Abstract
The circadian system is as an important regulator of immune function. Human inflammatory lung diseases frequently show time-of-day variation in symptom severity and lung function, but the mechanisms and cell types that are underlying these effects remain unclear. We show that pulmonary antibacterial responses are modulated by a circadian clock within epithelial club (Clara) cells. These drive circadian neutrophil recruitment to the lung via the chemokine CXCL5. Genetic ablation of the clock gene Bmal1 (also called Arntl or MOP3) in bronchiolar cells disrupts rhythmic Cxcl5 expression, resulting in exaggerated inflammatory responses to lipopolysaccharide and bacterial infection. Adrenalectomy blocks rhythmic inflammatory responses and the circadian regulation of CXCL5, suggesting a key role for the adrenal axis in driving CXCL5 expression and pulmonary neutrophil recruitment. Glucocorticoid receptor occupancy at the Cxcl5 locus shows circadian oscillations, but this is disrupted in mice with bronchiole-specific ablation of Bmal1, leading to enhanced CXCL5 expression despite normal corticosteroid secretion. In clock-gene disrupted mice the synthetic glucocorticoid dexamethasone loses anti-inflammatory efficacy. We now define a regulatory mechanism that links the circadian clock and glucocorticoid hormones to control both time-of-day variation and also the magnitude of pulmonary inflammation and responses to bacterial infection.
Human inflammatory lung diseases, including chronic obstructive pulmonary disease (COPD) and asthma, are responsible for 25% of premature deaths1. These conditions frequently show time-of-day variation in symptom severity and lung function, but the mechanisms and cell types that control this are unknown. The recruitment of immune cells to tissues is under strong circadian control2,3, and several immune cells, including macrophages and mast cells, exhibit robust oscillations of core clock genes and pro-inflammatory cytokines4–6. Animal studies show that susceptibility to endotoxic shock is clock dependent5,7–10. Further, the magnitude of response to pathogenic bacterial challenge varies with time of day 11. The immune response to bacterial infection is partly mediated by neutrophil recruitment to the affected site, which can also cause tissue damage and must therefore be tightly regulated12.
Inflammatory responses are inhibited by glucocorticoids, a suppressive mechanism that operates via an indirect ‘tethering’ action of the ligand-activated glucocorticoid receptor (GR)13. Glucocorticoid hormone secretion is clock-controlled14 and acts as a potent circadian-entrained stimulus in many peripheral tissues15. In addition, the function of the GR is determined by physical interactions with clock component proteins 16–18. It remains unclear how system-wide signals like glucocorticoids integrate with locally operating circadian oscillators to tune responses to environmental insults while avoiding excessive local tissue damage. The lung is uniquely exposed to environmental factors, and disordered amplitude and duration of inflammatory reactions in the lung underpin many lung diseases. We now define a regulatory mechanism that links the circadian clock and glucocorticoid hormones to control of both time-of-day variation and the magnitude of pulmonary inflammation and responses to infective agents.
RESULTS
Circadian control of pulmonary inflammation
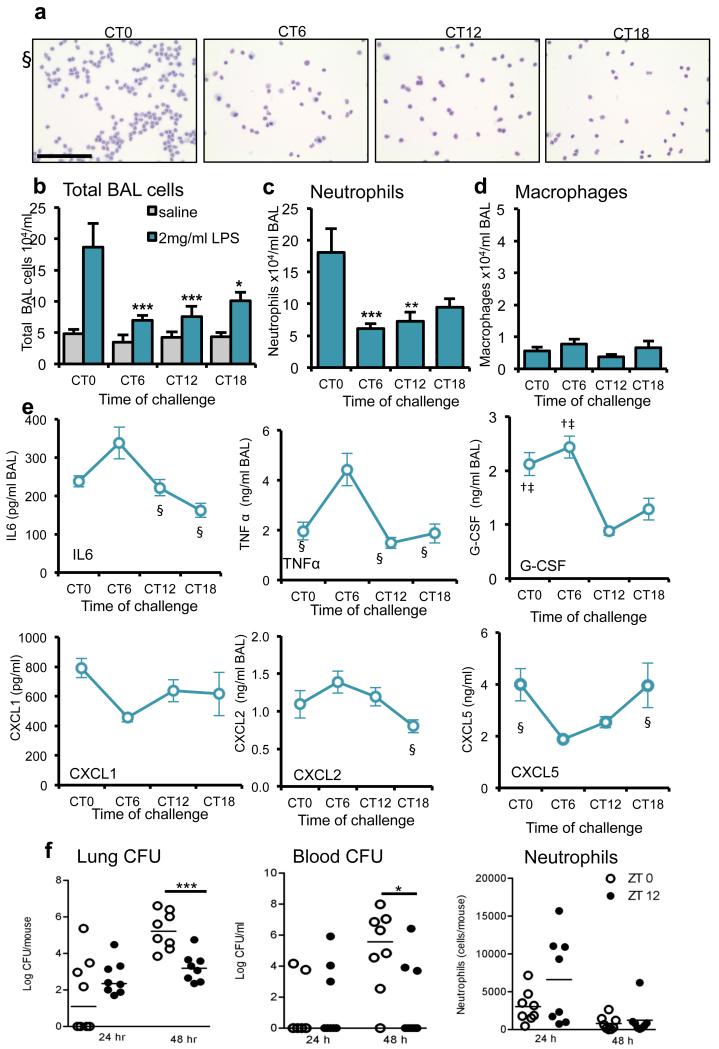
We assessed pulmonary responses to endotoxin challenge in C57/Bl6 mice across the circadian cycle using aerosolized lipopolysaccharide (LPS). Mice were entrained on 12:12hr light-dark cycles and housed in darkness for one circadian cycle before study. Animals were then subjected to LPS, and killed while maintained in darkness over the subsequent cycle (Aschoff type 2 protocol 19). This revealed a threefold variation in inflammatory responses measured in bronchoalveolar lavage (BAL) fluid, with peak infiltration of inflammatory cells, predominantly neutrophils, at circadian time 0 (CT0, dawn; Fig. 1a–c). Pulmonary macrophage numbers remained low throughout the cycle (Fig. 1d). Quantification of inflammatory and chemotactic cytokines in BAL fluid revealed strong circadian rhythmic changes in several cytokines (Fig. 1e). LPS binds to Toll-like receptor 4 (TLR4), activating the nuclear factor-κB (NF-κB) pathway and inducing pro-inflammatory cytokine transcription. Tlr4 is implicated in mediating LPS-induced phase shifts to light20. Tlr4 expression showed no variation across the circadian cycle (Supplementary Fig. 1), and thus circadian pulmonary LPS responses are unlikely to involve rhythmic regulation of Tlr4 expression.
Figure 1.
The pulmonary inflammatory response to LPS administration is gated by the circadian clock. (a) Staining of cytospins of BAL fluid from LPS-treated C57Bl/6 mice. Scale bar, 100 μm. (b) Total cell counts in BAL samples collected after LPS challenge at CT0 (n = 6), CT6 (n = 7) CT12 (n = 8) and CT18 (n = 6) or vehicle (n = 4/time point), two-way analysis of variance (ANOVA) and post hoc Bonferroni) (c) Neutrophil and (d) macrophage numbers in the same samples (one-way ANOVA and post hoc Bonferroni) (e) Cytokine levels in these BAL samples were compared between time points using one-way ANOVA and post hoc Bonferroni) § significantly different from CT6 (P≤0.05); † significantly different from CT12 (P≤0.05); ‡ significantly different from CT18 (P≤0.05).; G-CSF, granulocyte colony–stimulating factor. (f) Bacterial load in the lung and blood, and neutrophil counts in BAL, 48h after infection of C57Bl/6 mice with S. pneumoniae (n = 8/time point) at dawn (ZT0) and dusk (ZT12) (median values marked, Mann-Whitney U-test). Data are expressed as mean ± s.e.m and *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.005.
The observed time-of-day variation in LPS response suggested clock control of pulmonary immunity to bacterial infection. Accordingly, we infected mice at dawn (ZT0) and dusk (ZT12) with Streptococcus pneumoniae. At 24 h after ZT12 infection, there was increased pulmonary neutrophilia but no change in lung bacterial burden (Fig. 1f). By 48h, this early neutrophilia led to significant reductions in lung bacterial burden and dissemination to blood, which suggest a time-of-day mechanism for antibacterial defense in the lung. This apparent phase reversal in timing reflects the well-characterized kinetics of pneumococcal infection in mice, with an initial decline in bacteria over 8-12 h post infection followed by bacterial proliferation and tissue invasion 21.
