Abstract
Although there has been much success in identifying genetic variants associated with common diseases using genome-wide association studies (GWAS)1, it has been difficult to demonstrate which variants are causal and what role they play in disease. Moreover, the modest contribution these variants make to disease risk has raised questions regarding their medical relevance2. We have investigated a single nucleotide polymorphism (SNP) in the TNFRSF1A gene, that encodes TNF receptor 1 (TNFR1), which was discovered through GWAS to be associated with multiple sclerosis (MS)3,4, but not with other autoimmune conditions such as rheumatoid arthritis (RA)5, psoriasis6 and Crohn’s disease7. By analyzing MS GWAS3,4 data in conjunction with the 1000 Genomes Project data8 we provide genetic evidence that strongly implicates this SNP, rs1800693, as the causal variant in the TNFRSF1A region. We further substantiate this through functional studies showing that the MS risk allele directs expression of a novel, soluble form of TNFR1 that can block TNF. Importantly, TNF blocking drugs can promote onset or exacerbation of MS9-11, but they have proven highly efficacious in the treatment of autoimmune diseases for which there is no association with rs1800693. This indicates that the clinical experience with these drugs parallels the disease association of rs1800693, and that the MS-associated TNFR1 variant mimics the effect of TNF blocking drugs. Hence, our study demonstrates that clinical practice can be informed by comparing GWAS across common autoimmune diseases and by investigating the functional consequences of the disease-associated genetic variation.
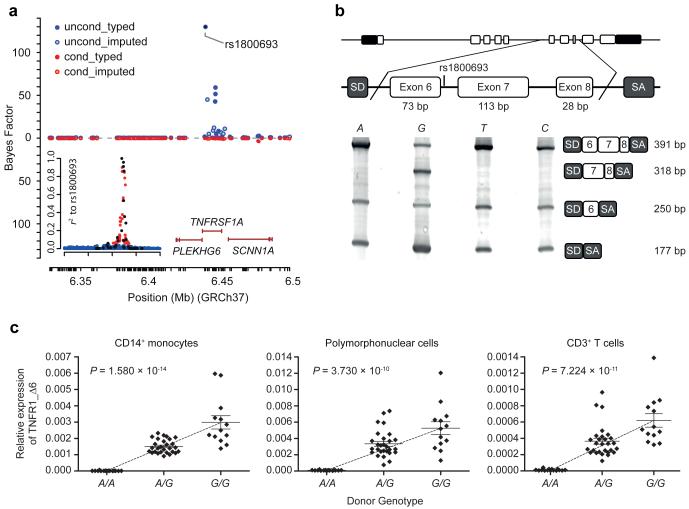
The largest MS GWAS4 reports rs1800693 as the most associated SNP in the TNFRSF1A region by over two orders of magnitude (odds ratio for risk allele = 1.12 (1.11-1.14); P = 4.1×10−14). To assess whether this SNP is mainly driving the association we examined the haplotype structure across the region in 379 individuals of European ancestry using whole genome sequence from the 1000 Genomes Project8 and we performed statistical imputation into a UK cohort of 1,853 MS patients and 5,174 controls4. Among genotyped SNPs the strongest signal is seen at rs1800693 and the variants in strongest association with this SNP were also genotyped in the study. Statistical imputation12 revealed no other variant with stronger association to MS within the region, including the previously reported3 nonsynonymous SNP rs4149584 (Supplementary Fig. 1), and analysis of association after controlling for the effect of rs1800693 removed almost all of the signal (Fig. 1a). These observations all support variation at rs1800693 as being primarily responsible for the MS association in the TNFRSF1A region.
Figure 1.
MS-associated TNFRSF1A region and rs1800693-dependent splicing. (a) MS association signal plot for the UK cohort4. Top: primary association with strongest signal at rs1800693 (blue: typed variants; light blue: variants imputed from 1000 Genomes Project). Bottom: signal conditional on rs1800693 (red: typed; orange: imputed). Inset: linkage disequilibrium to rs1800693 in 1000 Genomes Project (black: typed; untyped - red: r2≥0.05; blue: r2<0.05). (b) Rs1800693-dependent TNFR1 splicing using a minigene system. SD/SA: vector splice donor/acceptor site. (c) Relative expression of transcripts lacking exon 6 in primary human cells. Data = mean ±SEM; for A/A, A/G, G/G, n = 15, 28 and 13 donors.
To further substantiate the causality of rs1800693, we next sought to investigate the functional consequences of the variation at this SNP. As rs1800693 is proximal to the TNFRSF1A exon 6/intron 6 boundary, we hypothesized that this it may influence splicing of TNFR1 exon 6Ref13. In an in vitro minigene splicing assay, only the risk ‘G’ allele resulted in skipping of exon 6 (Fig. 1b). In primary human immune cells we found that the presence of the risk allele correlated with increased expression of transcripts lacking exon 6, with the gene dosage effect observed being consistent with the genetic effect on disease risk (Fig. 1c and Supplementary Fig. 2). Moreover, these transcripts are translated in the primary immune cells, as binding of polysomes to these transcripts demonstrated active translation (Supplementary Fig. 3). As full-length (FL-) TNFR1 expression did not vary in a genotype-dependent fashion (Supplementary Figs. 4 and 5), this indicates that the effect of rs1800693 may be specifically mediated by its influence on the generation of a functional protein isoform lacking exon 6 (Δ6-TNFR1), and we therefore investigated the properties of this molecule.
