Abstract
Purpose
To determine the genetic association of an inflammation-related gene, formyl peptide receptor 1 (FPR1), in exudative age-related macular degeneration (AMD) and polypoidal choroidal vasculopathy (PCV).
Methods
The coding region of FPR1 gene was sequenced in 554 unrelated Chinese individuals: 155 exudative AMD patients, 179 PCV patients, and 220 controls. Interactions and combined effects of FPR1 with complement factor H (CFH), high temperature requirement factor A1 (HTRA1), and smoking were also investigated.
Results
A total of 28 polymorphisms in FPR1 were identified. Single nucleotide polymorphisms (SNP) rs78488639 increased the risk to exudative AMD (P=0.043) and PCV (P=0.016), whereas SNP rs867229 decreased the risk to exudative AMD (P=0.0026), but not PCV. Homozygous G allele of rs1042229 was associated with exudative AMD (P=0.0394, odds ratio (OR)=2.27, 95% confident interval: 1.08–4.74), but not with PCV. Exudative AMD, but not PCV, was associated with the heterozygous genotypes of rs2070746 (P=0.019, OR=0.57) and rs867229 (P=0.0082, OR=0.54). Significantly, interactions were identified among FPR1 rs78488639, CFH rs800292, and HTRA1 rs11200638 in both exudative AMD and PCV. Combined heterozygous risk alleles of CFH rs800292 GA and FPR1 rs78488639 CA were posed to PCV (P=2.22 × 10−4, OR=10.47), but not exudative AMD. Furthermore, FPR1 rs78488639 CA combining with HTRA1 rs11200638 and smoking was also predisposed risks to exudative AMD and PCV.
Conclusion
FPR1 is associated with exudative AMD and PCV in a Hong Kong Chinese cohort. FPR1 rs78488639 interacted with CFH rs800292, HTRA1 rs11200638, and smoking, enhancing risk to exudative AMD and PCV.
Introduction
Age-related macular degeneration (AMD) is an irreversible, progressive, and sight-threatening macular disease in the elderly population, affecting over 50 million individuals worldwide.1 Clinically, the late stage of AMD can be divided into two subgroups: geographic atrophy and exudative/neovascular forms.2 Exudative AMD involves choroidal neovascularization (CNV), which is an ingrowth of new vascular vessels from choriocapillaries beneath retinal pigment epithelium (RPE) arising toward subretinal space and the neuroretina. These new vessels have greater tendency of leakage and bleeding.3 CNV invades into the retina via the Bruch's membrane and RPE, leading to serous or hemorrhagic detachments of either the RPE or neuroretina. Consequently, this leads to subretinal fibrosis4 and ultimately irreversible blindness. The exudative form accounts for >80% of significant rapid visual loss from AMD.5, 6
Inflammation has an influential role in exudative AMD.7 Drusen, the hallmark of AMD, contain inflammatory components, such as complement activators and inhibitors, membrane attack complex as well as inflammasome.8 Local RPE cell damage by aging activates and recruits choroidal dendritic cells, contributing to the formation of drusen and CNV.9 The choroidal infiltrate is also composed of macrophages and lymphocytes.10, 11 The activated inflammatory cells secrete collagenase and elastase, which erode the thinned Bruch's membrane and facilitate migration of choroidal capillaries.12 Moreover, interaction between inflammatory cells and vascular endothelial growth factor (VEGF) also indicates the role of inflammation in CNV development.13 In addition, significant association of inflammatory markers with AMD suggests that inflammation could participate in AMD pathogenesis.14, 15
As inflammation is involved in AMD pathogenesis, inflammation-related genes should be studied in AMD patients. The formyl peptide receptor 1 (FPR1) gene, located on chromosome 19q13.4,16 encodes a seven transmembrane domain G protein-coupled receptor. It is expressed mainly by mammalian phagocytic leukocytes and involved in inflammatory responses by activation of chemotatic peptides.17 This receptor allows phagocytic cells to recognize the presence of exogenous organisms, such as bacteria, and mediates the traffic of phagocytes to the sites of tissue damage.18 Therefore, FPR1 is a crucial molecule in innate immunity. Moreover, its ligand, the N-formyl peptides, is produced only by bacteria and mitochondrial proteins in nature.19 Mitochondrial dysfunction is also associated with AMD pathogenesis.20 Hence, we hypothesized that FPR1 could have a positive role in AMD pathogenesis.
Polypoidal choroidal vasculopathy (PCV) shares many similarities in clinical features with exudative AMD. But they differ in clinical course, imaging feature, response to treatment, and prognosis.21 In order to identify the role of FPR1 in exudative AMD, the whole-coding region of FPR1 gene was sequenced in this study. In addition, the FPR1 sequence was screened in PCV and compared with that of exudative AMD. Furthermore, the interactions and combined effects of FPR1 with complement factor H (CFH) and high temperature requirement factor A1 (HTRA1) were also determined as CFH and HTRA1 are associated with both exudative AMD and PCV.22, 23
Materials and methods
Study subjects
A total of 554 participants were recruited at the Prince of Wales Hospital Eye Centre, including 155 exudative AMD patients, 179 PCV patients and 220 age-matched control subjects (Supplementary Table 1). All study subjects were given complete ophthalmic examinations. AMD was graded according to an international classification and grading system.24 Patients with exudative AMD had non-drusenoid RPE detachment, choroidal neovascularization, serous or hemorrhagic retinal detachments, subretinal or sub-RPE hemorrhage, or fibrosis. The diagnosis of PCV was distinguished from exudative AMD by fluorescein angiography and indocyanine green angiography (ICGA).25 PCV patients had subretinal red or orange nodules and hemorrhagic pigment epithelial detachment and characteristic sacculated vascular abnormalities in the inner choroid as visualized on ICGA. The control subjects were recruited from elderly people >60 years of age. According to the complete ophthalmic examinations, the control subjects did not have any identifiable signs of AMD, PCV, or other retinal or optic nerve diseases except for mild senile cataract and refractive errors. Smoking habits were also recorded. A smoker was defined as a person who had smoked at least five cigarettes daily for >1 year. The study subjects were divided into two groups: those who had never smoked, those who were ex-smokers, and current smokers. The study protocol, approved by the Ethics Committee for Human Research at the Chinese University of Hong Kong, is in accordance with the tenets of the Declaration of Helsinki. Informed consent was obtained from each study subject.
