Abstract
Microbes attach to surfaces and form dense communities known as biofilms, which are central to how microbes live and influence humans. The key defining feature of biofilms is adhesion, whereby cells attach to one another and to surfaces, via attachment factors and extracellular polymers. While adhesion is known to be important for the initial stages of biofilm formation, its function within biofilm communities has not been studied. Here we utilise an individual-based model of microbial groups to study the evolution of adhesion. While adhering to a surface can enable cells to remain in a biofilm, consideration of within-biofilm competition reveals a potential cost to adhesion: immobility. Highly adhesive cells that are resistant to movement face being buried and starved at the base of the biofilm. However, we find that when growth occurs at the base of a biofilm, adhesion allows cells to capture substratum territory and force less adhesive, competing cells out of the system. This process may be particularly important when cells grow on a host epithelial surface. We test the predictions of our model using the enteric pathogen Vibrio cholerae, which produces an extracellular matrix important for biofilm formation. Flow cell experiments indicate that matrix-secreting cells are highly adhesive and form expanding clusters that remove non-secreting cells from the population, as predicted by our simulations. Our study shows how simple physical properties, such as adhesion, can be critical to understanding evolution and competition within microbial communities.
Introduction
Microbes secrete a range of products that aid in adhesion to surfaces and to other cells (Sauer et al., 2002; Hall-Stoodley et al., 2004; Petrova and Sauer, 2012), but even within a single species, microbes vary widely in their tendency to adhere (Tojo et al., 1988; Crociani et al., 1995; Vidal et al., 1998; Sutherland, 2001; Halme et al., 2004; Dranginis et al., 2007; Smukalla et al., 2008; Carter et al., 2011; Macklaim et al., 2011). Adhesion is critical for the formation of surface-attached microbial communities known as biofilms (Hall-Stoodley et al., 2004). In the standard model for biofilm formation, swimming or settling cells encounter surfaces and first attach reversibly, such that they can rapidly disperse if conditions are not favourable (O'Toole et al., 2000). When conditions are suitable, cells strengthen their attachment and divide to form a growing population on the surface (O'Toole et al., 2000). Attachment is facilitated by a range of secreted products, including polysaccharides, proteins, nucleic acids and amyloids (Absalon et al., 2011, Timmerman et al., 1991; Veenstra et al., 1996; Linke et al., 2006; Ma et al., 2006; Romero et al., 2010; Garcia et al., 2011; Shahid et al., 2012a, 2012b).
Microbes in biofilms adhere to each other in addition to surfaces (Heilmann et al., 1996; Conrady et al., 2013), suggesting a second possible function of adhesion: preferential attachment to cells of the same genotype, which can facilitate green-beard type co-operative interactions (Queller et al., 2003; Smukalla et al., 2008). Specifically, attaching to cells of the identical genotype allows groups to better exploit secondary phenotypes that improve final growth yield at an immediate cost to individual growth rate. An example is the secretion of enzymes that digest large nutrient sources that cannot be directly imported into cells (West et al., 2007; Drescher et al., 2014). If such secreting cells do not attach to their own kind, they can be exposed to genotypes that make use of the secreted products but do not themselves contribute to their production. These ‘cheater' genotypes can outcompete secreting cells and undermine the use of public goods over evolutionary time. Adhesion is therefore considered a factor that promotes the evolution of co-operation (Sachs, 2008).
Adhesion has almost exclusively been studied in the context of biofilm initiation and dispersal. Consequently, the evolutionary dynamics of adhesiveness over the course of biofilm growth are not well understood, even though adhesion may have a strong impact on competition between different strains and species. Here we extend an established individual-based computer model of bacterial biofilms to investigate the costs and benefits of adhesion within communities. We first consider the familiar idea that adhesion enables cells to form biofilms and avoid sloughing by shear forces. We then extend the model to examine how cells can use adhesion to resist displacement, which reveals roles for adhesion in competition within microbial communities. Finally, we perform experiments with the enteric pathogen V. cholerae to test the predictions of our model.
Materials and methods
Our model considers the growth of cells that passively push against each other as they grow and divide. Nutrient concentration gradients are calculated from consumption by cells and solute diffusion. Our simulations use periodic left-right boundary conditions and implement diffusion of nutrients into microbial colonies from the liquid above, the substratum below or both. The model extends an empirically tested, individual-based framework for the simulation of growth and division of unicellular organisms. The assumptions, justifications and implementations of these simulations are discussed at length elsewhere (Picioreanu et al., 1998; Kreft et al., 2001; Xavier et al., 2005; Xavier and Foster, 2007). In brief, the model implements a multigrid solver for two- or three-dimensional reaction diffusion partial differential equations. Using the standard assumption that diffusion occurs on a shorter timescale than bacterial growth, these equations are solved at each iteration to steady-state based on the flux at the boundaries and the production or consumption of diffusing solutes by biomass particles (cells). The biomass domain is separated from a homogeneous bulk liquid by a boundary layer in which only diffusion governs the concentration of solutes. The biofilm domain is derived from the second portion of our model, a simulation of cells in continuous space. Here, cells are represented as solid spheres. Cells grow according to the local steady-state concentration of nutrients (N) following the Monod equation
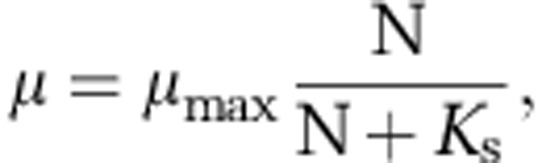 |
where the maximum growth rate μmax and the Monod constant Ks are modelling parameters (see Supplementary Tables S1 and S2).
Implementation of adhesion
Here, we extend the existing modelling framework to consider an additional modelling parameter σ, which implements adhesion. We model two effects of adhesion. The first is the improved ability of more adhesive cells to persist in biofilms. This property is captured by reducing biomass at the upper surface of biofilms to simulate cell sloughing by shear forces. In regions that are exposed to the liquid above the colony, biomass is reduced in proportion to the squared local thickness of the biofilm. Biomass is locally reduced at a rate dR according to the following equation
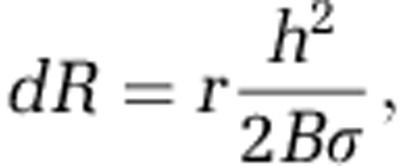 |
where r is the rate constant of detachment, h is the local thickness of the biofilm and B the amount of biomass in a computational voxel. This implementation of biofilm sloughing is a simple extension of previous erosion models (Xavier et al., 2005b).