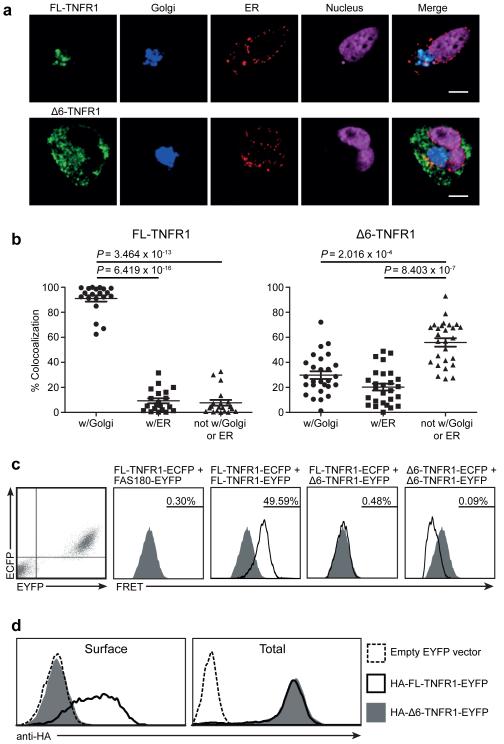
TNFR1 exon 6 skipping results in a frameshift and a premature stop codon, which translates into a protein comprising only the N-terminal 183 amino acids of FL-TNFR1, followed by a novel 45 amino acid sequence, as confirmed by tandem mass spectrometry (Supplementary Fig. 6). Consequently, Δ6-TNFR1 lacks the extracellular C-terminal portion of the fourth cysteine-rich domain (CRD) of FL-TNFR1, the transmembrane domain, and the intracellular region that is essential for appropriate subcellular localization14. While FL-TNFR1 localizes to the Golgi apparatus, Δ6-TNFR1 demonstrated a more diffuse intracellular distribution (Fig. 2), consistent with the absence of the Golgi-retention motif. Nevertheless, we observed partial subcellular colocalization of FL- and Δ6-TNFR1 (Supplementary Fig. 7), suggesting the potential for an interaction between the two isoforms. As Δ6-TNFR1 retains the pre-ligand assembly domain (PLAD/CRD1), required for TNFR1 trimerization at the cell surface14,15, it could exert a functional effect on FL-TNFR1 by associating with it to form heteromers that would therefore have modified properties. To investigate the existence of such an interaction, we used FL- and Δ6-TNFR1 proteins fused at their C-termini with fluorescent or luciferase proteins in fluorescence and bioluminescence resonance energy transfer (FRET and BRET, respectively). This demonstrated that Δ6-TNFR1 associates neither with the full-length protein nor with itself (Fig. 2c and Supplementary Fig. 8). Consistent with this and with the lack of transmembrane and cytoplasmic domains, Δ6-TNFR1 was not observed at the surface of transfected cells (Fig. 2d). Hence, as Δ6-TNFR1 cannot interact with FL-TNFR1, it is unlikely to have a direct impact on the latter, intracellularly or at the cell surface.
Figure 2.
Δ6-TNFR1 localization and analysis of isoform association. (a) TNFR1 subcellular localization. (b) FL/Δ6-TNFR1 localization analysis. Data = mean ±SEM; FL-TNFR1, n = 20, Δ6-TNFR1, n = 27 cells; scale bar: 1 μm. (c) FL/Δ6-TNFR1 association analysis. HEK 293T double-positive cells expressing ECFP/EYFP-tagged TNFR1 isoforms were analyzed for FRET to assess isoform association. FL-TNFR1-ECFP + FAS180-EYFP co-transfection was used for background signal definition with non-interacting proteins. Percentage of FRET-positive events (indicating protein association) is shown. (d) HEK 293T cells expressing HA-tagged FL- or Δ6-TNFR1-EYFP were analyzed for cell surface and total protein expression. ECFP/EYFP: enhanced cyan/yellow fluorescent protein.
To assess whether Δ6-TNFR1 has some intracellular function, regardless of its inability to associate with FL-TNFR1, we investigated TNFR1-mediated signaling in Δ6-TNFR1-transfected cells. As predicted by the absence of a death domain, which is necessary for both NF-κB-mediated signal transduction and apoptosis14,15, no significant spontaneous NF-κB signaling or TNFR1-mediated apoptosis were observed upon Δ6-TNFR1 expression (Supplementary Fig. 9 and 10). However, Δ6-TNFR1 could potentially retain some intracellular activity by accumulating in the endoplasmic reticulum (ER) and evoking a stress response15. Nevertheless, there was no evidence for increased ER-localization of Δ6-TNFR1 (Fig. 2a and Fig. 2b), or induction of the unfolded protein response in Δ6-compared with FL-TNFR1-transfected cells (Supplementary Fig. 11).