Sequence analysis
Genomic DNA was extracted (Qiagen QIAamp DNA Blood Mini kit, Qiagen, Hiden, Germany) from peripheral blood samples following the supplier's instructions. All samples were screened for sequence alterations in the entire coding region (exon 2) and intron–exon junctions of FPR1 (ENSG00000171051) by PCR with specific primer sets (Supplementary Table 2), followed by direct DNA sequencing (BigDye Terminator Cycle Sequencing Reaction Kit, v3.1, Applied Biosystems, Foster City, CA, USA) on a DNA sequencer (ABI 3130XL, Applied Biosystems). In addition, all samples were genotyped for CFH rs800292 and HTRA1 rs11200639 using direct DNA sequencing with specific primers.22, 23
Amino-acid substitution analysis
For the non-synonymous variants identified in FPR1, functional effects of the amino-acid substitutions were evaluated and predicted by two web-based analyzing programs: PolyPhen (polymorphism phenotyping, http://genetics.bwh.harvard.edu/pph/, provided by the Bork Group and the Sunyaev Lab, Brighamand Women's Hospital, Harvard Medical School, Boston, MA, USA) and SIFT (Sorting Intolerant from Tolerant, http://sift.jcvi.org/, provided in the public domain by the J Craig Venter Institute, Rockville, MD, USA). The PolyPhen score>1.0 would be considered as ‘probably damaging' and otherwise would be considered as ‘benign'. The SIFT score ≤0.05 is predicted to be damaging, and >0.05 is predicted to be tolerated.
Statistical analysis
All the identified polymorphisms were assessed for Hardy–Weinberg equilibrium in controls using χ2 analysis. Allelic and genotypic distributions were compared using the χ2-test or Fisher's exact test (SPSS, version 16.0; SPSS Science, Chicago, IL, USA). Bonferroni's correction was used for multiple testing corrections. Linkage disequilibrium (LD) plots and haplotype-based association analyses were generated by Haploview (v4.2, http://www.broadinstitute.org/).26 All associated polymorphisms in FPR1 were assessed by logistic regression (SPSS). Logistic regression analysis included only gender as indicator because the recruited control subjects were older, excluding the potential patients with major late-onset ocular diseases. SNPs that remained significant after adjusting for other individual SNPs were selected for interaction and combined effect analyses. The pair-wise association of FPR1 rs78488639 with CFH rs800292, HTRA1 rs11200638 or smoking was evaluated using the χ2-test or Fisher's exact test (SPSS).
Results
The gender distribution of PCV patients in our study was ∼2 : 1 (male:female, 123 : 51), whereas that of exudative AMD patients (84 : 70) and control subjects (101 : 119) was ∼1 : 1 (Supplementary Table 1). This was compatible to a previous study.27
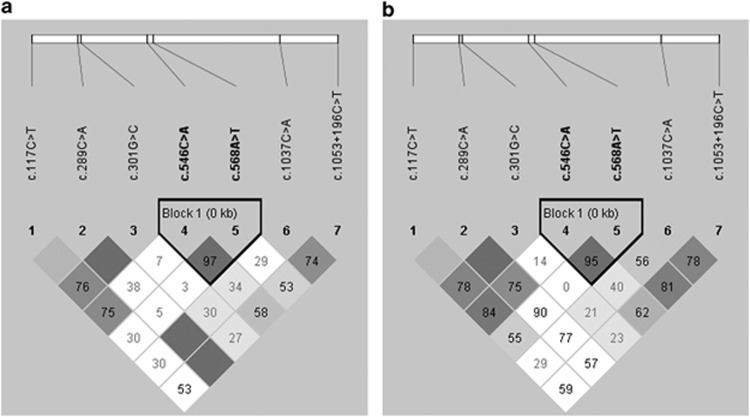
A total of 28 single nucleotide polymorphisms (SNPs) were identified in FPR1 (Table 1). Variants rs7253355 (c.1-342C>T) and c.513G>A (T171T) were excluded for further analysis as they violated Hardy–Weinberg equilibrium in the control subjects. Among the remaining 26 polymorphisms, 18 were rare SNPs (minor allele frequency <3%) and did not show significant association. They were also excluded for further analysis. The triallelic variant rs1042229 (c.576T>G/C, N192K/N) was analyzed independently. The homozygous of risk allele G was associated with exudative AMD (P=0.0394, odds ratio (OR)=2.27, 95% confident interval (CI): 1.08–4.74), but not with PCV (P=0.241) or in comparison between exudative AMD and PCV (P=0.137). Besides, the codon change caused by risk allele C was synonymous and did not show any association under either autosomal recessive or dominant model (data not shown). Thus, totally seven polymorphisms (c.117C>T, rs78488639, rs2070745, rs2070746, rs5030880, rs867228, and rs867229) were further analyzed (Table 1). SNP rs78488639 (c.289C>A, L97M) was associated with both exudative AMD (P=0.043) and PCV (P=0.029), whereas rs867229 (c.1053+196C>T) was associated with only exudative AMD (P=0.0026). Genotypic difference between exudative AMD and PCV was detected in rs867229 (P=0.014). Haplotype analysis revealed a LD block including two SNPs, rs2070746 and rs5030880, in both exudative AMD and PCV (Figure 1). However, haplotype-based association analysis could not identify any associated haplotype in exudative AMD or PCV (data not shown).