The second effect of adhesion is the ability to resist displacement. We assume that all cells reside in a matrix to which they adhere with strength proportional to their adhesion parameter σ. As a cell increases its diameter, or upon exceeding a maximum diameter (after which it divides into two cells), overlap between neighbouring cells occurs. Our algorithm probes all cells in the simulation in a random order and tests for such overlap. If overlap is found for a focal cell, a vector for cell displacement is calculated, and the focal cell is moved accordingly (Figure 1). A new cell is then chosen at random and moved. This process is repeated until no overlap remains in the system. The consequence is that during growth, an expansion of the biofilm domain occurs, and the top of the biofilm advances away from the substratum. Locally, if a focal cell overlaps with a cell of the same adhesiveness, the resulting movement vector is simply a function of the magnitude of their overlap. If the adhesion parameters of two cells differ, a weight is assigned to the resulting vector:
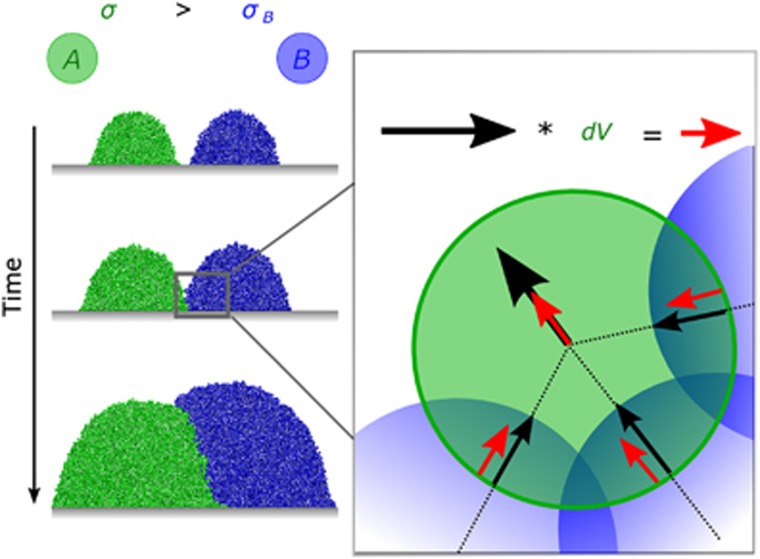
Figure 1.
Effect of adhesiveness on' cell–cell shoving. When cells that differ in their adhesiveness (σ) push each other, the more adhesive cell moves less than a less adhesive cell as it adheres more strongly to the biofilm matrix. More adhesive cells (a, green) then tend to localise below less adhesive cells (b, blue) when two growing colonies meet. The cartoon demonstrates our implementation of differential displacement. We move all overlapping cells, one cell at a time, until no overlap remains. The displacement vector (black) of a focal green cell resulting from overlap with blue cells is scaled by the factor dV, yielding the updated displacement vector (red); see also Supplementary Figure S1.
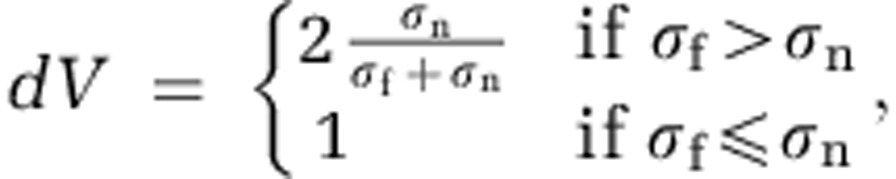 |
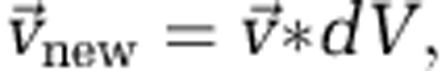 |
where σf is the adhesion parameter of the focal cell, σn is the adhesion parameter of the neighbouring cell and 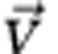 is the original movement vector prior to including the differential adhesion effect. Hence, a more adhesive cell (σf>σn) moves less upon meeting a cell of lower adhesiveness (Figure 1).
is the original movement vector prior to including the differential adhesion effect. Hence, a more adhesive cell (σf>σn) moves less upon meeting a cell of lower adhesiveness (Figure 1).
Our implementation of adhesiveness is an approximation of the true forces that cause highly adhesive cells to resist movement more strongly than less adhesive cells. In the Supplementary materials, we show that our implementation is consistent with a physical model of the drag forces spherical objects experience when they move through a viscous liquid, such as the biofilm matrix (Supplementary Figure S1). The result is that overlapping cells with the same adhesion displace equal distances on average, but differential movement occurs between cells that have different adhesion parameters. Finally, in some simulations, we explicitly simulate the secretion and accumulation of extracellular polymers, as detailed in Xavier and Foster (2007). These polymers are modelled as spheres, whose adhesiveness can be altered in the same manner as described above for cells.
Fitness calculation
To assess the evolutionary success of different genotypes, we calculate their relative fitnesses. The fitness, wX, of strain X is calculated as the average number of divisions per unit time achieved by that strain during a simulation (from t0 to tend).
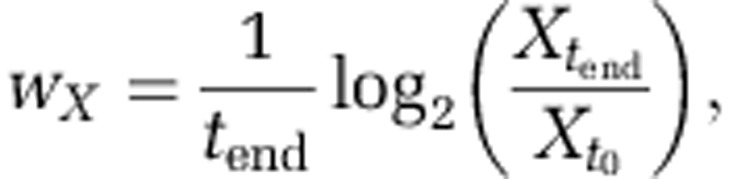 |
where Xt is the number of cells of species X at time t. The relative fitness, w, of species X in competition with Y is defined as 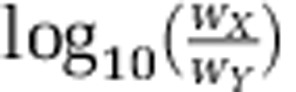 . We here consider only competition between genotypes in a local community and assume that biofilms persist on long timescales relative to cell division rates, such that the accumulated dispersal of cells from a focal biofilm to new habitats will follow the trend of local competition. Differences in the relative ability to disperse from biofilms are not considered here but can be important as is discussed in Nadell and Bassler (2011).
. We here consider only competition between genotypes in a local community and assume that biofilms persist on long timescales relative to cell division rates, such that the accumulated dispersal of cells from a focal biofilm to new habitats will follow the trend of local competition. Differences in the relative ability to disperse from biofilms are not considered here but can be important as is discussed in Nadell and Bassler (2011).
V. cholerae experiments
All experiments were conducted using strains derived from V. cholerae str. C6706 using standard molecular biology protocols. Briefly, a constitutive extracellular polymeric substances (EPS) producer (EPS+) was constructed by deleting genes encoding the quorum-sensing regulator (hapR) and the flagellin core protein (flaA). The EPS non-producer (EPS−) was built by further deletion of vpsL, which encodes a protein required for EPS biosynthesis. Further detail on strain construction and characterisation can be found in Nadell and Bassler (2011). Constitutive fluorescent protein expression constructs were inserted onto the chromosomes of each strain for visual distinction and quantification of our experiments. The two fluorescent genes used were mKate (Shcherbo et al., 2007) and mTFP1 (Ai et al., 2006), which were artificially coloured green and blue for comparison with our simulations. These expression constructs were previously shown to have negligible effects on bacterial growth rate (Nadell and Bassler, 2011). Biofilms were grown in simple straight-chamber microfluidic devices as previously described (Nadell and Bassler, 2011). To determine whether EPS+ cells displace EPS− cells by pushing against them, biofilms were initiated by inoculating the two strains in a 1:100 ratio (EPS+:EPS−). Biofilms were imaged along chamber substrata at regular time intervals for 48 h. For shorter sequences in which z-plane renderings were desired, biofilms were imaged throughout their volume at z-intervals of 1 μm. All microscopy was performed with a Nikon Eclipse fluorescence microscope (Tokyo, Japan) fitted with a Yokogawa CSU-X1 confocal scanning unit (Tokyo, Japan). Data were analysed using ImageJ and custom software written for MatLab (Natick, MA, USA).
