SUMMARY
The enteric pathogen enterohemorrhagic Escherichia coli (EHEC) causes severe diarrhea but the influence of the gut microbiota on EHEC infection is largely unknown. A predominant member of the microbiota, Bacteroides thetaiotaomicron (Bt), is resident at EHEC attachment sites. We show that Bt enhances EHEC virulence gene expression through the transcription factor, Cra, which is functionally sensitive to sugar concentrations. This enhanced virulence accompanies increased formation of attaching and effacing (AE) lesions requisite for EHEC colonization. Infection with Citrobacter rodentium, a natural mouse pathogen homologous to EHEC, in Bt-reconstituted mice results in increased gut permeability along with exacerbated host pathology and mortality compared to mice deplete of microflora. Bt modifies the metabolite environment at infection sites, increasing metabolites involved in gluconeogenesis, with stark increases in succinate, which can be sensed by Cra. Our findings suggest that microbiota composition affects disease outcome and may explain links between microbiota composition and disease susceptibility.
INTRODUCTION
Trillions of commensal bacteria inhabit the gastrointestinal tract, with the highest diversity and abundance of microbial species residing within the colon (Walter and Ley, 2011). The gut microbiota contribute to gut maturation, host nutrition, and pathogen resistance (Hooper et al., 2002; Sommer and Backhed, 2013), but recent findings also implicate a role for the microbiota in inflammatory bowel disease (IBD), cancer, obesity, diabetes, and heart disease (Honda and Littman, 2012; Spor et al., 2011). Multiple studies indicate that differences in microbial composition exist between healthy and diseased patients, however, it is not well understood if these shifts occur prior to disease or are a product of the disease (Gevers et al., 2014; Larsen et al., 2010; Peterson et al., 2008; Turnbaugh et al., 2009a; Turnbaugh et al., 2009b; Wang et al., 2009).
Transplantation of the microbiota from mouse strains resistant to Citrobacter rodentium infection rescued susceptible mice from a lethal challenge with C. rodentium, demonstrating that resistance could be transferred and that microbial compositions affect disease susceptibility (Willing et al., 2011). Additionally, successful treatment of recurrent Clostridium difficile infection through fecal transplantation further demonstrates that a balanced microbiota play an important role in preventing dysbiosis (Borody and Khoruts, 2012; Kelly et al., 2014). Specific taxa have been associated with increased disease susceptibility. A prospective study of the gut microbiota compositions in poultry workers before, during, and after exposure to Campylobacter found that those infected had elevated proportions of Bacteroides and Escherichia compared to those that remained uninfected despite repeated exposure (Dicksved et al., 2014).
A member of the Bacteroidetes phylum and major constituent of the microbiota, Bacteroides thetaiotaomicron encodes a number of glycoside hydrolases and polysaccharide lyases that enrich the availability of nutrients within the intestine (Sonnenburg et al., 2005). Within the intestine, Bt degrades complex polysaccharides into monosaccharides that can readily be used by non-glycophagic bacterial species such as E. coli and C. rodentium (Sonnenburg et al., 2005; Xu et al., 2003). Fluctuations in sugar concentrations modulate virulence gene expression and colonization of the human pathogen Enterohemorrhagic E. coli (EHEC) (Njoroge and Sperandio, 2012; Njoroge et al., 2012; Pacheco et al., 2012). EHEC colonizes the human colon, forming attaching and effacing (AE) lesions, invoking bloody diarrhea, hemorrhagic colitis, and hemolytic uremic syndrome (HUS) (Karmali et al., 1983). AE lesions result from extensive remodeling of the host cellular cytoskeleton that create a pedestal, or cup-like structure, beneath the bacteria (Kaper et al., 2004). EHEC also encodes a potent Shiga toxin (Stx) that is responsible for HUS (Kaper et al., 2004). EHEC intestinal colonization is dependent on the locus of enterocyte effacement (LEE) pathogenicity island (PAI) (McDaniel et al., 1995). The LEE region contains five major operons (LEE1–5) that encode a type III secretion system (T3SS), an adhesin (intimin) and its receptor (Tir), and effector proteins (Elliott et al., 1998; Kenny et al., 1997; Knutton et al., 1998). The ler gene encoded within LEE1 acts as the master regulator of the LEE genes (Elliott et al., 2000; Mellies et al., 1999). Glycolytic conditions, such as those found in the gut lumen, inhibit LEE1 expression while gluconeogenic conditions, akin to those found near the epithelial surface, activate LEE1 expression (Njoroge and Sperandio, 2012; Njoroge et al., 2012; Pacheco et al., 2012). We sought to understand if the metabolic changes induced by Bt affect susceptibility to infection and disease progression of EHEC.