Table 1. Sequence variants detected in FPR1.
| No. | Location | dbSNP ID | Sequence change | Codon change |
Genotype frequencya |
Genotype association P-value |
Minor allele frequency (%) |
|||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AMD | PCV | Control | AMD vs Control | PCV vs Control | AMD | PCV | Control | |||||
| 1 | Intron 1 | rs7253284 | c.1-514C>T | — | 1/4/150 | 0/9/161 | 0/16/203 | 0.068 | 0.533 | 6 (1.9) | 9 (2.6) | 16 (3.7) |
| 2 | Intron 1 | rs7253355 | c.1-342C>T | — | 11/54/90 | 14/68/88 | 5/85/129 | 0.070 | 0.020 | 76 (24.5) | 96 (28.2) | 95 (21.7) |
| 3 | Intron 1 | Novel | c.1-55C>T | — | 0/0/153 | 0/0/172 | 0/1/218 | 1.000 | 1.000 | 0 (0.0) | 0 (0.0) | 1 (0.2) |
| 4 | Intron 1 | Novel | c.1-18A>T | — | 0/0/153 | 0/2/170 | 0/0/219 | — | 0.193 | 0 (0.0) | 2 (0.6) | 0 (0.0) |
| 5 | Exon 2 | rs5030878 | c.32T>C | Ile11Thr | 0/6/147 | 0/12/160 | 0/12/207 | 0.491 | 0.540 | 6 (2.0) | 12 (3.5) | 12 (2.7) |
| 6 | Exon 2 | Novel | c.117C>T | Leu39Leu | 0/17/136 | 2/14/156 | 0/18/201 | 0.347 | 0.278 | 17 (5.6) | 18 (5.2) | 18 (4.1) |
| 7 | Exon 2 | Novel | c.279C>T | Phe93Phe | 0/0/153 | 0/0/170 | 0/1/218 | 1.000 | 1.000 | 0 (0.0) | 0 (0.0) | 1 (0.2) |
| 8 | Exon 2 | rs78488639 | c.289C>A | Leu97Met | 0/20/133 | 1/24/144 | 0/15/204 | 0.043 | 0.029 | 20 (6.5) | 26 (7.7) | 15 (3.4) |
| 9 | Exon 2 | rs2070745 | c.301G>C | Val101Leu | 45/75/33 | 60/71/39 | 54/102/63 | 0.261 | 0.066 | 165 (53.9) | 191(56.2) | 210 (47.9) |
| 10 | Exon 2 | rs28930680 | c.306T>C | Phe102Phe | 0/3/150 | 0/1/169 | 0/3/216 | 0.693 | 0.635 | 3 (0.7) | 1 (0.3) | 3 (0.7) |
| 11 | Exon 2 | rs5030879 | c.348C>T | Ile116Ile | 0/2/151 | 0/2/168 | 0/1/218 | 0.571 | 0.583 | 2 (0.7) | 2 (0.6) | 1 (0.2) |
| 12 | Exon 2 | Novel | c.368G>A | Arg123His | 0/1/152 | 0/0/170 | 0/0/219 | 0.411 | — | 1 (0.3) | 0 (0.0) | 0 (0.0) |
| 13 | Exon 2 | Novel | c.439A>T | Ile147Phe | 0/0/155 | 0/2/173 | 0/2/217 | 0.513 | 1.000 | 0 (0.0) | 2 (0.6) | 2 (0.5) |
| 14 | Exon 2 | Novel | c.513G>A | Thr171Thr | 0/0/155 | 0/0/175 | 1/1/217 | 0.491 | 0.448 | 0 (0.0) | 0 (0.0) | 3 (0.7) |
| 15 | Exon 2 | rs2070746 | c.546C>A | Pro182Pro | 37/60/58 | 33/95/47 | 52/108/59 | 0.065 | 0.462 | 134 (43.2) | 161 (46) | 212 (48.4) |
| 16 | Exon 2 | Novel | c.553A>G | Asn185Asp | 0/0/155 | 0/0/175 | 0/2/217 | 0.513 | 0.505 | 0 (0.0) | 0 (0.0) | 2 (0.5) |
| 17 | Exon 2 | rs5030880 | c.568A>T | Arg190Trp | 4/46/105 | 4/63/108 | 6/76/137 | 0.580 | 0.934 | 54 (17.4) | 81 (20.3) | 88 (20.1) |
| 18 | Exon 2 | rs1042229 | c.576T>Gb | Asn192Lys | — | — | — | — | — | — | — | — |
| — | — | — | c.576T>Cb | Asn192Asn | — | — | — | — | — | — | — | — |
| 19 | Exon 2 | Novel | c.721C>T | Arg241Trp | 0/0/155 | 0/0/175 | 0/1/218 | 1.000 | 1.000 | 0 (0.0) | 0 (0.0) | 1 (0.2) |
| 20 | Exon 2 | Novel | c.944 G >A | Arg309Gln | 0/0/153 | 0/2/176 | 0/0/220 | — | 0.199 | 0 (0.0) | 2 (0.6) | 0 (0.0) |
| 21 | Exon 2 | rs867228 | c.1037C>A | Glu346Ala | 17/63/73 | 16/81/81 | 17/108/95 | 0.249 | 0.750 | 97 (31.7) | 113 (31.7) | 142 (32.3) |
| 22 | 3′-UTR | Novel | c.1053+8G>T | — | 0/0/153 | 0/0/178 | 0/1/219 | 1.000 | 1.000 | 0 (0.0) | 0 (0.0) | 1 (0.2) |
| 23 | 3′-UTR | rs867341 | c.1053+75A>G | — | 1/7/145 | 0/5/173 | 0/7/213 | 0.378 | 0.829 | 9 (2.9) | 7 (1.4) | 7 (1.6) |
| 24 | 3′-UTR | Novel | c.1053+109G>C | — | 0/1/152 | 0/0/178 | 0/0/220 | 0.410 | — | 5 (1.6) | 0 (0.0) | 0 (0.0) |
| 25 | 3′-UTR | Novel | c.1053+162T>G | — | 0/0/152 | 0/1/170 | 0/0/220 | — | 0.437 | 0 (0.0) | 1 (0.3) | 0 (0.0) |
| 26 | Downstream | rs867229 | c.1053+196C>T | — | 29/50/75 | 18/79/74 | 23/107/87 | 0.0026 | 0.808 | 108 (35.1) | 115 (33.6) | 153 (35.2) |
| 27 | Downstream | rs1868943 | c.1053+205C>T | — | 0/2/152 | 0/0/171 | 0/8/209 | 0.205 | 0.010 | 2 (0.7) | 0 (0.0) | 8 (1.8) |
| 28 | Downstream | Novel | c.1053+219G>A | — | 0/1/153 | 0/3/168 | 0/1/216 | 1.000 | 0.325 | 1 (0.3) | 3 (0.9) | 1 (0.2) |
Genotype frequency presented as number of individual with homozygote/heterozygote/wild-type genotypes
Genotype and allele frequencies of c.576T>G/C is presented in a separate table.
