Abstract
The biosynthetic pathways for patellamide and related natural products have recently been studied by structural biology. These pathways produce molecules that have a complex framework and exhibit a diverse array of activity due to the variability of the amino acids that are found in them. As these molecules are difficult to synthesize chemically, exploitation of their properties has been modest. The patellamide pathway involves amino acid heterocyclization, peptide cleavage, peptide macrocyclization, heterocycle oxidation and epimerization; closely related products are also prenylated. Enzyme activities have been identified for all these transformations except epimerization, which may be spontaneous. This review highlights the recent structural and mechanistic work on amino acid heterocyclization, peptide cleavage and peptide macrocyclization. This work should help in using the enzymes to produce novel analogs of the natural products enabling an exploitation of their properties.
Introduction
Small molecules with potent biological properties are commonly isolated from bacteria. Analysis of well established rules for anthropogenic pharmaceuticals [1] indicates that in comparison, natural products tend to have more stereo centres and nitrogen atoms but are, in general, not wildly different [2 and 3]. Natural products that have some undesirable properties as medicines, but they can be improved by chemical modification. Natural products, including modified variants, are a major component in the pharmaceutical armory for treating diseases ranging from bacterial infection to cancer to immune suppression [4]. Synthetic biology promises, amongst other deliverables, the ability to tailor enzymes in biosynthetic pathways to create natural product variants with desirable properties in the quantities required for drug development [5].
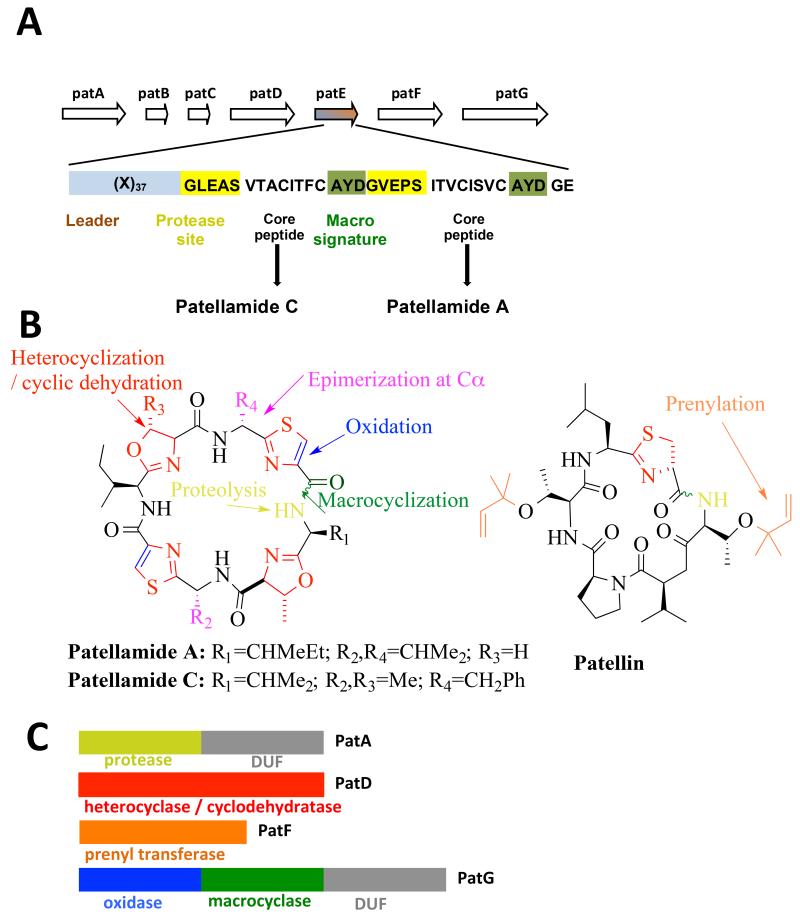
Peptide based natural products are particularly attractive from a chemical point of view; amino acids share a common standard in connectivity (the amide bond) with an almost infinitely configurable element (the side chain). One can thus ‘dial’ in chemical and structural properties into a shared basic design. Crucially, in the same way as no two proteins need to share any biological property, peptides by virtue of their different side chains can have divergent properties. The cyanobactin family of ribosomally synthesized and post-translationally modified peptides (RiPPs) are peptide macrocycles that exemplify this diversity, ranging from six to over 20 residues, with divergent sequences and very different biological properties including P-gp inhibition, cytotoxicity, immunomodulation, antifungal, antibacterial and antiviral properties [6]. Macrocyclic peptides are particularly appealing as they are intrinsically resistant to protease degradation and several cross membranes [7]. There is significant interest in their biosynthesis with a view to exploiting this class of molecule for novel drugs [8•, 9 and 10]. In this review, we use the example of the patellamide (an eight-residue macrocyclic cyanobactin) [11] to structure our discussion. In this pathway a single ribosomal seventy one-residue precursor peptide, PatE, gives rise to two eight-residue macrocycles; patellamide A and patellamide C [11] (Figure 1a). PatE contains two eight-residue core peptides and each is converted to the corresponding patellamides (Figure 1b) by enzyme action. Within PatE each core peptide is flanked by a conserved five-residue protease signature (N-terminal), a conserved three-residue macrocyclization signature (C-terminal) and in addition PatE has a thirty seven-residue leader peptide at N-terminus [11] (Figure 1a). During synthesis the peptide bonds between the core peptide and its flanking regions must be cut and the ends of the core peptide joined (Figure 1b). The final products contain oxazolines (derived from the heterocyclization of serine/threonine) and thiazoles (oxidized form of thiazoline which is derived from the heterocyclization of cysteine) (Figure 1b). The two residues adjacent to the thiazoles are epimerized from an l-configuration to d-configuration (Figure 1b). The functions of the enzymes that convert PatE into patellamides were first assigned by sequence analysis (Figure 1c) [11]. There are many closely related pathways in other marine organisms, such as the trunkamide and patellin (Figure 1b) biosynthetic pathway, that utilise biosynthetic enzymes which are very similar to those of the patellamide pathway [6, 9 and 11]. Moreover, many of the chemical reactions that are catalyzed by enzymes in the patellamide pathway occur in the biosynthesis of other natural products that are unrelated to cyclic peptides. We herein discuss insights from these other pathways under the corresponding chemical transformations observed in the patellamide system.
