SUMMARY
Infection of macaques with chimeric viruses based on SIVMAC but expressing the HIV-1 envelope (Env) glycoproteins (SHIVs) remains the most powerful model for evaluating prevention and therapeutic strategies against AIDS. Unfortunately, only a few SHIVs are currently available. Furthermore, their generation has required extensive adaptation of the HIV-1 Env sequences in macaques so they may not accurately represent HIV-1 Env proteins circulating in humans, potentially limiting their translational utility. We developed a strategy for generating large numbers of SHIV constructs expressing Env proteins from newly transmitted HIV-1 strains. By inoculating macaques with cocktails of multiple SHIV variants, we selected SHIVs that can replicate and cause AIDS-like disease in immunologically intact rhesus macaques without requiring animal-to-animal passage. One of these SHIVs could be transmitted mucosally. We demonstrate the utility of the SHIVs generated by this method for evaluating neutralizing antibody administration as a protection against mucosal SHIV challenge.
INTRODUCTION
The recent identification of antibodies that potently neutralize diverse HIV-1 strains has renewed enthusiasm for the development of antibody-based prevention and therapeutic strategies (Burton et al., 2012; Klein et al., 2013). Nonhuman primates such as macaques offer an immunologically intact model and can succumb to AIDS-like disease when infected with simian immunodeficiency viruses such as SIVMAC (Hatziioannou and Evans, 2012). However, the large genetic distance between SIV and HIV-1 is an important limitation of SIV/macaque models. Sequence variation obviously affects the nature and specificity of immunological responses, and neutralizing antibodies against HIV-1 envelope (Env) proteins are not generally crossreactive with SIVMAC Env proteins. In an effort to overcome such limitations, chimeric viruses based on SIV but expressing HIV-1 Env and certain HIV-1 accessory genes, (SHIVs) have been generated. Although there is concern that protection against some of the first SHIVs is too easily achieved with vaccine candidates, such viruses remain the best model to guide the design and in vivo testing of numerous vaccine candidates that raise immune responses against HIV-1 proteins (Hatziioannou and Evans, 2012).
Thus far, the generation of SHIVs that reflect the CCR5 coreceptor preference and pathogenicity of HIV-1 has been slow, and despite many years of research, there are currently only a few such SHIVs that are widely used. Moreover, their generation has required multiple animal-to-animal passages, resulting in extensive adaptation of the HIV-1 Env sequences (Harouse et al., 2001; Nishimura et al., 2010; Song et al., 2006), and thus they have significantly diverged from their HIV-1 counterparts circulating in humans.
Recent evidence shows that following sexual transmission of HIV-1 in humans, systemic infection is typically established by a single transmitted/founder (T/F) viral variant (Keele et al., 2008; Salazar-Gonzalez et al., 2008; Zhang et al., 1993; Zhu et al., 1993). T/F viruses may differ in their biological properties from viruses typically isolated later in HIV-1 infection (Liao et al., 2013; Parker et al., 2013). Importantly, T/F Env proteins represent clinically relevant targets that neutralizing antibodies and inhibitors need to engage to prevent infection. SHIV chimeras expressing T/F Env proteins would thus constitute preferred viruses for nonhuman primate studies of prophylactic and treatment interventions targeting HIV-1 Env proteins.
Herein, we have established a strategy for generating and screening large numbers of SHIVs expressing HIV-1 Env proteins from newly transmitted viruses. SHIVs selected by our approach were capable of replicating efficiently and causing AIDS-like disease in immunologically intact rhesus macaques. We further demonstrate that one of these SHIVs can be transmitted mucosally and can be used to evaluate immunoprophylactic interventions using broadly neutralizing antibodies.
RESULTS
Generation and In Vitro Characterization of a SHIV Library
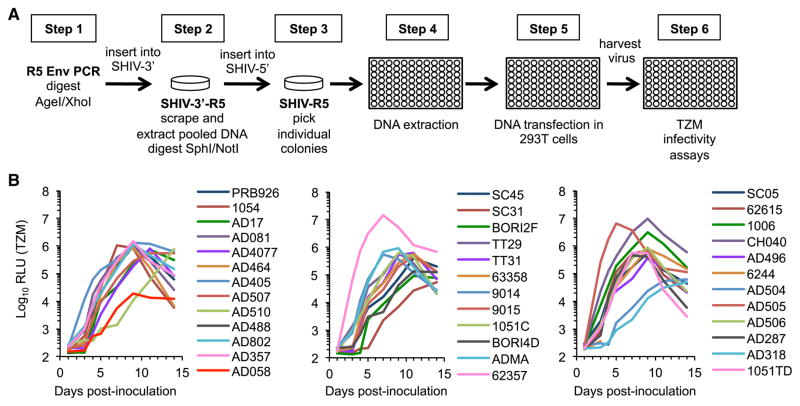
We optimized a cloning strategy that allowed the introduction of Env sequences from clade B HIV-1 strains into a proviral plasmid based on SHIVKB9, a virus clone derived after in vivo passage (Karlsson et al., 1997) (Figures 1A; Figure S1A available online). We pooled distinct clones generated using this approach into two groups of SHIVs. Group 1 consisted of 21 SHIV clones expressing Env proteins from R5-tropic T/F clade B viruses (Keele et al., 2008) (B.F.K. and G.M.S., unpublished data; Table S1). Group 2 consisted of 16 SHIVs expressing Env sequences obtained from patient lymphocyte samples taken early (28–63 days) after infection and prior to any antiretroviral treatment from a cohort of men who have sex with men. We confirmed that SHIVs expressing Env proteins that had not been previously tested were R5-tropic (Figure S1B) and that all SHIVs were replication competent in MT2 cells engineered to express CCR5 (Figure 1B).