Our results demonstrate that Bt increases virulence gene expression of EHEC and C. rodentium in vitro and during murine infection, respectively, and that Bt regulation of EHEC virulence occurs through the transcription factor Cra by sensing fluctuations in sugar concentrations. Mice reconstituted with Bt following antibiotics treatment lost weight and succumbed to infection more rapidly than mice deplete of microflora. Perturbations to a functional T3SS or to the presence of Shiga toxin attenuated disease progression, indicating that both a functional T3SS and Shiga toxin contribute to disease. Bt reconstitution augments the pathophysiology associated with C. rodentium infection, enhancing edema of the colonic epithelium, exacerbating crypt destruction, increasing immune infiltration, and impairing intestinal epithelial repair. Additionally, we identified metabolites at sites of infection specific to Bt colonization, C. rodentium infection, or a combination of Bt colonization and C. rodentium infection. The metabolites are primarily involved in oxidative stress, nucleotide synthesis, and gluconeogenesis. Furthermore, we linked succinate, a metabolite increased at the site of infection when Bt is present and that has previously been shown to act as a virulence factor (Rotstein et al., 1985; Rotstein et al., 1989), with augmented secretion of the EHEC T3SS translocon. Together, our findings indicate that a prominent member of the microbiota enhances virulence gene expression of an enteric pathogen and worsens the prognosis of an enteric infection.
RESULTS
Bt Increases Expression of One-Fifth of the E. coli Array Probe Sets
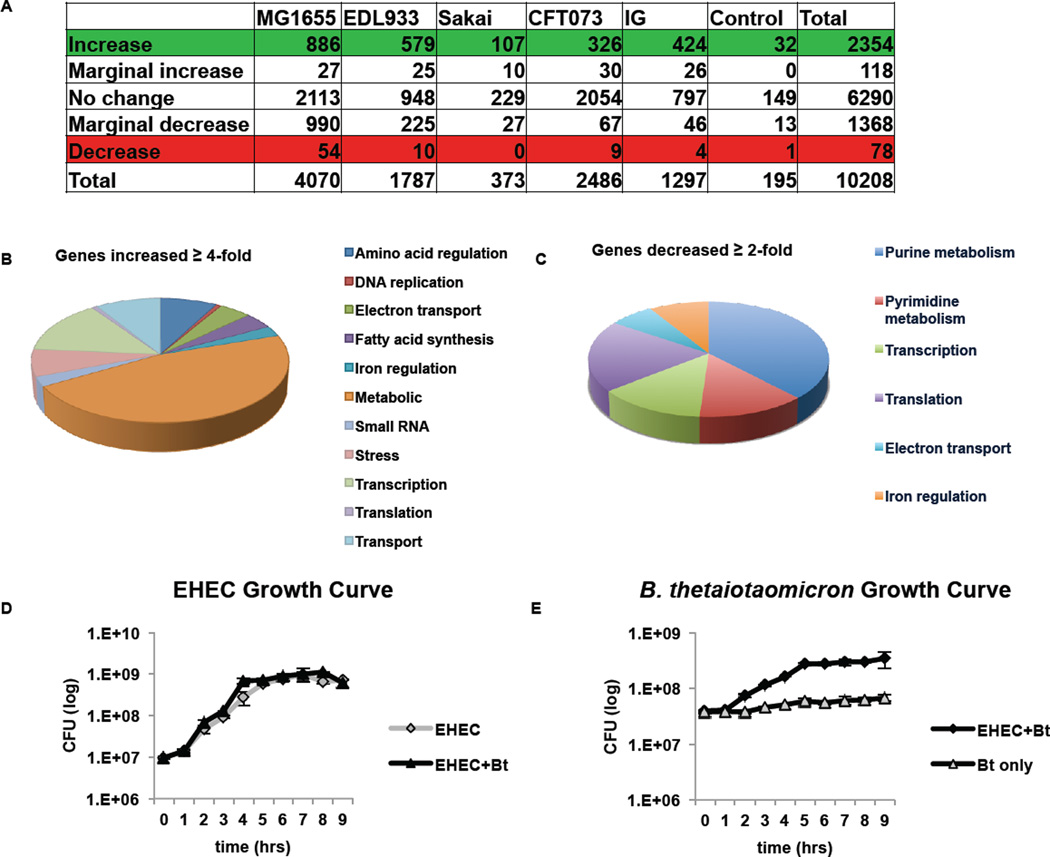