Figure 1.
Haplotype block structure for the seven common polymorphisms. The haplotype analysis revealed a LD block lying across rs2070746 and rs5030880 in (a) AMD and (b) PCV.
The heterozygous genotype of rs78488639 contributed a 2.05- and 2.27-fold of increased risk, respectively, to exudative AMD (P=0.043) and PCV (P=0.016; Table 2). Homozygous genotype of rs2070745 was associated with PCV (P=0.034, OR=1.80, 95% CI: 1.04–3.09). Heterozygous genotypes of rs2070746 and rs867229 were associated with a decreased risk in exudative AMD (P=0.019, OR=0.57, 95% CI: 0.35–0.91; P=0.0082, OR=0.54, 95% CI: 0.34–0.86, respectively). Heterozygous genotype of rs2070746 was also different between exudative AMD and PCV (P=0.0086, OR=0.51, 95% CI: 0.31–0.85). Moreover, five novel rare variants were identified. Amino-acid substitution prediction suggested that the protein function of c.368G>A (R123H) could be affected with a SIFT score of 0.01 (Table 3). This was further supported by the PolyPhen prediction with a score of 2.75. However, neither individual rare variant nor pooling of the variants were associated with exudative AMD or PCV (data not shown). The association became not significant after Bonferroni's correction (P=0.05/28=0.0018).
Table 2. Odds ratios of FPR1-associated polymorphisms.
| SNP ID | Sequence change |
Homo vs WT |
Hetro vs WT |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AMD–Control | OR (95% CI) | PCV–Control | OR (95% CI) | AMD–PCV | OR (95% CI) | AMD–Control | OR (95% CI) | PCV–Control | OR (95% CI) | AMD–PCV | OR (95% CI) | ||
| rs78488639 | c.289C>A | — | — | 0.415 | 1.01 (0.99–1.02) | 1.000 | 0.99 (0.98–1.01) | 0.043 | 2.05 (1.01–4.14) | 0.016 | 2.27 (1.15–4.47) | 0.752 | 0.90 (0.48 –1.71) |
| rs2070745 | c.301G>C | 0.114 | 1.59 (0.89–2.84) | 0.034 | 1.80 (1.04–3.09) | 0.695 | 0.89 (0.49–1.62) | 0.197 | 1.40 (0.84–2.35) | 0.646 | 1.12 (0.68–1.86) | 0.442 | 1.25 (0.71–2.20) |
| rs2070746 | c.546C>A | 0.254 | 0.72 (0.42–1.26) | 0.443 | 0.80 (0.45–1.42) | 0.757 | 0.91 (0.50–1.67) | 0.019 | 0.57 (0.35–0.91) | 0.681 | 1.10 (0.69–1.77) | 0.0086 | 0.51 (0.31–0.85) |
| rs867229 | c.1053+196C>T | 0.234 | 1.46 (0.78–2.74) | 0.813 | 0.92 (0.46–1.84) | 0.173 | 1.59 (0.81–3.11) | 0.0082 | 0.54 (0.34–0.86) | 0.514 | 0.87 (0.57–1.33) | 0.053 | 0.62 (0.39–1.0) |
Note: There was no homozygous genotype of rs78488639 identified in AMD and control group.
Table 3. Amino-acid substitution prediction of novel rare FPR1 variants.
| Sequence change | Codon change |
Genotypic frequency |
Predictions |
|||||
|---|---|---|---|---|---|---|---|---|
| AMD | PCV | Control | SIFT | Score | Polyphen | Score | ||
| c.368G>A | R123H | 0/1/152 | 0/0/170 | 0/0/219 | Affect protein function | 0.01 | Probably damaging | 2.753 |
| c.439A>T | I147F | 0/0/155 | 0/2/173 | 0/2/217 | Tolerated | 0.29 | Benign | 0 |
| c.553A>G | N185D | 0/0/155 | 0/0/175 | 0/2/217 | Tolerated | 0.69 | Benign | 0.525 |
| c.721C>T | R241W | 0/0/155 | 0/0/175 | 0/1/218 | Tolerated | 0.09 | Benign | 0.807 |
| c.944G >A | R309Q | 0/0/153 | 0/2/176 | 0/0/220 | Tolerated | 0.22 | Benign | 0.538 |
The G allele of CFH rs800292 increased the risk for both exudative AMD (homozygous: OR=2.60, 95% CI: 1.27–5.31, P=0.0074; heterozygous: OR=1.41, 95% CI: 0.68–2.92, P=0.36) and PCV (homozygous: OR=3.12, 95% CI: 1.42–6.86, P=0.0035; heterozygous: OR=2.50, 95% CI: 1.14–5.51, P=0.020). The risk was also increased by the A allele of HTRA1 rs11200638 in exudative AMD (homozygous: OR=12.48, 95% CI: 6.53–23.83, P=1.07 × 10−16; heterozygous: OR=2.46, 95% CI: 1.31–4.64, P=0.0046) and PCV (homozygous: OR=5.24, 95% CI: 2.89–9.50, P=1.79 × 10−8; heterozygous: OR=2.51, 95% CI: 1.47–4.29, P=5.94 × 10−4). Logistic regression analysis revealed that only FPR1 rs78488639 remained significant after adjusting for gender and other individual-associated SNPs in both exudative AMD (P=0.032) and PCV (P=0.022). Further interaction analysis identified significantly positive interactions among FPR1 rs78488639, CFH rs800292, and HTRA1 rs11200638 in exudative AMD (P=0.022) and PCV (P=0.023). In the combined effect analysis, ORs of combined FPR1 rs78488639 CA and CFH rs800292 GG genotypes were approximately twice greater than the individual ORs of FPR1 rs78488639 CA or CFH rs800292 GG in both exudative AMD (P=0.0062, OR=4.83, 95% CI: 1.51–15.51) and PCV (P=0.019, OR=4.03, 95% CI: 1.22–13.28; Table 4). A combined risk OR of 10.47 (P=2.22 × 10−4, 95% CI: 2.72–40.29) was identified in the heterozygous risk alleles of these two variants in the PCV patients, but not in exudative AMD (P=0.133). In addition, ORs of combined FPR1 rs78488639 CA and HTRA1 rs11200638 AA genotypes were at least 1.5-fold higher than the individual ORs of HTRA1 rs11200638 AA and >6-fold higher than FPR1 rs78488639 CA in both exudative AMD (P=1.02 × 10−4, OR=19.47, 95% CI: 3.75–100.97) and PCV (P=7.45 × 10−4, OR=14.19, 95% CI: 2.72–74.20). Furthermore, combined ORs of FPR1 rs78488639 CA and smoking were >5-fold higher than FPR1 rs78488639 CA in both exudative AMD (P=0.010, OR=10.93, 95% CI: 1.30–92.10) and PCV (P=9.96 × 10−4, OR=16.94, 95% CI: 2.06–139.38; Table 5).
