Abstract
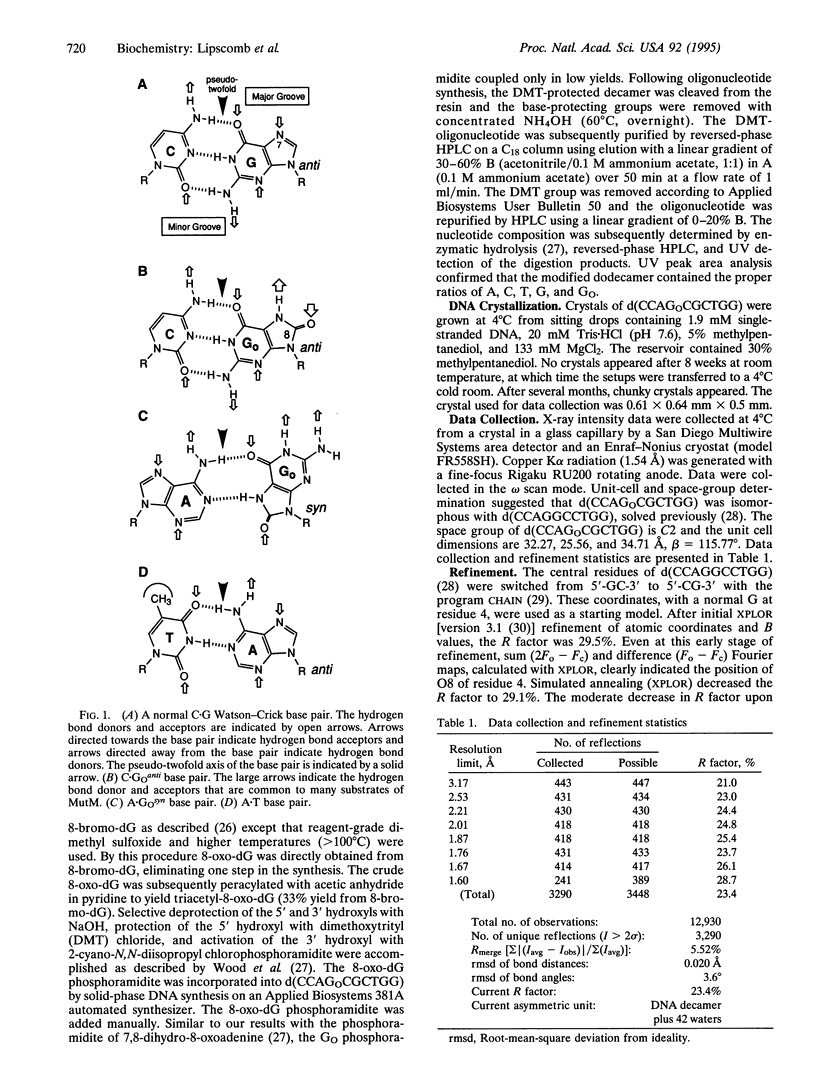
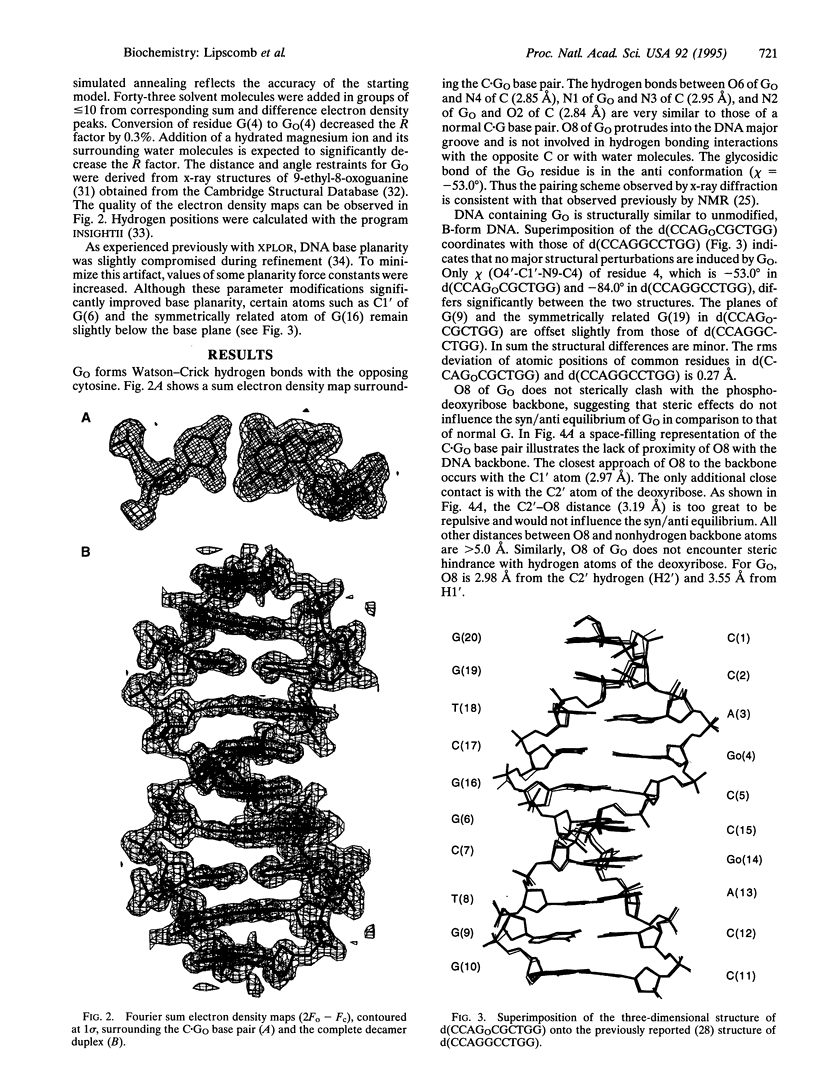
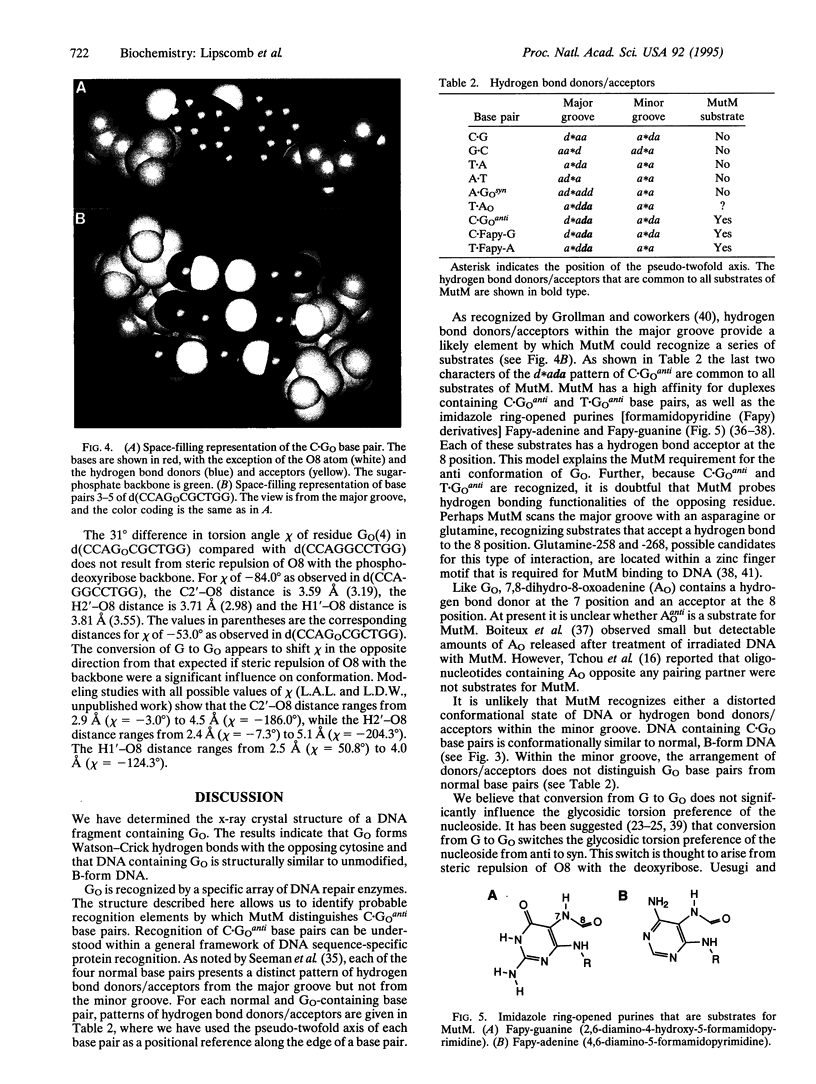
We have determined the x-ray structure of a DNA fragment containing 7,8-dihydro-8-oxoguanine (G(O)). The structure of the duplex form of d(CCAGOCGCTGG) has been determined to 1.6-A resolution. The results demonstrate that GO forms Watson-Crick base pairs with the opposite C and that G(O) is in the anti conformation. Structural perturbations induced by C.G(O)anti base pairs are subtle. The structure allows us to identify probable elements by which the DNA repair protein MutM recognizes its substrates. Hydrogen bond donors/acceptors within the major groove are the most likely element. In that groove, the pattern of hydrogen-bond donors/acceptors of C.G(O)anti is unique. Additional structural analysis indicates that conversion of G to G(O) would not significantly influence the glycosidic torsion preference of the nucleoside. There is no steric interaction of the 8-oxygen of G(O) with the phospho-deoxyribose backbone.
