Abstract
Increasing evidence points to a negative correlation between KRAS mutations and patients' responses to anti-EGFR monoclonal antibody treatment. Therefore, patients must undergo KRAS mutation detection to be eligible for treatment. High resolution melting analysis (HRM) is gaining increasing attention in KRAS mutation detection. However, its accuracy has not been systematically evaluated. We conducted a meta-analysis of published articles, involving 13 articles with 1,520 samples, to assess its diagnostic accuracy compared with DNA sequencing. The quality of included articles was assessed using the revised Quality Assessment for Studies of Diagnostic Accuracy (QUADAS-2) tools. Random effects models were applied to analyze the performance of pooled characteristics. The overall sensitivity and specificity of HRM were 0.99 (95% confidence interval [CI]: 0.98–1.00) and 0.96 (95%CI: 0.94–0.97), respectively. The area under the summary receiver operating characteristic curve was 0.996. High sensitivity and specificity, less labor, rapid turn-around and the closed-tube format of HRM make it an attractive choice for rapid detection of KRAS mutations in clinical practice. The burden of DNA sequencing can be reduced dramatically by the implementation of HRM, but positive results still need to be sequenced for diagnostic confirmation.
KRAS, a critical gene in the development of many cancers, is one of three members of a family of genes (KRAS, NRAS and HRAS) that encode small guanine nucleotide-binding proteins. It is a key element of cell-signaling pathways, such as mitogen-activated protein kinase (MAPK), Janus kinase/signal transducers and activators of transcription (JAK–STAT) and phosphatidylinositol-3 kinase (PI3K) pathways and plays an important role in a variety of cellular processes, including proliferation and apoptosis1. The protein encoded by KRAS is a key signal transducer for a variety of cellular receptors, including the epidermal growth factor receptor (EGFR). KRAS mutations have been associated with poor prognosis in different tumor types, including pancreatic cancer (~65%), colon cancer (~40%), lung cancer (~20%) and ovarian cancer (~15%)2. While having some utility as a genetic marker for diagnostic and prognostic purposes, KRAS mutation status has great value in assisting with EGFR-targeted therapy decisions because of its strong association as a negative predictor of responses to monoclonal antibody based therapies in colon cancer, and as a marker of resistance to small-molecule tyrosine kinase inhibitors in non-small-cell lung cancer (NSCLC)1,3,4. These findings not only make KRAS a strong predictor of clinical resistance to EGFR-targeted therapies but also demonstrate the significant diagnostic and prognostic implications of KRAS mutation status in a broad range of clinical settings. Therefore, the demand for KRAS mutational analysis as a predictive marker has increased rapidly. Prior to treatment with EGFR inhibitors in colorectal cancer (CRC), KRAS testing has become mandatory in the European Union5 and is recommended in the United States6.
A number of laboratory methods have been utilized to detect mutation status in the KRAS gene, most of which fall under the categories of DNA sequencing, single-strand conformation polymorphisms, allele-specific PCR, denaturing high performance liquid chromatography, denaturant gradient gel electrophoresis, array/strip analysis and high resolution melting analysis (HRM)1,7,8,9. All of these laboratory methods have been successfully applied to clinical KRAS mutation testing, and each has its unique feature. Although DNA sequencing, including direct DNA sequencing and pyrosequencing, is considered to be the “golden standard” for known/unknown mutation scanning10, its relatively low sensitivity or limits of detection may not be optimal for clinical settings.
HRM is a simple, PCR-based method for detecting DNA sequence variation by measuring changes in the melting of a DNA duplex11. To follow the transition of double-stranded DNA (dsDNA) to single-stranded (ssDNA), intercalating dyes such as LC Green and LC Green Plus, ResoLight, EvaGreen and SYTO 9 were employed12. These dyes emit more strongly when bound to dsDNA than ssDNA, namely, the fluorescence intensity decreases as two strands of the dsDNA melt apart. The level of fluorescence intensity vs. temperature is plotted, which is known as a melt curve. The melting temperature at which 50% of the DNA is in the double stranded state can be approximated by taking the derivative of the melting curve13. The distinctive melting curve can be used to detect DNA sequence variants without the need for any post-PCR handling. Advantages of the method include a rapid turn-around time, a closed-tube format that greatly reduces contamination risk, high sensitivity and specificity, low cost and, unlike other methods, no sample processing or separations after PCR14. Furthermore, HRM is a nondestructive method. Therefore, subsequent analysis of the sample by other methods, such as DNA sequencing or gel-electrophoresis can still be performed after HRM13. Due to the advantages mentioned above, HRM might be an attractive choice for the detection of KRAS mutations. However, the accuracy of HRM for the detection of KRAS mutations has not been systematically assessed. Thus, we conducted this meta-analysis to assess accuracy of HRM for the detection of KRAS mutations.