Results and discussion
We simulate a microbial community residing on a solid surface. Our model builds on an empirically tested individual-based framework designed to predict biofilm structure and composition (Picioreanu et al., 1997; Kreft et al., 2001; Xavier et al., 2005; Xavier and Foster, 2007; Nadell et al., 2008, 2010; Mitri et al., 2011; Schluter and Foster, 2012). Our simulations implement cells that consume nutrients, grow and divide, which can lead to the formation of spatial nutrient concentration gradients. In some regions, nutrients may become depleted below the point at which cells can grow (Xavier and Foster, 2007). We focus primarily on conditions under which biofilms are nutrient limited, which is likely to be common in many environments (Stewart and Franklin, 2008). In this scenario, competition for nutrients is high, and we measure fitness after a finite amount of nutrients has been consumed. We later relax these constraints and consider conditions of high or unlimited nutrient availability. In all cases, those genotypes that achieve the highest overall growth rate and retention within biofilms increase in frequency and outcompete other genotypes. To measure the effects of selection, we calculate a commonly used metric for relative fitness, w, which corresponds to the relative number of cell divisions (see Materials and methods) and consider a species competitively successful if w>0.
Adhesion as a mechanism to form biofilms
In this first model, nutrients diffuse into the biofilm from a bulk fluid above, and we assume that cell biomass is removed at a rate proportional to the square of the local biofilm height, a simple form of sloughing due to flow over the upper surface. Regions with more adherent cells accumulate biofilm more rapidly than other regions, because adhesiveness in this model reduces the rate at which cells are washed from the biofilm surface (Materials and methods). This analysis reveals the first and obvious potential advantage of being adhesive: thicker biofilm formation (Figure 2a). In biofilms containing more than one cell type, the more adhesive genotype dominates, as continual sloughing of cells preferentially removes the less adhesive genotype from the population (Figure 2b).
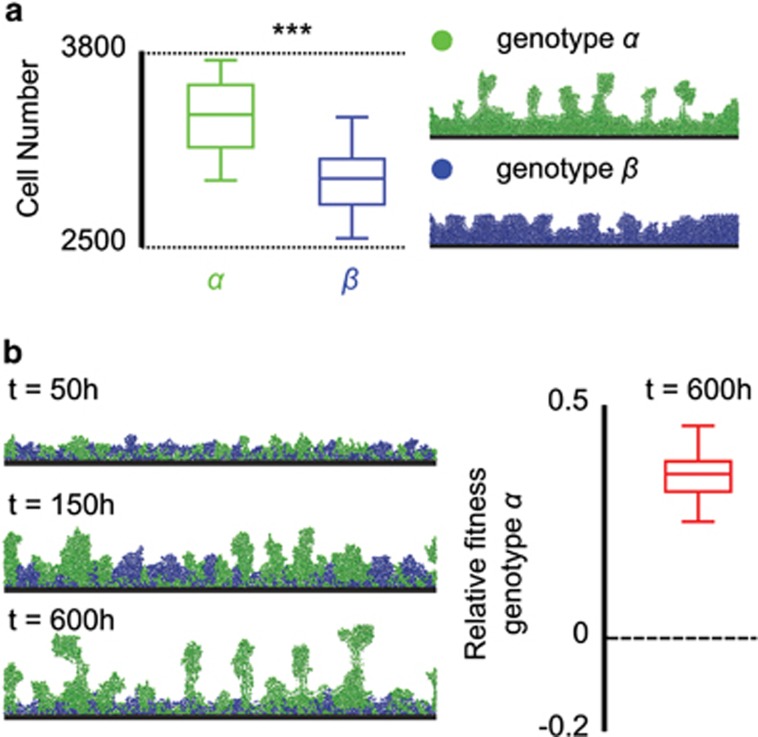
Figure 2.
Adhesion is beneficial when cells are sloughed from biofilms. (a) Monoculture biofilms in which equilibrium biofilm thickness is limited by cell detachment, which increases with the square of local biofilm height (bulk nutrient concentration: 2 × 10−3 (g l−1), detachment rate: r=0.1, see Materials and methods). Greater adhesion reduces this effect and therefore leads to thicker biofilms consisting of the more adhesive genotype α than those of the less adhesive genotype β (boxplots, Wilcoxon signed-rank test, ***P<10−8). (b) Greater adhesion allows cells of genotype α to outcompete genotype β when the two strains are co-inoculated. Snapshots show a representative simulation, with relative fitness calculated from 30 independent simulations.
Resisting displacement can be costly within biofilms
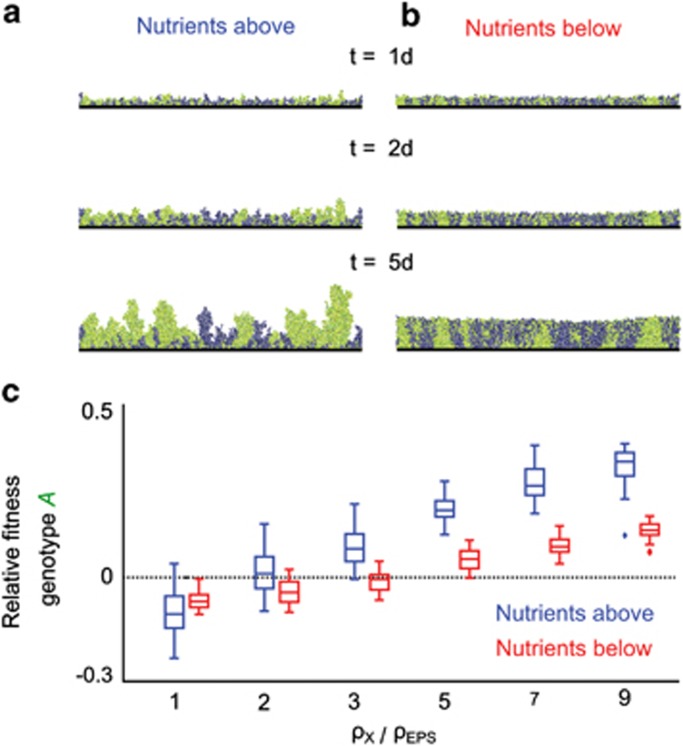
We next explored the competitive dynamics of strains with different abilities to resist displacement when they divide and push against each other. Specifically, when cells grow and divide, passive shoving occurs among neighbours to reveal space for new biomass. A more adhesive cell moves less when pushed by a less adhesive cell and vice versa, because the former attaches more securely to the surrounding extracellular matrix (see Materials and methods and Supplementary materials for details on the implementation of adhesion). We define an adhesion parameter σ, which determines the distance a cell moves aside when pushed (Supplementary Text S1). We simulate competition between two genotypes, with genotype A more adhesive than genotype B (σA=2, σB=1), and compare results to control simulations in which the two genotypes have equivalent adhesion parameters (σA=σB=1, see Materials and methods). This comparison reveals that the more adhesive genotype has lower relative fitness when in competition with the less adhesive genotype. The more adhesive genotype performs poorly when two cells of differential adhesiveness collide because more adhesive cells tend to localise underneath less adhesive cells, where nutrient concentrations are usually lower if the nutrient source is above the biofilm. Thus, the less adhesive genotype more frequently passes through population bottlenecks that often occur in biofilms for which nutrients are supplied from above (Hallatschek et al., 2007; Mitri and Foster, 2013; Figure 3b).