Figure 1.
(a) The patellamide gene cluster contains genes patA to patG, coding for proteins PatA to PatG. The PatE protein contains: (1) a 37 residue leader and (2) two core peptides, which are processed to give patellamides C and A [11]. The core peptides are flanked at their N and C termini
(b) the final natural product patellamides C and A showing the chemical transformations during post-translational processing. The closely related patellin which is prenylated is shown
(c) the enzymes which tailor the core peptide. PatG encodes two functions and in addition has a DUF. PatA encodes the protease and also posses the same DUF.
Heterocyclization
The heterocyclase (or cyclodehydratase) class of enzymes modify cysteine residues (and in some cases serine and threonine residues, too) within the context of the core peptide(s), to create thiazolines (or oxazolines) (Figure 1b) and eliminate water [12]. Patellamides A and C contain four heterocycles each and these five membered rings profoundly change the chemistry and flexibility of the peptide [13]. Their selective introduction is a powerful tool to tune the molecular shape and activity of not only peptide macrocycles, but also linear peptide natural product families such as the antibacterial microcin peptides [14]. The microcin ‘Trojan horse’ antibiotic peptide MccC7 has a C-terminal phosphoramidate that is linked to adenosine. During the biosynthesis of the peptide an intermediate with a C-terminal five membered succinamide ring is synthesized by heterocyclization of the C-terminal Asn residue [15]. The structure of the enzyme responsible, MccB, was revealed to have two domains. The larger of the two (the C-terminal) adopts a classic adenylase superfamily fold whilst the fold of the smaller (N-terminal) was novel [15]. MccB activates the substrate peptide carboxy terminus by first converting it to an adenylate then the amide of the side chain of Asn displaces AMP to give the succinamide five membered ring. MccB catalyzes a second adenylation reaction that adds adenosine to the peptide. Domain 1 of MccB acts as a clamp holding the peptide substrate for processing. More recently thiazole/thiazoline-containing (modified) microcins [16], collectively known as TOMMs have been studied [17•• and 18••]. Enzymatic characterization revealed that the catalytic activity required ATP and that full activity was only observed with two proteins BalhC and BalhD, although BalhD was shown to catalyze the reaction on its own, albeit very slowly [17••]. Biochemical data established that the protein functioned as a kinase, phosphorylating the hemiorthoamide intermediate formed by cysteine attack at the preceding carbonyl [17••]. The formation of hemiorthoamide intermediates is commonplace in food chemistry where thiazolines and other five membered rings occur in heated proteins [19]. By analogy then, it could be that the irreversible step is actually the removal of water (either by heat or by formation of a phosphorous oxygen bond followed by elimination of phosphate) and that the reversible formation of hemiorthoamides in a protein may in energetic terms be a relatively low barrier process.
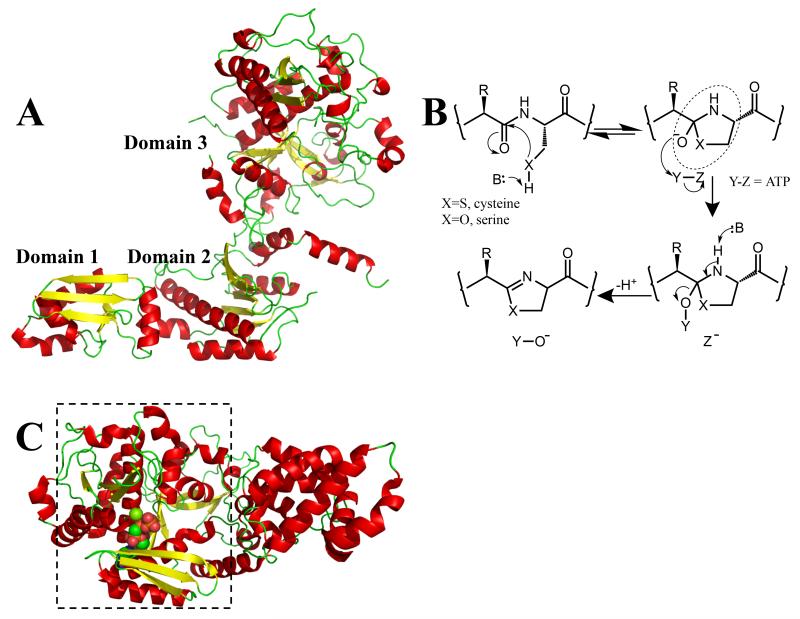
In the patellamide pathway, the enzyme PatD catalyzes the formation of multiple thiazolines (from cysteine) and oxazolines (from serine and threonine); the closely related enzyme TruD (from the trunkamide pathway) only processes cysteines with both enzymes operating in an ATP dependent and processive manner [20 and 21]. It has been shown that selenocysteine (a rare but known amino acid) is processed to a selenazoline by both PatD and TruD [22], making it possible that natural products could be discovered that contain selenazoline or selenazole modifications. The crystal structure of TruD [23••] shows it to be a three-domain protein (Figure 2a). Domains 1 and 2 are, in structural terms, essentially identical to the MccB protein [15 and 23••] with both proteins sharing a structural zinc bound in the second domain. BalhC has sequence homology to these two domains, whilst BalhD shares homology with the third domain of TruD, which is structurally novel [23••]. Thus PatD-like enzymes can be considered fusions of BalhC and BalhD.