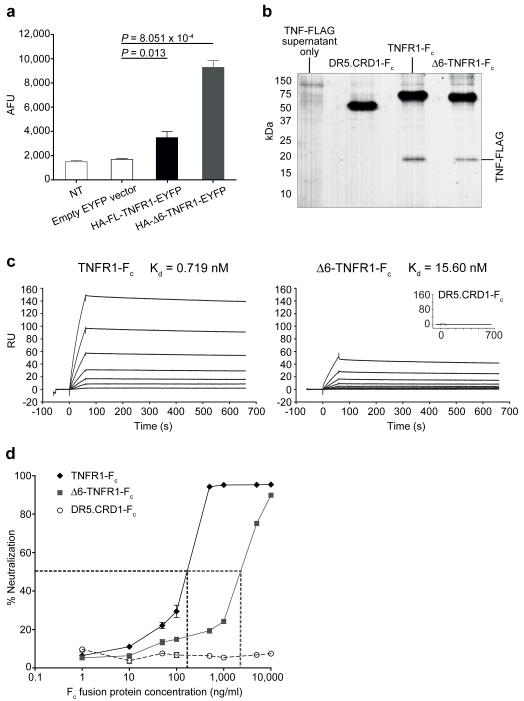
Given that no intracellular Δ6-TNFR1 activity was observed, and that this isoform has no transmembrane region and does not associate with FL-TNFR1, we hypothesized that Δ6-TNFR1 could exist as a soluble, functional molecule. Soluble TNFR1 generation has been previously described through exosomal release of full-length receptor and through metalloprotease-dependent cleavage of the FL-TNFR1 extracellular domain. Here, we demonstrate that skipping of exon 6 constitutes a novel mechanism of stable, soluble TNFR1 production: a higher level of soluble protein was found in supernatants of Δ6-compared to FL-TNFR1-transfected cells (Fig. 3a and Supplementary Fig. 12). Given the importance of determining the presence of Δ6-TNFR1 protein in primary human samples, an ELISA for this molecule was established. This required the generation of an anti-Δ6-TNFR1-specific monoclonal antibody, using a human combinatorial antibody library in conjunction with phage display, as commercially available anti-TNFR1 antibodies are not specific for the splice isoform (Supplementary Fig. 13). This ELISA allowed the detection of Δ6-TNFR1 in the sera of both healthy individuals (range = <1.25-14.8 ng/ml; n = 12) and MS patients (range = <1.25-37.3 ng/ml; n = 13) (Supplementary Fig. 14). Furthermore, soluble Δ6-TNFR1 can bind to TNF, as shown in TNF pull-down assays using TNFR1-Fc and Δ6-TNFR1-Fc fusion proteins (Fig. 3b). Using surface plasmon resonance the TNF binding ability of Δ6-TNFR1-Fc was determined to be in the nanomolar range (Kd = 15.6 nM), indicating that the isoform has a high affinity for TNF (Fig. 3c). Critically, Δ6-TNFR1-Fc proved to have a TNF antagonistic function as it can bind and neutralize the ability of TNF to signal through membrane-bound TNFR1 (Fig. 3d), with the neutralization capacity of Δ6-TNFR1-Fc being consistent with the surface plasmon resonance data.
Figure 3.
Δ6-TNFR1 solubility and TNF binding and neutralization. (a) Soluble TNFR1 detected by anti-HA ELISA using TNFR1-transfected HEK 293T supernatants. AFU: absolute fluorescence units; NT: non-transfected. (b) TNF-FLAG pulldown using TNFR1-Fc proteins. Inset: negative control for TNF binding (DR5.CRD1-Fc). (c) Surface plasmon resonance analysis of TNFR1-Fc protein TNF binding. Sensorgrams show resonance units (RU) over time and model fit for dissociation constant (Kd) derivation. (d) TNF neutralization by TNFR1-Fc proteins using HEK-Blue™ TNFα SEAP reporter cells. Dotted lines = IC50 for TNFR1-Fc and Δ6-TNFR1-Fc (158 and 3,035 ng/ml, respectively). Data = mean ±SEM; n = 3.
Our combined genetic and functional analyses strongly implicate rs1800693 as the causal SNP in the MS-associated TNFRSF1A region, with the risk allele directing an increased expression of Δ6-TNFR1. Since Δ6-TNFR1 protein is soluble and is capable of TNF antagonism, our evidence is consistent with the reported worsening of MS upon anti-TNF therapy10,11. TNF antagonists were originally used in MS patients partly because increased levels of TNF were detected in both active lesions and cerebrospinal fluid from MS patients16,17, and partly because of reports on animal models of MS where blockade of TNF prevented or delayed disease onset18,19. However, opposing findings have also been obtained from animal studies20, and the paradoxical outcomes from these pre-clinical models, as well as the precise mechanism of TNF antagonist action, are unexplained but remain an active area of research21,22.