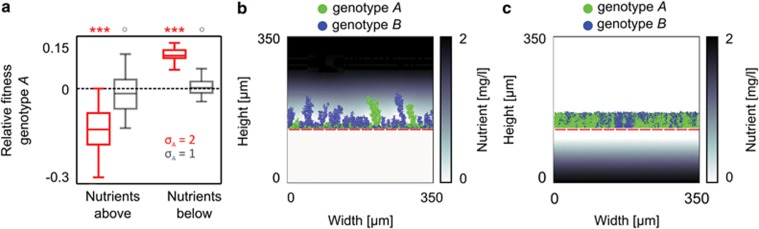
Figure 3.
The fitness effects of displacement by other cells. (a) Relative fitness of the more adhesive genotype A when nutrients diffuse from above and below the biofilm. Grey boxplots represent control simulations, in which competing genotypes are equally adhesive, and red boxplots summarise simulations for which A is more adhesive than B; the dashed line corresponds to no fitness difference. ***P<10−6 and O: P>0.4, sign test for difference from zero. Snapshots of simulations in which nutrients diffuse from above (b) or below (c) showing the nutrient gradients arising from consumption in biofilms growing on an impenetrable surface (dashed red line).
When nutrients are limited and diffuse from above, our model predicts that natural selection for adhesion will result in the following outcomes. First, provided that cells are sufficiently adherent to remain attached to a surface, we expect a general upper bound on the expression of genes promoting adhesion during biofilm growth, as high expression leads to substratum localisation away from areas of high nutrient concentration. Second, cells might be selected to regulate adhesion such that they are adherent during the initial stages of substratum attachment and then repress expression of genes encoding adhesins later during biofilm growth. Third, if there is genotypic variation in adhesin expression within a biofilm community, less adhesive genotypes might be increasingly favoured as biofilms mature because they deprive nutrients to highly adhesive cells in the basal layers.
Boles et al. (2004) describe a potential example of selection for less adhesive genotypes over time in growing laboratory biofilms of the opportunistic pathogen Pseudomonas aeruginosa. A recurring variant that arose in their experimental communities was weakly adhesive to the biofilm. The authors interpreted this reduction in adhesive capability to be part of an insurance policy for the biofilm as a whole: less adhesive cells can more easily detach and disperse to new potential habitats than could strongly adhesive cells. However, for less adhesive genetic variants to arise and increase in frequency by natural selection, their phenotype must also provide a direct short-term evolutionary advantage within the biofilm itself. Our model offers a putative advantage: compared with highly adhesive cells, weakly adhesive cells can more easily spread to the upper biofilm surface, where they will gain preferential access to nutrients diffusing from above.
An evolutionary advantage to resisting displacement
In the above model, due to limiting nutrients diffusing from above, only cells along the biofilm's advancing front can grow and divide, but this scenario captures only a subset of habitats that microbes occupy. First, if nutrients are abundant, growth occurs throughout a biofilm, and thus there is no cost to localising to the substratum. Second, nutrients can diffuse from the substratum itself, for example, in detrital particles in the ocean (Cordero et al., 2012; Shapiro et al., 2012) or the soil (Böckelmann et al., 2003). Similarly, many host-associated bacteria catabolise host-made products on the tissues on which they grow (Hooper et al., 1999; Meibom et al., 2004; Cash et al., 2006; Church et al., 2006; Derrien et al., 2010; Bevins and Salzman, 2011; Schluter and Foster, 2012).
Our model shows that high adhesiveness localises cells to the attachment surface. On this basis, we reasoned that the cost of adhesion observed in the preceding model could be converted to an advantage when nutrients are obtained from the substratum. To assess this idea, we modelled an environment in which nutrients diffuse from below the biofilm. Indeed, the adhesiveness that is detrimental in the above model becomes beneficial when access to limiting nutrients is highest at the substratum, because the more adhesive strain has preferential access to nutrients (Figure 3c) and rapidly dominates the attachment surface.
Resisting displacement when abundant nutrients are present
We have shown that adhesiveness can be strongly beneficial when it fosters access to a nutrient source on which cells are growing. In some circumstances, however, nutrients enter biofilms from above and below the basal substratum. The gut epithelium is an example of such an environment. We therefore evaluated the costs and benefits of resisting displacement when nutrients simultaneously diffuse from below and above, and when cells are sloughed from the biofilm's outermost surface (Schluter and Foster, 2012).
In this model, relative adhesiveness has no initial effect on fitness because the entire community is nutrient saturated, which allows all cells to grow at high rates (Figure 4). However, the more adhesive cells dominate on longer timescales. By localising to the base of the biofilm, the most adhesive cells gradually but reliably displace less adhesive cells away from the surface and into the sloughing region. This result is also obtained when nutrients diffuse only from above the biofilm but are not limiting, such that adhesive cells at the base of the colony are never starved (Supplementary Figure S1).
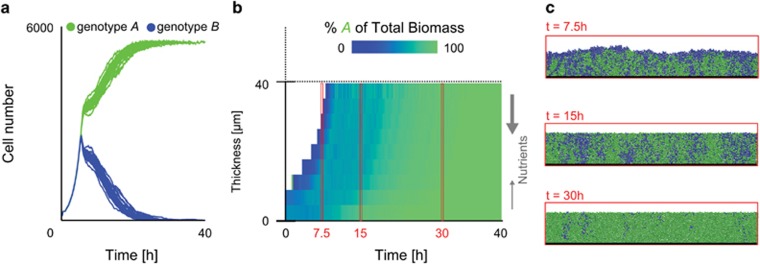
Figure 4.
Competition in a model of a nutrient-saturated biofilm with nutrients diffusing into the colony from above (N=4) and below (N=0.8). (a) Cell number over time for simulations initiated with the same number of each genotype; genotype A outcompetes genotype B over time. (b) The heat map shows the biomass distribution of the two genotypes averaged over the width of a single simulated biofilm community. (c) Snapshots expand these average values. Cells grow protected from sloughing in a 40-μm-thick layer beyond which cells are lost from the biofilm. Because genotype A is more adhesive, it displaces genotype B into the biofilm's sloughing region, leading to dominance of A. Identical results are obtained when nutrients exclusively diffuse into the biofilm from above, but remain at high enough concentration for growth to occur throughout the biofilm.
Two potential effects of secreted polymers: adhesion and expansion
The models explored above assume that cells adhere with different strengths to secreted EPS, which were not explicitly implemented. We next consider simulations in which EPS is explicitly implemented: strains can differ in their investment into EPS, and EPS may itself vary in adhesiveness. We do this because previous theoretical and empirical work suggests that polymer secretion, in addition to mediating adhesion, can also produce an inexpensive expansion in cell cluster volume relative to pure biomass production (Xavier and Foster, 2007; Kim et al., 2014). Such volume expansion positions EPS-secreting strains into nutrient-rich areas of the biofilm at the expense of strains that do not produce EPS, a competitive advantage that could counteract the costs of adhesiveness. Previous work on EPS has only considered the case where limiting nutrients diffuse into the biofilm from above (Xavier and Foster, 2007; Nadell et al., 2008; Kim et al., 2014), which raises the question of whether volume expansion via EPS is advantageous when nutrients are obtained from the substratum.