Table 4. Combined effects of FPR1 rs78488639 with CFH rs800292 and HTRA1 rs11200638 in exudative AMD and PCV.
|
CFH rs800292 |
|||||||
|---|---|---|---|---|---|---|---|
| FPR1 rs78488639 |
AA |
GA |
GG |
||||
| ORFPR1(95% CI) | P | OR (95% CI) | P | OR (95% CI) | P | OR (95% CI) | |
| Exudative AMD | |||||||
| CC | 1.00 (Ref) | 1.00 (Ref) | 0.317 | 1.52 (0.66–3.49) | 0.0083 | 2.90 (1.29–6.54) | |
| CA | 2.05 (1.01–4.14) | 0.321 | 3.22 (0.55–18.85) | 0.196 | 3.22 (0.67–15.56) | 0.0062 | 4.83 (1.51–15.51) |
| PCV | |||||||
| CC | 1.00 (Ref) | — | 1.00 (Ref) | 0.090 | 2.01 (0.89–4.55) | 0.020 | 2.58 (1.14–5.84) |
| CA | 2.27 (1.15–4.47) | 1.000 | 0.91 (0.81–1.01) | 2.22 × 10−4 | 10.47 (2.72–40.29) | 0.019 | 4.03 (1.22–13.28) |
| AA | 1.01 (0.99–1.02) | — | — | — | — | 0.256 | 1.11 (0.90–1.37) |
|
HTRA1 rs11200638 |
|||||||
|---|---|---|---|---|---|---|---|
| FPR1 rs78488639 |
GG |
GA |
AA |
||||
| ORFPR1(95% CI) | P | OR (95% CI) | P | OR (95% CI) | P | OR (95% CI) | |
| Exudative AMD | |||||||
| CC | 1.00 (Ref) | — | 1.00 (Ref) | 0.083 | 1.81 (0.82–3.57) | 1.94 × 10−13 | 11.02 (5.55–21.90) |
| CA | 2.05 (1.01–4.14) | 1.000 | 0.70 (0.08–6.08) | 3.83 × 10−4 | 8.11 (2.56–25.74) | 1.02 × 10−4 | 19.47 (3.75–100.97) |
| PCV | |||||||
| CC | 1.00 (Ref) | — | 1.00 (Ref) | 0.0018 | 2.59 (1.41–4.76) | 3.15 × 10−8 | 6.20 (3.16–12.16) |
| CA | 2.27 (1.15–4.47) | 0.037 | 4.06 (1.26–13.04) | 0.0023 | 6.08 (1.92–19.30) | 7.45 × 10−4 | 14.19 (2.72–74.20) |
| AA | 1.01 (0.99–1.02) | — | — | — | — | 0.207 | 1.06 (0.95–1.17) |
Note: Allele A is the risk allele for FPR1 rs78488639, allele G is risk allele for CFH rs800292, and allele A is the risk allele for HTRA1 rs11200638.
Table 5. Combined effects of FPR1 rs78488639 with smoking in exudative AMD and PCV.
|
Smoking |
|||||
|---|---|---|---|---|---|
| FPR1 rs78488639 |
No |
Yes |
|||
| ORFPR1(95% CI) | P | OR (95% CI) | P | OR (95% CI) | |
| Exudative AMD | |||||
| CC | 1.00 (Ref) | — | 1.00 (Ref) | 0.038 | 1.93 (1.04–3.58) |
| CA | 2.05 (1.01–4.14) | 0.739 | 0.67 (0.16–2.74) | 0.010 | 10.93 (1.30–92.10) |
| PCV | |||||
| CC | 1.00 (Ref) | — | 1.00 (Ref) | 0.992 | 1.00 (0.48–2.10) |
| CA | 2.27 (1.15–4.47) | 0.164 | 2.15 (0.72–6.44) | 9.96 × 10−4 | 16.94 (2.06–139.38) |
| AA | 1.01 (0.99–1.02) | — | — | — | — |
Note: Allele A is the risk allele for FPR1 rs78488639.
Discussion
Inflammation is an important component in AMD etiology. Chronic localized inflammation initiated and stimulated by drusen, together with the accumulation of lipofuscin, compromise RPE function.28, 29 The injured RPE actively recruits choroidal dendritic cells,29 microglia, macrophages,30 and leukocytes,31 which express VEGF and stimulate CNV growth,32 the hallmark of exudative AMD. The initiation of AMD by inflammation and infections has been supported by clinical observations of Chlamydia pneumonia and cytomegalovirus infections in AMD patients.33, 34 As FPR1 is a receptor for phagocytic cells recognizing the presence of exogenous organisms and mediating the traffic of phagocytes to the sites of tissue damage,18 FPR1 could have an important role in AMD initiation by infections.
The involvement of FPR1 in innate immunity is supported by multiple evidences. First, the release of formylated peptides from mitochondria secondary to cell death might attract phagocytic leukocytes through the receptor.35 Second, the Fpr1−/− mice model had accelerated mortality and increased bacterial burden in the liver and spleen early after infection,36 showing a role of Fpr1 in the host defense. Moreover, rs5030879 (c.348C>T, I116I) in FPR1 is associated with aggressive periodontitis,37 which is an infectious–inflammatory disease. In addition, c.329T>C (F110S), c.378C>G (C126W), and rs5030878 (c.32C>T, I11T) of FPR1 are significantly correlated with defective polymorphonuclear neutrophil (PMN) function and C-reactive protein.38, 39 However, limited data reported the association of FPR1 with AMD, and also PCV. In this study, the results of FPR1 sequencing in both exudative AMD and PCV revealed five associated polymorphisms (Table 1), indicating a role of FPR1 in the inflammation mechanisms of exudative AMD and PCV.