TNF antagonists are largely beneficial in the treatment of other autoimmune conditions, including RA, psoriasis, Crohn’s disease, and ankylosing spondylitis (AS)23. In accordance with this, common genetic variants in the TNFRSF1A region have not been identified for these conditions5-7 with the exception of AS24, that has a distinct association signal compared to MS (Supplementary Fig. 15), indicating that Δ6-TNFR1 is unlikely to influence the pathology of these conditions. Notably, the MS-associated rs1800693 SNP is also associated with primary biliary cirrhosis25, but there is no controlled clinical study of TNF antagonists in this disease. Interestingly, side effects associated with the use of TNF antagonists in treating non-MS autoimmune diseases include clinical onset of MS and isolated demyelinating diseases, such as optic neuritis, which is often an early manifestation of MS9,26. These side effects are relatively rare, indicating that they may only arise in individuals with a propensity for demyelinating disease that is unmasked upon treatment.
We have identified a disease-associated genetic variant that directs increased expression of a molecule that is analogous to drugs whose adverse effects can promote or exacerbate disease. This finding has broader implications. A criticism against the potential clinical value of GWAS findings has been the modest effect on disease risk conferred by most associated genetic variants2. However, this notion does not consider that drugs targeting the same pathways as the genetic variants are likely to have a larger functional impact and thus may be clinically relevant27. We show that naturally-occurring variation and therapeutic agents can have the same target, but their effect differs in magnitude: Δ6-TNFR1 and a TNF antagonist both neutralized TNF, but the antagonist had a greater ability to do so. Therefore, our result suggests that a signal of disease association for rs1800693 could be predictive of an adverse effect of TNF antagonist treatment. In conclusion, our study shows that, through genetic and functional follow-up investigations, GWAS can serve to directly inform therapeutic choice in the treatment of a common disease.
METHODS
Statistical analysis of disease association
SNP genotype data from the UK cohort of the IMSGC and WTCCC2 MS GWAS study4 were used to localize primary association signals. SNP and sample exclusion lists were as in the original study. SNPs that were not in Hardy-Weinberg equilibrium (HWE) within either control cohort (P<0.001) were also excluded. Inferred haplotypes for 379 samples of European ancestry (EUR) from the March 2012 release of the 1000 Genomes Project were used for analysis of haplotype structure and imputation of untyped variants. Only SNPs typed successfully in case and control cohorts were used for imputation with IMPUTE Version 2Ref12, removing SNPs whose minor allele frequencies (MAF) differed by more than 0.1 between control cohorts. Association analysis with a logistic model accounting for uncertainty in genotype imputation, including gender as a covariate, was carried out using SNPTEST28, on SNPs with MAF>0.01 that had an imputation information score greater than 0.9. We used a Bayesian, additive test with the score method to measure evidence for association and we report the Bayes factor comparing the models of association and no association. We assumed the default priors of SNPTEST for the analysis, namely the standard normal for the intercept and a normal distribution with mean = 0 and standard deviation = 0.2 for the genotypes (which were coded as 0, 1 or 2 for the minor homozygous, heterozygous and major homozygous sites respectively). Gender had a normal prior as well, with mean = 0 and variance = 1000. Under the assumption of a single associated variant in the region, the Bayes factor for a variant is a proportional (under weak assumptions) to the posterior probability that it is driving the association. The conditional analysis was performed in the same way, including the genotype at rs1800693 as an additional covariate (with an additive effect).
Minigene assays
TNFRSF1A minigene constructs were made using a ~1.5 kb genomic region comprising rs1800693 and spanning the region from introns 5 to 8. Site-directed mutagenesis was performed using the QuikChange II XL Site-Directed Mutagenesis Kit (Stratagene). RNA was isolated from HEK 293T cells (ATCC) using the RNeasy Micro Kit (Qiagen) and cDNA was synthesized using the High Capacity RNA-to-cDNA Kit (Applied Biosystems), according to the manufacturer’s instructions. Minigene splicing was analyzed by PCR amplification of cDNA using primers specific to the SD and SA sites. All resulting amplification products were sequenced.
Peripheral blood donors
With informed consent and ethics committee (NRES Committee South Central – Oxford B; REC Ref. No.: 10/H0605/5;v.1) approval, blood samples were obtained from healthy white European volunteers (without self-reported autoimmune disorders); their ages ranged from 20 – 61 years (mean = 33.38 years) and 70% were females. Relapsing-remitting MS patient serum samples were obtained in accordance with the Ruhr-University Bochum (Germany) ethics committee.
Genotyping
Rs1800693 was genotyped using a TaqMan® SNP Genotyping Assay (Applied Biosystems) according to the manufacturer’s instructions. Genotyping data were scored independently by two operators to minimize error.
Isolation of human blood cell subsets
Peripheral blood mononuclear cells (PBMCs) were separated on Lymphprep™ (Axis-Shield). Polymorphonuclear cells were obtained following erythrocyte pellet lysis. CD14-positive monocytes were isolated from PBMCs using anti-CD14 MicroBeads (Miltenyi Biotec); CD3-positive T-cells were isolated using anti-CD3 MicroBeads (Miltenyi Biotec), according to the manufacturer’s instructions.