We investigated whether EPS-based volume expansion can confer a competitive advantage when cells acquire nutrients from the surface on which they reside, independently of the potential for adhesive effects. In contrast to the results described above, Figure 5 shows that volume expansion via EPS enables cells to gain preferential nutrient access by rapidly colonising the surface when nutrients diffuse from below. Likewise, when nutrients diffuse from above, EPS-producing cell lineages can expand upwards and deny competitors access to nutrients, as shown by Xavier and Foster (2007). In the supplement, we explore the interactions between the two properties of EPS, volume expansion, and adhesion, and find that volume expansion can compensate for the costs of adhesion when limiting nutrients diffuse into biofilms from above (Supplementary Figures S2 and S3).
Figure 5.
The fitness effect of volume expansion. EPS-producing genotype A (investment in EPS 25% of total growth, green) is competed against a non-producing genotype (blue), in which EPS (yellow) is not adhesive and only increases cell lineage volume. (a and b) Snapshots from representative simulations with EPS modelled to be five times less costly to produce than cell biomass. Simulations are shown with nutrients diffusing from above (a) or below (b). (c) Fitness of the EPS-producing genotype A for different costs of EPS production (ρEPS) relative to the cost of biomass production (ρX). Decreasing the cost of EPS relative to the cost of producing new cells (increasing ρX/ρEPS) leads to increased fitness of the EPS-producing genotype A, irrespective of whether nutrients come from above (blue) or below (red); N=2 × 10−3 (g l−1).
The benefits of an adhesive polymer in V. cholerae
We performed an empirical test of our model predictions using biofilms produced by the enteric pathogen V. cholerae (Nalin et al., 1979; Huq et al., 1983; Meibom et al., 2004; Alam et al., 2006). Previous research has demonstrated that EPS-producing cells (EPS+) produce larger biofilms when alone and have a substantial competitive advantage over an isogenic non-producing strain (EPS−) in mixed genotype biofilms. These results support the first putative advantage of adhesiveness proposed above: EPS-producing cells accumulate more biomass within biofilms by virtue of their adhesion to each other and the substratum. Nadell and Bassler (2011) also found that the presence of EPS+ cells reduces the ability of EPS−cells to accumulate biofilm biomass by 80%, while the presence of EPS− cells has no effect on EPS+ biomass accumulation, indicating direct between-strain interference competition. The mechanistic basis for this result, however, remains unexplained (Nadell and Bassler, 2011). Here we explored whether the two other benefits identified by our model—active displacement of competing strains and cell cluster volume expansion—also contribute to the success of EPS-producing cells.
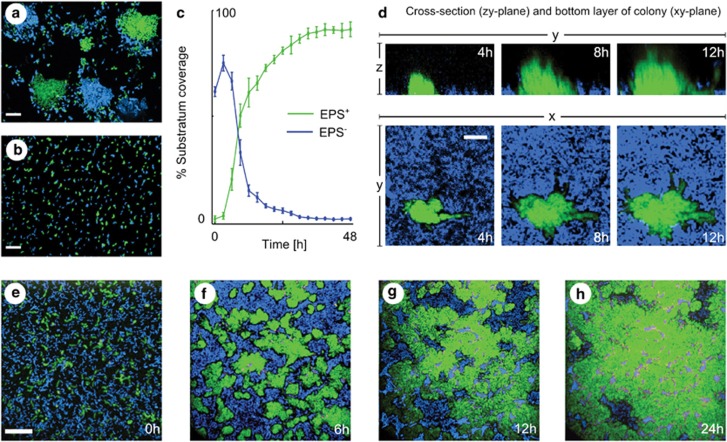
As observed previously, the EPS+ strain forms more robust biofilms (10–20 cell lengths thick) relative to the EPS− strain (1–2 lengths thick), suggesting increased adhesion among EPS+ cells. To further assess this inference, we grew gently shaken cultures of 1:1 blue EPS+: green EPS+ cells and observed that they form dense mono-colour groups. This result indicates strong mother–daughter cell adherence (Figure 6a). In contrast, EPS− cells do not adhere to one another (Figure 6B). To examine the competitive interaction of EPS+ and EPS− cells in biofilms, we inoculated the two strains together on the glass substrata of straight-chamber microfluidic devices composed of polydimethysiloxane bonded to microscope slides and imaged the resulting biofilms at regular intervals for 48 h. Visual inspection and quantification of fluorescence intensities of the basal biofilm layer show that EPS– cells are predominantly cleared from the substratum along the expanding front of EPS+ clusters, though they are occasionally trapped underneath EPS+ clusters. Ultimately, EPS+ cells occupy nearly 100% of the substratum (Figures 6c and e–h and Supplementary Figure S4).
Figure 6.
EPS-producing cells displace non-producing cells from the substratum in a V. cholerae experimental system. (a) A 1:1 mixture of green and blue EPS+ cells form clonal clusters in liquid culture. (b) A 1:1 mixture of blue and green EPS− cells remain dispersed. (c–h) EPS+ (green) in competition with EPS− cells (blue) at different time points. (c) Quantification of surface area coverage by the two strains over time. Bars denote s.e.m., n=3 replicates. (d) EPS+ cells (green) displace V. cholerae EPS− cells (blue) by burrowing under the EPS− strain along the attachment surface in co-culture, even though EPS− cells can remain attached when grown alone (Supplementary Figure S5). (e–h) A time series for the bottom layer of EPS+ (green) competing against EPS− cells (blue). The scale bars in panels a, b and e, f denote 20 μm. The scale bar in panel d denotes 8 μm.
The mother–daughter cell adhesion of EPS+ cells and rapid reduction in surface occupation by EPS− cells during biofilm competition is consistent with our second proposed advantage of adhesion, namely active displacement of competitors by EPS+ cells. However, if EPS− cells are intrinsically more likely to detach from the substratum than EPS+ cells, it could be that EPS− cells are simply revealing space that is subsequently occupied by EPS+ cells without any direct interaction between the two genotypes. Two pieces of evidence contradict this interpretation. First, EPS−cells are equally capable as EPS+ cells of forming and maintaining confluent monolayers on the glass surface, even when subjected to 20-fold higher flow velocities than those used in the competition experiment (Supplementary Figure S5). Second, three-dimensional imaging of co-cultured biofilms illustrates a sloughing effect, by which expanding EPS+ clusters displace EPS− cells from the glass along the boundary at which the two strains meet (Figure 6d).
The third predicted benefit of EPS-mediated adhesion is that matrix-secreting cells expand volumetrically more rapidly than non-secreting competitors, allowing the secreting strain to occupy more space from which to draw growth substrate and thereby deny neighbouring cells access to nutrients. Though nutrients were supplied from above the biofilm in our system, cells near open spaces on the substratum grew throughout the experiment, showing that they were not being denied nutrients by EPS+ clusters. In addition, microscopy did not indicate significantly increased spacing between EPS+ cells compared with EPS− cells (Supplementary Figures S4 and S5). Thus, volume expansion does not appear to be a dominant mechanism underpinning the competitive advantage of the EPS+ strain over the EPS− strain in this system.