Figure 1. Cloning Strategy and In Vitro Replication of R5-SHIVs.
(A) Flowchart of the strategy used for cloning and screening R5-SHIVs. Restriction sites used to introduce the Env PCR products and ligate the two genome halves are noted.
(B) Replication of SHIVs encoding the indicated Env proteins in MT2 cells engineered to express CCR5. Infectious virus in supernatant samples collected from MT2-R5 cells at the indicated times p.i. was measured using TZM-bl cells. Results from a representative experiment (of two performed) are shown. See also Figure S1 and Table S1.
In Vivo Replication and Selection of R5-SHIVs
To select SHIVs that replicated best in vivo, two rhesus macaques were each inoculated intravenously with a cocktail containing 2.5 × 104 i.u. (as measured on TZM-bl cells) of each Group 1 virus. A third animal was inoculated with a cocktail containing 5 × 104 i.u of each Group 2 virus. Animals were depleted of CD8+ cells prior to and 1 week postinoculation (p.i.) to enhance initial virus replication. All animals developed high levels of acute viremia, reaching >108 viral RNA copies/ml (Figures 2A and 2B). Pronounced peripheral and gut-associated lymphoid tissue (GALT) CD4+ T cell depletion was observed in all animals (Figure S2). Notably, one animal inoculated with Group 1 viruses and the animal inoculated with Group 2 viruses rapidly succumbed; the Group 1 animal was sacrificed at 16 weeks p.i. suffering from extensive AIDS-defining extranodal B cell lymphoma, and the Group 2 animal was sacrificed at 13 weeks p.i. suffering from profound anemia, hypoproteinemia as well as weight loss, and sustained low CD4+ T cell counts characteristic of AIDS.
Figure 2. In Vivo Selection of R5-SHIVs.
(A and B) Plasma viral load in macaques inoculated with cocktails of SHIVs from Group 1 (A) or Group 2 (B). Animals were depleted of CD8+ cells at the time of and 1 week post inoculation. Red x indicates euthanasia due to AIDS. (C and D) Phylogenetic analysis of Env sequences found in the plasma of Group 1 (C) or Group 2 (D) SHIV-infected macaques. Black lines represent unmodified Env sequences from Group 1 or Group 2 viruses inoculated into animals and colored lines Env sequences obtained from infected animals at the indicated times p.i. Predominant Env sequences are labeled. See also Figure S2.
Analysis of Env sequences in plasma revealed that only one Env clone, 1054, was detectable at week 3 p.i. in both animals infected with Group 1 viruses (Figure 2C). At later time points, other Env sequences were also detected, such as AD17, that had recombined with 1054 sequences to various extents. Similarly, in the animal inoculated with Group 2 viruses, a single Env clone, AD081, predominated throughout all sampling time points (Figure 2D). Overall, these data suggest that the 1054 and AD081 Env proteins conferred an advantage over other Env proteins in the same cocktail, supporting more efficient SHIV replication in macaques.