Human TNFR1 transcript quantification
TNFR1 transcript levels were measured by real-time quantitative PCR (qRT-PCR) with an ABI Prism 7900HT Sequence Detection System and analyzed using SDS v.2.4 software (Applied Biosystems). GAPDH house-keeping gene transcripts and transcripts containing exons 6 and 7 were detected using Taqman® Gene Expression Assays Hs02758991_g1 and Hs00533565_g1, respectively (Applied Biosystems). A custom primer/probe set spanning the exon 5-7 boundary was designed for specific detection of transcripts lacking exon 6. Relative expression is expressed as 2−dCT, where dCT = (cycle threshold for the transcript of interest) – (cycle threshold for the house-keeping gene transcript).
Polysome profiling assay
PBMCs obtained from peripheral blood samples of healthy volunteers were cultured in RPMI containing 10% FCS, 4 ng/ml IL-2, 50 ng/ml IL-4 and 50 ng/ml GM-CSF for 72 h, followed by stimulation with 10 ng/ml PMA and 100 ng/ml LPS for 16h. Cells were then treated with 10 μg/ml of cycloheximide, lysed and post-nuclear supernatants were centrifuged at 40,000 rpm for 2 h 15 min through 12 ml linear 20-47% (w/v) sucrose gradients containing 20 mM Tris-HCl pH 8.0, 5 mM MgCl2, 140 mM KCl. EDTA control gradients contained 20 mM Tris-HCl pH 8.0, 10 mM EDTA, 140 mM KCl. 1 ml fractions were collected and absorbance at 260 nm was used to generate chromatograms of sedimentation profiles. RNA was extracted from the fractions and cDNA was analysed by qRT-PCR.
Human immune cell TNFR1 surface expression
The anti-human monoclonal antibodies used for immunostaining were Alexa-Fluor®647 anti-CD120a/TNFR1 (clone H398; AbD Serotec), Alexa-Fluor®700 anti-CD11c, e-Fluor®450 anti-CD123 (clones 3.9 and 6h6 respectively; eBioscience), FITC and PE anti-HLA-DR (clone L243, BioLegend), PerCP-Cy5.5 anti-CD14 and Pacific Blue™ anti-CD16 (clones HCD14 and 3G8 respectively; BioLegend) and FITC Lin-1 cocktail (BD Biosciences). Alexa-Fluor®647 mouse IgG2a (AbD Serotec) was used as an isotype control for CD120a/TNFR1 staining. Data obtained were analyzed using FlowJo (Tree Star, Inc.). TNFR1 surface expression was calculated by dividing the median fluorescence intensity (MedFI) of the anti-TNFR1 staining by the MedFI of the isotype control for each sample. Each day that donor samples were evaluated, FluoroSpheres were also analyzed (Blank Beads and Calibration Beads, Dakocytomation). Data normalization was performed using the Calibration Beads to control for day-to-day variation in flow cytometer function.
Confocal microscopy
HeLa cells (ATCC) were transfected with constructs expressing, TNFR1 fluorescent fusion proteins, a DsRed2-tagged beta subunit of the signal particle receptor (SRβ) as a marker of the ER, and an ECFP-tagged trans-Golgi-resident protein N-acetylgalactosaminyltransferase-2 (GalNAc-T2) stalk region as a marker of the Golgi. TO-PRO®-3 iodide (Molecular Probes, Invitrogen) was used for nuclear staining, with staining being performed 48 h after transfection. Images were taken with an LSM510 confocal microscope (Zeiss) and processed using Zeiss LSM510 v.3.2 and Image J software. Colocalization was quantified using MetaMorph software (Molecular Devices).
FRET
FRET analysis by flow cytometry was performed as described previously29. Briefly, HEK 293T cells were transfected with the indicated ECFP and EYFP fusion protein constructs and harvested 24 h later for FRET analysis. FRET signals were examined on viable, ECFP/EYFP double-positive cells.
BRET
BRET assays were performed and analyzed as described by previously30. GFP2 and RLuc fusion protein constructs for BRET studies were generated by cloning TNFR1 sequences into pGFP2-N3 and pRLuc-N3 vectors (BioSignal Packard).
Flow cytometric analysis of TNFR1-transfected cells
HEK 293T cells were transfected with the indicated N-terminally HA-tagged TNFR1-EYFP fusion protein constructs. TNFR1 expression was measured in EYFP-positive cells using anti-HA.11 (clone 16B12; Covance) and APC-polyclonal goat anti-mouse IgG secondary antibody (BD Pharmingen™). For total anti-HA.11 staining, cells were permeabilized with ice-cold 70% ethanol for 30 min before incubation with the primary antibody.
Spontaneous NFκB signaling analysis
HEK 293T cells were transfected with the indicated TNFR1 construct along with a Renilla luciferase construct (pGL4.74, Promega) and the luc2/NFκB/RE reporter (pGL4.32, Promega). Cells were analyzed for firefly normalized to Renilla luciferase activity using the Dual Luciferase Reporter Assay System (Promega). IL-8 levels in culture supernatant were measured by immunoassay using the Human CXCL8/IL-8 Quantikine® ELISA kit (R & D Systems), according the manufacturer’s instructions.