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bessho T., Roy R., Yamamoto K., Kasai H., Nishimura S., Tano K., Mitra S. Repair of 8-hydroxyguanine in DNA by mammalian N-methylpurine-DNA glycosylase. Proc Natl Acad Sci U S A. 1993 Oct 1;90(19):8901–8904. doi: 10.1073/pnas.90.19.8901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bessho T., Tano K., Kasai H., Ohtsuka E., Nishimura S. Evidence for two DNA repair enzymes for 8-hydroxyguanine (7,8-dihydro-8-oxoguanine) in human cells. J Biol Chem. 1993 Sep 15;268(26):19416–19421. [PubMed] [Google Scholar]
- Bhatnagar S. K., Bullions L. C., Bessman M. J. Characterization of the mutT nucleoside triphosphatase of Escherichia coli. J Biol Chem. 1991 May 15;266(14):9050–9054. [PubMed] [Google Scholar]
- Boiteux S., Gajewski E., Laval J., Dizdaroglu M. Substrate specificity of the Escherichia coli Fpg protein (formamidopyrimidine-DNA glycosylase): excision of purine lesions in DNA produced by ionizing radiation or photosensitization. Biochemistry. 1992 Jan 14;31(1):106–110. doi: 10.1021/bi00116a016. [DOI] [PubMed] [Google Scholar]
- Boiteux S., O'Connor T. R., Lederer F., Gouyette A., Laval J. Homogeneous Escherichia coli FPG protein. A DNA glycosylase which excises imidazole ring-opened purines and nicks DNA at apurinic/apyrimidinic sites. J Biol Chem. 1990 Mar 5;265(7):3916–3922. [PubMed] [Google Scholar]
- Castaing B., Geiger A., Seliger H., Nehls P., Laval J., Zelwer C., Boiteux S. Cleavage and binding of a DNA fragment containing a single 8-oxoguanine by wild type and mutant FPG proteins. Nucleic Acids Res. 1993 Jun 25;21(12):2899–2905. doi: 10.1093/nar/21.12.2899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho B. P., Evans F. E. Structure of oxidatively damaged nucleic acid adducts. 3. Tautomerism, ionization and protonation of 8-hydroxyadenosine studied by 15N NMR spectroscopy. Nucleic Acids Res. 1991 Mar 11;19(5):1041–1047. doi: 10.1093/nar/19.5.1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epe B. Genotoxicity of singlet oxygen. Chem Biol Interact. 1991;80(3):239–260. doi: 10.1016/0009-2797(91)90086-m. [DOI] [PubMed] [Google Scholar]
- Fiala E. S., Nie G., Sodum R., Conaway C. C., Sohn O. S. 2-Nitropropane-induced liver DNA and RNA base modifications: differences between Sprague-Dawley rats and New Zealand white rabbits. Cancer Lett. 1993 Oct 15;74(1-2):9–14. doi: 10.1016/0304-3835(93)90037-a. [DOI] [PubMed] [Google Scholar]
- Fuciarelli A. F., Wegher B. J., Blakely W. F., Dizdaroglu M. Yields of radiation-induced base products in DNA: effects of DNA conformation and gassing conditions. Int J Radiat Biol. 1990 Sep;58(3):397–415. doi: 10.1080/09553009014551761. [DOI] [PubMed] [Google Scholar]
- Grollman A. P., Moriya M. Mutagenesis by 8-oxoguanine: an enemy within. Trends Genet. 1993 Jul;9(7):246–249. doi: 10.1016/0168-9525(93)90089-z. [DOI] [PubMed] [Google Scholar]
- Hahn M., Heinemann U. DNA helix structure and refinement algorithm: comparison of models for d(CCAGGCm5CTGG) derived from NUCLSQ, TNT and X-PLOR. Acta Crystallogr D Biol Crystallogr. 1993 Sep 1;49(Pt 5):468–477. doi: 10.1107/S0907444993004858. [DOI] [PubMed] [Google Scholar]
- Heinemann U., Alings C. Crystallographic study of one turn of G/C-rich B-DNA. J Mol Biol. 1989 Nov 20;210(2):369–381. doi: 10.1016/0022-2836(89)90337-9. [DOI] [PubMed] [Google Scholar]