Results
Literature search outcome
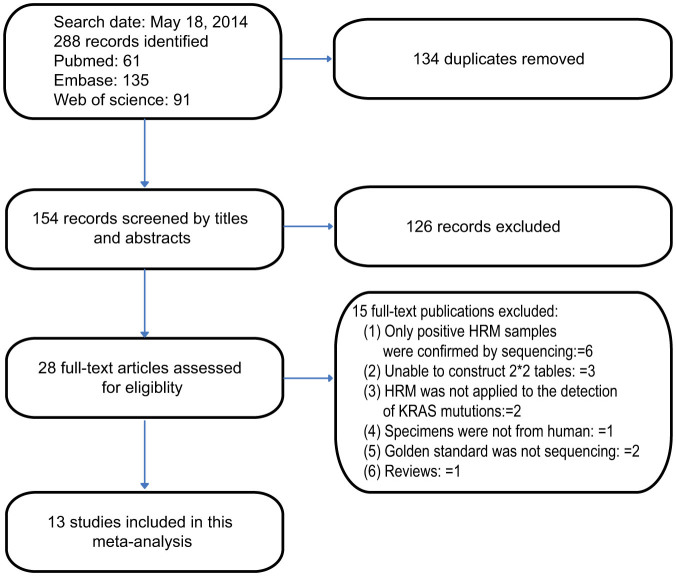
A total of 288 records were retrieved after an independent search of the scientific literature by reviewers. One hundred and thirty-four records were excluded because of duplicates and 126 records were excluded after reviewing of the title and abstract. Twenty-eight full-text papers were deemed to be potentially relevant and were examined in detail. Fifteen full-text papers were excluded for the reasons described in Figure 1. Finally, 13 studies5,10,15,16,17,18,19,20,21,22,23,24,25 met the inclusion criteria and were included in this meta-analysis. These articles were divided into 15 ‘units' for statistical analysis according to the specimen source.
Figure 1. Flowchart describing the systematic literature search and study selection process.
Study characteristics and quality assessment
The main characteristics of the eligible studies are summarized in Table 1. A total of 13 studies with 1,520 samples were included in our meta-analysis study. Disease types included colorectal cancer (CRC) or colon cancer (CC; n = 9), non-small cell lung cancer (NSCLC; n = 3) and pancreatic cancer (PA; n = 1). The average sample number was 101 (range 28 to 200). The most common specimen source was formalin-fixed and paraffin-embedded tissues (FFPE; n = 7), followed by fresh frozen tissues (FF; n = 6). All studies used DNA extraction kits from Qiagen. The most frequently used dye was Syto9 (S9; n = 7), followed by Resolight (RL; n = 7). Amplicon lengths varied from 59 bp to 247 bp, with an average length of 143 bp. The total volume (μL) of HRM was 9 μL (n = 1), 10 μL (n = 8), 20 μL (n = 3), 25 μL (n = 1) and 50 μL (n = 1), while one study did not report the total volume of HRM. All the eligible studies used DNA direct sequencing as the reference standard. The number of false positives and false negatives obtained from each eligible study varied greatly. The number of false positives ranged from 0 to 22 while the number of false negatives ranged from 0 to 2.
Table 1. Characteristics of the 13 eligible studies in the meta-analysis.