To determine the global impact of Bt on EHEC gene expression, we performed a microarray from EHEC cultures grown in the presence or absence of Bt using the GeneChip® E. coli Genome 2.0 array. The GeneChip® E. coli Genome 2.0 array includes approximately 10,000 probe sets for all genes present in the following four strains of E. coli: K-12 lab strain MG1655, uropathogenic strain CFT073, O157:H7 enterohemorrhagic strain EDL933, and O157:H7 enterohemorrhagic strain Sakai. The microarray data represents a single replicate from each condition; therefore, no statistical analysis or multiple hypotheses testing correction was performed. Expression of one-fifth of the array probe sets increased when EHEC was cultured with Bt (Fig. 1A). The affected genes reflect an increase in competition for nutrients, with 47% of the increased genes participating in metabolism, 8% in amino acid regulation, and 9% in cellular transport (Fig. 1B). Expression of less than 1% of the EHEC genome decreased, with the most affected genes playing a role in purine and pyrimidine metabolism (Fig. 1C). Despite the increase in metabolic genes, EHEC displayed no growth advantage when cultured with Bt (Fig. 1D). However, EHEC provided a distinct growth advantage to Bt, increasing Bt generation time from 282 min/gen when grown alone to 97 min/gen when grown with EHEC (Fig. 1E).
Figure 1. B. thetaiotaomicron (Bt) increases expression of one-fifth of the E. coli array probe sets.
A, Summary of microarray results comparing EHEC grown with Bt to EHEC grown alone (EHEC+Bt/EHEC only). B, C, Pie graph categorizing the E. coli genes by function b, that increased ≥ 4-fold or c, that decreased ≥ 2-fold in the presence of Bt D, E, Growth curves (of 6 biological samples, experiments were repeated 2 times, each with 3 independent biological samples) of in vitro growth of D, EHEC or E, Bt. The error bars indicate the standard deviation of the mean. Doubling time for EHEC grown alone and in the presence of Bt is 50 min/gen and 53 min/gen, respectively. Doubling time for Bt grown alone and in the presence of EHEC is 282 min/gen and 97 min/gen, respectively.