Previous studies have highlighted the role of the macrophage clock in circadian responses to endotoxin4,5. Bronchoalveolar macrophages from mice expressing a Per2-luciferase fusion gene (Per2-luc mice) 22 exhibited circadian bioluminescent rhythms and clock gene expression (Supplementary Fig. 2a,b). Mice lacking the core clock gene Bmal1 (also known as Arntl or MOP3) within myelomonocytic (macrophages and monocytes) and neutrophils (LysM-Bmal1−/−; generated by crossing LysM-cre+/− with Bmal1fl/fl)5 showed suppressed clock gene expression; Nr1d1, and Nr1d2 (also known as rev-erbα and rev-erbβ respectively). in bronchoalveolar macrophages (Supplementary Fig. 2b). Nonetheless, after administration of aerosolized LPS, they demonstrated amplitude and phasing of neutrophil and cytokine responses similar to those of wild-type mice (Bmal1fl/fl)(Supplementary Fig. 2c,d and Supplementary Table 1), suggesting that cells of monocytic origin are not involved in the circadian gating we observed.
Targeting the circadian clock in pulmonary epithelial cells
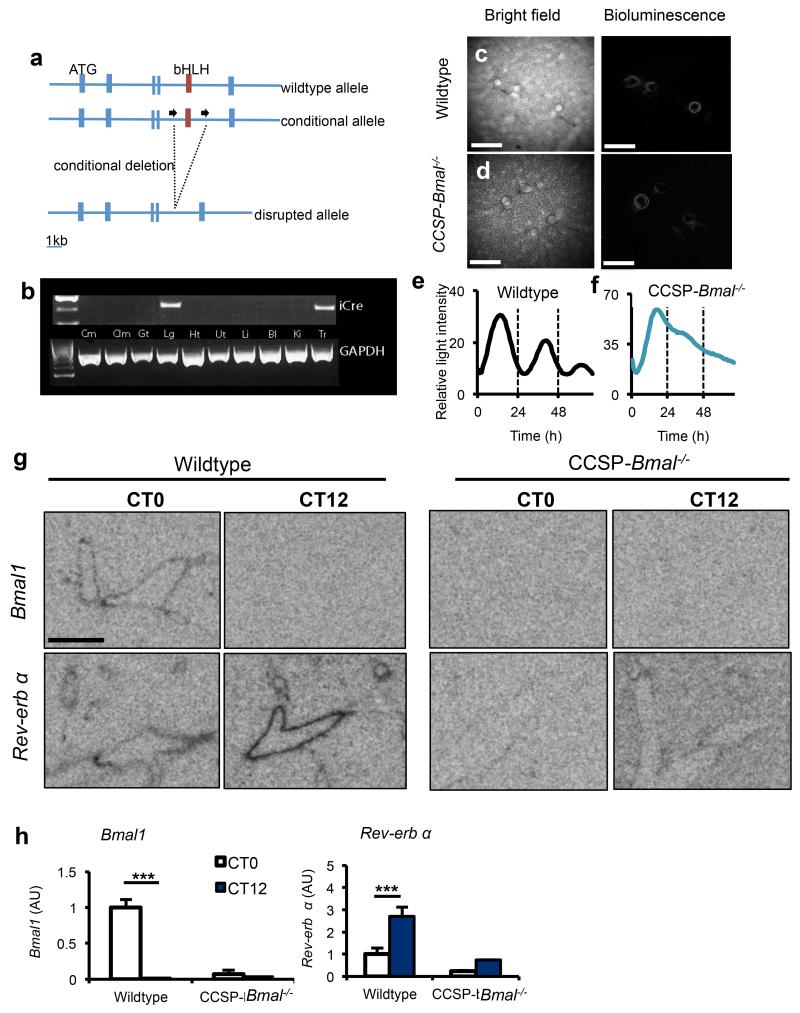
The non-ciliated epithelial cells lining the bronchioles have well-defined roles in pulmonary immune function23,24. Our earlier studies showed that these club cells express clock proteins and are essential for the maintenance of circadian rhythmicity in lung25. We crossed Bmal1fl/fl mice with mice expressing a codon-improved Cre recombinase (iCre) under the control of the club cell secretory protein (CCSP; also known as Scgb1a1) promoter26 (Fig. 2a) to produce BMAL1 loss in club cells and compared these with littermate controls carrying Bmal1fl/fl. In CCSP-icre+/− mice, expression of iCre was limited to the lung and trachea (Fig. 2b).
Figure 2.
Targeted ablation of Bmal1 in CCSP expressing cells disrupts circadian rhythmicity in whole lung. (a) Schematic illustrating the region surrounding the basic helix-loop-helix (bHLH) domain of the mouse Bmal1 locus, the conditional floxed allele and the disrupted allele. (b) Expression of iCre (and the housekeeping gene Gapdh) in tissues harvested from CCSP-Bmal−/− (Bmal1fl/fl; CCSP-icre+/−) mice (Cm, cerebrum; Clm, cerebellum; Gt, gut; Lg, lung; Ht, heart; Ut, uterus; Li, liver; Bl, bladder; Ki, kidney; Tr, trachea). (c,d) Ectopic lung slices from Bmal1fl/fl; CCSP-icre+/− (CCSP-Bmal−/−) or CCSP-icre negative littermate controls (wildtype) on a Per2-luc background were placed under a bioluminescence camera. Scale bars, 500 μm. (e,f) Bioluminescence intensity from bronchioles quantified and plotted as a function of time. Data are representative of 3 independent trials. (g, h) Quantification of clock genes Bmal1 and Nr1d1 (rev-erb α) expression in the bronchioles of lungs at CT0 (n=4/genotype) and CT12 (n=3/genotype) (scale bar, 5 mm, two-way ANOVA and post hoc Bonferroni. Data are expressed as mean ± s.e.m and *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.005.
Using mice expressing the PER2-luc transgene as a background strain, we imaged ectopic lung slices from Bmal1fl/fl; CCSP-icre −/−, and Bmal1fl/fl; CCSP-icre+/-25,27 (Fig. 2c,d and Supplementary Videos 1 and 2). This revealed circadian oscillations in Per2 in the bronchioles of Bmal1fl/fl; CCSP-icre −/− mice (25.8 ± 0.17 h mean ± s.e.m. Fig. 2e) that were disrupted in Bmal1fl/fl; CCSP-icre+/−mice (32.5 ± 0.8 h, Fig. 2f). Loss of rhythmicity was confined to bronchioles, as clock gene (Bmal1, Nr1d1, Per2 and Cry1) oscillations were similar in both genotypes in the whole lung (Supplementary Fig. 3a). Gene targeting had no impact on oscillations of clock genes in the liver (Supplementary Fig. 3b) or behavioral rhythms (Supplementary Fig. 3c). In situ hybridization revealed expression of Bmal1 and Nr1d1, at CT0 and CT12, respectively, in bronchial epithelia (Fig. 2g,h and Supplementary Fig. 4). As expected, targeted loss of Bmal1 in the bronchioles suppressed expression of its target gene, Nr1d1.
Bronchiolar epithelial cells drive circadian responses
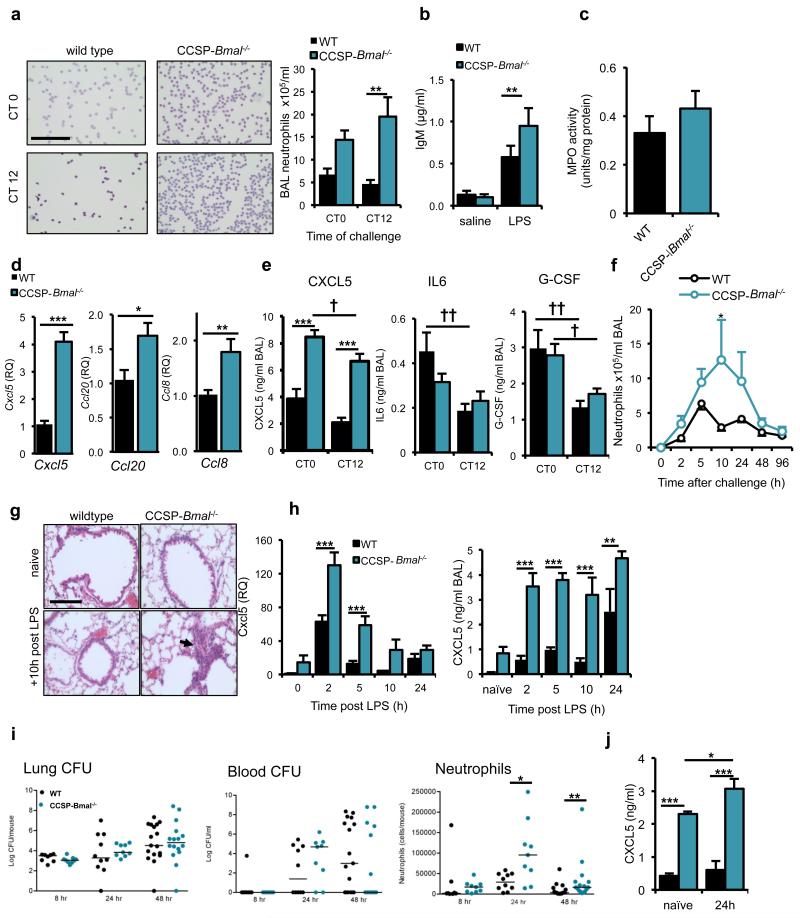
Bmal1fl/fl; CCSP-icre+/− mice exhibited disrupted pulmonary responses to LPS, with a three- to six-fold increase in neutrophilia and overall loss of circadian gating (ie the difference in neutrophilia between CT0 and CT12) (Fig. 3a). This elevated neutrophilia was associated with loss of barrier function as indicated by elevated BAL fluid IgM (Fig. 3b). Despite increased neutrophil recruitment to the lungs, there was no change in myeloperoxidase activity (Fig. 3c). Flow cytometry revealed a low-amplitude time-of-day change in neutrophil numbers in naive wild-type mice, whereas in Bmal1fl/fl; CCSP-icre+/− mice, neutrophil levels were elevated but not altered by time of day (Supplementary Fig. 5a). Although these data showed slightly elevated pulmonary neutrophilia under naive conditions, we did not observe marked gross pathological alterations (alveolar volume or ultrastructure) in tissues collected from aged naive mice (Supplementary Fig. 5b).