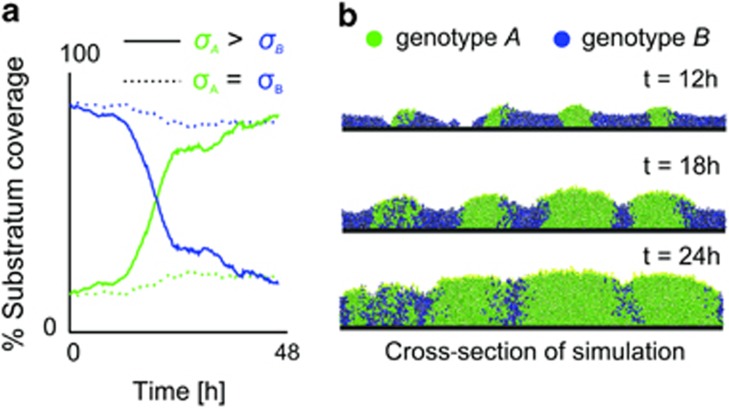
In summary, our experimental data suggest that the cell cluster formation that occurs due to the adhesiveness of V. cholerae EPS producers provides a competitive advantage to secreting cells primarily via the accumulation of thick biofilm and active displacement of non-secreting cells from the glass substratum. These dynamics can be recapitulated in simulations that simultaneously incorporate increased resistance to sloughing and displacement by EPS-producing cells (Figure 7). Importantly, the parameters used for these simulations were the same as those in the above sections and were chosen prior to experimentation (Figures 2, 3, 4 and Supplementary Table S1) with two exceptions. First, the rate of sloughing from biofilms was chosen to obtain a biofilm thickness comparable with that of the V. cholerae EPS+ strain. Second, the fraction of biomass devoted to EPS production by EPS+ cells was reduced from 25% to 5% to reflect that little EPS is observed between cells in micrographs obtained from the experiments (Supplementary Figure S5B).
Figure 7.
Model of EPS+ versus EPS− competition in V. cholerae. Simulations were performed in which genotype A produces adhesive EPS (investment 5% of growth rate), whereas genotype B does not. Here, nutrients diffuse from above and are saturating (N=0.8 (g l−1)) to enable growth throughout the biofilm, which is limited in thickness through sloughing of biomass proportional to the square of local biofilm height. Rate of sloughing (r=20, see Materials and methods), and investment into EPS are parameters chosen based upon the experimental system (see text), but all other parameters are the same as those used in previous simulations (Supplementary Table S1). Areas containing EPS exhibit reduced sloughing of cells (see Materials and methods) and EPS-producing cells are more resistant to displacement by non-producing cells (initial frequency of A=0.1, 35 cells were seeded in total). (a) The graph shows coverage of the substratum by the two genotypes and indicates displacement of B by A when A is more adhesive than genotype B (solid line); no displacement occurs in an otherwise identical control simulation in which the two strains are equally adhesive dashed line). (b) Simulation snapshots σA>σB.
Conclusions
Our work highlights the potential importance of adhesion for the evolutionary fate of microbes in biofilms, beyond simple attachment to surfaces. Resistance to displacement can be costly when nutrients do not reach the base of a biofilm. Under these conditions, highly adhesive cells can be overgrown by less adhesive genotypes, which results in starvation of the more adhesive genotype. However, when cells are able to grow at the base of a biofilm, adhesion is beneficial, as it enables cell lineages to persist during biofilm growth and to expand across the substratum by displacing other cells in their paths. Therefore, in biofilms residing on host tissues, adhesiveness is likely to be important for the outcome of competition for space and nutrients.
In vivo studies have demonstrated the importance of adhesiveness for initial surface colonisation (Liu et al., 2008); however, we do not know of studies that examine whether adhesion provides a competitive advantage within host-associated communities. We do know that both pathogenic and commensal bacteria produce molecules that anchor them to epithelial cells or to the associated mucus layer (MacKenzie et al., 2009; Chattopadhyay et al., 2012). Moreover, bacteria have apparently evolved environment-specific adhesion molecules. For example, Lactobacilli living in the gut possess adhesins that differ from those of Lactobacilli strains residing in the vagina (Macklaim et al., 2011), and vaginal Lactobacilli display increased adhesiveness relative to their counterparts that exist in other environments (Boris et al., 1998; Malik et al., 2013). The hypothesis that adhesion is a key competitive strategy raises the interesting possibility that arms races occur in which strains evolve methods to decrease adhesion of their competitors. While speculative, some evidence exists for this notion. For example, the soil bacterium Lysinibacillus fusiformis secretes non-bactericidal biosurfactants that can act as wetting agents that reduce attachment and biofilm formation by competitors (Pradhan et al., 2014).
Evolutionary competition in biofilms is often intense, and factors that tip the competitive balance in favour of a particular genotype can be rapidly selected. The literature emphasises roles for secreted toxins and rapid growth in competition. Our work demonstrates the importance of strategies that modify endogenous physical properties of microbial communities. A genotype can dominate a community simply being more adhesive than its competitors.
Acknowledgments
We thank Michael A Bentley, Katharine Coyte, William Durham, Knut Drescher, Eamonn Gaffney, Wook Kim, Sara Mitri, Rene Niehus, Nuno M Oliveira, Akira Sasaki, Sam Brown, Stuart West and Roman Popat for helpful discussions and comments on the manuscript. We also thank Will Smith for his help with the analysis and comparison of our model with a physical model of viscous draft.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Absalon C, Van Dellen K, Watnick PI. A communal bacterial adhesin anchors biofilm and bystander cells to surfaces. PLoS Pathog. 2011;7:e1002210. doi: 10.1371/journal.ppat.1002210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ai H-W, Henderson JN, Remington SJ, Campbell RE. Directed evolution of a monomeric, bright and photostable version of Clavularia cyan fluorescent protein: structural characterization and applications in fluorescence imaging. Biochem J. 2006;400:531–540. doi: 10.1042/BJ20060874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alam M, Sultana M, Nair G. Toxigenic Vibrio cholerae in the aquatic environment of Mathbaria, Bangladesh. Appl Environ Microbiol. 2006;72:2849–2855. doi: 10.1128/AEM.72.4.2849-2855.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bevins CL, Salzman NH. The potter's wheel: the host's role in sculpting its microbiota. Cell Mol Life Sci. 2011;68:3675–3685. doi: 10.1007/s00018-011-0830-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Böckelmann U, Szewzyk U, Grohmann E. A new enzymatic method for the detachment of particle associated soil bacteria. J Microbiol Methods. 2003;55:201–211. doi: 10.1016/s0167-7012(03)00144-1. [DOI] [PubMed] [Google Scholar]