Bt Augments EHEC Virulence Via the Catabolite Repressor/Activator Protein
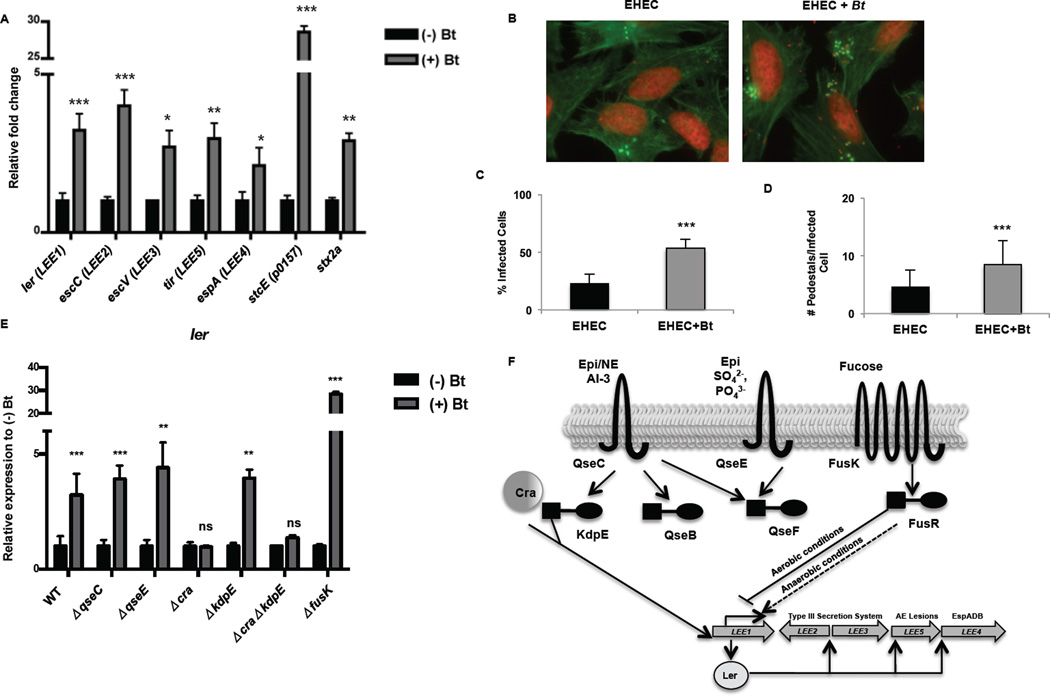
Expression of a myriad of EHEC virulence genes is increased in the presence of Bt, including the LEE, stx2a (encoding Stx) and stcE (encoding a mucinase) genes (Fig. 2A). The LEE pathogenicity island consists of 41 genes organized into five major operons (Kaper et al., 2004). Transcription of all LEE operons is increased when EHEC is cultured in the presence of Bt. In the presence of Bt, the master regulator of the LEE, ler, increased more than 3-fold (p < 0.001). Transcription of the T3SS structural components escC and escV increased approximately 4-fold and 3-fold, respectively (escC: p < 0.0003; escV: p = 0.0319) in the presence of Bt while the translocated intimin receptor tir increased 3-fold (p = 0.0021) and the T3SS filament espA increased 2-fold (p = 0.05) (Fig. 2A). In accordance, AE lesion formation in EHEC significantly increased in the presence of Bt. In the presence of Bt, AE lesion formation on HeLa cells increased from 23% to 54% (p < 0.0001), with an increase of 8.5 pedestals per infected cell in the presence of Bt compared to 4.6 pedestals per cell in the absence of Bt (Figs. 2B–D).
Figure 2. Bt augments EHEC virulence via the catabolite repressor/activator protein.
A, qRT-PCR of LEE, stcE and stx2A genes in EHEC grown alone (−) Bt or in the presence of Bt (+) Bt (n= 9; error bars, s.d.; ***P < 0.001, **P < 0.01, *P < 0.05). B, Fluorescent actin staining assay of HeLa cells infected with EHEC alone or in the presence of Bt, stained with fluorescein isothiocyanate-phalloidin (actin, green) and propidium iodide (bacterial and HeLa DNA, red). Original magnification, 63X. C, Quantification of fluorescent actin staining assay of the percentage of HeLa cells infected, as defined by pedestal formation by EHEC. D, Number of pedestals/infected cell. (n = 350 cells; error bars, s.d.; *** P<0.001). E, qRT-PCR of ler in WT, ΔqseC, ΔqseE, Δcra, ΔkdpE, ΔΔcrakdpE, and ΔfusK grown alone or in the presence of Bt (n = 9–15; error bars, s.d.; ***P < 0.001, **P < 0.01, P > 0.05 = ns). Each mutant has been normalized to 1 to show the fold-increase of the mutant when grown in the presence of Bt (+) Bt F, Representation of LEE pathogenicity island regulation.