Figure 3.
Loss of the club cell clock results in enhanced neutrophil responses to LPS. (a) Neutrophil recruitment after aerosolized LPS challenge in CCSP-Bmal−/− and wild-type littermates at CT0 (n=8/genotype) and CT12 (n=8 wild-type, n=10 CCSP-Bmal−/−) (two-way ANOVA and post hoc Bonferroni; scale bar, 100 μm). (b) IgM levels in BAL fluid after saline (n = 6/genotype) or LPS exposure (n=11 CCSP-Bmal−/−, n = 9 wild-type littermates) (two-way ANOVA and post hoc Bonferroni). (c) Myeloperoxidase (MPO) activity levels in lung tissue from CCSP-Bmal−/− (n = 10) and wild-type littermates (n = 8) mice challenged with LPS at CT12. (d) Confirmation of a quantitative PCR (qPCR) array, measuring transcription of Cxcl5, Ccl20 and Ccl8 in CCSP-Bmal−/− (n = 12) and wild-type littermate (n = 7) lung tissue 2 h after LPS challenge, qPCR data normalized to wild-type littermate controls and values expressed as relative quantification (RQ) to wild-type levels, Student’s t-test). (e) Diurnal variation in LPS-induced cytokine secretion in CCSP-Bmal−/− mice (n = 8 at CT0, n = 10 at CT12) and wild-type littermates (n = 7/time point) (two-way ANOVA and post hoc Bonferroni; genotype differences ***P ≤ 0.005, time-of-day differences †P ≤ 0.05 and ††P ≤ 0.01,). (f) Quantification of BAL neutrophils at timed intervals after aerosolized LPS at ZT4 in CCSP-Bmal−/− mice (0h: n = 3 and 2h-96h: n = 4) and wild-type littermates (0h–10h: n = 4 and 24-96h: n=5); two-way ANOVA and post hoc Bonferroni). (g) H&E staining of histological sections of lung collected 10h after LPS illustrating neutrophil infiltration (arrow) (representative of n = 4; scale bar, 100 μm). (h) Cxcl5 transcript and CXCL5 protein levels in LPS-stimulated lungs from CCSP-Bmal−/− mice (0h: n = 3 and 2h-24h: n = 4)) and wild-type litttermates (n = 4); qPCR data normalized to wild-type naive lung; two-way ANOVA and post hoc Bonferroni). (i) Bacterial load in the lung and blood, and neutrophil counts in BAL after infection with S. pneumonia in CCSP-Bmal−/− mice (8h – 24h: n = 9; 48h: n = 19) and wild-type littermates (8h: n = 8; 24h: n = 10; and 48h: n = 14), (median values marked, Mann-Whitney U-test). (j) CXCL5 levels in BAL fluid from naïve, and 24h post bacterial infection in CCSP-Bmal−/− mice (naïve: n = 4 and infected: n = 6) and wild-type littermates (n = 5), two-way ANOVA and post hoc Bonferonni). Data are expressed as mean ± s.e.m and *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.005.
We next quantified a wide range of inflammatory mediators (Supplementary Table 2) and found that six mediators were substantially up-regulated in Bmal1fl/fl; CCSP-icre+/− mice. We subsequently confirmed that three of these six (Cxcl5, Ccl20 and Ccl8) were significantly different in a repeat cohort of LPS-treated mice (Fig. 3d and Supplementary Fig. 6a,b), and, as the only neutrophil chemo-attractant, CXCL5 emerged as a candidate effector molecule. CXCL5 expression was also in phase with the peak of LPS-induced neutrophilia (Fig. 1e). LPS-treated Bmal1fl/fl; CCSP-icre+/− mice secreted significantly more (two- to threefold) CXCL5 in BAL fluid compared to Bmal1fl/fl; CCSP-icre−/− at both CT0 and CT12 (Fig. 3e), unlike any other cytokine (Fig. 3e and Supplementary Table 3). Neutrophilic inflammation in response to an initial challenge in Bmal1fl/fl; CCSP-icre+/− mice compared to Bmal1fl/fl; CCSP-icre−/− persisted at a high level and for over several days after challenge (Fig. 3f,g), accompanied by elevated CXCL5 (Fig. 3h).
We next assessed responses to S. pneumoniae infection. As predicted from the LPS challenge, we saw marked elevation of neutrophil recruitment in Bmal1fl/fl; CCSP-icre+/− mice, probably in response to elevated CXCL5 production (Fig. 3i,j). However, this augmented neutrophilia occurred without a difference in bacterial CFU recovered from lung, or circulation. This contrasts with the time-of-day protective effect seen in wild-type animals and demonstrates an unexpected and counterintuitive consequence of circadian disruption, which results in augmented recruitment of neutrophils apparently lacking full antibacterial efficacy. This is compatible with earlier observations identifying no commensurate induction in pulmonary myeloperoxidase activity with augmented neutrophilia in response to LPS.
CXCL5 drives pulmonary rhythmic inflammatory responses
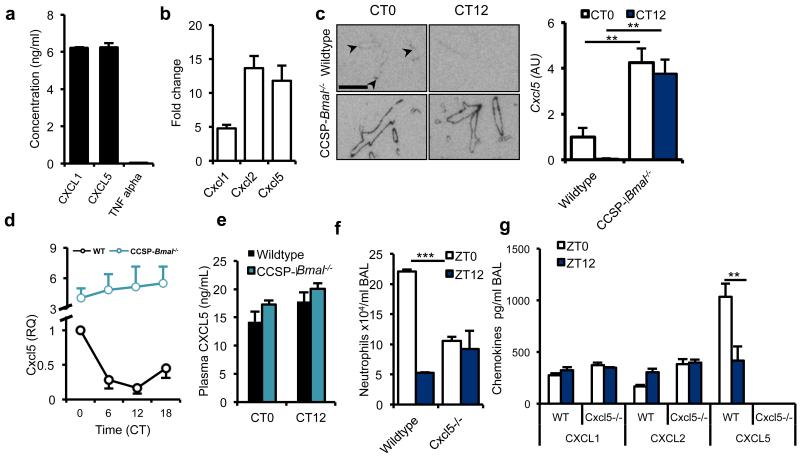
Using LPS-stimulated purified mouse bronchial epithelial cells, we observed expression of CXCL5 and CXCL1, but not tumor necrosis factor-α (TNF-α) (Fig. 4a). CXCL5 acts as a potent neutrophil chemo-attractant, as assessed by migratory responses of mouse bone marrow–derived neutrophils (Supplementary Fig. 7). Primary human bronchial epithelial cells stimulated with interleukin-1β (IL-1β) also expressed CXCL5 (Fig. 4b); in contrast, pulmonary macrophages do not28. In situ hybridization demonstrated Cxcl5 expression in naive Bmal1fl/fl littermate controls, that was confined to the bronchioles and elevated at CT0, while in Bmal1fl/fl; CCSP-icre+/− mice expression was elevated at both time points within these tissues (Fig. 4c). Expression of Cxcl5 mRNA co-localized with CCSP expression within bronchiolar epithelial cells (Supplementary Fig. 8). Profiling over 24 h in wild-type mice revealed that Cxcl5 mRNA expression was rhythmic, peaking in the early light phase, coincident with the elevation in protein (Fig. 4d). In Bmal1fl/fl; CCSP-icre+/− mice, mRNA expression was elevated (fourfold), compared to Bmal1fl/fl; CCSP-icre−/− and non-rhythmic (Fig. 4d). These elevated baseline CXCL5 levels in naive CCSP-Bmal−/− mice are lung-specific, as serum CXCL5 levels showed no time-of-day or genotype difference (Fig. 4e). Thus, our results suggest that CCSP-expressing cells are the critical cell type mediating circadian responses to endotoxin challenge.
Figure 4.