- Kouchakdjian M., Bodepudi V., Shibutani S., Eisenberg M., Johnson F., Grollman A. P., Patel D. J. NMR structural studies of the ionizing radiation adduct 7-hydro-8-oxodeoxyguanosine (8-oxo-7H-dG) opposite deoxyadenosine in a DNA duplex. 8-Oxo-7H-dG(syn).dA(anti) alignment at lesion site. Biochemistry. 1991 Feb 5;30(5):1403–1412. doi: 10.1021/bi00219a034. [DOI] [PubMed] [Google Scholar]
- Lin T. S., Cheng J. C., Ishiguro K., Sartorelli A. C. 8-Substituted guanosine and 2'-deoxyguanosine derivatives as potential inducers of the differentiation of Friend erythroleukemia cells. J Med Chem. 1985 Sep;28(9):1194–1198. doi: 10.1021/jm00147a012. [DOI] [PubMed] [Google Scholar]
- Maki H., Sekiguchi M. MutT protein specifically hydrolyses a potent mutagenic substrate for DNA synthesis. Nature. 1992 Jan 16;355(6357):273–275. doi: 10.1038/355273a0. [DOI] [PubMed] [Google Scholar]
- McAuley-Hecht K. E., Leonard G. A., Gibson N. J., Thomson J. B., Watson W. P., Hunter W. N., Brown T. Crystal structure of a DNA duplex containing 8-hydroxydeoxyguanine-adenine base pairs. Biochemistry. 1994 Aug 30;33(34):10266–10270. doi: 10.1021/bi00200a006. [DOI] [PubMed] [Google Scholar]
- Michaels M. L., Cruz C., Grollman A. P., Miller J. H. Evidence that MutY and MutM combine to prevent mutations by an oxidatively damaged form of guanine in DNA. Proc Natl Acad Sci U S A. 1992 Aug 1;89(15):7022–7025. doi: 10.1073/pnas.89.15.7022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaels M. L., Miller J. H. The GO system protects organisms from the mutagenic effect of the spontaneous lesion 8-hydroxyguanine (7,8-dihydro-8-oxoguanine). J Bacteriol. 1992 Oct;174(20):6321–6325. doi: 10.1128/jb.174.20.6321-6325.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaels M. L., Pham L., Cruz C., Miller J. H. MutM, a protein that prevents G.C----T.A transversions, is formamidopyrimidine-DNA glycosylase. Nucleic Acids Res. 1991 Jul 11;19(13):3629–3632. doi: 10.1093/nar/19.13.3629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaels M. L., Pham L., Nghiem Y., Cruz C., Miller J. H. MutY, an adenine glycosylase active on G-A mispairs, has homology to endonuclease III. Nucleic Acids Res. 1990 Jul 11;18(13):3841–3845. doi: 10.1093/nar/18.13.3841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaels M. L., Tchou J., Grollman A. P., Miller J. H. A repair system for 8-oxo-7,8-dihydrodeoxyguanine. Biochemistry. 1992 Nov 17;31(45):10964–10968. doi: 10.1021/bi00160a004. [DOI] [PubMed] [Google Scholar]
- Mo J. Y., Maki H., Sekiguchi M. Hydrolytic elimination of a mutagenic nucleotide, 8-oxodGTP, by human 18-kilodalton protein: sanitization of nucleotide pool. Proc Natl Acad Sci U S A. 1992 Nov 15;89(22):11021–11025. doi: 10.1073/pnas.89.22.11021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moriya M., Grollman A. P. Mutations in the mutY gene of Escherichia coli enhance the frequency of targeted G:C-->T:a transversions induced by a single 8-oxoguanine residue in single-stranded DNA. Mol Gen Genet. 1993 May;239(1-2):72–76. doi: 10.1007/BF00281603. [DOI] [PubMed] [Google Scholar]