In Vivo Replication of Individual “Winner” SHIVs
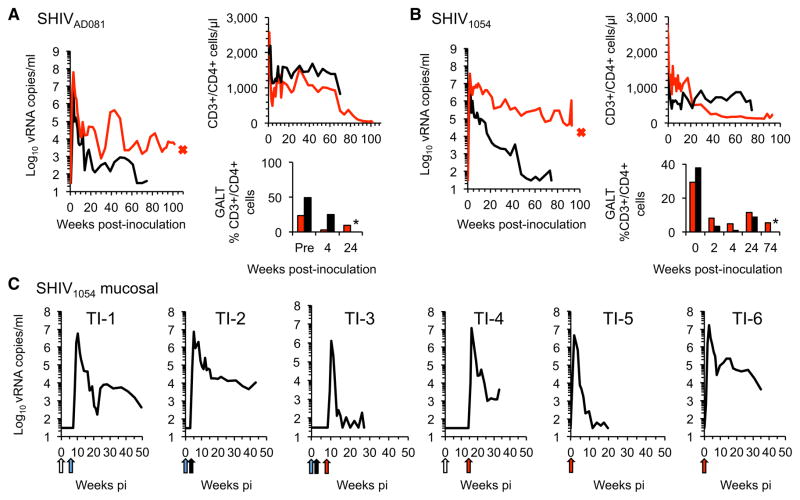
We next determined the ability of individual SHIVs encoding unadapted AD081 or 1054 Env proteins to replicate in macaques without CD8+ cell depletion. SHIVAD081 and SHIV1054 were each intravenously inoculated into two macaques, all of which developed high levels of acute viremia (Figures 3A and 3B). Specifically, peak viremia approached 107–108 viral RNA copies/ml in SHIVAD081-inoculated animals and was accompanied by marked GALT CD4+ T cell depletion (Figure 3A). Peripheral CD4+ T cell numbers decreased substantially during acute infection and continued to progressively decline. Importantly, one SHIVAD081-infected animal developed severe Pneumocystis pneumonia and was euthanized at 25 months p.i. for this AIDS-defining clinical endpoint. In SHIV1054-inoculated animals peak viremia reached 107 viral RNA copies/ml and remained high in one animal (Figure 3B). Although both SHIV1054-inoculated animals showed significant CD4+ T cell depletion in the GALT, peripheral CD4+ T cells in the animal with the highest viremia decreased dramatically during acute infection and continued to decline during the course of infection (Figure 3B). This animal succumbed to an AIDS-defining opportunistic infection, severe Pneumocystis pneumonia, and was euthanized at 21.5 months p.i. Thus, both unadapted R5-SHIVs selected by our strategy were capable of causing AIDS in macaques, a noteworthy property for R5-SHIVs that had not been adapted by serial passage in macaques.
Figure 3. Replication of Individual R5-SHIVs in Macaques.
(A) Plasma viremia, CD4+ T cell counts in blood, and CD4+ T cell frequency in the GALT for 2 macaques (one in red and one in black) inoculated intravenously with 2.9 × 105 i.u. of SHIVAD081. Asterisks in bar graphs indicate unavailable samples.
(B) As in (A), except that two macaques were inoculated intravenously with 4.8 × 105 i.u. of SHIV1054.
(C) Plasma viremia for six macaques inoculated intrarectally with varying doses of SHIV1054. Arrows indicate time and colors the dose of viral challenge; white arrow is 3 × 103 i.u., blue arrow is 3 × 104 i.u., black arrow is 7.4 × 104 i.u., and red arrow is 2.1x105 i.u. See also Figure S3.
Evaluation of Immunoprophylaxis Approaches against SHIVs Expressing Authentic, Transmitted HIV-1 Env Proteins
Analysis of the sensitivity of SHIV1054, SHIVAD081, and each of the other T/F-SHIV variants to several neutralizing antibodies revealed that, like most transmitted HIV-1 strains (Seaman et al., 2010), all exhibited Tier 2 neutralization properties (Table S2). We elected to perform a passive immunoprophylaxis experiment with SHIV1054, since it encodes a precise Env sequence that initiated a virus transmission event in humans.
For these studies, we first determined whether SHIV1054 could be transmitted to macaques via mucosal challenge, since HIV-1 transmission in humans occurs predominantly at mucosal sites. Six macaques were inoculated with varying doses of SHIV1054 intrarectally to establish an optimal challenge dose at which all animals became infected. All animals developed high acute viremia of between 106 and 107 viral RNA copies/ml (Figure 3C). Viral replication gradually declined thereafter. Although a substantial decrease in GALT CD4+ T cells was observed in five out of six infected animals, peripheral CD4+ T cell decrease was generally modest and transient (Figure S3). These findings demonstrate the ability of SHIV1054 to infect macaques via intrarectal transmission and established the optimal virus dose for such mucosal challenges.
We then attempted to protect macaques from infection using PGT121, a potent, broadly neutralizing candidate prophylactic and therapeutic monoclonal antibody that targets a glycan-dependent V3 loop epitope (Julien et al., 2013; Walker et al., 2011) and has been previously demonstrated to protect macaques against mucosal challenges by a macaque-adapted SHIV (Moldt et al., 2012). PGT121 neutralized SHIV1054 in vitro (Figure S4A).
We infused six macaques with PGT121 and six with a control antibody, both at a dose of 10 mg/kg. Serum antibody levels peaked 1 day postinfusion (Figure 4A), and plasma obtained from PGT121-infused animals up to 6 days postinfusion potently neutralized SHIV1054 in vitro (Figure S3B). One day after antibody infusion, all macaques were challenged intrarectally with 2.1 × 105 i.u. of SHIV1054. All control animals developed high levels of acute viremia, reaching peaks of 106–107 viral RNA copies/ml. In contrast, animals that received PGT-121 infusions did not develop viremia (Figure 4B). A single plasma viral load “blip” was detected at week 3 or 4 postchallenge in five of six PGT121-infused animals. However, this likely reflects trace contamination of the plasma samples since (1) all “blips” occurred in samples from the same collection date, rather than at the same time after infection, and (2) there was no evidence of established infection in these animals using other assays, including quantitative RT-PCR (qRT-PCR) assays of matched serum samples, qRT-PCR/PCR assays for PBMC-associated viral RNA and DNA, and immunoblot analysis (Figures S4C and S4D; Table S3). Overall, our data demonstrate that PGT121 is capable of protecting macaques against mucosal challenge with a SHIV encoding an HIV-1 envelope that mediated HIV-1 transmission in humans.