BMAL1 regulates CXCL5 expression in CCSP positive cells. (a) Secretion of CXCL1, CXCL5 and TNF-α by cultured mouse club cells (representative of 3 independent trials). (b) Expression of Cxcl1, Cxcl2 and Cxcl5 by primary human normal bronchial epithelial cells in response to IL-1β stimulation (values normalized to expression levels in unstimulated epithelial cells, n = 3). (c) Localization of Cxcl5 expression in the lungs of CCSP-Bmal−/− and wild-type littermates at CT0 (n = 4) and CT12 (n = 3); two-way ANOVA and post hoc Bonferroni; arrowheads mark bronchioles where expression is low; scale bar, 5 mm). (d) Expression of Cxcl5 mRNA over circadian time in lung from CCSP-Bmal−/− mice (n = 4) and wild-type littermates (n = 4), normalized to wild-type littermate value at CT0. (e) Circulating levels of CXCL5 in plasma determined at two opposing time points by ELISA in CCSP-Bmal−/− (CT0: n = 3 and CT12: n = 4) and wild-type littermates (CT0: n = 4 and CT12: n = 5). (f,g) Neutrophil counts and chemokine levels in BAL from Cxcl5−/− mice exposed to LPS at two opposing time points, n = 3, one-way ANOVA and post hoc Bonferroni. Data are expressed as mean ± s.e.m and *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.005.
To determine the role of CXCL5 in conferring clock control to pulmonary innate immunity, we studied global CXCL5-knockout mice. We observed an attenuated dawn (Zeitgeber time 0, ZT0) neutrophilic response to nebulized LPS, consistent with our previous results29 (Fig. 4f). Together with the observation that there were no significant time-of-day differences in expression of other CXCL chemokines (Fig. 4g), our findings suggest that CXCL5 regulates pulmonary responses to infection and plays a central role in conferring clock control of inflammation.
Role of glucocorticoids in the rhythmic regulation of CXCL5
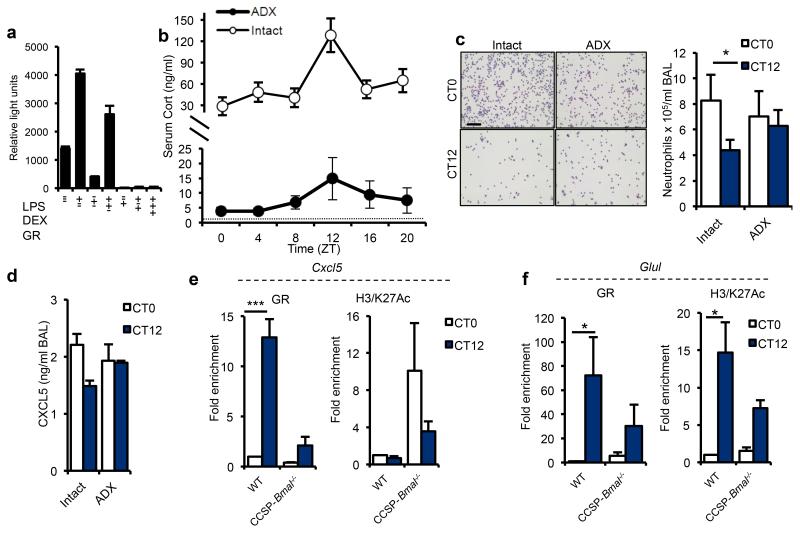
The isolated Cxcl5 proximal promoter did not show intrinsic circadian rhythmicity (Supplementary Fig. 9a) or respond to BMAL1 and CLOCK either alone or together (Supplementary Fig. 9b), but it was strongly repressed by dexamethasone-activated GR, in cells transfected with a GR expression vector 30 (Fig. 5a). We next explored whether systemic adrenal glucocorticoids activating GR act as a circadian repressor of CXCL5 expression. We assessed circadian variation in pulmonary neutrophilic inflammation in adrenalectomized mice. The circadian rhythm in circulating corticosterone in these mice was suppressed but still detectable (Fig. 5b), due to an extra-adrenal source31. Adrenalectomy caused loss of rhythmic CXCL5, and pulmonary neutrophilia in response to nebulized LPS but no general increase in lung inflammation (Fig. 5c,d). In contrast, intraperitoneal injection of LPS to adrenalectomised mice caused a significant increase in circulating IL-6 (Supplementary Fig. 10a). Measurement of pulmonary Per2, Bmal1 and Nr1d1 expression revealed similar high-amplitude, time-of-day–dependent changes in both adrenalectomized and intact animals, indicating that altered pulmonary responses did not arise as a consequence of internal de-synchronization of the local clock (Supplementary Fig. 10b).
Figure 5.
Endogenous glucocorticoid rhythms regulate rhythmic repression of Cxcl5. (a) The effects of LPS, glucocorticoids and GR over-expression on activity of the Cxcl5-luciferase promoter in RAT-1 cells, as assessed by luciferase activity (representative of 4 independent trials). (b) Concentration of circulating glucocorticoids (Cort) in intact (n = 5/time point) and adrenalectomized (ADX, n = 8/time point) C57Bl/6 mice across the circadian day; dotted line represents lower limit of detection. (c) LPS-induced neutrophilia in ADX mice (CT0: n = 11; CT12: n = 13) and intact counterparts (n = 12) (scale bar, 100 μm Student’s t-test with Bonferroni correction for multiple comparisons) (d) LPS induced CXCL5 production in ADX mice (CT0: n = 6; CT12: n = 7) and intact counterparts (CT0: n = 6; CT12: n = 5). (e,f) Chromatin immunoprecipitation (ChIP) analysis in CCSP-Bmal1−/− and littermate controls (WT) of GR binding and H3/K27Ac at CT0 and CT12 to a hypersensitive region on the Cxcl5 promoter (e) and the Glul promoter (f) (n = 3; two-way ANOVA and post hoc Bonferroni). Data are expressed as mean ± s.e.m and *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.005.
Genome-wide mapping of GR occupancy in relation to chromatin architecture and target gene transcription has previously revealed binding sites in the proximal CXCL5 promoter that overlap with regions of DNase1 hypersensitivity, indicating enhancers30. We identified strong diurnal regulation of GR binding to the Cxcl5 glucocorticoid response element in whole lung of Bmal1fl/fl; CCSP-icre−/− mice (Fig. 5e), with augmented binding coincident with the night-time rise in endogenous corticosterone (Fig. 5b). In Bmal1fl/fl; CCSP-icre+/− mice, GR recruitment to Cxcl5 was reduced and no longer rhythmic (Fig 5e). Acetylation of lysine 27 on histone H3 (H3/K27Ac) is a robust epigenetic mark of active enhancers32,33. We measured H3/K27Ac to determine enhancer activity and gauge the consequence of GR recruitment. We found increased H3/K27Ac in targeted mice at both time points, consistent with the constitutive increase in Cxcl5 gene expression (Fig. 5e). These data are therefore compatible with a model in which disrupted GR-mediated repression leads to up-regulation of Cxcl5 mRNA in clock gene-targeted mice.
Extending these observations to a GR-transactivated gene, glutamine synthetase (Glul), we observed diurnal variation in GR binding, and gene expression in wild-type tissue (Fig. 5f, Supplementary Figure 11). Histone H3 acetylation followed GR recruitment, as predicted, as a marker of enhancer activity (Fig. 5f). In Bmal1fl/fl; CCSP-icre+/− mice, we saw reduced GR enrichment and reduced H3/K27Ac (Fig. 5f) with loss of diurnal Glul expression in bronchioles but not in other lung structures (Supplementary Figure 11)., Thus, club cell disruption of Bmal1 leads to altered dynamics of GR activity specifically in bronchiolar epithelial cells for both GR-repressed and GR-activated genes. The differences we observed were not attributable to altered expression of GR in the lung (Supplementary Fig. 12a,b).