Figure 2.
(a) The structure of TruD (cartoon with α-helices in red; β-sheets in yellow; β-turns in green) shows it has three domains [23••]. Domains 1 and 2 are very similar to a known adenylase MccB [15] and to BalhC from the TOMM pathway. Domain 3 has a novel structure and is related in sequence to BalhD in the TOMM pathway [17••]
(b) heterocyclization of amino acids (cysteine X = S, selenocysteine X = Se, serine/threonine X = O) to yield azoline rings. TOMMS were first predicted to go through a hemiorthamide intermediate (circled). It is likely that in the patellamide biosynthetic pathway, heterocyclization goes through the same central intermediate. In the TOMM system [17••] ATP is used to phosphorylate (Y = ADP, Z = PO43−) the intermediate, whereas in the patellamide pathway the intermediate was proposed to be adenylated (Y = AMP, Z = P2O74−) [23••]. Either mechanism would be irreversible and would promote elimination yielding the azoline. There is a third possibility, pyrophosphorylation (Y = P2O74−, Z = AMP), which although rare, has precedent and is consistent with much of the published data;
(c) a cartoon representation of the structure of the YcaO domain [18••] from E. coli has located the nucleotide binding site. This domain is boxed and is structurally similar to domain 3 of TruD [23••], shown in Figure 2a. The binding of ATP to domain 3 disfavors the adenylase mechanism previously proposed [23••]. The protein is colored as Figure 2a, the nucleotide analog AMP-PNP is shown in space fill with carbon atoms in green, nitrogen atoms in blue and oxygen atoms in red.
NMR and assay data show that TruD produces AMP and pyrophosphate (PPi), the hall mark of an adenylase or less commonly a pyrophosphorylase mechanisms [23••] (Figure 2b). However, the loops that form the ATP binding site in domain 2 of MccB [15] are absent in the TruD structure (and in PatD-like sequences more generally) [23••], precluding ATP binding in the manner seen in other adenylating enzymes [23••]. An adenylase rather than a kinase mechanism was proposed as the most likely for TruD with the data in hand [23••].
TruD was shown to have a preferred order of reactivity, proceeding from the C-terminus [23••]. The processivity of the enzyme required the leader peptide to be attached to the core peptide and similar observations had previously been made for the BalhC/D system [24• and 25•]. All the heterocyclase enzymes are promiscuous and tolerate diversity in chemical and structural nature of amino acids, which flank the target cysteine (or threonine, serine) [20, 21, 24• and 25•].
A very recent paper has reported the structure of the YcaO domain from Escherichia coli with a bound nucleotide [18••] (Figure 2c). The product and substrate of YcaO are unknown but the YcaO domain has the same structure as the third domain of TruD and shares sequence homology with BalhD [18••]. The residues that ligate the nucleotide are also conserved, essentially ruling out the adenylase domain of TruD as the nucleotide binding site [18••] and disfavouring an adenylase mechanism for TruD. The key chemical step in both TruD and BalhD is the irreversible modification of the hemiorthoamide and it is possible, but perhaps unlikely, that one operates by a pyrophosporylation and the other by a kinase mechanism (Figure 2b). It has been pointed out that adenylation and phosphorylation share chemical similarities [26] and in one case similar enzyme folds [27]. It is worth noting that in the absence of substrate, YcaO produced AMP and PPi [18••], suggesting more work is needed to resolve the mechanism(s) in detail.
Peptide cleavage and macrocyclization
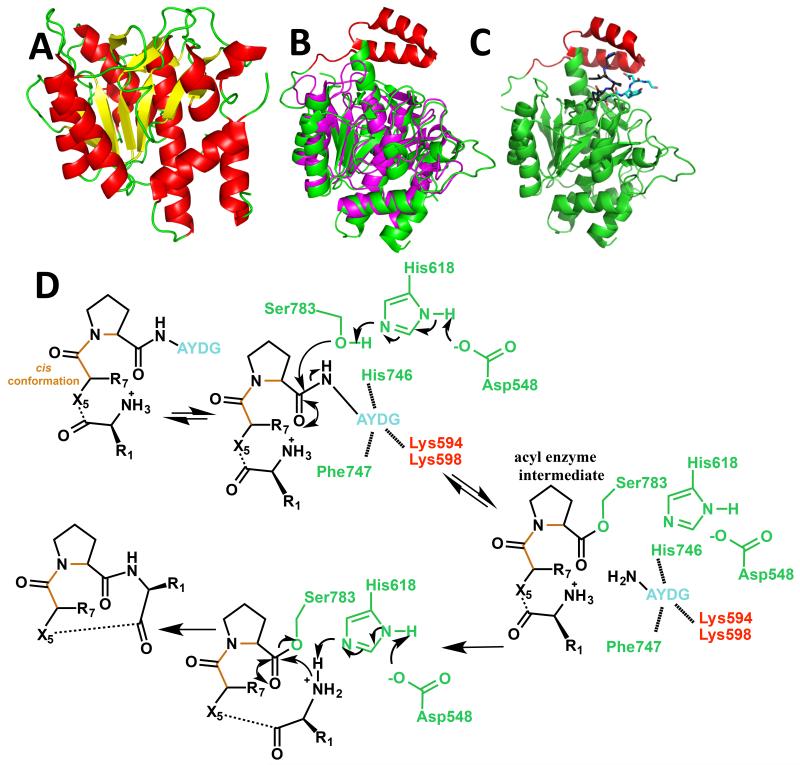
In the patellamide pathway, two domains, one from the PatG protein and one from the PatA protein, possess sequences matching the classic subtilisin fold [11]. However, PatA catalyzes the cleavage of the precursor peptide at the N-terminal protease site, removing the leader peptide (Figure 1a). The PatG domain catalyzes the formation of the macrocyclic peptide (simultaneously removing the C-terminal protease signature) [11]. The structure of the protease domain from PatA without substrate (Fig. 3ai) was determined independently by two groups [28• and 29••]. The apo structure of a close PatA homolog, PagA, was also reported [29••]. PatA possesses a classic serine protease fold but in both native PatA structures, the catalytic triad is perturbed from the classic arrangement [28• and 29••]. In a PatA variant (catalytic serine mutated to alanine) [28•] and in PagA [29••] the key histidine of the triad adopts its conventional conformation. It would seem unlikely that mis-setting of the catalytic triad in PatA is a consequence of the lack of the C-terminal domain, as similar protease activity is observed in both forms [29••]. This leaves two explanations for the misalignment: either crystal packing or missing protein-protein interactions (from the substrate). Unfortunately the PagA structure, which has the correct constellation of active site residues, does not resolve the argument, as it has extensive crystal contacts around its active site which could mimic some protein-protein interactions [29••]. Cleavage of PatE by PatA occurs at a very slow rate (orders of magnitude slower than subtilisin) [28• and 29••] and it has been suggested a slow protease might be important in helping regulate the timing of the multiple chemical steps during processing [29••].