Figure 4. HIV-1 Monoclonal Antibody Protects against T/F Env Virus Challenge.
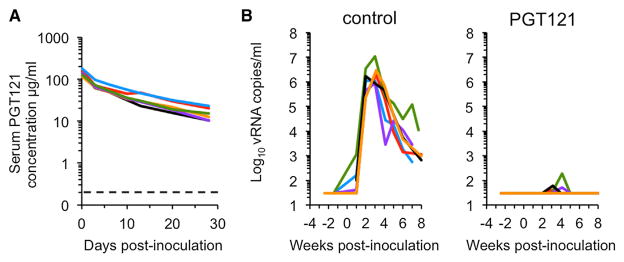
(A) PGT121 antibody concentration in serum of animals infused with the antibody on day −1. Day 0 is the day of challenge with SHIV1054. Dotted line indicates antibody concentration that achieves 90% neutralization of SHIV1054 in vitro.
(B) Plasma viremia in macaques receiving pre-exposure transfusion of PGT121 or control monoclonal antibodies at 10 mg/kg, followed by an intrarectal SHIV1054 challenge using 2.1 × 105 i.u. of virus. Animals in both groups are shown in #1 blue, #2 green, #3 purple, #4 red, #5 black, and #6 orange. See also Figure S4 and Table S3.
DISCUSSION
Two major limitations that plague current SHIV models are the paucity in Env diversity represented and the inconsistency in pathogenicity exhibited by SHIVs developed to date. To overcome these limitations, we have designed an approach that (a) facilitates the generation of a large number of SHIVs by introducing many different HIV-1 Env proteins into a SHIV backbone and (b) allows in vivo competition to select Env sequences that confer the highest replicative capacity in macaques. Using this approach, we generated and screened 37 new SHIVs and selected two HIV-1 Env proteins that can support efficient replication and cause AIDS-like disease in macaques without animal-to-animal passage.
The fact that each of the SHIVs selected from the two different groups of viruses is pathogenic in immunologically intact macaques establishes this approach for identifying HIV-1 Env proteins best suited for generation of SHIVs. It also suggests that the CD8+ cell depletion performed during the initial selection of these “winners” did not grossly affect the outcome. Even though the number of animals used in these experiments is very small, the fact that SHIV1054 dominated in 2/2 animals infected with 21 different SHIVs is statistically significant (p = 0.0059). At present, it is unclear what determinants allowed the “winner” Env proteins to outperform others in their respective groups, and we did not observe any correlation between “winning” and (1) sensitivity to various antibodies or soluble CD4 (Table S2) or (2) unusual characteristics of the patients from which they were derived (see Supplemental Experimental Procedures). It is also unclear how many of the other SHIVs would have replicated efficiently in non-CD8+-depleted animals. However, the goal of our approach was to identify HIV-1 Env proteins best suited for SHIV development, and as such, it was successful.
All our SHIV variants express HIV-1 Env proteins from viruses that have successfully mediated human-to-human transmission. The characteristics that render these Env proteins capable of being preferentially transmitted between humans from a swarm of other viruses remain uncertain (Liao et al., 2013; Parker et al., 2013; Parrish et al., 2012; Sagar et al., 2009; Salazar-Gonzalez et al., 2009; Wilen et al., 2011). Nevertheless, it is conceivable that the properties that allow them to successfully infect new human hosts may improve their ability to perform better in the context of SHIVs in animals.
Importantly, these viruses should be useful and relevant tools in determining the efficacy of prophylactic interventions targeting HIV-1 Env, because they accurately represent the sequences that such strategies must target to prevent HIV-1 infection in humans. Indeed, we show that a single neutralizing monoclonal antibody against HIV-1 Env can provide protection against mucosal challenges with a SHIV expressing an authentic T/F HIV-1 Env, underscoring the potential utility of such interventions in humans.
EXPERIMENTAL PROCEDURES
Cloning Strategy
Two plasmid vectors, containing the 5′ and 3′ halves of the SHIVKB9 genome (Karlsson et al., 1997), were generated using a low copy number plasmid backbone (pXf3). Full-length proviral plasmids encoding the different HIV-1 Env proteins were generated as outlined in Figure 1A and described in detail in Supplemental Experimental Procedures.
Cell Lines
MT2 cells were obtained from NIH AIDS Reagent Program. Following transduction with an MLV-derived vector based on LPCX (Clontech) that expressed human CCR5 and selection with puromycin, a single cell clone expressing high levels of CCR5, as determined by FACS using a monoclonal anti-CCR5 antibody conjugated to PE (clone 3A9, BD Pharmigen), was selected for use in all experiments.