| Study | Country | Year | Disease | Number | Specimen source | Instrument | Dye | AL | Total volume (μL) | TP | FP | FN | TN |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Krupuy et al.15 | Australia | 2006 | NSCLC | 30 | FF | RG | S9 | 92, 189 | 20 | 9 | 0 | 0 | 21 |
| Do et al.16 | Australia | 2008 | NSCLC | 200 | FFPE | RG | S9 | 92 | 20 | 25 | 0 | 0 | 175 |
| Simi et al.17 | Italy | 2008 | CRC | 116 | FF | RG | S9 | 167 | 10 | 50 | 0 | 0 | 66 |
| Fassina et al.18 | Italy | 2009 | NSCLC | 77 | CS | LC480 | RL | 172 | NR | 9 | 0 | 0 | 68 |
| Ma. et al.19 | China | 2009 | CRC | 100 | FFPE | LC480 | RL | 170 | 10 | 61 | 0 | 1 | 38 |
| Whitehall et al.20 | Australia | 2009 | CRC | 160 | FFPE | LC480 | S9 | 80–92 | 10 | 54 | 0 | 2 | 104 |
| Whitehall et al.20 | Australia | 2009 | CRC | 140 | FF | RG | S9 | 80–92 | 10 | 40 | 4 | 0 | 96 |
| van Eijk, R. et al.23 | Netherlands | 2010 | CRC | 92 | FFPE | LS96 | S9 | 166 | 10 | 52 | 5 | 0 | 35 |
| van Eijk, R. et al.23 | Netherlands | 2010 | CRC | 28 | FF | LS96 | S9 | 166 | 10 | 14 | 1 | 0 | 13 |
| Franklin et al.22 | USA | 2010 | CC | 118 | FFPE | LC480 | RL | NA | 10 | 42 | 22 | 0 | 54 |
| Bennani et al.21 | Morocco | 2010 | CRC | 56 | PE | LC480 | RL | 143 | 10 | 17 | 7 | 0 | 32 |
| Zhang et al.25 | China | 2011 | PC | 50 | FFPE | LC480 | RL | 59, 163 | 20 | 19 | 0 | 0 | 31 |
| Solassol et al.24 | France | 2011 | CRC | 131 | FF | LC480 | RL | 247 | 50 | 47 | 0 | 0 | 84 |
| Krol. et al.5 | Netherlands | 2012 | CRC | 125 | FF | LC480 | RL | 189 | 25 | 39 | 2 | 0 | 84 |
| Akiyoshi et al.10 | Japan | 2013 | CRC | 97 | FFPE | LS96 | LS | 92 | 9 | 51 | 3 | 2 | 41 |
AL: Amplicon length; NSCLC: non-small cell lung cancer; CRC: colorectal cancer; CC: colon cancer; PC: pancreatic cancer; FF: fresh frozen tissue; FFPE: formalin-fixed and paraffin-embedded; CS: cytologic slides; PE: paraffin-embedded; RG: Rotorgene6000; LS96:lightScanner96; S9: Syto 9; RL: Resolight; LS: LightScanner Master Mix; NR: not reported; TP: true-positive; FP: false-positive; FN: false-negative; TN: true-negative.
We assessed the quality of the eligible studies using the wildly accepted revised Quality Assessment for Studies of Diagnostic Accuracy (QUADAS–2) tools26. The domains of patient selection, index test and reference standard, both in risks of bias and applicability concerns, were labeled as “low risk”. The flow and timing domain was labeled as “unclear risk”.
Diagnostic accuracy and threshold analysis
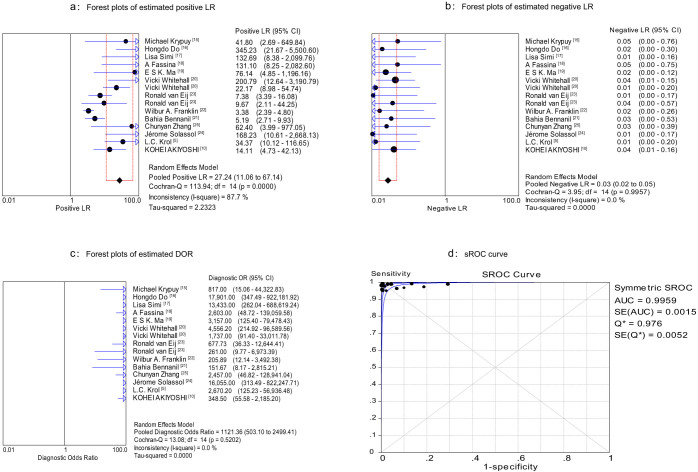
The Spearman correlation coefficient was 0.150 (P = 0.593), which indicated an absence of a threshold effect. Therefore, we pooled the sensitivity, specificity, positive likelihood ratio (PLR), negative likelihood ratio (NLR) and diagnostic odds ratio (DOR) by the data from the eligible studies. The overall diagnostic sensitivity was 0.99 (95% CI: 0.98–1.00) and the overall specificity was 0.96 (95%CI: 0.94–0.97; Figure 2). PLR, NLR and DOR were three other commonly used test indicators for diagnostic tests27. The greater the value of PLR, the more likely the test result was associated with the disease, while the lower the value of NLR, the more likely the result was associated with absence of the disease. DOR is defined as the odds of true positives vs. that of false positives. The value of DOR ranged from 0 to infinity, higher values indicating better discriminatory test performances28. Figures 3a–b show that HRM has a high PLR (27.24, 95%CI: 11.06–67.14) and a low NLR (0.03, 95% CI: 0.02–0.05), indicating HRM's excellent ability to both confirm and exclude the presence of KRAS mutations. In addition, the value of DOR was 1,121.36 (95%CI: 503.10–2,499.41), further indicating HRM is an effective method for KRAS mutation scanning (Figure 3c). The I2 test for heterogeneity demonstrated greater heterogeneity for specificity (I2 = 88.3%, P = 0.00) and PLR (I2 = 87.7%, P = 0.00). The summary receiver operating characteristic (sROC) curve, which is intended to present the relationship between sensitivity and specificity across all included studies in the context of a meta-analysis29, is shown in Figure 3d. The sROC curve from our data showed that the Q value was 0.97, while the area under the curve (AUC) was 0.996.