Figure 3.
(a) Crystal structure of the PatA protease domain [28• and 29••] in cartoon representation (colored as in Figure 2a);
(b) structural alignment of the PatG macrocyclase domain [30••] (subtilisin fold in green and ‘capping’ or ‘macrocyclization’ insert in red) with Akt1, a subtilisin protease (magenta);
(c) crystal structure of the PatG macrocyclase domain in complex with substrate peptide VPAPIPFPAYDG (only PIPFPAYDG ordered) [30••]. The cartoon of the protein is colored as 2b, the peptide is shown in stick with carbon atoms of the core peptide in black, carbon atoms of the macrocyclization signature in cyan, nitrogen atoms blue and oxygen atoms red; and
(d) mechanism for PatG macrocyclase domain catalyzed C-terminal cleavage of the core peptide to form an acyl enzyme intermediate and subsequent decomposition of this intermediate by the incoming amino terminus of the core peptide to form the macrocycle [30••]. The core peptide is shown in black, the macrocyclisation signature of the substrate is shown in magenta. The cis peptide is shown in orange. The protein residues are colored in green for those in the subtilisin fold and red for those in the insert.
The crystal structure of the macrocyclase domain of PatG (Figure 3aii), determined by two laboratories [29•• and 30••], shows it does indeed possess a subtilisin fold (thus structurally similar to PatA [28• and 29••]). PatG does however possess a unique and additional structural feature: two helices that are inserted between a helix and a strand of the subtilisin fold (Figure 3b). These helices, which have been termed as the ‘capping’ or ‘macrocyclization’ insert, sit directly above the catalytic site. Sequence alignment shows that the entire macrocyclase superfamily has an insertion in this region but neither length nor the sequence of the insertions have any obvious conservation. For example, the insertion in one ortholog (TenG) is eight residues longer whilst another (AnaB90) is four residues shorter than PatG [30••]. As yet, no relationship between the insertion and the size or composition of the macrocycle has been reported. Protease substrate residues are conventionally labelled from the N to C terminus as P5 P4 P3 P2 P1–P1′ P2′ P3′ P4′ P5′. In general, subtilisins bind their substrates as a β-strand with the protein–protein recognition occurring N-terminal to the scissile bond (P1–P1′), that is, residues P5 to P1 of the substrate peptide; this is exemplified by PatA (Figure 1a). This P5 to P1 binding cleft is blocked in PatG by changes in the loop structure and insertion of large hydrophobic residues; these changes are conserved in PatG homologs [30••]. As a result PatG actively prevents substrates from binding with an extended β-strand conformation for residues P5 to P1 [30••]. The crystal structure of a complex between an inactive PatG mutant (Figure 3c) and a peptide substrate (P8-VPAPIPFPAYDG-P4′) has been determined [30••]. The N-terminal region of the substrate curves away from the protein active site (P8–P6 (VPA) are disordered). The P5 (P) and P4 (I) residues do not make any contact with the protein, the P3 (P) residue makes only a few contacts and the aromatic side chain of residue P2 (F) sits in a shallow pocket. The lack of contact is consistent with the promiscuity of the macrocyclase domain since it is largely insensitive to the identity of residues within the core peptide [31]. The complex reveals a number of very specific interactions between the C-terminal region of the substrate P1′ P2′ P3′ (AYD) and residues from the macrocyclization/capping insert [30••]. PatG stands as a mirror opposite of subtilisin where recognition is N-terminal and promiscuity C-terminal. For a PatG substrate, there is one additional requirement: a proline residue or a thiazoline at P1 [31]. The PatG substrate complex shows that the proline of the substrate adopts a cis configuration of the P2–P1 substrate bond [30••]. A trans peptide (β-strand) configuration would result in clashes with the macrocyclase due to the conserved insertions and thus only substrates capable of adopting a cis (proline) or cis-like (thiazoline) configuration will bind to PatG, rationalizing the requirement at P1. PatG does form an acyl enzyme intermediate with its peptide substrate [30••] and a mechanism in which this intermediate is decomposed by attack of the free amino terminus of the core peptide has been proposed (Figure 3d). An elegant study has shown that the PatG acyl enzyme intermediate can be decomposed with short peptides [29••]; in essence demonstrating that PatG can function as a true transpeptidase. The rate of macrocyclization by PatG is slow [29•• and 30••], but this does not necessarily imply the enzyme is a poor catalyst; the uncatalyzed rate for peptide macrocyclization has not been reported and in our hands no spontaneous macrocyclization was observed [30••].