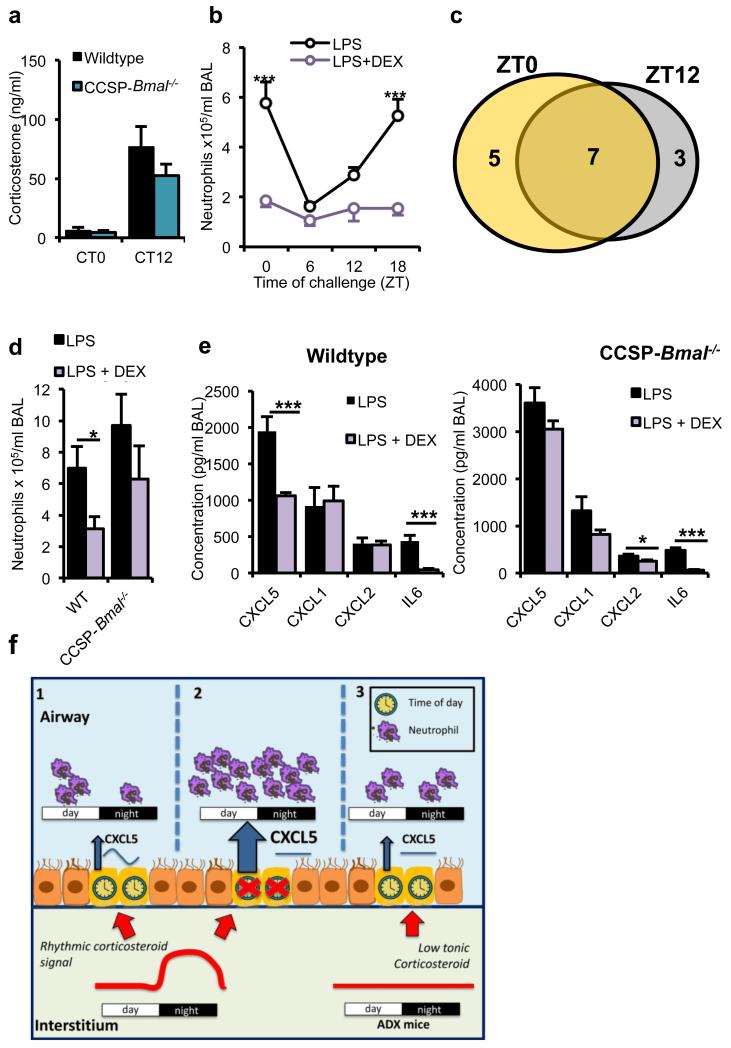
Bmal1fl/fl; CCSP-icre+/− mice showed the same diurnal variation in hypothalamic-pituitary-adrenal axis activity, as determined by serum corticosterone, as wild-type mice (Fig. 6a). This reveals an impact on GR promoter occupancy, consequent to targeting Bmal1 in club cells, which is independent of rhythmic adrenal activity. To determine whether differences in GR recruitment to cis elements in the lung were solely dependent on circadian variation in corticosterone or whether a local cellular control system was operating, we treated Bmal1fl/fl; CCSP-icre−/− mice with systemic dexamethasone (DEX) before induction of pulmonary inflammation. This activated known GR-responsive genes within the lung, confirming action on target tissue (Supplementary Fig. 13). LPS treatment induced circadian-gated neutrophilic responses, which was reduced by DEX treatment, irrespective of challenge time (Fig. 6b). Of 15 cytokines exhibiting sensitivity to DEX, 7 were responsive at both time points tested (Fig. 6c and Supplementary Fig. 14). In contrast, in Bmal1fl/fl; CCSP-icre+/− mice, suppression of LPS-induced neutrophilia by DEX was lost (Fig. 6d), and this was associated with failure to suppress CXCL5 (Fig. 6e).
Figure 6.
Anti-inflammatory effects of glucocorticoids depend on a bronchiolar clock. a) Endogenous circadian rhythms of circulating corticosterone in CCSP-Bmal−/− mice (CT0: n = 11; CT12: n = 10) and wild-type littermates (CT0: n = 10; CT12: n = 11). (b) Effects of pretreatment with DEX (1 mg per kg body weight) on pulmonary neutrophilia in C57Bl/6 mice across the circadian day (n = 6 per time point; two-way ANOVA, post hoc Bonferonni;). (c) Venn diagram illustrating the numbers of cytokines significantly repressed by DEX at ZT0 (yellow), ZT12 (gray) or both, n = 6 per time point. (d,e) BAL neutrophil numbers after indicated treatments in CCSP-Bmal−/− (LPS: n = 6; + DEX: n = 7) and littermate (WT) controls (LPS: n = 3; + DEX: n = 6).; Student’s t-test; *P ≤ 0.05) (d) and local cytokine and chemokine production in the same study; one-way ANOVA and post hoc Bonferroni (e). (f) Schematic illustrating how a local bronchiolar clock and circulating rhythmic glucocorticoids regulate the response to timed endotoxin and bacterial challenge. In the normal state, CXCL5 is regulated by interaction of a local circadian bronchiolar clock and systemic repressive glucocorticoid signals of adrenal origin, resulting in clock-regulated responses to endotoxin and bacterial infection (1). Ablation of Bmal1 in the epithelial club cells leads to disrupted GR signaling, non-rhythmic expression of CXCL5 and neutrophilia, despite a persistent glucocorticoid rhythm (2). In this state, neutrophil chemotaxis may be impaired, leading to reduced efficiency of bacterial clearance. In adrenalectomized mice, loss of the rhythmic repressive adrenal signal leads to loss of circadian gating of CXCL5 and an increased neutrophilic response to endotoxin (3). Data are expressed as mean ± s.e.m and *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.005.
DISCUSSION
Neutrophilic pulmonary inflammation is a feature of adult respiratory distress syndrome and COPD and is an important common feature of severe steroid-resistant asthma34–36. Little is known of the signals regulating neutrophil trafficking and antibacterial action in the lung. We show that a local pulmonary clock specific to bronchiolar CCSP-expressing cells regulates dynamic changes in response to in vivo bacterial challenge. Furthermore, targeting Bmal1 in these cells leads to an exaggerated but apparently ineffective neutrophilic inflammatory response to bacterial infection. This highlights the importance of the bronchiolar epithelium as part of a cell-specific timing mechanism regulating overall homeostasis of pulmonary immunological responses.
Our results show that CXCL5, a club-cell produced neutrophil chemokine, is clock-controlled, and mediates circadian variation in lung inflammation. In support of this notion, CXCL5-null mice showed a loss of circadian rhythmic responses to LPS. The specific role of CXCL5 is notable, as several members of this chemokine family regulate neutrophil homeostasis (CXCL1, CXCL2, CXCL5 and CXCL15)29,37–40, but only CXCL5 was circadian. CXCL5 is also associated with several autoimmune diseases, including rheumatoid arthritis, that have circadian phenotypes 41.
Neutrophils are proposed to use chemokines as initial signals to guide localization but then switch to bacterial-derived signals for final homing42. If excess CXCL5 is secreted, this may impair final neutrophil targeting of bacteria and hence killing. However, the augmented neutrophilia is predicted to lead to excessive pulmonary damage. Therefore, our results support a role for the pulmonary circadian clock in tissue homeostasis and responses to infection. Furthermore, aberrant control of neutrophilic lung inflammation, an important feature in COPD, may arise as a consequence of disruption of normal circadian function. In this context, it is notable that cigarette smoke suppresses pulmonary clock gene expression43,44.
Our studies, using whole-lung tissue, reveal strong time-of-day changes in GR occupancy on Cxcl5. This may indicate an underlying change in chromatin architecture, resulting in enhancer activation and thereby impaired GR recruitment; alternatively, it may indicate that impaired GR recruitment, due to altered chromatin architecture, results in loss of GR enhancer inhibition (or it could be a combination of both mechanisms). Similarly, analysis of Glul revealed impairment of GR function with less GR recruitment and, on this transactivation target, consequently reduced H3/K27Ac. Taken together, this implies a broader dysregulation of GR function in the Bmal1-disrupted cells. Furthermore, adrenalectomy, which removes endogenous glucocorticoid timing signals, also abolished time-of-day variation in CXCL5 and neutrophil infiltration without de-repressing neutrophilia. Thus, loss of normal circadian GR function in club cells is dominant over a rhythmic corticosteroid signal in the phenotype we observed.
Using systemic DEX administration, which models therapeutic intervention to treat human pulmonary inflammation, we found loss of glucocorticoid responses in CCSP-Bmal1−/− mice. This provided further evidence for a local circadian circuit regulating bronchiolar GR function. Our studies were conducted under tightly matched time-controlled conditions, and one possibility not tested is whether glucocorticoid treatment shifted the phase of target cells in addition to the impact on the amplitude of response.
In conclusion, we identify resonance between an oscillating circadian systemic glucocorticoid signal and locally operating circadian control of GR function to confer clock control of pulmonary innate immunity. We define a mechanism that couples the circadian clockwork and bronchiolar GR to pulmonary innate immunity (Fig. 6f). The bronchiolar club cell, acting through a single neutrophil chemokine, is the final effector pathway. The efficacy of therapeutically administered glucocorticoids depends on intact clock function in the airway. Therefore, human conditions of persisting pulmonary neutrophilic inflammation, which characteristically fail to resolve in response to glucocorticoid therapy (COPD, adult respiratory distress syndrome and glucocorticoid-resistant asthma), may share a common underlying mechanism of clock disruption, either de novo or in response to particular inflammatory signals.
ONLINE METHODS
Transgenic mouse lines
Conditional club cell Bmal1-knockout mice (CCSP-Bmal1−/−) were generated by breeding Bmal1fl/fl mice (Jackson Labs 007668) with CCSP-icre mice, which are an improvement on the CCSP-Cre line26,47. Both lines were on a C57BL/6 background. Tissue expression of iCre in CCSP-Bmal−/− mice was assessed by RT-PCR (forward primer AGATGCCAGGACATCAGGAAC CTG; reverse primer ATCAGCCACACCAGACACAGAGATC). CCSP-Bmal−/− mice were subsequently crossed onto a PER2-luc background22. Mice lacking Bmal1 in cells of a monocyte lineage (LysM-Bmal1−/−) were utilized as described elsewhere5. In studies utilizing conditional targeted mice, wild-type littermates (Bmal1fl/fl) were used as control; these mice carried no copies of the Cre (LysM) or iCre (CCSP). Transgenic and littermate control (C57BL/6) mice were routinely housed in 12:12 light/dark (L:D) cycles with ad libitum access to food and water. All experiments were carried out in accordance with the Animals (Scientific Procedures) Act 1986 (UK) or the Guide for the Care and Use of Laboratory Animals and with approval of the Children’s Hospital of Philadelphia Institutional Animal Care and Use Committee (USA). In all studies, male mice were used age 6-24 weeks). Analysis of wheel-running activity determined behavioral circadian period under different lighting conditions.