- Boles BR, Thoendel M, Singh PK. Self-generated diversity produces ‘insurance effects' in biofilm communities. Proc Natl Acad Sci USA. 2004;101:16630–16635. doi: 10.1073/pnas.0407460101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boris S, Suárez JE, Vázquez F, Barbés C. Adherence of human vaginal lactobacilli to vaginal epithelial cells and interaction with uropathogens. Infect Immun. 1998;66:1985–1989. doi: 10.1128/iai.66.5.1985-1989.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter MQ, Brandl MT, Louie JW, Kyle JL, Carychao DK, Cooley MB, et al. Distinct acid resistance and survival fitness displayed by Curli variants of enterohemorrhagic Escherichia coli O157:H7. Appl Environ Microbiol. 2011;77:3685–3695. doi: 10.1128/AEM.02315-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cash HL, Whitham CV, Behrendt CL, Hooper LV. Symbiotic bacteria direct expression of an intestinal bactericidal lectin. Science. 2006;80:313–1126. doi: 10.1126/science.1127119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chattopadhyay S, Tchesnokova V, McVeigh A, Kisiela DI, Dori K, Navarro A, et al. Adaptive evolution of class 5 fimbrial genes in enterotoxigenic Escherichia coli and its functional consequences. J Biol Chem. 2012;287:6150–6158. doi: 10.1074/jbc.M111.303735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Church D, Elsayed S, Reid O, Winston B, Lindsay R. Burn wound infections. Clin Microbiol Rev. 2006;19:403–434. doi: 10.1128/CMR.19.2.403-434.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrady DG, Wilson JJ, Herr AB. Structural basis for Zn 2+-dependent intercellular adhesion in staphylococcal biofilms. Proc Natl Acad Sci USA. 2013;110:E202–E211. doi: 10.1073/pnas.1208134110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordero OX, Ventouras L-A, DeLong EF, Polz MF. Public good dynamics drive evolution of iron acquisition strategies in natural bacterioplankton populations. Proc Natl Acad Sci USA. 2012;109:20059–20064. doi: 10.1073/pnas.1213344109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crociani J, Grill JP, Huppert M, Ballongue J. Adhesion of different bifidobacteria strains to human enterocyte-like Caco-2 cells and comparison with in vivo study. Lett Appl Microbiol. 1995;21:146–148. doi: 10.1111/j.1472-765x.1995.tb01027.x. [DOI] [PubMed] [Google Scholar]
- Derrien M, van Passel MW, van de Bovenkamp JH, Schipper RG, de Vos WM, Dekker J. Mucin-bacterial interactions in the human oral cavity and digestive tract. Gut Microbes. 2010;1:254–268. doi: 10.4161/gmic.1.4.12778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dranginis AM, Rauceo JM, Coronado JE, Lipke PN. A biochemical guide to yeast adhesins: glycoproteins for social and antisocial occasions. Microbiol Mol Biol Rev. 2007;71:282–294. doi: 10.1128/MMBR.00037-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drescher K, Nadell CD, Stone HA, Wingreen NS, Bassler BL. Solutions to the public goods dilemma in bacterial biofilms. Curr Biol. 2014;24:50–55. doi: 10.1016/j.cub.2013.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia MC, Lee JT, Ramsook CB, Alsteens D, Dufrêne YF, Lipke PN. A role for amyloid in cell aggregation and biofilm formation. PLoS One. 2011;6:e17632. doi: 10.1371/journal.pone.0017632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallatschek O, Hersen P, Ramanathan S, Nelson DR. Genetic drift at expanding frontiers promotes gene segregation. Proc Natl Acad Sci USA. 2007;104:19926–19930. doi: 10.1073/pnas.0710150104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall-Stoodley L, Costerton JW, Stoodley P. Bacterial biofilms: from the natural environment to infectious diseases. Nat Rev Microbiol. 2004;2:95–108. doi: 10.1038/nrmicro821. [DOI] [PubMed] [Google Scholar]
- Halme A, Bumgarner S, Styles C, Fink GR. Genetic and epigenetic regulation of the FLO gene family generates cell-surface variation in yeast. Cell. 2004;116:405–415. doi: 10.1016/s0092-8674(04)00118-7. [DOI] [PubMed] [Google Scholar]
- Heilmann C, Schweitzer O, Gerke C, Vanittanakom N, Mack D, Götz F. Molecular basis of intercellular adhesion in the biofilm-forming Staphylococcus epidermidis. Mol Microbiol. 1996;20:1083–1091. doi: 10.1111/j.1365-2958.1996.tb02548.x. [DOI] [PubMed] [Google Scholar]
- Hooper LV, Xu J, Falk PG, Midtvedt T, Gordon JI. A molecular sensor that allows a gut commensal to control its nutrient foundation in a competitive ecosystem. Proc Natl Acad Sci USA. 1999;96:9833–9838. doi: 10.1073/pnas.96.17.9833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huq A, Small EB, West PA, Huq MI, Rahman R, Colwell RR. Ecological relationships between Vibrio cholerae and planktonic crustacean copepods. Appl Environ Microbiol. 1983;45:275–283. doi: 10.1128/aem.45.1.275-283.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim W, Racimo F, Schluter J, Levy SB, Foster KR. Importance of positioning for microbial evolution. Proc Natl Acad Sci USA. 2014;111:E1639–E1647. doi: 10.1073/pnas.1323632111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreft JU, Picioreanu C, Wimpenny JW, van Loosdrecht MC. Individual-based modelling of biofilms. Microbiology. 2001;147:2897–2912. doi: 10.1099/00221287-147-11-2897. [DOI] [PubMed] [Google Scholar]
- Linke D, Riess T, Autenrieth IB, Lupas A, Kempf VAJ. Trimeric autotransporter adhesins: variable structure, common function. Trends Microbiol. 2006;14:264–270. doi: 10.1016/j.tim.2006.04.005. [DOI] [PubMed] [Google Scholar]
- Liu CH, Lee SM, Vanlare JM, Kasper DL, Mazmanian SK. Regulation of surface architecture by symbiotic bacteria mediates host colonization. Proc Natl Acad Sci USA. 2008;105:3951–3956. doi: 10.1073/pnas.0709266105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L, Jackson KD, Landry RM, Parsek MR, Wozniak DJ. Analysis of Pseudomonas aeruginosa conditional psl variants reveals roles for the psl polysaccharide in adhesion and maintaining biofilm structure postattachment. J Bacteriol. 2006;188:8213–8221. doi: 10.1128/JB.01202-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacKenzie DA, Tailford LE, Hemmings AM, Juge N. Crystal structure of a mucus-binding protein repeat reveals an unexpected functional immunoglobulin binding activity. J Biol Chem. 2009;284:32444–32453. doi: 10.1074/jbc.M109.040907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macklaim J, Gloor G, Anukam K, Cribby S, Reid G. At the crossroads of vaginal health and disease, the genome sequence of Lactobacillus iners AB-1. Proc Natl Acad Sci USA. 2011;108:4688–4695. doi: 10.1073/pnas.1000086107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik S, Petrova MI, Claes IJJ, Verhoeven TLA, Busschaert P, Vaneechoutte M, et al. The high auto-aggregative and adhesive phenotype of the vaginal Lactobacillus plantarum strain CMPG5300 is sortase-dependent. Appl Environ Microb. 2013;79:4576–4585. doi: 10.1128/AEM.00926-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meibom KL, Li XB, Nielsen AT, Wu C-Y, Roseman S, Schoolnik GK. The Vibrio cholerae chitin utilization program. Proc Natl Acad Sci USA. 2004;101:2524–2529. doi: 10.1073/pnas.0308707101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitri S, Foster KR. The genotypic view of social interactions in microbial communities. Annu Rev Genet. 2013;47:247–273. doi: 10.1146/annurev-genet-111212-133307. [DOI] [PubMed] [Google Scholar]