Prenyl transferases, DUF and oxidase
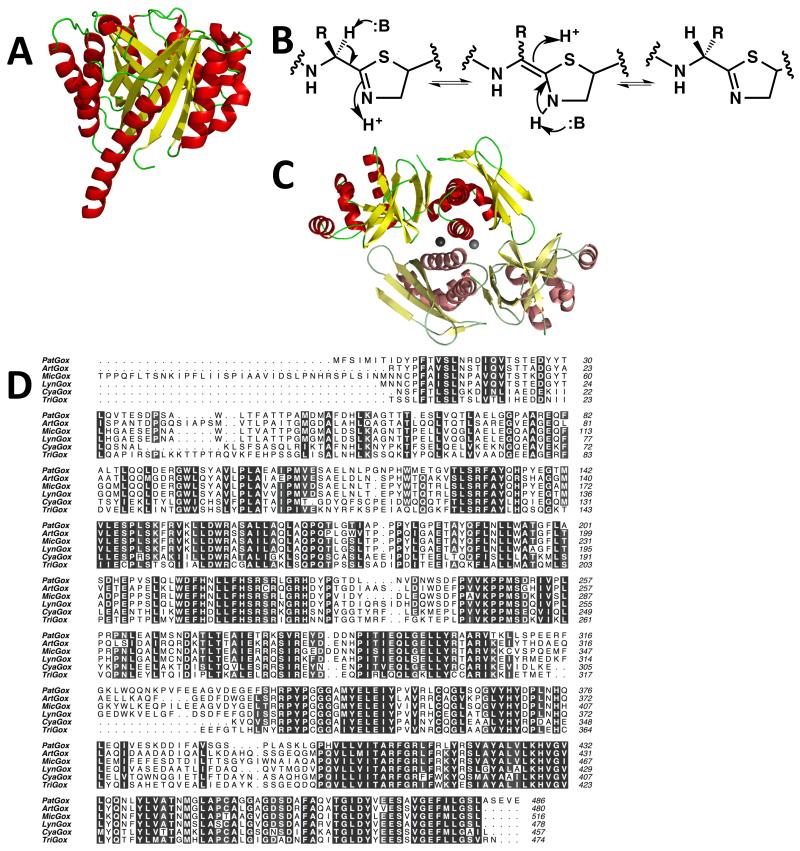
Although patellamides A and C are not prenylated, the related patellin (Figure 1b) and trunkamide natural products are (Figure 1b) [32, 33, 34 and 35]. The prenyl transferases from patellamide like pathways in Lyngbya aestuarii (LynF) and Prochloron spp. (TruF1) have been characterized biochemically [35 and 36]. LynF prenylates a tyrosine residue on the phenol group to generate the corresponding allyl phenyl ether intermediate which then spontaneously undergoes a Claisen re-arrangement to yield the ortho-substituted phenol [36]. On the other hand, TruF prenylates serine and threonine residues on the hydroxyl side chain [35] (Figure 1b). The structure of PatF from the patellamide pathway reveals that it does indeed adopt the classic TIM barrel fold (Figure 4a) seen in other prenyl transferases but no enzymatic activity was detected [37], consistent with the lack of prenylation of patellamides A and C. A detailed analysis of PatF reveals that two residues, His125 and Met136, are located at positions that are usually occupied by conserved and catalytically active Asp and Lys residues in a known prenyl transferase (dimethylallyl tryptophan synthase from Aspergillus fumigatus) [38], rationalizing the inactivity of PatF. Other cyanobactin prenyl transferases possess the Asp and either a Lys or Arg at these key positions [37]. Whether PatF has another role in the synthesis of patellamides A and C, and if not why PatF is conserved in the gene cluster, remains unknown.
Figure 4.
(a) The structure of PatF [37] represented as Figure 2a. Although the structure is that of a prenyl transferase, the protein is inactive. In other biosynthetic pathways, homologs of PatF have been shown to be active [35 and 36]
(b) a mechanism for the epimerization at the carbon adjacent to a thiazoline. The, study of a chemically related compound has led to the suggestion that in the patellamide pathway this is a spontaneous reaction [40]. However, an enzyme catalyzed processes has not been formally ruled out nor has epimerization adjacent to a thiazole been excluded
(c) cartoon representation of the structure of the PatG C-terminal DUF [39] reveals it to be a dimer, linked by zinc atoms. One monomer is colored as Figure 2a, the second monomer has duller colors
(d) the oxidase domain of PatG is highly conserved in other patellamide-like biosynthetic pathways with PatG homologs. Clustal Omega sequence alignment of the N-terminal oxidase domains of cyanobactin proteins PatG (Prochloron didemini), ArtG (Arthrospira platensis), MicG (Microcystis aeruginosa NIES-298), LynG (Lyngbya sp. PCC 8106), CyaG (Cyanothece sp. PCC7822) and TriG (Trichodesmium erythraeum IMS101). Figure created using ALINE.
Both PatA and PatG are multi-domain proteins and both contain a C-terminal domain of unknown function (DUF) [11] (Figure 1c). The sequences of the two DUF domains are homologous and are found in PatA and PatG homologs of other patellamide like biosynthetic clusters [39]. A number of roles for the DUF domain have been considered including epimerization of the two residues in patellamide A and C (Figure 1b). The pKa of the Cα proton will be lowered by the adjacent thiazoline due to resonance stabilization (Figure 4b); the loss of aromaticity in the thiazole that would occur upon stabilization of the negative charge would be expected to raise the pKa. For this reason, it is commonly assumed epimerization follows heterocyclization and precedes oxidation, but this has not been established. Epimerization has been proposed to be chemically spontaneous [40], if true, DUF domains may have no role in patellamide biosynthesis. The question then would be why the DUF domains are so well conserved in patellamide like biosynthetic gene clusters? The structure of the DUF domain from PatG has been determined and the protein is found as a dimer with two zinc ions at the interface [39] (Figure 4c). The zinc ion binding residues are not conserved in other DUF domains and since the dimerization depends on the zinc, it is unclear whether other domains are dimers [39]. The fold of the DUF domain is novel but gave no clue as to a potential function [39]. It was established that the DUF domain does not bind linear precursor peptides, nor linear precursor peptides with four heterocycles [39]. Further experiments are needed to establish whether the DUF domain binds the macrocycle (or perhaps the core peptide alone). Secondly, an evaluation of the chemical spontaneity or otherwise of epimerization in patellamide C is also needed before a final assessment of the DUF domain activity can be made.