Formyl peptide receptors (FPRs) are expressed mainly in PMNs and monocytes, but also found in neurons, microglial, and glial cells.40 In the inflammatory response, phagocytes are induced to move directionally through chemotaxis.41 When the ligands (N-formyl peptides) bind to FPR1, FPR1 will direct the activated phagocyte migration through the PI3k/PTEN pathways. This implies an important role of FPR1 in inflammatory cell migration. FPR1 also induces superoxide production, and this oxidative stress could further damage the RPE cells.42, 43 Furthermore, FPR1 expression can be enhanced by inflammation in Alzheimer's disease,44 which is associated with age as AMD and involves accumulation of amyloid β (Aβ) in senile plaques. Aβ has been found in AMD drusen.45 Therefore, FPR1 likely has an etiological role in AMD.
CFH and HTRA1 are well recognized as susceptibility genes for AMD.22, 23 CFH is an inhibitor in alternative complement pathway, controlling complement activation.46 Abnormal regulation of complement activation by CFH might contribute to AMD pathogenesis.47 HTRA1 is also involved in inflammatory responses.48, 49 In this study, FPR1 significantly interacted with CFH and HTRA1, with respective combined ORs of 10.47 and 19.47 for increasing disease risk (Table 4). Moreover, the risks of FPR1 to exudative AMD and PCV were also increased by smoking, with respective combined ORs of 10.93 and 16.94 (Table 5). The combined effect with smoking has been demonstrated in our previous studies on CFH and HTRA1.22, 23 Therefore, the role of FPR1 as well as the contribution of inflammation genes and smoking to AMD and PCV are further implicated.
SNP rs1042229 is a triallelic (T, G or C) variant, whereas the majority of SNPs are biallelic. Its G allele specifies lysine and T/C specifies asparagines at codon 192. The N192 locates in the center of the second extracellular loop, which the lysine substitution may alter the ligand-binding specificity and binding affinity through the binding pocket for N-formyl peptides.50 In this study, the G allele is the risk allele of rs1042229, and homozygous G genotype of rs1042229 was associated with 2.27-fold of increasing risk in exudative AMD, but not in PCV. Another variant (L97M; rs78488639), located in the first extracellular loop, markedly decreases in the affinity of ligand binding.51 This variant showed significant association with both exudative AMD and PCV, contributing an approximately twofold increasing disease risk (Table 2). Differential associations of various FPR1 SNPs with AMD and PCV are also noted, suggesting that exudative AMD and PCV might have different gene variant association patterns.
In summary, significant interactions and combined effects of FPR1 with CFH, HTRA1, and smoking in exudative AMD and PCV were revealed in this study. FPR1 could have a role in AMD and PCV pathogenesis, and its biological functions in the disease mechanisms need to be elucidated.
Acknowledgments
We express our greatest appreciation to all the study participants. This study was supported in part by the Endowment Fund for Lim Por-Yen Eye Genetics Research Centre, the Chinese University of Hong Kong and the General Research Fund from the Research Grants Council (grant number 473410), Hong Kong.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on Eye website (http://www.nature.com/eye)
Supplementary Material
References
- Pascolini D, Mariotti SP, Pokharel GP, Pararajasegaram R, Etya'ale D, Négrel AD, et al. 2002 global update of available data on visual impairment: a compilation of population-based prevalence studies. Ophthalmic Epidemiol. 2004;11:67–115. doi: 10.1076/opep.11.2.67.28158. [DOI] [PubMed] [Google Scholar]
- Jager RD, Mieler WF, Miller JW. Age-related macular degeneration. N Engl J Med. 2008;358:2606–2617. doi: 10.1056/NEJMra0801537. [DOI] [PubMed] [Google Scholar]
- Fine SL, Berger JW, Maguire MG, Ho AC. Age-related macular degeneration. N Engl J Med. 2000;342:483–492. doi: 10.1056/NEJM200002173420707. [DOI] [PubMed] [Google Scholar]
- Ambati J, Ambati BK, Yoo SH, Ianchulev S, Adamis AP. Age-related macular degeneration: etiology, pathogenesis, and therapeutic strategies. Surv Ophthalmol. 2003;48:257–293. doi: 10.1016/s0039-6257(03)00030-4. [DOI] [PubMed] [Google Scholar]
- Age-Related Eye Disease Study Research Group A randomized, placebo-controlled, clinical trial of high-dose supplementation with vitamins C and E, beta carotene, and zinc for age-related macular degeneration and vision loss: AREDS report no. 8. Arch Ophthalmol. 2001;119:1417–1436. doi: 10.1001/archopht.119.10.1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skaf AR, Mahmoud T. Surgical treatment of age-related macular degeneration. Semin Ophthalmol. 2011;26:181–191. doi: 10.3109/08820538.2011.577133. [DOI] [PubMed] [Google Scholar]
- Ding X, Patel M, Chan CC. Molecular pathology of age-related macular degeneration. Prog Ret Eye Res. 2009;28:1–18. doi: 10.1016/j.preteyeres.2008.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanton CM, Wright AF. Inflammatory biomarkers for AMD. Adv Exp Med Biol. 2014;801:251–257. doi: 10.1007/978-1-4614-3209-8_32. [DOI] [PubMed] [Google Scholar]
- Moshfeghi DM, Blumenkranz MS. Role of genetic factors and inflammation in age-related macular degeneration. Retina. 2007;27:269–275. doi: 10.1097/IAE.0b013e31802e3e9b. [DOI] [PubMed] [Google Scholar]