- Mitri S, Xavier JB, Foster KR. Social evolution in multispecies biofilms. Proc Natl Acad Sci USA. 2011;108:10839–10846. doi: 10.1073/pnas.1100292108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadell CD, Bassler BL. A fitness trade-off between local competition and dispersal in Vibrio cholerae biofilms. Proc Natl Acad Sci USA. 2011;108:14181–14185. doi: 10.1073/pnas.1111147108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadell CD, Foster KR, Xavier JB. Emergence of spatial structure in cell groups and the evolution of cooperation. PLoS Comput Biol. 2010;6:e1000716. doi: 10.1371/journal.pcbi.1000716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadell CD, Xavier JB, Levin SA, Foster KR. The evolution of quorum sensing in bacterial biofilms. PLoS Biol. 2008;6:e14. doi: 10.1371/journal.pbio.0060014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nalin DR, Daya V, Reid a, Levine MM, Cisneros L. Adsorption and growth of Vibrio cholerae on chitin. Infect Immun. 1979;25:768–770. doi: 10.1128/iai.25.2.768-770.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Toole G, Kaplan H, Kolter R. Biofilm formation as microbial development. Annu Rev Microbiol. 2000;54:49–79. doi: 10.1146/annurev.micro.54.1.49. [DOI] [PubMed] [Google Scholar]
- Petrova OE, Sauer K. Sticky situations: key components that control bacterial surface attachment. J Bacteriol. 2012;194:2413–2425. doi: 10.1128/JB.00003-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picioreanu C, van Loosdrecht MC, Heijnen JJ. Mathematical modeling of biofilm structure with a hybrid differential-discrete cellular automaton approach. Biotechnol Bioeng. 1998;58:101–116. doi: 10.1002/(sici)1097-0290(19980405)58:1<101::aid-bit11>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- Picioreanu C, van Loosdrecht MCM, Heijnen JJ. Modelling the effect of oxygen concentration on nitrite accumulation in a biofilm airlift suspension reactor. Water Sci Tech. 1997;36:147–156. [Google Scholar]
- Pradhan AK, Pradhan N, Sukla LB, Panda PK, Mishra BK. Inhibition of pathogenic bacterial biofilm by biosurfactant produced by Lysinibacillus fusiformis S9. Bioprocess Biosyst Eng. 2014;37:139–149. doi: 10.1007/s00449-013-0976-5. [DOI] [PubMed] [Google Scholar]
- Queller DC, Ponte E, Bozzaro S, Strassmann JE. Single-gene greenbeard effects in the social amoeba Dictyostelium discoideum. Science. 2003;299:105–106. doi: 10.1126/science.1077742. [DOI] [PubMed] [Google Scholar]
- Romero D, Aguilar C, Losick R, Kolter R. Amyloid fibers provide structural integrity to Bacillus subtilis biofilms. Proc Natl Acad Sci USA. 2010;107:2230–2234. doi: 10.1073/pnas.0910560107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sachs JL. Resolving the first steps to multicellularity. Trends Ecol Evol. 2008;23:245–248. doi: 10.1016/j.tree.2008.02.003. [DOI] [PubMed] [Google Scholar]
- Sauer K, Camper AK, Ehrlich GD, William J, Davies DG, Costerton JW. Pseudomonas aeruginosa displays multiple phenotypes during development as a biofilm. J Bacteriol. 2002;184:1140–1154. doi: 10.1128/jb.184.4.1140-1154.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schluter J, Foster KR. The evolution of mutualism in gut microbiota via host epithelial selection. PLoS Biol. 2012;10:e1001424. doi: 10.1371/journal.pbio.1001424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahid S, Markovic S, Linke D, Rossum B. van. Assignment and secondary structure of the YadA membrane protein by solid-state MAS NMR. Sci Rep. 2012;2:803. doi: 10.1038/srep00803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahid SA, Bardiaux B, Franks WT, Krabben L, Habeck M, van Rossum B-J, et al. Membrane-protein structure determination by solid-state NMR spectroscopy of microcrystals. Nat Methods. 2012;9:1212–1217. doi: 10.1038/nmeth.2248. [DOI] [PubMed] [Google Scholar]
- Shapiro BJ, Friedman J, Cordero OX, Preheim SP, Timberlake SC, Szabo G, et al. Population genomics of early events in the ecological differentiation of bacteria. Science. 2012;336:48–51. doi: 10.1126/science.1218198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shcherbo D, Merzlyak EM, Chepurnykh TV, Fradkov AF, Ermakova GV, Solovieva EA, et al. Bright far-red fluorescent protein for whole-body imaging. Nat Methods. 2007;4:741–746. doi: 10.1038/nmeth1083. [DOI] [PubMed] [Google Scholar]
- Smukalla S, Caldara M, Pochet N, Beauvais A, Guadagnini S, Yan C, et al. FLO1 is a variable green beard gene that drives biofilm-like cooperation in budding yeast. Cell. 2008;135:726–737. doi: 10.1016/j.cell.2008.09.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart PS, Franklin MJ. Physiological heterogeneity in biofilms. Nat Rev Microbiol. 2008;6:199–210. doi: 10.1038/nrmicro1838. [DOI] [PubMed] [Google Scholar]
- Sutherland IW. Biofilm exopolysaccharides: a strong and sticky framework. Microbiology. 2001;147:3–9. doi: 10.1099/00221287-147-1-3. [DOI] [PubMed] [Google Scholar]
- Timmerman CP, Fleer A, Besnier JM, De Graaf L, Cremers F, Verhoef J. Characterization of a proteinaceous adhesin of Staphylococcus epidermidis which mediates attachment to polystyrene. Infect Immun. 1991;59:4187–4192. doi: 10.1128/iai.59.11.4187-4192.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tojo M, Yamashita N, Goldmann DA, Pier GB. Isolation and Characterization of a Capsular Polysaccharide Adhesin from Staphylococcus epidermidis. J Infect Dis. 1988;157:713–722. doi: 10.1093/infdis/157.4.713. [DOI] [PubMed] [Google Scholar]
- Veenstra G, Cremers F, van Dijk H, Fleer A. Ultrastructural organization and regulation of a biomaterial adhesin of Staphylococcus epidermidis. J Bacteriol. 1996;178:537–541. doi: 10.1128/jb.178.2.537-541.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidal O, Longin R, Prigent-Combaret C, Dorel C, Hooreman M, Lejeune P. Isolation of an Escherichia coli K-12 mutant strain able to form biofilms on inert surfaces: involvement of a new ompR allele that increases curli expression. J Bacteriol. 1998;180:2442–2449. doi: 10.1128/jb.180.9.2442-2449.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West SA, Diggle SP, Buckling A, Gardner A, Griffin AS. The social lives of microbes. Annu Rev Ecol Evol S. 2007;38:53–77. [Google Scholar]
- Xavier JB, Foster KR. Co-operation and conflict in microbial biofilms. Proc Natl Acad Sci USA. 2007;104:876–881. doi: 10.1073/pnas.0607651104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xavier JB, Picioreanu C, van Loosdrecht MCM. A framework for multidimensional modelling of activity and structure of multispecies biofilms. Environ Microbiol. 2005;7:1085–1103. doi: 10.1111/j.1462-2920.2005.00787.x. [DOI] [PubMed] [Google Scholar]
- Xavier JB, Picioreanu C, van Loosdrecht MCM. A general description of detachment for multidimensional modelling of biofilms. Biotechnol Bioeng. 2005;91:651–669. doi: 10.1002/bit.20544. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.