The only domain in the patellamide biosynthetic pathway without a structure is the oxidase (dehydrogenase) domain of PatG (Figure 1c). The domain is conserved in PatG homologs from patellamide-like pathways (Figure 4d). Sequence analysis identifies the domain as FMN dependent and a recent biochemical analysis of the related thiazoline oxidase (which is a separate protein) in the microcin pathway has been reported [41]. This study showed FMN was bound and identified residues crucial for activity [41]. The domain belongs to the class of FMN oxidases most closely resembling the structure 3EO7 (determined by www.jcsg.org). Although the TOMM and patellamide enzymes are related in sequence, the basis of substrate recognition remains unclear since microcin is linear and patellamides are macrocycles. A recent study has shown that one homolog of the PatG oxidase domain can oxidize both linear and macrocyclic thiazoline containing substrates [42••], whereas another homolog appeared only to operate on a macrocyclic substrate [42••].
Future prospects.
The patellamide biosynthetic pathway is comprised of multiple different chemical steps (thus enzymes) and this has made for particularly interesting mechanistic and structural studies. The interest in this specific biosynthetic pathway is driven by the potential utility of the compounds that can be made. Several studies have reported the in vitro and in vivo biotechnological application of the pathway to make novel compounds [42••, 43• and 44•]. In one in vitro approach the slow protease step catalyzed by PatA has been replaced by engineering of the PatE so that trypsin can be used instead; this step has greatly accelerated the production and increased the yield of patellamide analogs [42••]. Despite all these efforts, the order of the epimerization, macrocyclization and oxidation remains to be experimentally verified. Important questions also remain to be answered about the relationship between the macrocyclase sequence with respect to macrocycle ring size, the mechanism of heterocyclization, the precise substrate for oxidation and the nature of epimerization. Answering these questions will not only enhance our knowledge of the chemistry of the enzymatic reactions found in the patellamide pathway (and other natural product biosynthetic pathways), but should allow a greater diversity of analogs to be created both in vivo and in vitro. Two principal bottlenecks in the in vitro production of analogs remain [42••]. Firstly the macrocyclase is slow and is the rate-limiting step in the whole process. Secondly the requirement for the leader peptide for introduction of heterocycles means that PatE can only be economically produced by expression in a heterologous host, which limits the chemical diversity within the final patellamide analog. Resolving these issues would transform the prospects for large-scale production of very diverse analogs. PatD has been shown to be activated in trans by addition of exogenous leader [43•] and this suggests there is scope to overcome at least one of the bottlenecks.
Highlights.
Substantial progress has been made in the structural biology of patellamide biosynthesis
The enzyme activities are widely distributed in the cyanobactin superfamily.
Activities include macrocyclization and heterocyclization.
These enzymes have potential for biotechnology through engineering.
Acknowledgements
This work was supported by grants from the ERC339367 (JHN and MJ) and BBSRCBB/K015508/1 (JHN and MJ).
Footnotes
No conflict
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
• of special interest
•• of outstanding interest
- 1.Lipinski CA, Lombardo F, Dominy BW, Feeney PF. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv Drug Deliv Rev. 2001;46:3–26. doi: 10.1016/s0169-409x(00)00129-0. [DOI] [PubMed] [Google Scholar]
- 2.Lee M-L, Schneider G. Scaffold architecture and pharmacophoric properties of natural products and trade drugs: application in the design of natural product-based combinatorial libraries. Journal of combinatorial chemistry. 2001;3:284–289. doi: 10.1021/cc000097l. [DOI] [PubMed] [Google Scholar]
- 3.Ganesan A. The impact of natural products upon modern drug discovery. Curr Opin Chem Biol. 2008;12:306–317. doi: 10.1016/j.cbpa.2008.03.016. [DOI] [PubMed] [Google Scholar]
- 4.Ngo LT, Okogun JI, Folk WR. 21st century natural product research and drug development and traditional medicines. Nat Prod Rep. 2013;30:584–592. doi: 10.1039/c3np20120a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zadran S, Levine RD. Perspectives in metabolic engineering: understanding cellular regulation towards the control of metabolic routes. Appl Biochem Biotechnol. 2013;169:55–65. doi: 10.1007/s12010-012-9951-x. [DOI] [PubMed] [Google Scholar]
- 6.Sivonen K, Leikoski N, Fewer DP, Jokela J. Cyanobactins-ribosomal cyclic peptides produced by cyanobacteria. Appl Microbiol Biotechnol. 2010;86:1213–1225. doi: 10.1007/s00253-010-2482-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Joo SH. Reviews: Cyclic Peptides as Therapeutic Agents and Biochemical Tools. Biomolecules & Therapeutics. 2012;20:19–26. doi: 10.4062/biomolther.2012.20.1.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tianero MD, Donia MS, Young TS, Schultz PG, Schmidt EW. Ribosomal route to small-molecule diversity. J Am Chem Soc. 2012;134:418–425. doi: 10.1021/ja208278k. [• This paper is a powerful illustration of how the patellamide pathway can be harnessed to make novel and diverse molecules.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schmidt EW, Donia MS. Life in cellulose houses: symbiotic bacterial biosynthesis of ascidian drugs and drug leads. Curr Opin Biotechnol. 2010;21:827–833. doi: 10.1016/j.copbio.2010.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kotz J. Bringing macrocycles full circle. SciBX: Science-Business eXchange. 2012;5:11. [Google Scholar]