Aerosolized lipopolysaccharide
Mice were placed into a Perspex chamber and exposed to aerosolized lipopolysaccharide (0127:B8; 2 mg/ml) or vehicle (saline) for 20 min. Animals were returned back to their cages for 5 h (or 2–96 h in time-course studies) before killing (pentobarbital i.p.). For circadian timed studies, animals were maintained in constant darkness (D:D) 24 h before and during LPS exposure at CT0, CT6, CT12 or CT18. To assess the effects of glucocorticoids on the pulmonary inflammatory response, mice were pretreated with dexamethasone (1 mg per kg body weight, i.p.) 1 h before LPS exposure. After LPS exposure, the lungs were lavaged using 1 ml BAL fluid (10 mM EDTA and 1% BSA), instilled and removed via a tracheal cannula. The left lobe of the lung was collected for RNA analysis. Lavage fluid was centrifuged and the supernatant utilized for cytokine and chemokine analysis using the Bioplex Suspension Array System (BioRad) or ELISA (R&D Systems). The pellet was resuspended in fresh BAL fluid to allow quantification of total cell numbers using a Casy Cell Counter (Schärfe System, Germany), and subsequently cytospins were produced that were stained with Leishman’s eosin methylene blue (VWR) to enable detection of macrophages and neutrophils48. Images were captured (Leica DM2000 microscope and Leica DFC296 camera), and macrophages and neutrophils were counted from 5 individual fields to determine the relative percentage of each. By combining total cell counts and relative cell counts, total numbers of macrophages and neutrophils were calculated.
Intraperitoneal lipopolysaccharide
Mice (ADX or intact controls) were treated with intraperitoneal LPS (1 mg per kg body weight, Sigma Aldrich) at CT0, and the concentration of IL-6 in the serum was analyzed by ELISA (R&D Systems) 4 h later5.
Primary cell culture
Macrophages were isolated from the bronchoalveolar space of naive mice by lavage with sterile saline (2 × 1 ml). The retrieved fluid was pooled from 5–10 mice, centrifuged and the pellet harvested immediately for RNA (RNeasy, Qiagen) or resuspended in RPMI and plated out for cell culture. After 2 h, nonadherent cells were removed by gentle washing. For photomultiplier tube (PMT) analysis25, RPMI was replaced by recording medium containing luciferin (Promega). Club cells were isolated as described previously25, stimulated with LPS (100 ng/ml, Sigma Aldrich) for 24 h and the supernatant assayed for cytokines (ELISA, R&D Systems).
Primary normal human bronchial epithelial cells
Primary NHBE cells were purchased from Lonza (Walkersville, MD) and cultured as monolayers in serum-free bronchial epithelium growth medium supplemented with growth factors (BEGM, Lonza). Subculture reagents (Reagentpack, Lonza) were used according to the supplier’s instructions. Passage 3 cells were seeded in a 24-well plate (6 × 105 cells/well) and allowed to attach overnight before synchronization with 50% FBS in BEGM (1 h). Cells were then treated 22 h after synchronization with 1 ng/ml IL-1β. 2 h later, RNA was extracted.
RNA analysis
RNA was extracted from tissue using Trizol Reagent (tissue) or the RNeasy method (Cells; Qiagen). After DNase treatment, cDNA was prepared (RNA to cDNA kit, Applied Biosystems). qPCR was performed using commercial primer/probe mixes (Applied Biosystems) or primer and probe sets as listed in Supplementary Table 4. Genes encoding β-actin and 36B4 were utilized as housekeeping genes for TAQMAN and SYBR green qPCR, respectively.
Chemotaxis assay
Chemotaxis assays using bone marrow–derived neutrophils were performed using a modified Boyden chamber with a 5-μm polycarbonate filter (NeuroProbe, USA). Bone marrow was flushed from femurs and tibias, and neutrophils were negatively selected out using the EasySep Mouse Neutrophils Enrichment Kit (STEMCELL Technologies). 1.25 × 105 cells were applied to the upper compartment and recombinant mouse CXCL5 (0.5ng/ml - 5μg/ml) (R&D systems) to the lower compartment. After 45 min, neutrophil migration across the membrane was quantified using CellTiter Glo (Promega, UK).
Ectopic lung slices
Precision-cut ectopic lung slices (275 μm) were prepared25. After washes to remove residual agarose, slices were placed onto cell culture inserts (Millicell) within 35-mm dishes containing 1 ml recording medium, sealed with a coverslip and placed under a self-contained Olympus LV200 luminescence microscopy system fitted with a cooled Hamamatsu C9100-13 EM-CCD camera (Olympus, Japan) as previously described27. Individual regions of interest were delineated using ImageJ software (v1.41o).
Histology and immunohistochemistry
Lungs were inflated with 4% PFA and fixed overnight before processing and paraffin embedding. 5-μm sections were cut and mounted onto slides. Immunohistochemical staining was performed using an antibody to GR (1:400, clone M-20, sc-1004, Santa Cruz) and standard procedures (including 20 minute antigen retrieval with Citrate Buffer pH6.0). After DAB staining, sections were counterstained with Toludine Blue. H&E staining and Masson’s trichrome staining of lung sections were carried out using standard protocols.
Corticosterone measurements
Levels of corticosterone in serum samples were measured using enzyme immunoassay as per kit instructions (ENZO Life Sciences Ltd). Samples were diluted 1:40.
Adrenalectomy
Adrenalectomies were performed via a dorsal approach under isoflurane anesthesia. Incisions (1 cm) were made parallel to the spinal cord, and the muscle was pulled away before removal of the adrenal glands via blunt dissection. Analgesic was given peri- and postoperatively. Adrenalectomized (ADX) mice were provided 1% saline as drinking water to maintain salt balance and were monitored for at least 1 week before additional procedures. Assessment of successful surgery was achieved by quantifying corticosterone in serum samples.
PCR array
A PCR-based array (PAMM 011, SABiosciences) was utilized to detect differences in expression of inflammatory genes in lung tissue harvested from mice 2 h after aerosolized LPS challenge. RNA was extracted (Trizol), cleaned up (RNeasy) and the integrity checked (Agilent 2100 Bioanalyser). The array and analysis was carried out as per kit instructions.
Cxcl5 promoter analysis
Murine Cxcl5 promoter was cloned (−2090 to −43), inserted into an insulated PGL4 vector49 and expressed in RAT1 cells. Stably transfected cells were placed under PMTs, and transiently transfected cells were treated with LPS (100 ng/ml) in the absence or presence of DEX (100 nM) or overexpression of the GR, BMAL or CLOCK.
Chromatin immunoprecipitation
Mouse lung was harvested at CT0 or CT12, diced into small pieces, fixed and homogenized. ChIP assays were performed as per standard protocols50 using anti-GR (3μg/ml, clone M-20, sc-1004, Santa Cruz) and anti–acetylated H3/K27 (3μg/ml, ab4729, Abcam). DNA fragmentation was confirmed by agarose gel electrophoresis, with a size distribution of less than 500 bp. Nonspecific rabbit IgG (Santa Cruz Biotechnology) was used as a control. Chromatin was cleaned up using MinElute kit (Qiagen). Real-time quantitative PCR was performed using primers specific to the glucocorticoid hypersensitive sites on the Cxcl5 and Glul promoters30. Experiments were repeated on three separate occasions.
In situ hybridization
Lung tissue was collected from CCSP-Bmal−/− and wild-type littermates at CT0 and CT12. The trachea was cannulated postmortem and lungs inflated with 1 ml PBS/OCT compound medium mixture (1:1). The trachea was tied off, and the lungs and heart were removed en bloc and snap frozen. Frozen sections (12 μm) of lung were mounted onto slides and stored at −80 °C. The mouse Nr1d1, Bmal1, Cxcl5 and Glul plasmids were prepared by cloning a fragment of cDNA using the following primers: Glul: forward GCGACATGTACCTCCATCCT and reverse ACC CGATGCAAGATAAAACG; Cxcl5: forward TAGAGCCCCAATCTCCACAC and reverse GTGCATTCCGCTTAGCTTTC; Nr1d1: forward AGGGCACAAGCAACATTACC and reverse CTGAGAGAAGCCCACCAAAG; Bmal1: forward GTAGATCAGAGGGCGACA GC and reverse GGGAGGCGTACTTGTGATGT). cDNA fragments were cloned into pGEMT-easy vector (Promega), which was linearized using appropriate restriction enzymes, to generate templates for sense and antisense transcripts. Antisense and sense riboprobes were synthesized in the presence of [33P]UTP. Probes were hybridized overnight at 60 °C, and hybridization signals were visualized by film autoradiography. Films were scanned using a CoolSNAP-Pro camera (Photometrics, Marlow, UK) while on a light box. Signal intensity was quantified by densitometry analysis of autoradiographs. Optical density (relative units, RU) within the target region was calculated on each section with a minimum of 3 sections per animal, 3 or 4 animals per group.