To determine the pathway through which Bt regulates EHEC, we analyzed virulence gene expression in a panel of EHEC mutants grown in the presence or absence of Bt. In the presence of Bt, ler expression increased more than 3-fold in wild-type EHEC. Similarly, in the qseC, qseE, and kdpE EHEC mutants, ler expression increased approximately 3 to 4-fold when the mutants were cultured in the presence of Bt. In contrast, no enhancement of ler expression occurred in the cra EHEC mutant, demonstrating that Bt regulation of the LEE occurs through Cra, a transcription factor that positively regulates EHEC virulence by sensing fluctuations in sugar concentrations indicative of a gluconeogenic environment (Njoroge et al., 2013). Bt regulation does not occur through the histidine sensor kinases QseC, QseE, or FusK or through the response regulator KdpE that play other important signaling roles in EHEC virulence gene expression (Njoroge et al., 2013; Pacheco et al., 2012; Reading et al., 2007; Sperandio et al., 2002) (Figs. 2E, F). Fluctuations in the concentrations of carbon metabolites are a major mechanism to modulate virulence gene expression and colonization of EHEC (Njoroge et al., 2013; Pacheco et al., 2012). Analysis of our microarray data corresponds with Bt regulation of EHEC gene expression through Cra (Supplementary Fig. 1) (Chin et al., 1989; Feldheim et al., 1990; Saier and Ramseier, 1996).
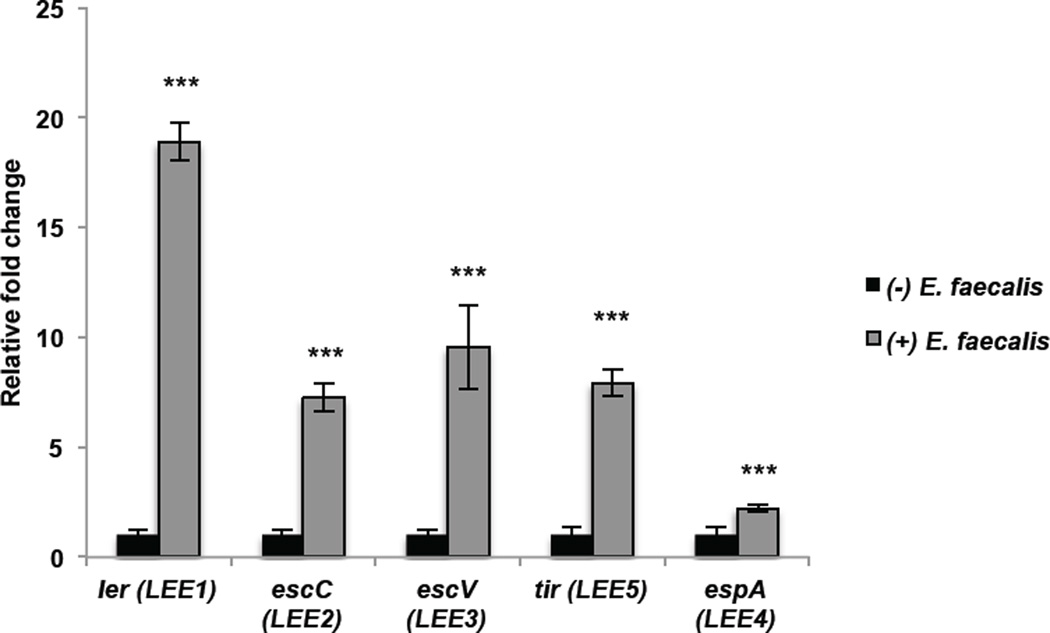
To determine if augmentation of the LEE extends beyond Bt to other members of the intestinal microflora, we cultured EHEC in the presence of Enterococcus faecalis, a prominent member of the Firmicutes phyla. In the presence of E. faecalis, an opportunistic Firmicutes, transcription of all LEE operons was significantly increased (Fig. 3). Transcription of ler increased 19-fold (p < 0.0001), and escC, escV, and tir increased 7–10 fold in the presence of Bt (escC: p < 0.0001, escV: p < 0.0001, tir: p < 0.0001), indicating that members of the two major intestinal microbiota phyla, Bacteroidetes and Firmicutes, induce the LEE in EHEC.