Figure 2. Forest plots of estimated sensitivity (a) and specificity (b) for HRM with 95%CI.
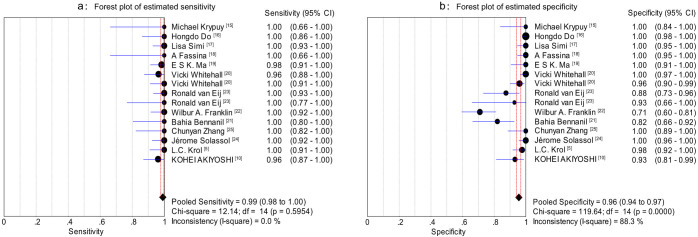
Each solid circle represents an eligible study. The size of the solid circle reflects the sample size of each eligible study. Error bars represents 95%CI.
Figure 3. Forest plots of estimated PLR (a), NLR (b) and DOR (c) for HRM with 95%CI, and sROC (d).
Each solid circle represents an eligible study. The size of the solid circle reflects the sample size of each eligible study. Error bars represents 95%CI.
Meta-regression analysis and publication bias
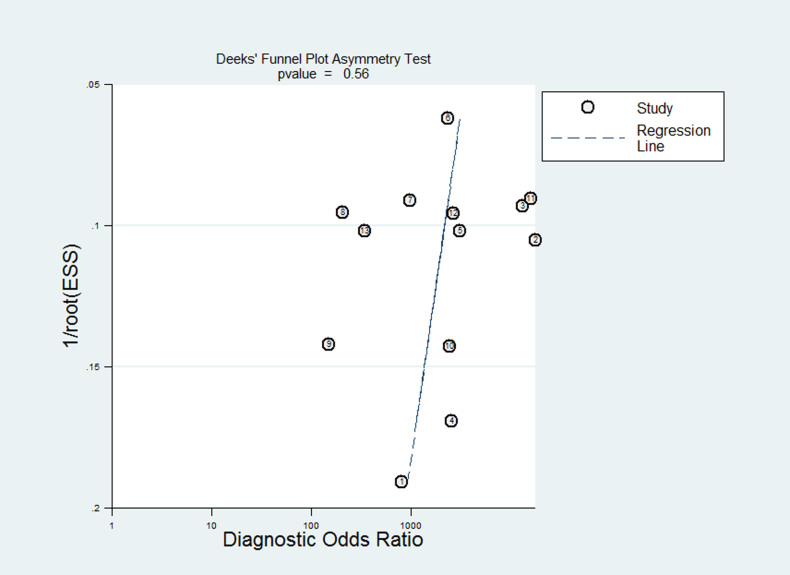
The results of multivariate meta-regression analysis (Table 2) demonstrated that there was no statistical significance between eligible studies. The subgroup analysis was not executed because the source of heterogeneity was not found. In our study, the Deek's Funnel Plot Asymmetry Test demonstrated that publication bias was not significant (P = 0.56; Figure 4).
Table 2. Results of the multivariable meta-regression model for the characteristics with backward regression analysis (Inverse variance weights; variables were retained in the regression model if P<0.05).
| Variables | Coeff. | Std.Err. | P-value | RDOR | [95%CI] |
|---|---|---|---|---|---|
| Cte. | 4.238 | 6.4547 | 0.5405 | --- | --- |
| S | −0.472 | 0.3554 | 0.2417 | --- | --- |
| Disease | 0.612 | 1.1813 | 0.6262 | 1.85 | 0.09–38.44 |
| Number | 0.011 | 0.0180 | 0.5818 | 1.01 | 0.96–1.06 |
| Specimen source | −0.151 | 0.6377 | 0.8222 | 0.86 | 0.17–4.43 |
| Instrument | −0.702 | 1.0401 | 0.5299 | 0.50 | 0.03–7.19 |
| Dye | −0.640 | 0.5565 | 0.2728 | 0.53 | 0.16–1.77 |
| Lengths | 0.009 | 0.0107 | 0.4266 | 1.01 | 0.98–1.04 |
| Total volume | 0.018 | 0.0648 | 0.7903 | 1.02 | 0.86–‘1.20 |
Cte: Constant Coefficient; S: Statistic S; RDOR: Relative diagnostic odds ratio.
Figure 4. Deek's Funnel Plot Asymmetry Test for the assessment of potential publication bias.
Each solid circle represents a study in this meta-analysis. The publication bias was not significant (P = 0.56).