Lentivirus preparation
The lentiviral expression plasmid pHRsinUbEm was a gift from Paul J. Lehner (University of Cambridge). TNFR1 expression plasmids were co-transfected with the vesicular stomatitis virus-G envelope plasmid pMD2.G and packaging plasmid psPAX2 (containing HIV-1 Gag and Rev) into HEK 293T cells to package lentiviral particles. Viral titers were determined by serial dilution and transduction of HEK 293T cells.
Spontaneous apoptosis analysis
For spontaneous apoptosis measurement by mitochondrial integrity analysis, HeLa cells were transfected with the indicated TNFR1-ECFP construct. Mitochondrial membrane potentials (ΔψM) were analyzed by staining cells with the mitochondrion-selective probe tetramethylrhodamine methyl ester (TMRM). The TMRM signal was measured by flow cytometry in ECFP-positive cells. For spontaneous apoptosis measurement by cell cycle analysis, HEK 293T cells were transduced with the indicated TNFR1 lentivirus at a multiplicity of infection of 50. Cell DNA content was measured by flow cytometry using propidium iodide (PI) staining. Following exclusion of debris and cell doublets, cells with a sub-diploid (G0/G1) DNA content were considered apoptotic.
UPR analysis
The XBP1 luciferase reporter assay was performed as for the NFκB/RE luciferase reporter assay but with the use of an XBP1 luciferase reporter construct and HT1080 cells (ATCC). ER stress-induced splicing of the 26-bp intron of human XBP1 was analyzed.
Cell culture supernatant TNFR1 measurement
HEK 293T cells were transfected with the indicated N-terminally HA-tagged TNFR1-EYFP fusion protein constructs. Culture supernatants or anti-TNFR1 antibodies (R&D Systems; Santa Cruz Biotechnology, Inc.) were used to coat Nunc™ microtitre plates. HA-tagged TNFR1 proteins were detected using biotinylated anti-HA.11 (clone 16B12; Covance) or biotinylated anti-TNFR1 antibodies (R&D Systems; Santa Cruz Biotechnology, Inc.) and Europium-conjugated Streptavidin (Perkin Elmer). For Western blots, HA-tagged TNFR1 proteins detected using anti-HA.11 (clone 16B12; Covance) and IRDye® 800CW goat-anti-mouse IgG secondary antibody, and visualized using an Odyssey Infrared Imaging System (LI-COR Biosciences).
Δ6-TNFR1 measurement in human serum
Endogenous Δ6-TNFR1 was measured in control and patient sera by ELISA. An anti-Δ6-TNFR1-specific monoclonal Fab antibody was generated using HuCAL® antibody technology (AbD Serotec) that employs a human combinatorial antibody library and the CysDisplay® method of phage display. The anti-Δ6-TNFR1-specific antibody was coated onto Nunc™ microtitre plates at a concentration of 2 μg/ml overnight. The anti-human TNFR1 mouse monoclonal antibody (clone 16805; R&D Systems) was used at 2 μg/ml for detection, in conjunction with Europium-conjugated anti-mouse IgG. The sensitivity limit of the assay was determined to be ~1.25 ng/ml.
Recombinant protein expression, purification and TNF immunoprecipitation
HEK 293T cells were transiently transfected with TNF-FLAG or Fc fusion constructs. Fc fusion proteins were purified from culture supernatant using protein A Sepharose (Sigma). Following Fc-fusion protein pre-incubation with TNF-FLAG, immunoprecipitation was performed using protein A Sepharose. An Fc fusion of the first cysteine-rich domain of DR5 (DR5.CRD1-Fc) was used as a negative control for TNF binding.
Tandem mass spectrometry
Purified Δ6-TNFR1-Fc protein and transfected cell supernatants were solubilized and digested with chymotrypsin (Roche). The digested protein mixture was separated with a 140 min gradient from 5 to 60 % acetonitrile (uHPLC, Proxeon) and loaded onto a LTQ-Orbitrap or a Q-Exactive mass spectrometer (Thermo Fisher). The instrument was operated in a data-dependent top10 acquisition modus. Raw data were searched using the MaxQuant software suite (version 1.2.2.0) against the complete IPI human database (v3.68, 87061 entries) with an additional entry for the Δ6-TNFR1 isoform. The fragmentation spectra were plotted and annotated by the Viewer interface of MaxQuant.
Surface plasmon resonance
Surface plasmon resonance spectroscopy was performed using the BIAcore T100 system. TNF-FLAG was directly immobilized onto anti-FLAG-coated chips from culture supernatants. Experiments were carried out using serial dilutions of the indicated purified Fc-fusion proteins. Binding curves were analyzed using BIAcore T100 Evaluation software to fit the data and determine dissociation constants (Kd), assuming 1:1 binding.