- Cao X, Shen D, Patel MM, Patel MM, Tuo J, Johnson TM, et al. Macrophage polarization in the maculare of age-related macular degeneration: a pilot study. Pathol Int. 2011;61:528–535. doi: 10.1111/j.1440-1827.2011.02695.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B, Wei L, Meverle C, Tuo J, Sen HN, Li Z, et al. Complement component C5a promotes expression of IL-22 and IL-17 from human T cells and its implication in age-related macular degeneration. J Transl Med. 2011;9:1–21. doi: 10.1186/1479-5876-9-111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Schaft TL, Mooy CM, de Bruijn WC, de Jong PT. Early stages of age-related macular degeneration: an immunofluorescence and electron microscopy study. Br J Ophthalmol. 1993;77:657–661. doi: 10.1136/bjo.77.10.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kijlstra A, La Heij E, Hendrikse F. Immunological factors in the pathogenesis and treatment of age-related macular degeneration. Ocul Immunol Inflamm. 2005;13:3–11. doi: 10.1080/09273940590909185. [DOI] [PubMed] [Google Scholar]
- Machalińska A, Dziedziejko V, Mozolewska-Piotrowska K, Karczewicz D, Wiszniewska B, Machaliński B. Elevated plasma levels of C3a complement compound in the exudative form of age-related macular degeneration. Ophthalmic Res. 2009;42:54–59. doi: 10.1159/000219686. [DOI] [PubMed] [Google Scholar]
- Hong T, Tan AG, Mitchell P, Wang JJ. A review and meta-analysis of the association between C-reactive protein and age-related macular degeneration. Surv Ophthalmol. 2011;56:184–194. doi: 10.1016/j.survophthal.2010.08.007. [DOI] [PubMed] [Google Scholar]
- Haviland DL, Borel AC, Fleischer DT, Haviland JC, Wetsel RA. Structure, 5'-flanking sequence, and chromosome location of the human N-formyl peptide receptor gene. A single-copy gene comprised of two exons on chromosome 19q.13.3 that yields two distinct transcripts by alternative polyadenylation. Biochemistry. 1993;32:4168–4174. doi: 10.1021/bi00067a003. [DOI] [PubMed] [Google Scholar]
- Gao JL, Murphy PM. Species and subtype variants of the N-formyl peptide chemotactic receptor reveal multiple important functional domains. J Biol Chem. 1993;268:25395–25401. [PubMed] [Google Scholar]
- Le Y, Murphy PM, Wang JM. Formyl-peptide receptors revisited. Trends Immunol. 2002;23:541–548. doi: 10.1016/s1471-4906(02)02316-5. [DOI] [PubMed] [Google Scholar]
- Bhattacharya M, Wang J, Ribeiro FM, Dixon SJ, Feldman RD, Hegele RA, et al. Analysis of a missense variant of the human N-formyl peptide receptor that is associated with agonist-independent beta-arrestin association and indices of inflammation. Pharmacogenomics J. 2007;7:190–199. doi: 10.1038/sj.tpj.6500416. [DOI] [PubMed] [Google Scholar]
- Karunadharma PP, Nordgaard CL, Olsen TW, Ferrington DA. Mitochondrial DNA damage as a potential mechanism for age-related macular degeneration. Invest Ophthalmol Vis Sci. 2010;51:5470–5479. doi: 10.1167/iovs.10-5429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laude A, Cackett PD, Vithana EN, Yeo IY, Wong D, Koh AH, et al. Polypoidal choroidal vasculopathy and neovascular age-related macular degeneration: same or different disease. Prog Retin Eye Res. 2010;29:19–29. doi: 10.1016/j.preteyeres.2009.10.001. [DOI] [PubMed] [Google Scholar]
- Ng TK, Chen LJ, Liu DT, Tam PO, Chan WM, Liu K, et al. Multiple gene polymorphisms in the complement factor h gene are associated with exudative age-related macular degeneration in chinese. Invest Ophthalmol Vis Sci. 2008;49:3312–3317. doi: 10.1167/iovs.07-1517. [DOI] [PubMed] [Google Scholar]
- Tam PO, Ng TK, Liu DT, Chan WM, Chiang SW, Chen LJ, et al. HTRA1 variants in exudative age-related macular degeneration and interactions with smoking and CFH. Invest Ophthalmol Vis Sci. 2008;49:2357–2365. doi: 10.1167/iovs.07-1520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird AC, Bressler NM, Bressler SB, Chisholm IH, Coscas G, Davis MD, et al. An international classification and grading system for age-related maculopathy and age-related macular degeneration. The International ARM Epidemiological Study Group. Surv Ophthalmol. 1995;39:367–374. doi: 10.1016/s0039-6257(05)80092-x. [DOI] [PubMed] [Google Scholar]
- Cackett P, Wong D, Yeo I. A classification system for polypoidal choroidal vasculopathy. Retina. 2009;29:187–191. doi: 10.1097/IAE.0b013e318188c839. [DOI] [PubMed] [Google Scholar]
- Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- Seitsonen SP, Onkamo P, Peng G, Xiong M, Tommila PV, Ranta PH, et al. Multifactor effects and evidence of potential interaction between complement factor H Y402H and LOC387715 A69S in age-related macular degeneration. PLoS One. 2008;3:e3833. doi: 10.1371/journal.pone.0003833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundelin S, Wihlmark U, Nilsson SE, Brunk UT. Lipofuscin accumulation in cultured retinal pigment epithelial cells reduces their phagocytic capacity. Curr Eye Res. 1998;17:851–857. [PubMed] [Google Scholar]
- Hageman GS, Luthert PJ, Victor Chong NH, Johnson LV, Anderson DH, Mullins RF. An integrated hypothesis that considers drusen as biomarkers of immune-mediated processes at the RPE-Bruch's membrane interface in aging and age-related macular degeneration. Prog Retin Eye Res. 2001;20:705–732. doi: 10.1016/s1350-9462(01)00010-6. [DOI] [PubMed] [Google Scholar]
- Killingsworth MC, Sarks JP, Sarks SH. Macrophages related to Bruch's membrane in age-related macular degeneration. Eye (Lond) 1990;4:613–621. doi: 10.1038/eye.1990.86. [DOI] [PubMed] [Google Scholar]