Discussion
Since it was first introduced for genotyping in 200330, HRM has been used to detect mutations such as EGFR31, KIT32, BRAF33, BRCA34, TP5335 and KRAS15. The present meta-analysis found 13 published studies on the diagnostic accuracy of HRM for detection of KRAS mutations. The results of the overall sensitivity and specificity indicate a very high level of overall diagnostic accuracy for HRM. In addition, no publication bias was found in the present meta-analysis, indicating that the results of the present meta-analysis are reliable. Therefore, we concluded that HRM is a very sensitive and specific method for scanning KRAS mutations. The AUC under the sROC curve is a global indicator for assessing the diagnostic performance of an index test36. The present meta-analysis found that the AUC was close to 1 (0.996), also indicating that HRM is an effective method for KRAS mutation scanning. Similar results were obtained in a recently published study28 designed to evaluate overall accuracy of HRM for rapid detection of BRAF mutations. Overall sensitivity and specificity values in that study were 0.99 (95%CI = 0.99–1.00) and 0.99 (95%CI = 0.96–1.00), respectively. Results from the two studies showed that HRM is a robust method for mutation scanning for both KRAS and BRAF.
However, there are discordant results between HRM and DNA sequencing in seven5,10,19,20,21,22,23 of the 13 included studies. The total number of false positives was 44, which may be explained by different limits of detection or sensitivity for the two methods. HRM can detect mutations in samples containing ~1% to 10% cells with mutated DNA, as opposed to direct sequencing, which requires the presence of mutated DNA in at least 10% to 30% of the sample37. Some clinical samples that are low in tumor cells or DNA may be labeled as positive by HRM but negative by direct sequencing, which results in a false positive. The total number of false negatives was five, which may be explained by greater amplicon length and lower GC content38.
Considerable heterogeneity was observed in the pooled value of specificity and PLR. Though a multivariate meta-regression analysis was performed, we could not find the source of heterogeneity. This heterogeneity may be related to additional factors, such as the sequence length, GC content and the presence of substances such as DMSO or betaine38,39,40. However, this information is hard to collect and quantitatively analyze.
Though the accuracy of HRM is very high, several factors have to be taken into consideration when implementing HRM techniques into routine clinical practice, such as PCR specificity, sample type, length of the amplicon, GC content, dye, instrument, and melting analysis software. Melting is performed directly after PCR and different heterozygotes may produce melting curves so similar to each other that, although they clearly vary from homozygous variants, they are not differentiated from each other. Therefore, specific amplification of the interested targets is critical, requiring careful choice of primers and optimized temperature cycling41.
Both FF and FFPE can be used for KRAS mutation scanning; however, a higher sensitivity was obtained when fresh tissues were used for HRM, because DNA may degenerate during sample processing and storage14. A head-to-head comparison study24 showed that the agreement for the mutational status of KRAS between FF and matched FFPE specimens was low. Therefore, the authors suggested that fresh frozen specimens (FF) are favored when possible. When only FFPE samples are available, the risk of DNA degeneration should be prevented by using large amounts of template DNA or by performing multiple amplifications. Mutations in a long amplicon complicate the analysis. Hence, shortening the length of the amplicons can make melting curve differences between mutant and wild-type alleles more obvious and, in turn, easier to distinguish42. For example, for PCR products below 400 bp, sensitivity and specificity were 100%, decreasing to 96.1% sensitivity and 99.4% specificity if the PCR products were between 400 bp and 1000 bp14. Some studies showed that a lower GC content might have an association with false negative results11,38. Besides, different dyes are variably effective. For example, LCGreen Plus detects heterozygotes better than Syto 9, which is better than EvaGreen, which is better than SYBR Green I9. Instruments also have an impact on the sensitivity and specificity of HRM, because instruments have differences in their ability to resolve shape and absolute temperature differences, explained by their signal-to-noise ratios, acquisition rates, data density and software8,12,14. Also, it should be noted that although HRM may prove to be a helpful screening method for KRAS mutations, the requirement for confirmation by direct sequencing or other methods is necessary, especially in a diagnostic context.
Several limitations must be pointed out in our present meta-analysis despite the analysis showing high sensitivity and specificity. Firstly, though a meta-regression had been performed, we still could not find the source of the heterogeneity. Secondly, the sample size (1,520) included in our meta-analysis is relatively small, which might have some effect on the overall accuracy of the HRM. Lastly, sufficient information about other factors was not collected, which may affect the accuracy of HRM, such as the sequence length, GC content and the presence of substances such as DMSO or betaine.
In summary, high sensitivity and specificity, less labor, a rapid turn-around and the closed-tube format of HRM make it an attractive choice for rapid detection of KRAS mutations in clinical practice. The DNA sequencing burden can be reduced dramatically by the implementation of HRM, but positive results still need to be sequenced for diagnostic confirmation.