Western blotting
Lung tissue was collected at ZT12 and protein prepared in FastPrep-24 lysing matrix tubes (MP Biomedicals) then lysed using Radio-Immunoprecipitation Assay (RIPA) buffer (50mM TricCl pH 7.4, 1% NP40, 0.25% Na-deoxycholate, 150mM NaCl, 1 mM EDTA) containing protease (Calbiochem, San Diego, CA, USA) and phosphatase inhibitors (Sigma-Aldrich Corp.). 15μg protein was run on an SDS 8-12% Tris-Glycine gel (Novex, Life Technologies) then transferred onto a 0.2μm nitrocellulose membrane (BioRad) overnight, blocked with 1% milk and probed for GR (1:1000, clone M-20, sc-1004, Santa Cruz) 25. Immunoreactivity was visualized using enhanced chemiluminescence (GE Healthcare).
Flow cytometry
Resident cells were isolated from naive lung at CT0 and CT12 via lavage with BAL fluid. After staining with a live/dead marker (Life Technologies #L10119, 1:1000) in PBS, Fc receptors were blocked (1:100 anti-CD16/32, eBioscience #14-0161) before application of the following antibodies in 30 μl FACS buffer (PBS, 1% BSA and 0.1% sodium azide): CD11b-PerCP-Cy5.5 (1:200, clone M1/70, #45-0112), CD11c-APC (1:400, clone N418, #17-0114), Ly6G-FITC (1:100, clone RB6-8C5, #11-5931) and F4/80-PE-Cy7 (1:200, clone BM8, #25-4801) (all purchased from eBioscience). After washing, cells were resuspended in 50 μl FACS buffer and fixed by addition of an equal volume of 3.6% formaldehyde for 20 min. Cells were resuspended in FACS buffer, and analysis was carried out on a BD LSR II flow cytometer. Neutrophils were identified as F4/80−CD11b+Ly6G+. Alveolar macrophages were identified as F4/80+CD11c+CD11blo.
Streptococcus pneumonia infection
Mice were infected at ZT0 (C57BL/6 only) or ZT12 (C57BL/6 and CCSP-Bmal1−/−) with Streptococcus pneumoniae (serotype 2), strain D39 (National Collection of Type Cultures [NCTC] 7466), intranasally (2 × 104 CFU per mouse) under isoflurane anesthesia. Animals were killed 8, 24 or 48 h later via terminal anesthesia. Blood was collected from the femoral artery (into 1:20 heparin), and the lungs were lavaged with BAL fluid and then dissected out. The BAL fluid was spun (300g, 5 min) to isolate cellular infiltrates. A lung tissue homogenate was prepared by digesting minced tissue in a solution of 0.154 mg/ml collagenase I and II (Liberase TM, Roche #05401127001) and 50 μg/ml DNase I (Roche #11284932001). After 30 min at 37 °C, the reaction was stopped with EDTA, and the digest passed through a 70 μM filter. Whole blood and lung homogenate were plated out on Columbia blood-agar plates in serial dilution overnight at 37°C and CFUs counted the next day. After red blood cell lysis (in 0.15 M ammonium chloride, 1 M potassium hydrogen carbonate, and 0.01mM EDTA, pH 7.2), BAL cells were counted, and flow cytometry was used to quantify neutrophil numbers (as above).
IgM ELISA
IgM levels in BAL samples were quantified using an ELISA kit for mouse IgM as per kit instructions (Immunology Consultants Laboratory Inc, USA) using blood free BAL samples diluted 1:2 with the provided diluent.
Myeloperoxidase (MPO) assay
Lung tissue was homogenized in 5 mM potassium phosphate buffer. After centrifugation (16,000g, 30 min 4 °C), the pellet was resuspended in extraction buffer (50 mM potassium phosphate, 0.5% hexadecyl trimethylammonium bromide) at a concentration of 800 μg protein/ml. The sample was freeze-thawed thrice, and after centrifugation (13,000g, 15 min, 4 °C), the supernatant was diluted 1 in 10. This was then assayed for MPO activity by measuring the change in optical density at 460 nm using kinetic readings over 3 min. MPO activity is expressed as an arbitrary unit (MPO units; change in absorbance over 60 s) made relative to protein concentrations.
Statistical analyses
Values are expressed as mean ± s.e.m. unless otherwise stated. Data were analyzed using SPSS 16.0 or GraphPad Prism 5.0. Parametric statistical analyses were applied when data showed normal distribution, otherwise non-parametric tests were utilized. ***P ≤ 0.005, **P ≤ 0.01, *P ≤ 0.05. qPCR data are expressed relative to the housekeeping gene encoding β-actin (or 36B4) and relative to a control sample, as stated. PMT recordings were normalized to a rolling average before plotting as a function of time; period analysis was carried out using RAP software51; and Cosinor analysis was carried out using Cosinor.exe v2.3 (R. Refinetti).
Statistical estimates are based on achieving a statistical significance (based on achieving 80% power with an estimated 30-50% magnitude of effect.
All animals were randomized to treatment and control groups and investigators were blinded to genotype and treatment.
Supplementary Material
ACKNOWLEDGMENTS
We thank J.S. Takahashi, Southwestern University, for the gift of the PER2::luc mice, F. Scott and A. Hughes for help with imaging lung tissue slices and J. Woodburn for technical help. We thank M.H. Hastings for comments on the manuscript. The work was supported by research grants to A.L. from GlaxoSmithKline and the Biotechnology and Biological Sciences Research Council, UK (BB/D004357/1, BB/K003119/1 and BB/K003097/1), to D.R. from the Wellcome Trust and the UK National Institute for Health Research Musculoskeletal Biomedical Research Unit Manchester, to T.B. from the Wellcome Trust (093612/Z/10/Z), as well as the US National Institute of Diabetes and Digestive and Kidney Diseases grant P01 DK59820 to F.D. and US National Institutes of Health grants 1RO1HL105834 and 1RO1AI099479 to G.S.W.
Footnotes
Note: Any Supplementary Information and Source Data files are available in the online version of the paper.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
References
- 1.Martinez FJ, Donohue JF, Rennard SI. The future of chronic obstructive pulmonary disease treatment–difficulties of and barriers to drug development. Lancet. 2011;378:1027–1037. doi: 10.1016/S0140-6736(11)61047-7. Medline CrossRef</jrn>. [DOI] [PubMed] [Google Scholar]
- 2.Scheiermann C, et al. Adrenergic nerves govern circadian leukocyte recruitment to tissues. Immunity. 2012;37:290–301. doi: 10.1016/j.immuni.2012.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Scheiermann C, Kunisaki Y, Frenette PS. Circadian control of the immune system. Nat. Rev. Immunol. 2013;13:190–198. doi: 10.1038/nri3386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Keller M, et al. A circadian clock in macrophages controls inflammatory immune responses. Proc. Natl. Acad. Sci. USA. 2009;106:21407–21412. doi: 10.1073/pnas.0906361106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gibbs JE, et al. The nuclear receptor REV-ERBα mediates circadian regulation of innate immunity through selective regulation of inflammatory cytokines. Proc. Natl. Acad. Sci. USA. 2012;109:582–587. doi: 10.1073/pnas.1106750109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang X, Reece SP, Van Scott MR, Brown JM. A circadian clock in murine bone marrow-derived mast cells modulates IgE-dependent activation in vitro. Brain Behav. Immun. 2011;25:127–134. doi: 10.1016/j.bbi.2010.09.007. [DOI] [PubMed] [Google Scholar]
- 7.Shackelford PG, Feigin RD. Periodicity of susceptibility to pneumococcal infection: influence of light and adrenocortical secretions. Science. 1973;182:285–287. doi: 10.1126/science.182.4109.285. [DOI] [PubMed] [Google Scholar]
- 8.Silver AC, Arjona A, Walker WE, Fikrig E. The circadian clock controls Toll-like receptor 9-mediated innate and adaptive immunity. Immunity. 2012;36:251–261. doi: 10.1016/j.immuni.2011.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Halberg F, Johnson NEA, Brown BW, Bittner JJ. Susceptibility rhythm to E. coli endotoxin and bioassay. Proc. Soc. Exp. Biol. Med. 1960;103:142–144. doi: 10.3181/00379727-103-25439. [DOI] [PubMed] [Google Scholar]
- 10.Castanon-Cervantes O, et al. Dysregulation of inflammatory responses by chronic circadian disruption. J. Immunol. 2010;185:5796–5805. doi: 10.4049/jimmunol.1001026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bellet MM, et al. Circadian clock regulates the host response to Salmonella. Proc. Natl. Acad. Sci. USA. 2013;110:9897–9902. doi: 10.1073/pnas.1120636110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Witko-Sarsat V, Rieu P, Descamps-Latscha B, Lesavre P, Halbwachs-Mecarelli L. Neutrophils: molecules, functions and pathophysiological aspects. Lab. Invest. 2000;80:617–653. doi: 10.1038/labinvest.3780067. [DOI] [PubMed] [Google Scholar]