Tropism Assays
TZM-bl cells seeded at 8 × 103 cells/well in 96-well plates the day before were incubated for 30 min in 100 μl of medium containing Maraviroc (16 μM) or AMD3100 (2 μM) or no inhibitor. Subsequently, dilutions of SHIVs expressing different Env proteins in a total volume of 100 μl were added to wells so that the final concentration of Maraviroc was 8 μM and AMD3100 1 μM. Medium was changed 24 hr postinfection and 48 hr postinfection β-galactosidase activity was determined using GalactoStar reagent (Applied Biosystems).
Animal Experiments
Rhesus macaques (Macaca mulatta) were housed and cared for in accordance with American Association for Accreditation of Laboratory Animal Care (AAALAC) standards in an AAALAC-accredited facility, and all animal procedures were performed according to a protocol approved by the Institutional Animal Care and Use Committee of the National Cancer Institute (see also Supplemental Experimental Procedures). For infections with pooled viral inocula, two macaques were inoculated with a cocktail of 21 SHIVs expressing group 1 Envs at a dose of 2.5 × 104 i.u. (as measured on TZM-bl) for each virus (Figure 1A), and one macaque was inoculated with a cocktail of 16 SHIVs expressing group 2 Envs, at a dose of 5 × 104 i.u. for each virus (Figure 1B). Inoculations were performed intravenously (saphenous vein), and the animals were subjected to CD8+ cell depletion by subcutaneous administration of a chimeric “rhesusized” anti-CD8 mAb, M-T807R1 (obtained from Keith Reimann, NIAID NHP Reagent Program, Harvard/Beth Israel Deaconess Medical Center) at a dose of 25 mg/kg, 30 min prior to virus inoculation and again 1 week later.
Virus inoculations with individual SHIVs were performed intravenously in macaques that were not CD8+ cell depleted. Two animals each received 2.9 × 105 i.u. of SHIVAD081 (Figure 3A), and two animals each received 4.8 × 105 i.u. of SHIV1054 (Figure 3B).
Six animals were inoculated intrarectally using increasing doses of SHIV1054 (Figure 3C). The specific doses used were 3 × 103 i.u. at week 0 and 3 × 104 i.u. at week 6 for TI-1, 3 × 104 i.u. at week 0 and 7.3 × 104 i.u. at week 3 for TI-2, 3 × 104 i.u. at week 0, 7.3 × 104 i.u. at week 3 and 2.1 × 105 i.u. at week 8 for TI-3, 3 × 103 i.u. at week 0 and 2.1 × 105 i.u. at week 14.4 for TI-4, and 2.1 × 105 i.u. at week 0 for TI-5 and TI-6.
For immunoprophylactic studies, six macaques received 10 mg/kg of PGT121 and six the control antibody DEN3 (Moldt et al., 2012) via intravenous infusion. All animals were subsequently inoculated intrarectally with 2.1 × 105 i.u. of SHIV1054 at 24 hr post-antibody infusion.
GALT Biopsies
Pinch biopsies of lower duodenal/upper jejunal tissue (up to 15 pinches per procedure, approximately 2 to 3 mm3) were obtained using biopsy forceps under direct endoscopic visualization from macaques under isoflurane anesthesia. For flow cytometry analysis, mononuclear cells were obtained from biopsy specimens processed using mechanical disruption and enzymic digestion of connective tissues, as previously described (Lifson et al., 2003; Veazey et al., 2001).
Viral Load Measurements
Virions were pelleted from plasma and virion-associated RNA extracted, as described previously (Cline et al., 2005). Briefly, plasma viral load was quantified using a two-step real-time qRT-PCR based on amplification of an SIV-mac239-derived sequence located in the Gag coding region.
Single-Genome Amplification/Sequencing of SHIV env
A viral DNA fragment that includes the entire env gene was amplified from the inoculum stock and each macaque at peak viremia using limiting dilution, single-genome amplification PCR approach. For details, see Supplemental Experimental Procedures.
Flow Cytometry
Antibodies and reagents were obtained from BD Biosciences, unless indicated otherwise, and data analysis was performed using FCS Express (De Novo Software). Samples were prepared and absolute cell counting and lymphocyte immunophenotyping were performed as previously described (Del Prete et al., 2012; Hatziioannou et al., 2009; Tabb et al., 2013). For details, see Supplemental Experimental Procedures.
Cell-Associated Viral RNA and DNA
A hybrid real-time/digital PCR format and analysis approach, previously described (Hansen et al., 2011), was applied for determination of cell-associated viral loads in isolated PBMCs. Quantitative hybrid real-time/digital nested PCR were performed as previously described (Hansen et al., 2011). A total of 12 replicates of each DNA or RNA sample were tested with two of the replicates containing a spike of DNA or RNA standard, as appropriate, to monitor assay performance and to guide retest requirements. Determined SIV DNA and RNA values were normalized to cell equivalents determined from coamplification of a target sequence in the Rhesus CCR5 gene sequence present at single haploid copy per genome.