- 11.Schmidt EW, Nelson JT, Rasko DA, Sudek S, Eisen JA, Haygood MG, Ravel J. Patellamide A and C biosynthesis by a microcin-like pathway in Prochloron didemni, the cyanobacterial symbiont of Lissoclinum patella. Proc Natl Acad Sci U S A. 2005;102:7315–7320. doi: 10.1073/pnas.0501424102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Arnison PG, Bibb MJ, Bierbaum G, Bowers AA, Bugni TS, Bulaj G, Camarero JA, Campopiano DJ, Challis GL, Clardy J, et al. Ribosomally synthesized and post-translationally modified peptide natural products: overview and recommendations for a universal nomenclature. Nat Prod Rep. 2013;30:108–160. doi: 10.1039/c2np20085f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.In Y, Doi M, Inoue M, Ishida T, Hamada Y, Shioir T. Molecular conformation of patellamide A, a cytotoxic cyclic peptide from the ascidian Lissoclinum patella, by X-ray crystal analysis. Chem Pharm Bull. 1993;41:1686–1690. doi: 10.1248/cpb.41.1686. T. [DOI] [PubMed] [Google Scholar]
- 14.Duquesne S, Destoumieux-Garzon D, Peduzzi J, Rebuffat S. Microcins, gene-encoded antibacterial peptides from enterobacteria. Nat Prod Rep. 2007;24:708–734. doi: 10.1039/b516237h. [DOI] [PubMed] [Google Scholar]
- 15.Regni CA, Roush RF, Miller DJ, Nourse A, Walsh CT, Schulman B. How the MccB bacterial ancestor of ubiquitin E1 initiates biosynthesis of the microcin C7 antibiotic. EMBO J. 2009;28:1953–1964. doi: 10.1038/emboj.2009.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Melby JO, Nard NJ, Mitchell DA. Thiazole/oxazole-modified microcins: complex natural products from ribosomal templates. Curr Opin Chem Biol. 2011;15:369–378. doi: 10.1016/j.cbpa.2011.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dunbar KL, Melby JO, Mitchell DA. YcaO domains use ATP to activate amide backbones during peptide cyclodehydrations. Nat Chem Biol. 2012;8:569–575. doi: 10.1038/nchembio.944. [•• This is a detailed enzymatic characterization of a cysteine heterocyclase, the paper suggests that BahlD operates by kinase like mechanism. The use of 18O labeled substrate coupled to 31P NMR to probe mechanism showed the oxygen is transferred to phosphate.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dunbar KL, Chekan JR, Cox CL, Burkhart BJ, Nair SK, Mitchell DA. Discovery of a new ATP-binding motif involved in peptidic azoline biosynthesis. 2014:823–829. doi: 10.1038/nchembio.1608. [•• The structure of the YcaO domain (Figure 2c) shows a novel ATP binding site and since the structure of this domain matches domain 3 of TruD (Figure 2A), it seems ATP will bind here also. This structure disfavors the adenylation mechanism proposed for TruD.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Paulus T, Riemer C, Beck-Sickinger AG, Henle T, Klostermeyer H. Protein-backbone-modifications: Formation of imidazolines. European Food Research and Technology. 2006;222:242–249. [Google Scholar]
- 20.McIntosh JA, Donia MS, Schmidt EW. Insights into heterocyclization from two highly similar enzymes. J Am Chem Soc. 2010;132:4089–4091. doi: 10.1021/ja9107116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McIntosh JA, Schmidt EW. Marine molecular machines: heterocyclization in cyanobactin biosynthesis. Chembiochem. 2010;11:1413–1421. doi: 10.1002/cbic.201000196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Koehnke J, Morawitz F, Bent AF, Houssen WE, Shirran SL, Fuszard MA, Smellie IA, Botting CH, Smith MCM, Jaspars M, Naismith JH. An enzymatic route to selenazolines. Chembiochem. 2013;14:564–567. doi: 10.1002/cbic.201300037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Koehnke J, Bent AF, Zollman D, Smith K, Houssen WE, Zhu X, Mann G, Lebl T, Scharff R, Shirran SL, et al. The cyanobactin heterocyclase enzyme: a processive adenylase that operates with a defined order of reaction. Angew Chem Int Ed Engl. 2013;52:13991–13996. doi: 10.1002/anie.201306302. [•• This is the first structural report of csyeine heterocyclase. In contrast to the TOMM system this enzyme was reported to produce AMP and P2O74− and this lead to the suggestion of an adenylation mechanism.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Melby JO, Dunbar KL, Trinh NQ, Mitchell DA. Selectivity, directionality, and promiscuity in peptide processing from a Bacillus sp. Al Hakam cyclodehydratase. J Am Chem Soc. 2012;134:5309–5316. doi: 10.1021/ja211675n. [• This papers demonstrates there is an order to heterocyclization of multiple cysteines in microcins.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dunbar KL, Mitchell DA. Insights into the mechanism of peptide cyclodehydrations achieved through the chemoenzymatic generation of amide derivatives. J Am Chem Soc. 2013;135:8692–8701. doi: 10.1021/ja4029507. [• This paper builds upon reference 17 and further explores the mechanism of the cyclodehydratase in the microcin pathway.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schmelz S, Naismith JH. Adenylate-forming enzymes. Curr Opin Struct Biol. 2009;19:666–671. doi: 10.1016/j.sbi.2009.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schmelz S, Kadi N, McMahon SA, Song L, Oves-Costales D, Oke M, Liu M, Johnson KA, Carter LG, Botting CH, et al. AcsD catalyzes enantioselective citrate desymmetrization in siderophore biosynthesis. Nat Chem Biol. 2009;5:174–182. doi: 10.1038/nchembio.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Houssen WE, Koehnke J, Zollman D, Vendome J, Raab A, Smith MCM, Naismith JH, Jaspars M. The Discovery of New Cyanobactins from Cyanothece PCC 7425 Defines a New Signature for Processing of Patellamides. Chem Biochem. 2012;13:1–13. doi: 10.1002/cbic.201200661. [•The paper reports the structure of the protease domain of PatA. Modelling was used to rationalize the substrate specificity of the enzyme.] [DOI] [PubMed] [Google Scholar]