- 13.Perkins ND. Integrating cell-signalling pathways with NF-κB and IKK function. Nat. Rev. Mol. Cell Biol. 2007;8:49–62. doi: 10.1038/nrm2083. [DOI] [PubMed] [Google Scholar]
- 14.Ota T, Fustin JM, Yamada H, Doi M, Okamura H. Circadian clock signals in the adrenal cortex. Mol. Cell. Endocrinol. 2012;349:30–37. doi: 10.1016/j.mce.2011.08.010. [DOI] [PubMed] [Google Scholar]
- 15.Dickmeis T. Glucocorticoids and the circadian clock. J. Endocrinol. 2009;200:3–22. doi: 10.1677/JOE-08-0415. [DOI] [PubMed] [Google Scholar]
- 16.Nader N, Chrousos GP, Kino T. Circadian rhythm transcription factor CLOCK regulates the transcriptional activity of the glucocorticoid receptor by acetylating its hinge region lysine cluster: potential physiological implications. FASEB J. 2009;23:1572–1583. doi: 10.1096/fj.08-117697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lamia KA, et al. Cryptochromes mediate rhythmic repression of the glucocorticoid receptor. Nature. 2011;480:552–556. doi: 10.1038/nature10700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Han DH, Lee YJ, Kim K, Kim CJ, Cho S. Modulation of glucocorticoid receptor induction properties by core circadian clock proteins. Mol. Cell. Endocrinol. 2014;383:170–180. doi: 10.1016/j.mce.2013.12.013. [DOI] [PubMed] [Google Scholar]
- 19.Aschoff J. In: Circadian Clocks. Aschoff J, editor. Amsterdam, North-Holland: 1965. pp. 95–111. [Google Scholar]
- 20.Paladino N, Leone MJ, Plano SA, Golombek DA. Paying the circadian toll: the circadian response to LPS injection is dependent on the Toll-like receptor 4. J. Neuroimmunol. 2010;225:62–67. doi: 10.1016/j.jneuroim.2010.04.015. [DOI] [PubMed] [Google Scholar]
- 21.Kadioglu A, et al. Host cellular immune response to pneumococcal lung infection in mice. Infect. Immun. 2000;68:492–501. doi: 10.1128/iai.68.2.492-501.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yoo SH, et al. PERIOD2:LUCIFERASE real-time reporting of circadian dynamics reveals persistent circadian oscillations in mouse peripheral tissues. Proc. Natl. Acad. Sci. USA. 2004;101:5339–5346. doi: 10.1073/pnas.0308709101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Elizur A, et al. Clara cells impact the pulmonary innate immune response to LPS. Am. J. Physiol. Lung Cell. Mol. Physiol. 2007;293:L383–L392. doi: 10.1152/ajplung.00024.2007. [DOI] [PubMed] [Google Scholar]
- 24.Hirota T, et al. Identification of small molecule activators of cryptochrome. Science. 2012;337:1094–1097. doi: 10.1126/science.1223710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gibbs JE, et al. Circadian timing in the lung; a specific role for bronchiolar epithelial cells. Endocrinology. 2009;150:268–276. doi: 10.1210/en.2008-0638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li S, et al. Foxp1/4 control epithelial cell fate during lung development and regeneration through regulation of anterior gradient 2. Development. 2012;139:2500–2509. doi: 10.1242/dev.079699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Guilding C, et al. Suppressed cellular oscillations in after-hours mutant mice are associated with enhanced circadian phase-resetting. J. Physiol. (Lond.) 2013;591:1063–1080. doi: 10.1113/jphysiol.2012.242198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jeyaseelan S, et al. Induction of CXCL5 during inflammation in the rodent lung involves activation of alveolar epithelium. Am. J. Respir. Cell Mol. Biol. 2005;32:531–539. doi: 10.1165/rcmb.2005-0063OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mei J, et al. CXCL5 regulates chemokine scavenging and pulmonary host defense to bacterial infection. Immunity. 2010;33:106–117. doi: 10.1016/j.immuni.2010.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.John S, et al. Interaction of the glucocorticoid receptor with the chromatin landscape. Mol. Cell. 2008;29:611–624. doi: 10.1016/j.molcel.2008.02.010. [DOI] [PubMed] [Google Scholar]
- 31.Dalm S, Brinks V, van der Mark MH, de Kloet ER, Oitzl MS. Non-invasive stress-free application of glucocorticoid ligands in mice. J. Neurosci. Methods. 2008;170:77–84. doi: 10.1016/j.jneumeth.2007.12.021. [DOI] [PubMed] [Google Scholar]
- 32.The ENCODE Project Consortium et al. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vyas S, et al. Insights into negative regulation by the glucocorticoid receptor from genome-wide profiling of inflammatory cistromes. Mol. Cell. 2013;49:1–3. doi: 10.1016/j.molcel.2012.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Grommes J, Soehnlein O. Contribution of neutrophils to acute lung injury. Mol. Med. 2011;17:293–307. doi: 10.2119/molmed.2010.00138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nakagome K, Matsushita S, Nagata M. Neutrophilic inflammation in severe asthma. Int. Arch. Allergy Immunol. 2012;158(Suppl 1):96–102. doi: 10.1159/000337801. [DOI] [PubMed] [Google Scholar]
- 36.Quint JK, Wedzicha JA. The neutrophil in chronic obstructive pulmonary disease. J. Allergy Clin. Immunol. 2007;119:1065–1071. doi: 10.1016/j.jaci.2006.12.640. [DOI] [PubMed] [Google Scholar]
- 37.Eash KJ, Greenbaum AM, Gopalan PK, Link DC. CXCR2 and CXCR4 antagonistically regulate neutrophil trafficking from murine bone marrow. J. Clin. Invest. 2010;120:2423–2431. doi: 10.1172/JCI41649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Suratt BT, et al. Role of the CXCR4/SDF-1 chemokine axis in circulating neutrophil homeostasis. Blood. 2004;104:565–571. doi: 10.1182/blood-2003-10-3638. [DOI] [PubMed] [Google Scholar]
- 39.Kobayashi Y. Neutrophil infiltration and chemokines. Crit. Rev. Immunol. 2006;26:307–316. doi: 10.1615/critrevimmunol.v26.i4.20. [DOI] [PubMed] [Google Scholar]
- 40.Mei J, et al. Cxcr2 and Cxcl5 regulate the IL-17/G-CSF axis and neutrophil homeostasis in mice. J. Clin. Invest. 2012;122:974–986. doi: 10.1172/JCI60588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tuller T, Atar S, Ruppin E, Gurevich M, Achiron A. Common and specific signatures of gene expression and protein-protein interactions in autoimmune diseases. Genes Immun. 2013;14:67–82. doi: 10.1038/gene.2012.55. [DOI] [PubMed] [Google Scholar]
- 42.Heit B, Tavener S, Raharjo E, Kubes P. An intracellular signaling hierarchy determines direction of migration in opposing chemotactic gradients. J. Cell Biol. 2002;159:91–102. doi: 10.1083/jcb.200202114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hwang JW, Sundar IK, Yao H, Sellix MT, Rahman I. Circadian clock function is disrupted by environmental tobacco/cigarette smoke, leading to lung inflammation and injury via a SIRT1-BMAL1 pathway. FASEB J. 2014;28:176–194. doi: 10.1096/fj.13-232629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vasu VT, Cross CE, Gohil K. Nr1d1, an important circadian pathway regulatory gene, is suppressed by cigarette smoke in murine lungs. Integr. Cancer Ther. 2009;8:321–328. doi: 10.1177/1534735409352027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Smith JB, Herschman HR. Glucocorticoid-attenuated response genes encode intercellular mediators, including a new C-X-C chemokine. J. Biol. Chem. 1995;270:16756–16765. doi: 10.1074/jbc.270.28.16756. [DOI] [PubMed] [Google Scholar]
- 46.Miranda TB, Morris SA, Hager GL. Complex genomic interactions in the dynamic regulation of transcription by the glucocorticoid receptor. Mol. Cell. Endocrinol. 2013;380:16–24. doi: 10.1016/j.mce.2013.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li H, et al. Cre-mediated recombination in mouse Clara cells. Genesis. 2008;46:300–307. doi: 10.1002/dvg.20396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.McGrath EE, et al. TNF-related apoptosis-inducing ligand (TRAIL) regulates inflammatory neutrophil apoptosis and enhances resolution of inflammation. J. Leukoc. Biol. 2011;90:855–865. doi: 10.1189/jlb.0211062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Meng QJ, et al. Ligand modulation of REV-ERBα function resets the peripheral circadian clock in a phasic manner. J. Cell Sci. 2008;121:3629–3635. doi: 10.1242/jcs.035048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lee TI, Johnstone SE, Young RA. Chromatin immunoprecipitation and microarray-based analysis of protein location. Nat. Protoc. 2006;1:729–748. doi: 10.1038/nprot.2006.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Okamoto K, Onai K, Ishiura M. RAP, an integrated program for monitoring bioluminescence and analyzing circadian rhythms in real time. Anal. Biochem. 2005;340:193–200. doi: 10.1016/j.ab.2004.11.007. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.