Neutralization Tier Phenotyping and Neutralization Assays
Neutralization tier phenotyping of the R5-SHIVs was performed using TZM-bl cells as described previously (Li et al., 2005). Neutralization titers are the sample dilution (for plasma) or antibody concentration (for sCD4, purified IgG preparations and monoclonal antibodies) at which relative luminescence units (RLUs) were reduced by 50% compared to RLU in virus control wells after subtraction of background RLU in cell control wells.
Serum neutralizing titers against the challenge SHIV1054 stock of animals infused with PGT121 were determined in TZM-bl cells, as previously described (Li et al., 2005). Briefly, virus was mixed with serial dilution of the serum samples starting at 100-fold dilution. Following 1 hr of incubation, the virus/serum mix was used to infect TZM-bl cells Forty-eight hours after infection, the cells were lysed, and luciferase expression was evaluated using the Bright-Glo luciferase assay (Promega).
ELISA Assays
PGT121 serum concentration was determined by a recombinant HIVJRFL gp120-specific (Progenics Pharmaceuticals) ELISA, as previously described (Parren et al., 2001). Briefly, microtiter plates were coated overnight with 2 μg/ml HIVJRFL gp120. Following a wash step, the plates were blocked for 1 hr with 3% (vol/vol) BSA and incubated for 2 hr with serial dilution of the serum samples starting at 20-fold dilution or control antibodies. Binding was detected with a goat anti-human IgG F(ab)2 fragment coupled to alkaline phosphatase (Pierce) and visualized with p-nitrophenyl phosphate substrate (Sigma-Aldrich).
Immunoblot Analyses
SHIV1054 virions generated in transfected 293T cells were purified by centrifugation through sucrose and resuspended in PBS. Following initial densitometry determination of capsid (CA) protein concentration in samples run on Coomassie-stained gels, 200 ng CA/lane were loaded and separated on 4%–20% acrylamide gradient gels. Proteins were then transferred onto a PVDF membrane (Millipore Immobilon-P, IPVH00010) using a semi-dry electroblotter (Ellard Instrumentation), cut into strips, and probed with heat-inactivated plasma from infected macaques clarified by centrifugation and diluted 1:1,000, followed by a goat anti-human IgG-peroxidase conjugate (Sigma cat. #A8667) diluted 1:10,000. Blots were developed using chemiluminescent detection reagents (Pierce).
Supplementary Material
Acknowledgments
We are grateful to Vicky Coalter, Adam Wiles, Rodney Wiles, and Donald Johnson for assistance with study coordination and processing of nonhuman primate blood and tissues and to Florian Douam and Melinda Ng for assistance in optimizing SHIV cloning procedures. This work was supported by grants from the NIH; R21AI093255 (to T.H.), R37AI64003, and R01AI50111 (to P.D.B.); by the Howard Hughes Medical Institute; by NIAID-NIH Contract No. HHSN27201100016C (to D.C.M.); and in part with federal funds from the National Cancer Institute, National Institutes of Health, under Contract No. HHSN261200800001E (to J.D.L.). Histology support was provided by the Pathology/Histotechnology Laboratory core service located at the Frederick National Laboratory for Cancer Research, Leidos Biomedical Research, Inc., Frederick, MD. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government.
Footnotes
ACCESSION NUMBERS
All sequences generated were deposited in GenBank under accession numbers KJ372039–KJ372207.
Supplemental Information includes four figures, three tables, and Supplemental Experimental Procedures and can be found with this article online at http://dx.doi.org/10.1016/j.chom.2014.08.003.
AUTHOR CONTRIBUTIONS
P.D.B., J.D.L., V.N.K., and T.H. conceived the study and designed the experiments. G.Q.D., B.A., B.M., B.F.K., J.D.E., A.R., M.S., K.O., R.F., C.M.T., E.C., J.S., C.C.L., M.P., J.D.L., and T.H. performed experiments. G.Q.D., B.F.K., P.D.B., J.D.L., V.N.K., and T.H. analyzed the data. D.C.M., D.R.B., G.M.S., and M.M. provided reagents. P.D.B. and T.H. wrote the paper with contributions from all authors.