- 29.Agarwal V, Pierce E, McIntosh J, Schmidt EW, Nair SK. Structures of cyanobactin maturation enzymes define a family of transamidating proteases. Chem Biol. 2012;19:1411–1422. doi: 10.1016/j.chembiol.2012.09.012. [••This paper reports structures of both the protease and macrocyclase domains from the patellamide pathway. The paper demonstrates that the acyl enzyme formed by macrocyclase can be intercepted by small peptides.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Koehnke J, Bent AF, Houssen WE, Zollman D, Morawitz F, Shirran SL, Vendome J, Nneoyiegbe AF, Trembleau L, Botting CH, et al. The mechanism of patellamide macrocyclization revealed by the characterization of the PatG macrocyclase domain. Nat Struct Mol Biol. 2012;19:767–772. doi: 10.1038/nsmb.2340. [•• This is the first structure of this class of macrocyclizing enzyme revealing the conserved insertion that is the hallmark of the family (Figure 3b). A structure complexed to a substrate peptide rationalizes the role of the macrocyclization signature (Figure 3c).] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McIntosh JA, Robertson CR, Agarwal V, Nair SK, Bulaj GW, Schmidt EW. Circular logic: nonribosomal peptide-like macrocyclization with a ribosomal peptide catalyst. J. Am. Chem. Soc. 2010:15499–15501. doi: 10.1021/ja1067806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Donia MS, Fricke WF, Ravel J, Schmidt EW. Variation in tropical reef symbiont metagenomes defined by secondary metabolism. PLoS One. 2011;6:e17897. doi: 10.1371/journal.pone.0017897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McIntosh JA, Donia MS, Schmidt EW: Ribosomal peptide natural products: bridging the ribosomal and nonribosomal worlds. Nat Prod Rep. 2009;26:537–559. doi: 10.1039/b714132g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Donia MS, Hathaway BJ, Sudek S, Haygood MG, Rosovitz MJ, Ravel J, Schmidt EW. Natural combinatorial peptide libraries in cyanobacterial symbionts of marine ascidians. Nat Chem Biol. 2006;2:729–735. doi: 10.1038/nchembio829. [DOI] [PubMed] [Google Scholar]
- 35.McIntosh JA, Donia MS, Nair SK, Schmidt EW. Enzymatic basis of ribosomal peptide prenylation in cyanobacteria. J Am Chem Soc. 2011;133:13698–13705. doi: 10.1021/ja205458h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Majmudar JD, Gibbs RA. Pericyclic prenylation: peptide modification through a Claisen rearrangement. Chembiochem. 2011;12:2723–2726. doi: 10.1002/cbic.201100612. [DOI] [PubMed] [Google Scholar]
- 37.Bent AF, Koehnke J, Houssen WE, Smith MCM, Jaspars M, Naismith JH. Structure of PatF from Prochloron didemni. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2013;69:618–623. doi: 10.1107/S1744309113012931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Metzger U, Schall C, Zocher G, Unsold I, Stec E, Li SM, Heide L, Stehle T. The structure of dimethylallyl tryptophan synthase reveals a common architecture of aromatic prenyltransferases in fungi and bacteria. Proc Natl Acad Sci U S A. 2009;106:14309–14314. doi: 10.1073/pnas.0904897106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Milne BF, Long PF, Starcevic A, Hranueli D, Jaspars M. Spontaneity in the patellamide biosynthetic pathway. Org Biomol Chem. 2006;4:631–638. doi: 10.1039/b515938e. D. [DOI] [PubMed] [Google Scholar]
- 40.Mann G, Koehnke J, Bent AF, Graham R, Houssen WE, Jaspars M, Schwarz-Linek U, Naismith JH. The structure of the cyanobactin domain of unknown function from PatG in the patellamide gene cluster. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2014 doi: 10.1107/S2053230X1402425X. Accepted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Melby JO, Li X, Mitchell DA. Orchestration of enzymatic processing by thiazole/oxazole-modified microcin dehydrogenases. Biochemistry. 2014;53:413–422. doi: 10.1021/bi401529y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Houssen W, Bent AF, Naismith JH, Jaspars M. An Efficient Method for the in vitro Production of Azol(in)e-based Cyclic Peptides. Angew Chem Int Ed Engl. 2014 doi: 10.1002/anie.201408082. [•• The paper demonstrates the power of the in vitro approach to production of analogues of patellamide. By re-engineering PatE (substrate) the slow PatA can be eliminated from the process.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Goto Y, Ito Y, Kato Y, Tsunoda S, Suga H. One-pot synthesis of azoline-containing peptides in a cell-free translation system integrated with a posttranslational cyclodehydratase. Chem Biol. 2014:766–774. doi: 10.1016/j.chembiol.2014.04.008. [• This paper reports a cell free system to produce novel patellamide analogues. Interestingly the heterocyclase was shown to be activated in trans by addition of exogenous leader peptide.] [DOI] [PubMed] [Google Scholar]
- 44.Rufffner DE, Schmidt EW, Heemstra JR. Assessing the combinatorial potential of the RiPP cyanobactin tru pathway. ACS Synth Biol. 2014 doi: 10.1021/sb500267d. Online. [• This paper extends in vivo processing efforts to make diverse analogues using the trunkamide enzymes.] [DOI] [PMC free article] [PubMed] [Google Scholar]