- Oda Y., Uesugi S., Ikehara M., Nishimura S., Kawase Y., Ishikawa H., Inoue H., Ohtsuka E. NMR studies of a DNA containing 8-hydroxydeoxyguanosine. Nucleic Acids Res. 1991 Apr 11;19(7):1407–1412. doi: 10.1093/nar/19.7.1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park E. M., Shigenaga M. K., Degan P., Korn T. S., Kitzler J. W., Wehr C. M., Kolachana P., Ames B. N. Assay of excised oxidative DNA lesions: isolation of 8-oxoguanine and its nucleoside derivatives from biological fluids with a monoclonal antibody column. Proc Natl Acad Sci U S A. 1992 Apr 15;89(8):3375–3379. doi: 10.1073/pnas.89.8.3375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavlov Y. I., Minnick D. T., Izuta S., Kunkel T. A. DNA replication fidelity with 8-oxodeoxyguanosine triphosphate. Biochemistry. 1994 Apr 19;33(15):4695–4701. doi: 10.1021/bi00181a029. [DOI] [PubMed] [Google Scholar]
- Pflaum M., Boiteux S., Epe B. Visible light generates oxidative DNA base modifications in high excess of strand breaks in mammalian cells. Carcinogenesis. 1994 Feb;15(2):297–300. doi: 10.1093/carcin/15.2.297. [DOI] [PubMed] [Google Scholar]
- Seeman N. C., Rosenberg J. M., Rich A. Sequence-specific recognition of double helical nucleic acids by proteins. Proc Natl Acad Sci U S A. 1976 Mar;73(3):804–808. doi: 10.1073/pnas.73.3.804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibutani S., Takeshita M., Grollman A. P. Insertion of specific bases during DNA synthesis past the oxidation-damaged base 8-oxodG. Nature. 1991 Jan 31;349(6308):431–434. doi: 10.1038/349431a0. [DOI] [PubMed] [Google Scholar]
- Tchou J., Bodepudi V., Shibutani S., Antoshechkin I., Miller J., Grollman A. P., Johnson F. Substrate specificity of Fpg protein. Recognition and cleavage of oxidatively damaged DNA. J Biol Chem. 1994 May 27;269(21):15318–15324. [PubMed] [Google Scholar]
- Tchou J., Kasai H., Shibutani S., Chung M. H., Laval J., Grollman A. P., Nishimura S. 8-oxoguanine (8-hydroxyguanine) DNA glycosylase and its substrate specificity. Proc Natl Acad Sci U S A. 1991 Jun 1;88(11):4690–4694. doi: 10.1073/pnas.88.11.4690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tchou J., Michaels M. L., Miller J. H., Grollman A. P. Function of the zinc finger in Escherichia coli Fpg protein. J Biol Chem. 1993 Dec 15;268(35):26738–26744. [PubMed] [Google Scholar]
- Uesugi S., Ikehara M. Carbon-13 magnetic resonance spectra of 8-substituted purine nucleosides. Characteristic shifts for the syn conformation. J Am Chem Soc. 1977 May 11;99(10):3250–3253. doi: 10.1021/ja00452a008. [DOI] [PubMed] [Google Scholar]
- Vidmar J. J., Cupples C. G. MutY repair is mutagenic in mutT- strains of Escherichia coli. Can J Microbiol. 1993 Sep;39(9):892–894. doi: 10.1139/m93-133. [DOI] [PubMed] [Google Scholar]
- Wood M. L., Dizdaroglu M., Gajewski E., Essigmann J. M. Mechanistic studies of ionizing radiation and oxidative mutagenesis: genetic effects of a single 8-hydroxyguanine (7-hydro-8-oxoguanine) residue inserted at a unique site in a viral genome. Biochemistry. 1990 Jul 31;29(30):7024–7032. doi: 10.1021/bi00482a011. [DOI] [PubMed] [Google Scholar]
- Wood M. L., Esteve A., Morningstar M. L., Kuziemko G. M., Essigmann J. M. Genetic effects of oxidative DNA damage: comparative mutagenesis of 7,8-dihydro-8-oxoguanine and 7,8-dihydro-8-oxoadenine in Escherichia coli. Nucleic Acids Res. 1992 Nov 25;20(22):6023–6032. doi: 10.1093/nar/20.22.6023. [DOI] [PMC free article] [PubMed] [Google Scholar]