- Dastgheib K, Green WR. Granulomatous reaction to Bruch's membrane in age-related macular degeneration. Arch Ophthalmol. 1994;112:813–818. doi: 10.1001/archopht.1994.01090180111045. [DOI] [PubMed] [Google Scholar]
- Lopez PF, Sippy BD, Lambert HM, Thach AB, Hinton DR. Transdifferentiated retinal pigment epithelial cells are immunoreactive for vascular endothelial growth factor in surgically excised age-related macular degeneration-related choroidal neovascular membranes. Invest Ophthalmol Vis Sci. 1996;37:855–868. [PubMed] [Google Scholar]
- Robman L, Mahdi O, McCarty C, Dimitrov P, Tikellis G, McNeil J, et al. Exposure to Chlamydia pneumoniae infection and progression of age-related macular degeneration. Am J Epidemiol. 2005;161:1013–1019. doi: 10.1093/aje/kwi130. [DOI] [PubMed] [Google Scholar]
- Miller DM, Espinosa-Heidmann DG, Legra J, Dubovy SR, Sũner IJ, Sedmak DD, et al. The association of prior cytomegalovirus infection with neovascular age-related macular degeneration. Am J Ophthalmol. 2004;138:323–328. doi: 10.1016/j.ajo.2004.03.018. [DOI] [PubMed] [Google Scholar]
- Migeotte I, Communi D, Parmentier M. Formyl peptide receptors: a promiscuous subfamily of G protein-coupled receptors controlling immune responses. Cytokine Growth Factor Rev. 2006;17:501–519. doi: 10.1016/j.cytogfr.2006.09.009. [DOI] [PubMed] [Google Scholar]
- Gao JL, Lee EJ, Murphy PM. Impaired antibacterial host defense in mice lacking the N-formylpeptide receptor. J Exp Med. 1999;189:657–662. doi: 10.1084/jem.189.4.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maney P, Walters JD. Formylpeptide receptor single nucleotide polymorphism 348T>C and its relationship to polymorphonuclear leukocyte chemotaxis in aggressive periodontitis. J Periodontol. 2009;80:1498–1505. doi: 10.1902/jop.2009.090103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gwinn MR, Sharma A, De Nardin E. Single nucleotide polymorphisms of the N-formyl peptide receptor in localized juvenile periodontitis. J Periodontol. 1999;70:1194–1201. doi: 10.1902/jop.1999.70.10.1194. [DOI] [PubMed] [Google Scholar]
- Benachour H, Zaiou M, Herbeth B, Lambert D, Lamont JV, Pfister M, et al. Human formyl peptide receptor 1 (FPR1) c.32C>T SNP is associated with decreased soluble E-selectin levels. Pharmacogenomics. 2009;10:951–959. doi: 10.2217/pgs.09.29. [DOI] [PubMed] [Google Scholar]
- Becker EL, Forouhar FA, Grunnet ML, Boulay F, Tardif M, Bormann BJ, et al. Broad immunocytochemical localization of the formylpeptide receptor in human organs, tissues, and cells. Cell Tissue Res. 1998;292:129–135. doi: 10.1007/s004410051042. [DOI] [PubMed] [Google Scholar]
- Janetopoulos C, Firtel RA. Directional sensing during chemotaxis. FEBS Lett. 2008;582:2075–2085. doi: 10.1016/j.febslet.2008.04.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boxer LA, Yoder M, Bonsib S, Schmidt M, Ho P, Jersild R, et al. Effects of a chemotactic factor, N-formylmethionyl peptide, on adherence, superoxide anion generation, phagocytosis, and microtubule assembly of human polymorphonuclear leukocytes. J Lab Clin Med. 1979;93:506–514. [PubMed] [Google Scholar]
- Lehmeyer JE, Snyderman R, Johnston RB., Jr Stimulation of neutrophil oxidative metabolism by chemotactic peptides: influence of calcium ion concentration and cytochalasin B and comparison with stimulation by phorbol myristate acetate. Blood. 1979;54:35–45. [PubMed] [Google Scholar]
- Salminen A, Ojala J, Kauppinen A, Kaarniranta K, Suuronen T. Inflammation in Alzheimer's disease: amyloid-beta oligomers trigger innate immunity defence via pattern recognition receptors. Prog Neurobiol. 2009;87:181–194. doi: 10.1016/j.pneurobio.2009.01.001. [DOI] [PubMed] [Google Scholar]
- Ohno-Matsui K. Parallel findings in age-related macular degeneration and Alzheimer's disease. Prog Retin Eye Res. 2011;30:217–238. doi: 10.1016/j.preteyeres.2011.02.004. [DOI] [PubMed] [Google Scholar]
- Chen LJ, Liu DT, Tam PO, Chan WM, Liu K, Chong KK, et al. Association of complement factor H polymorphisms with exudative age-related macular degeneration. Mol Vis. 2006;12:1536–1542. [PubMed] [Google Scholar]
- Anderson DH, Radeke MJ, Gallo NB, Chapin EA, Johnson PT, Curletti CR, et al. The pivotal role of the complement system in aging and age-related macular degeneration: hypothesis re-visited. Prog Retin Eye Res. 2010;29:95–112. doi: 10.1016/j.preteyeres.2009.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng TK, Yam GH, Chen WQ, Lee VY, Chen H, Chen LJ, et al. Interactive expressions of HtrA1 and VEGF in human vitreous humors and fetal RPE cells. Invest Ophthalmol Vis Sci. 2011;52:3706–3712. doi: 10.1167/iovs.10-6773. [DOI] [PubMed] [Google Scholar]
- Ng TK, Liang XY, Pang CP. HTRA1 in age-related macular degeneration. Asia Pac J Ophthalmol. 2012;1:51–63. doi: 10.1097/APO.0b013e31823e57fe. [DOI] [PubMed] [Google Scholar]
- Perez HD, Holmes R, Vilander LR, Adams RR, Manzana W, Jolley D, et al. Formyl peptide receptor chimeras define domains involved in ligand binding. J Biol Chem. 1993;268:2292–2295. [PubMed] [Google Scholar]
- Le Y, Ye RD, Gong W, Li J, Iribarren P, Wang JM. Identification of functional domains in the formyl peptide receptor-like 1 for agonist-induced cell chemotaxis. FEBS J. 2005;272:769–778. doi: 10.1111/j.1742-4658.2004.04514.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.