TNF neutralization assay
Varying concentrations of purified Fc fusion proteins ranging from 1-10,000 ng/ml were incubated with 5 × 104 HEK-Blue™ TNFα/IL-1β cells (Invivogen) in the presence of 25 ng/ml of TNF. The assay was performed according to the manufacturer’s instructions.
Statistical analysis of functional data
All statistical tests were performed using GraphPad Prism, GraphPad StatMate 2.0, and the R statistical software package. Regression analysis was used to test for correlations between rs1800693 genotype and TNFR1 transcript levels and TNFR1 cell surface expression in human immune cell subsets, assuming a linear genotype-to-phenotype relationship (1 d.f. F-test); age and sex were not included as covariates as for all data sets no age or sex effects were observed (P>0.05). For the TNFR1_Δ6 transcripts the slope calculated by the linear regression analysis for the CD14+ monocytes, polymorphonuclear cells and CD3+ T cells was 0.0014, 0.0026 and 0.0003, respectively. Using a Bonferroni correction for multiple testing (taking P = 0.05 and considering three independent hypotheses for the three different human immune cell subsets for RNA-level analyses), the significance threshold estimated was P = 0.017. At this significance threshold we obtained >90% power to detect the differences observed with our sample size. The percentage of TNFR1_Δ6 transcript level variation accounted for by genotype at rs1800693 was estimated by least-squares regression analysis at 67, 52 and 55% for the CD14+ monocytes, polymorphonuclear cells and CD3+ T cells, respectively. Linear regression analysis was used to quantify the correlation in TNFR1_Δ6 transcript levels between the different immune cell subsets. For all regression analyses there was no significant departure from linearity (P>0.05) as determined using a runs test. For the immune cell TNFR1 surface expression, using a Bonferroni correction (considering six independent hypotheses for six independent immune cell subsets), the significance threshold was P = 0.008; at this threshold we had >80% power to detect a 50% difference between the homozygous groups, given our sample size. For the statistical analysis of the confocal colocalization quantification, two-tailed, paired Student’s T tests were performed. For all other analyses, two-tailed, unpaired Student’s T tests were performed. For all Student’s T tests a 5% significance threshold was used.
Supplementary Material
Acknowledgements
We thank the volunteers who donated blood samples, the IMSGC and WTCCC2 for data access, A. Auton for help with association analysis data preparation, G.R. Screaton for providing TNFR1 constructs, and A. Vincent and N. Willcox for critical reading of the manuscript. Work in the authors’ laboratories is supported by the UK Medical Research Council (MRC), the European Union through grant FP7/2007-2013 (SYBILLA), the Naomi Bramson Trust (L. Fugger), and the Wellcome Trust (090532/Z/09/Z and 086084/Z/08/Z; G. McVean). A.P.G., A.H., L.L., O.A.L., and D.P. are supported by an MRC studentship, the Deutsche Forschungsgemeinschaft, a Christopher Welch Scholarship, funding from the MRC and the MS Society, and a Dorothy Hodgkin Postgraduate Award, respectively.
Footnotes
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
Author Information
The authors declare no competing financial interests.
References
- 1.Lander ES. Initial impact of the sequencing of the human genome. Nature. 2011;470:187–197. doi: 10.1038/nature09792. [DOI] [PubMed] [Google Scholar]
- 2.Goldstein DB. Common genetic variation and human traits. N Engl J Med. 2009;360:1696–1698. doi: 10.1056/NEJMp0806284. [DOI] [PubMed] [Google Scholar]
- 3.De Jager PL, et al. Meta-analysis of genome scans and replication identify CD6, IRF8 and TNFRSF1A as new multiple sclerosis susceptibility loci. Nature Genet. 2009;41:776–782. doi: 10.1038/ng.401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sawcer S, et al. Genetic risk and a primary role for cell-mediated immune mechanisms in multiple sclerosis. Nature. 2011;476:214–219. doi: 10.1038/nature10251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stahl EA, et al. Genome-wide association study meta-analysis identifies seven new rheumatoid arthritis risk loci. Nature Genet. 2010;42:508–514. doi: 10.1038/ng.582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Strange A, et al. A genome-wide association study identifies new psoriasis susceptibility loci and an interaction between HLA-C and ERAP1. Nature Genet. 2010;42:985–990. doi: 10.1038/ng.694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Franke A, et al. Genome-wide meta-analysis increases to 71 the number of confirmed Crohn’s disease susceptibility loci. Nature Genet. 2010;42:1118–1125. doi: 10.1038/ng.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.The 1000 Genomes Project Consortium A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bosch X, Saiz A, Ramos-Casals M. Monoclonal antibody therapy-associated neurological disorders. Nature Rev Neurol. 2011;7:165–172. doi: 10.1038/nrneurol.2011.1. [DOI] [PubMed] [Google Scholar]