References
- Burton DR, Poignard P, Stanfield RL, Wilson IA. Broadly neutralizing antibodies present new prospects to counter highly antigenically diverse viruses. Science. 2012;337:183–186. doi: 10.1126/science.1225416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cline AN, Bess JW, Piatak M, Jr, Lifson JD. Highly sensitive SIV plasma viral load assay: practical considerations, realistic performance expectations, and application to reverse engineering of vaccines for AIDS. J Med Primatol. 2005;34:303–312. doi: 10.1111/j.1600-0684.2005.00128.x. [DOI] [PubMed] [Google Scholar]
- Del Prete GQ, Kearney MF, Spindler J, Wiegand A, Chertova E, Roser JD, Estes JD, Hao XP, Trubey CM, Lara A, et al. Restricted replication of xenotropic murine leukemia virus-related virus in pigtailed macaques. J Virol. 2012;86:3152–3166. doi: 10.1128/JVI.06886-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen SG, Ford JC, Lewis MS, Ventura AB, Hughes CM, Coyne-Johnson L, Whizin N, Oswald K, Shoemaker R, Swanson T, et al. Profound early control of highly pathogenic SIV by an effector memory T-cell vaccine. Nature. 2011;473:523–527. doi: 10.1038/nature10003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harouse JM, Gettie A, Eshetu T, Tan RC, Bohm R, Blanchard J, Baskin G, Cheng-Mayer C. Mucosal transmission and induction of simian AIDS by CCR5-specific simian/human immunodeficiency virus SHIV(SF162P3) J Virol. 2001;75:1990–1995. doi: 10.1128/JVI.75.4.1990-1995.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatziioannou T, Evans DT. Animal models for HIV/AIDS research. Nat Rev Microbiol. 2012;10:852–867. doi: 10.1038/nrmicro2911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatziioannou T, Ambrose Z, Chung NP, Piatak M, Jr, Yuan F, Trubey CM, Coalter V, Kiser R, Schneider D, Smedley J, et al. A macaque model of HIV-1 infection. Proc Natl Acad Sci USA. 2009;106:4425–4429. doi: 10.1073/pnas.0812587106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Julien JP, Sok D, Khayat R, Lee JH, Doores KJ, Walker LM, Ramos A, Diwanji DC, Pejchal R, Cupo A, et al. Broadly neutralizing antibody PGT121 allosterically modulates CD4 binding via recognition of the HIV-1 gp120 V3 base and multiple surrounding glycans. PLoS Pathog. 2013;9:e1003342. doi: 10.1371/journal.ppat.1003342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlsson GB, Halloran M, Li J, Park IW, Gomila R, Reimann KA, Axthelm MK, Iliff SA, Letvin NL, Sodroski J. Characterization of molecularly cloned simian-human immunodeficiency viruses causing rapid CD4+ lymphocyte depletion in rhesus monkeys. J Virol. 1997;71:4218–4225. doi: 10.1128/jvi.71.6.4218-4225.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keele BF, Giorgi EE, Salazar-Gonzalez JF, Decker JM, Pham KT, Salazar MG, Sun C, Grayson T, Wang S, Li H, et al. Identification and characterization of transmitted and early founder virus envelopes in primary HIV-1 infection. Proc Natl Acad Sci USA. 2008;105:7552–7557. doi: 10.1073/pnas.0802203105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein F, Mouquet H, Dosenovic P, Scheid JF, Scharf L, Nussenzweig MC. Antibodies in HIV-1 vaccine development and therapy. Science. 2013;341:1199–1204. doi: 10.1126/science.1241144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Gao F, Mascola JR, Stamatatos L, Polonis VR, Koutsoukos M, Voss G, Goepfert P, Gilbert P, Greene KM, et al. Human immunodeficiency virus type 1 env clones from acute and early subtype B infections for standardized assessments of vaccine-elicited neutralizing antibodies. J Virol. 2005;79:10108–10125. doi: 10.1128/JVI.79.16.10108-10125.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao HX, Tsao CY, Alam SM, Muldoon M, Vandergrift N, Ma BJ, Lu X, Sutherland LL, Scearce RM, Bowman C, et al. Antigenicity and immunogenicity of transmitted/founder, consensus, and chronic envelope glycoproteins of human immunodeficiency virus type 1. J Virol. 2013;87:4185–4201. doi: 10.1128/JVI.02297-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lifson JD, Piatak M, Jr, Cline AN, Rossio JL, Purcell J, Pandrea I, Bischofberger N, Blanchard J, Veazey RS. Transient early post-inoculation anti-retroviral treatment facilitates controlled infection with sparing of CD4+ T cells in gut-associated lymphoid tissues in SIVmac239-infected rhesus macaques, but not resistance to rechallenge. J Med Primatol. 2003;32:201–210. doi: 10.1034/j.1600-0684.2003.00026.x. [DOI] [PubMed] [Google Scholar]