Methods
Literature search strategy
Medline (using PubMed as the search engine), Web of science and Excerpta Medica Database (EMBASE) were searched to identify relevant publications published in English until May 18, 2014. The following search words (in Title/Abstract fields) were used: ‘high resolution melting analysis' or ‘high resolution melting analyses' or ‘high resolution fluorescent melting analysis' or ‘high resolution amp icon melting analysis' or ‘HRM' or ‘HRMA' or ‘HRMCA' AND ‘mutation' or ‘mutations' AND ‘KRAS' or ‘K-RAS' AND ‘sequence' or ‘sequencing'. We also performed a manual search for additional eligible studies in the reference lists of the articles identified.
Inclusion and exclusion criteria
The following inclusion criteria were used: 1) HRM was applied to the detection of KRAS mutations in humans; 2) DNA sequencing was used as a reference standard; and 3) sensitivity and specificity of HRM were reported or could be calculated from the provided data. The exclusion criteria were as follows: 1) studies were performed using only HRM; 2) only positive HRM samples were confirmed by sequencing; 3) the reference standard was not DNA sequencing; 4) the samples were cell lines or artificially created sequences; 5) they were unable to construct 2×2 tables; 6) the articles were reviews, letters, comments, and conference abstracts because of limited data; and 7) publications were identified as duplicates.
Data extraction and quality assessment
Two reviewers (YPL and HYW) independently extracted relevant data from each eligible study. The following data were collected: author's name, country, disease type, specimens source, reference standard, instruments used, dye used, lengths of amplicon, number of samples, and number of samples with the indicated results (TP, FP, FN, and TN), and 0.5 was added to all cells to handle studies with empty cells. We assessed the quality of the eligible studies using the widely accepted revised Quality Assessment for Studies of Diagnostic Accuracy (QUADAS–2) tools. Disagreements between reviewers were resolved by discussion or by consensus including a third author.
Statistical analysis
The outcome parameters were overall sensitivity, specificity, positive likelihood ratio (PLR), negative likelihood ratio (NLR), diagnostic odds ratio (DOR) and their corresponding 95% confidence intervals (CIs) by a random-effects model. Forest plots were used to describe the pooled sensitivity and specificity, as well as heterogeneity of eligible studies.
The degrees of heterogeneity were explored with a chi-square test of heterogeneity (Q Cochran's Q statistical) and inconsistency index (I-square), which was determined using Meta-Disc (version 1.4) software43. Taking into account the low statistical power of these tests, significant heterogeneity was defined as a Q test P value of <0.10, or an I2 measure >30%.
The summary receiver operating characteristic (sROC) curves and area under the curve (AUC) were applied to demonstrate the overall diagnostic performance of HRM. In addition, the Spearman correlation coefficient was used to verify if the heterogeneity in meta-analysis could be explained by a threshold effect; a threshold effect was defined as a positive correlation (P<0.05). Deek's Funnel Plot Asymmetry Test was applied to determine the presence of publication bias using STATA 12.1 software (Stata Corp., College Station, Texas, USA.)44.
Meta-regression analysis
Meta-regression analysis was performed to explore the factors of heterogeneity using Meta-Disc (version 1.4) software. We applied a multivariable regression model and used a backward stepwise algorithm with the covariates including disease type, specimen source, instrument, dye, length of PCR products, and total volume of HRM; variables were retained in the regression model if P<0.05.The subgroup analysis was performed if factors of heterogeneity could be found.
Author Contributions
Conceived and designed the experiments: Y.P.L., W.L.F., Q.H. Performed the experiments: H.Y.W., X.Y., Q.H.X. Analyzed the data: D.C., Y.P.L. Wrote the manuscript: Y.P.L., H.Y.W.
Acknowledgments
This work was supported in part by grants from The National High Technology Research and Development Program of China (National 863 Program, No.2011AA02A121, and 2013AA020204), the Project Sponsored by the Scientific Research Foundation for the Returned Overseas Chinese Scholars of State Education Ministry, Chongqing Application and Development Projects (cstc2014yykfA110029).