- 10.van Oosten BW, et al. Increased MRI activity and immune activation in two multiple sclerosis patients treated with the monoclonal anti-tumor necrosis factor antibody cA2. Neurology. 1996;47:1531–1534. doi: 10.1212/wnl.47.6.1531. [DOI] [PubMed] [Google Scholar]
- 11.The Lenercept Multiple Sclerosis Study Group. The University of British Columbia MS/MRI Analysis Group TNF neutralization in MS: results of a randomized, placebo-controlled multicenter study. Neurology. 1999;53:457–465. [PubMed] [Google Scholar]
- 12.Howie BN, Donnelly P, Marchini J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet. 2009;5:e1000529. doi: 10.1371/journal.pgen.1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen M, Manley JL. Mechanisms of alternative splicing regulation: insights from molecular and genomics approaches. Nature Rev Mol Cell Biol. 2009;10:741–754. doi: 10.1038/nrm2777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schutze S, Tchikov V, Schneider-Brachert W. Regulation of TNFR1 and CD95 signalling by receptor compartmentalization. Nature Rev Mol Cell Biol. 2008;9:655–662. doi: 10.1038/nrm2430. [DOI] [PubMed] [Google Scholar]
- 15.Kimberley FC, Lobito AA, Siegel RM, Screaton GR. Falling into TRAPS - receptor misfolding in the TNF receptor 1-associated periodic fever syndrome. Arthritis Res Ther. 2007:9. doi: 10.1186/ar2197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hofman FM, Hinton DR, Johnson K, Merrill JE. Tumor necrosis factor identified in multiple sclerosis brain. J Exp Med. 1989;170:607–612. doi: 10.1084/jem.170.2.607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sharief MK, Hentges R. Association between tumor necrosis factor-alpha and disease progression in patients with multiple sclerosis. N Engl J Med. 1991;325:467–472. doi: 10.1056/NEJM199108153250704. [DOI] [PubMed] [Google Scholar]
- 18.Probert L, et al. TNFR1 signalling is critical for the development of demyelination and the limitation of T-cell responses during immune-mediated CNS disease. Brain. 2000;123:2005–2019. doi: 10.1093/brain/123.10.2005. [DOI] [PubMed] [Google Scholar]
- 19.Baker D, et al. Control of established experimental allergic encephalomyelitis by inhibition of tumor-necorsis-factor (TNF) activity within the central-nervous-system using monoclonal-antibodies and TNF receptor immunoglobulin fusion proteins. Eur J Immunol. 1994;24:2040–2048. doi: 10.1002/eji.1830240916. [DOI] [PubMed] [Google Scholar]
- 20.Liu JL, et al. TNF is a potent anti-inflammatory cytokine in autoimmune-mediated demyelination. Nature Med. 1998;4:78–83. doi: 10.1038/nm0198-078. [DOI] [PubMed] [Google Scholar]
- 21.Taoufik E, et al. Transmembrane tumour necrosis factor is neuroprotective and regulates experimental autoimmune encephalomyelitis via neuronal nuclear factor- {kappa}B. Brain. 2011;134:2722–2735. doi: 10.1093/brain/awr203. [DOI] [PubMed] [Google Scholar]
- 22.Brambilla R, et al. Inhibition of soluble tumour necrosis factor is therapeutic in experimental autoimmune encephalomyelitis and promotes axon preservation and remyelination. Brain. 2011;134:2736–2754. doi: 10.1093/brain/awr199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Feldmann M, Steinman L. Design of effective immunotherapy for human autoimmunity. Nature. 2005;435:612–619. doi: 10.1038/nature03727. [DOI] [PubMed] [Google Scholar]
- 24.Evans DM, et al. Interaction between ERAP1 and HLA-B27 in ankylosing spondylitis implicates peptide handling in the mechanism for HLA-B27 in disease susceptibility. Nature Genet. 2011;43:761–767. doi: 10.1038/ng.873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mells GF, et al. Genome-wide association study identifies 12 new susceptibility loci for primary biliary cirrhosis. Nature Genet. 2011;43:329–332. doi: 10.1038/ng.789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Compston A, Coles A. Multiple sclerosis. Lancet. 2008;372:1502–1517. doi: 10.1016/S0140-6736(08)61620-7. [DOI] [PubMed] [Google Scholar]
- 27.Altshuler D, Daly MJ, Lander ES. Genetic mapping in human disease. Science. 2008;322:881–888. doi: 10.1126/science.1156409. [DOI] [PMC free article] [PubMed] [Google Scholar]
For ONLINE METHODS
- 28.Marchini J, Howie B, Myers S, McVean G, Donnelly P. A new multipoint method for genome-wide association studies by imputation of genotypes. Nature Genet. 2007;39:906–913. doi: 10.1038/ng2088. [DOI] [PubMed] [Google Scholar]
- 29.Chan FK, et al. Fluorescence resonance energy transfer analysis of cell surface receptor interactions and signaling using spectral variants of the green fluorescent protein. Cytometry. 2001;44:361–368. doi: 10.1002/1097-0320(20010801)44:4<361::aid-cyto1128>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- 30.James JR, Oliveira MI, Carmo AM, Iaboni A, Davis SJ. A rigorous experimental framework for detecting protein oligomerization using bioluminescence resonance energy transfer. Nature Methods. 2006;3:1001–1006. doi: 10.1038/nmeth978. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.