- Moldt B, Rakasz EG, Schultz N, Chan-Hui PY, Swiderek K, Weisgrau KL, Piaskowski SM, Bergman Z, Watkins DI, Poignard P, Burton DR. Highly potent HIV-specific antibody neutralization in vitro translates into effective protection against mucosal SHIV challenge in vivo. Proc Natl Acad Sci USA. 2012;109:18921–18925. doi: 10.1073/pnas.1214785109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura Y, Shingai M, Willey R, Sadjadpour R, Lee WR, Brown CR, Brenchley JM, Buckler-White A, Petros R, Eckhaus M, et al. Generation of the pathogenic R5-tropic simian/human immunodeficiency virus SHIVAD8 by serial passaging in rhesus macaques. J Virol. 2010;84:4769–4781. doi: 10.1128/JVI.02279-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker ZF, Iyer SS, Wilen CB, Parrish NF, Chikere KC, Lee FH, Didigu CA, Berro R, Klasse PJ, Lee B, et al. Transmitted/founder and chronic HIV-1 envelope proteins are distinguished by differential utilization of CCR5. J Virol. 2013;87:2401–2411. doi: 10.1128/JVI.02964-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parren PW, Marx PA, Hessell AJ, Luckay A, Harouse J, Cheng-Mayer C, Moore JP, Burton DR. Antibody protects macaques against vaginal challenge with a pathogenic R5 simian/human immunodeficiency virus at serum levels giving complete neutralization in vitro. J Virol. 2001;75:8340–8347. doi: 10.1128/JVI.75.17.8340-8347.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parrish NF, Wilen CB, Banks LB, Iyer SS, Pfaff JM, Salazar-Gonzalez JF, Salazar MG, Decker JM, Parrish EH, Berg A, et al. Transmitted/founder and chronic subtype C HIV-1 use CD4 and CCR5 receptors with equal efficiency and are not inhibited by blocking the integrin α4β7. PLoS Pathog. 2012;8:e1002686. doi: 10.1371/journal.ppat.1002686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sagar M, Laeyendecker O, Lee S, Gamiel J, Wawer MJ, Gray RH, Serwadda D, Sewankambo NK, Shepherd JC, Toma J, et al. Selection of HIV variants with signature genotypic characteristics during heterosexual transmission. J Infect Dis. 2009;199:580–589. doi: 10.1086/596557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salazar-Gonzalez JF, Bailes E, Pham KT, Salazar MG, Guffey MB, Keele BF, Derdeyn CA, Farmer P, Hunter E, Allen S, et al. Deciphering human immunodeficiency virus type 1 transmission and early envelope diversification by single-genome amplification and sequencing. J Virol. 2008;82:3952–3970. doi: 10.1128/JVI.02660-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salazar-Gonzalez JF, Salazar MG, Keele BF, Learn GH, Giorgi EE, Li H, Decker JM, Wang S, Baalwa J, Kraus MH, et al. Genetic identity, biological phenotype, and evolutionary pathways of transmitted/founder viruses in acute and early HIV-1 infection. J Exp Med. 2009;206:1273–1289. doi: 10.1084/jem.20090378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seaman MS, Janes H, Hawkins N, Grandpre LE, Devoy C, Giri A, Coffey RT, Harris L, Wood B, Daniels MG, et al. Tiered categorization of a diverse panel of HIV-1 Env pseudoviruses for assessment of neutralizing antibodies. J Virol. 2010;84:1439–1452. doi: 10.1128/JVI.02108-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song RJ, Chenine AL, Rasmussen RA, Ruprecht CR, Mirshahidi S, Grisson RD, Xu W, Whitney JB, Goins LM, Ong H, et al. Molecularly cloned SHIV-1157ipd3N4: a highly replication- competent, mucosally transmissible R5 simian-human immunodeficiency virus encoding HIV clade C Env. J Virol. 2006;80:8729–8738. doi: 10.1128/JVI.00558-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabb B, Morcock DR, Trubey CM, Quiñones OA, Hao XP, Smedley J, Macallister R, Piatak M, Jr, Harris LD, Paiardini M, et al. Reduced inflammation and lymphoid tissue immunopathology in rhesus macaques receiving anti-tumor necrosis factor treatment during primary simian immunodeficiency virus infection. J Infect Dis. 2013;207:880–892. doi: 10.1093/infdis/jis643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veazey RS, Gauduin MC, Mansfield KG, Tham IC, Altman JD, Lifson JD, Lackner AA, Johnson RP. Emergence and kinetics of simian immunodeficiency virus-specific CD8(+) T cells in the intestines of macaques during primary infection. J Virol. 2001;75:10515–10519. doi: 10.1128/JVI.75.21.10515-10519.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker LM, Huber M, Doores KJ, Falkowska E, Pejchal R, Julien JP, Wang SK, Ramos A, Chan-Hui PY, Moyle M, et al. Broad neutralization coverage of HIV by multiple highly potent antibodies. Nature. 2011;477:466–470. doi: 10.1038/nature10373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilen CB, Parrish NF, Pfaff JM, Decker JM, Henning EA, Haim H, Petersen JE, Wojcechowskyj JA, Sodroski J, Haynes BF, et al. Phenotypic and immunologic comparison of clade B transmitted/founder and chronic HIV-1 envelope glycoproteins. J Virol. 2011;85:8514–8527. doi: 10.1128/JVI.00736-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang LQ, MacKenzie P, Cleland A, Holmes EC, Brown AJ, Simmonds P. Selection for specific sequences in the external envelope protein of human immunodeficiency virus type 1 upon primary infection. J Virol. 1993;67:3345–3356. doi: 10.1128/jvi.67.6.3345-3356.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu T, Mo H, Wang N, Nam DS, Cao Y, Koup RA, Ho DD. Genotypic and phenotypic characterization of HIV-1 patients with primary infection. Science. 1993;261:1179–1181. doi: 10.1126/science.8356453. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.