References
- Anderson S. M. Laboratory methods for KRAS mutation analysis. Expert Rev Mol Diagn 11, 635–642 (2011). [DOI] [PubMed] [Google Scholar]
- Wang H. L., Lopategui J., Amin M. B. & Patterson S. D. KRAS mutation testing in human cancers: The pathologist's role in the era of personalized medicine. Adv Anat Pathol 17, 23–32 (2010). [DOI] [PubMed] [Google Scholar]
- Riely G. J., Marks J. & Pao W. KRAS mutations in non-small cell lung cancer. Proc Am Thorac Soc 6, 201–205 (2009). [DOI] [PubMed] [Google Scholar]
- Riely G. J. & Ladanyi M. KRAS mutations: an old oncogene becomes a new predictive biomarker. J Mol Diagn 10, 493–495 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krol L. C. et al. Concordance in KRAS and BRAF mutations in endoscopic biopsy samples and resection specimens of colorectal adenocarcinoma. Eur J Cancer 48, 1108–1115 (2012). [DOI] [PubMed] [Google Scholar]
- Allegra C. J. et al. American Society of Clinical Oncology provisional clinical opinion: testing for KRAS gene mutations in patients with metastatic colorectal carcinoma to predict response to anti-epidermal growth factor receptor monoclonal antibody therapy. J Clin Oncol 27, 2091–2096 (2009). [DOI] [PubMed] [Google Scholar]
- Shackelford R. E., Whitling N. A., McNab P., Japa S. & Coppola D. KRAS Testing: A Tool for the Implementation of Personalized Medicine. Genes Cancer 3, 459–466 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montgomery J., Wittwer C. T., Palais R. & Zhou L. Simultaneous mutation scanning and genotyping by high-resolution DNA melting analysis. Nat Protoc 2, 59–66 (2007). [DOI] [PubMed] [Google Scholar]
- Wittwer C. T. High-resolution DNA melting analysis: advancements and limitations. Hum Mutat 30, 857–859 (2009). [DOI] [PubMed] [Google Scholar]
- Akiyoshi K. et al. KRAS mutations in patients with colorectal cancer as detected by high-resolution melting analysis and direct sequencing. Anticancer Res 33, 2129–2134 (2013). [PubMed] [Google Scholar]
- Taylor C. F. Mutation scanning using high-resolution melting. Biochem Soc Trans 37, 433–437 (2009). [DOI] [PubMed] [Google Scholar]
- Herrmann M. G., Durtschi J. D., Bromley L. K., Wittwer C. T. & Voelkerding K. V. Amplicon DNA melting analysis for mutation scanning and genotyping: cross-platform comparison of instruments and dyes. Clin Chem 52, 494–503 (2006). [DOI] [PubMed] [Google Scholar]
- Li B. S. et al. Is high resolution melting analysis (HRMA) accurate for detection of human disease-associated mutations? A meta analysis. PLoS One 6, e28078 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montgomery J. L., Sanford L. N. & Wittwer C. T. High-resolution DNA melting analysis in clinical research and diagnostics. Expert Rev Mol Diagn 10, 219–240 (2010). [DOI] [PubMed] [Google Scholar]
- Krypuy M., Newnham G. M., Thomas D. M., Conron M. & Dobrovic A. High resolution melting analysis for the rapid and sensitive detection of mutations in clinical samples: KRAS codon 12 and 13 mutations in non-small cell lung cancer. BMC Cancer 6 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Do H., Krypuy M., Mitchell P. L., Fox S. B. & Dobrovic A. High resolution melting analysis for rapid and sensitive EGFR and KRAS mutation detection in formalin fixed paraffin embedded biopsies. BMC Cancer 8 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simi L. et al. High-resolution melting analysis for rapid detection of KRAS, BRAF, and PIK3CA gene mutations in colorectal cancer. Am J Clin Pathol 130, 247–253 (2008). [DOI] [PubMed] [Google Scholar]
- Fassina A. et al. Detection of EGFR and KRAS mutations on transthoracic needle aspiration of lung nodules by high resolution melting analysis. J Clin Pathol 62, 1096–1102 (2009). [DOI] [PubMed] [Google Scholar]
- Ma E. S., Wong C. L., Law F. B., Chan W. K. & Siu D. Detection of KRAS mutations in colorectal cancer by high-resolution melting analysis. J Clin Pathol 62, 886–891 (2009). [DOI] [PubMed] [Google Scholar]
- Whitehall V. et al. A Multicenter Blinded Study to Evaluate KRAS Mutation Testing Methodologies in the Clinical Setting. J Mol Diagn 11, 543–552 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennani B. et al. Mutation analysis of BRAF exon 15 and KRAS codons 12 and 13 in Moroccan patients with colorectal cancer. Int J Biol Markers 25, 179–184 (2010). [PubMed] [Google Scholar]
- Franklin W. A. et al. KRAS mutation: comparison of testing methods and tissue sampling techniques in colon cancer. J Mol Diagn 12, 43–50 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Eijk R. et al. Sensitive and specific KRAS somatic mutation analysis on whole-genome amplified DNA from archival tissues. J Mol Diagn 12, 27–34 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solassol J. et al. KRAS mutation detection in paired frozen and formalin-fixed paraffin-embedded (FFPE) colorectal cancer tissues. Int J Mol Sci 12, 3191–3204 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C. et al. Differential High-Resolution Melting Analysis for the Detection of K-ras Codons 12 and 13 Mutations in Pancreatic Cancer. Pancreas 40, 1283–1288 (2011). [DOI] [PubMed] [Google Scholar]
- Whiting P. F. et al. QUADAS-2: a revised tool for the quality assessment of diagnostic accuracy studies. Ann Intern Med 155, 529–536 (2011). [DOI] [PubMed] [Google Scholar]
- Cleophas T. J. & Zwinderman A. H. Meta-analyses of diagnostic studies. Clin Chem Lab Med 47, 1351–1354 (2009). [DOI] [PubMed] [Google Scholar]
- Chen D. et al. High-resolution melting analysis for accurate detection of BRAF mutations: a systematic review and meta-analysis. Sci Rep 4, 4168 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter S. D. Properties of the summary receiver operating characteristic (SROC) curve for diagnostic test data. Stat Med 21, 1237–1256 (2002). [DOI] [PubMed] [Google Scholar]
- Wittwer C. T., Reed G. H., Gundry C. N., Vandersteen J. G. & Pryor R. J. High-resolution genotyping by amplicon melting analysis using LCGreen. Clin Chem 49, 853–860 (2003). [DOI] [PubMed] [Google Scholar]
- Fukui T. et al. Prospective study of the accuracy of EGFR mutational analysis by high-resolution melting analysis in small samples obtained from patients with non-small cell lung cancer. Clin Cancer Res 14, 4751–4757 (2008). [DOI] [PubMed] [Google Scholar]
- Fuster O. et al. Rapid detection of KIT mutations in core-binding factor acute myeloid leukemia using high-resolution melting analysis. J Mol Diagn 11, 458–463 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willmore-Payne C., Holden J. A., Tripp S. & Layfield L. J. Human malignant melanoma: detection of BRAF- and c-kit-activating mutations by high-resolution amplicon melting analysis. Hum Pathol 36, 486–493 (2005). [DOI] [PubMed] [Google Scholar]
- van der Stoep N. et al. Diagnostic guidelines for high-resolution melting curve (HRM) analysis: an interlaboratory validation of BRCA1 mutation scanning using the 96-well LightScanner. Hum Mutat 30, 899–909 (2009). [DOI] [PubMed] [Google Scholar]
- Garritano S. et al. Determining the effectiveness of High Resolution Melting analysis for SNP genotyping and mutation scanning at the TP53 locus. BMC Genet 10, 5 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Z. D., Liu X. F., Liu X. C., Ding C. M. & Hu C. J. Diagnostic accuracy of osteopontin for malignant pleural mesothelioma: a systematic review and meta-analysis. Clin Chim Acta 433, 44–48 (2014). [DOI] [PubMed] [Google Scholar]
- Bozzao C. et al. Survey of KRAS, BRAF and PIK3CA mutational status in 209 consecutive Italian colorectal cancer patients. Int J Biol Markers 27, e366–374 (2012). [DOI] [PubMed] [Google Scholar]
- Reed G. H. & Wittwer C. T. Sensitivity and specificity of single-nucleotide polymorphism scanning by high-resolution melting analysis. Clin Chem 50, 1748–1754 (2004). [DOI] [PubMed] [Google Scholar]
- Liew M. et al. Genotyping of single-nucleotide polymorphisms by high-resolution melting of small amplicons. Clin Chem 50, 1156–1164 (2004). [DOI] [PubMed] [Google Scholar]
- von Ahsen N., Oellerich M. & Schutz E. Limitations of genotyping based on amplicon melting temperature. Clin Chem 47, 1331–1332 (2001). [PubMed] [Google Scholar]
- Er T. K. & Chang J. G. High-resolution melting: applications in genetic disorders. Clin Chim Acta 414, 197–201 (2012). [DOI] [PubMed] [Google Scholar]
- Gundry C. N. et al. Amplicon melting analysis with labeled primers: a closed-tube method for differentiating homozygotes and heterozygotes. Clin Chem 49, 396–406 (2003). [DOI] [PubMed] [Google Scholar]
- Zamora J., Abraira V., Muriel A., Khan K. & Coomarasamy A. Meta-DiSc: a software for meta-analysis of test accuracy data. BMC Med Res Methodol 6, 31 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deeks J. J., Macaskill P. & Irwig L. The performance of tests of publication bias and other sample size effects in systematic reviews of diagnostic test accuracy was assessed. J Clin Epidemiol 58, 882–893 (2005). [DOI] [PubMed] [Google Scholar]