Figure 3. E. faecalis, a member of the Firmicutes phylum, augments EHEC virulence gene expression.
qRT-PCR of LEE genes in EHEC grown alone (−) E. faecalis or in the presence of E. faecalis (+) E.faecalis (n= 9; error bars, s.d.; ***P < 0.001).
C. rodentium as an Infection Model
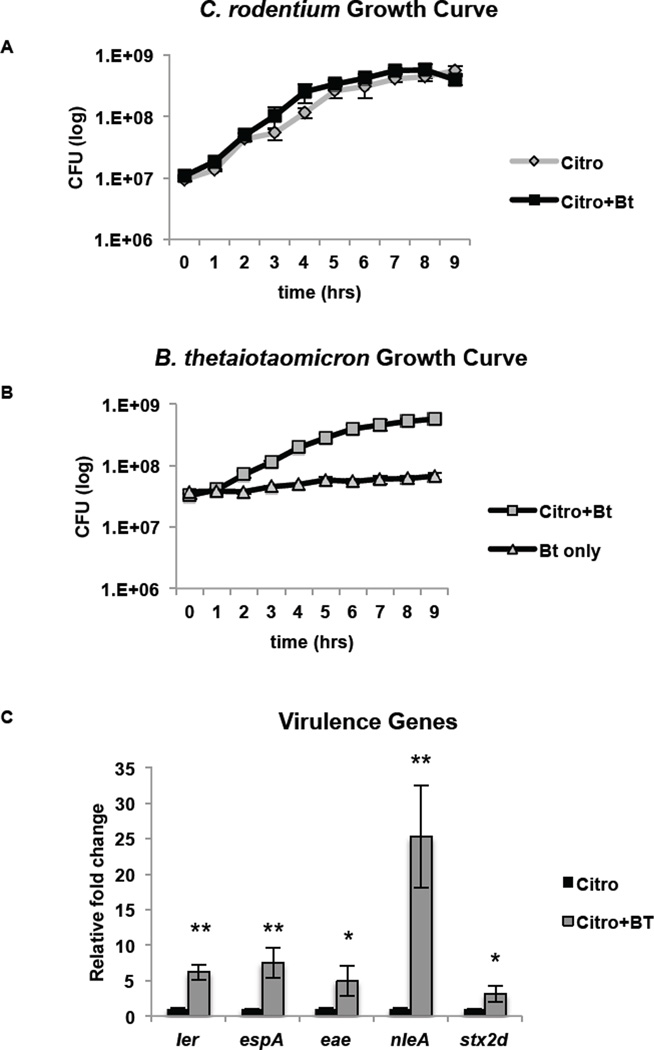
EHEC is a natural human pathogen; however, EHEC poorly infects mice, and mice do not develop key features of the disease such as AE lesions, intestinal damage, and systemic illness (Mallick et al., 2012; Mundy et al., 2005). To study the characteristics of EHEC infection, we employed an infection model using a Stx-producing C. rodentium strain (DBS770) constructed by Schauer and colleagues (Mallick et al., 2012). Mice infected with this strain develop AE lesions on the intestinal epithelium and Stx-dependent damage to the intestinal epithelium and kidneys (Mallick et al., 2012). In vitro analysis confirmed that Bt affects C. rodentium in a similar manner to EHEC. Similarly to EHEC, C. rodentium growth was unaffected by the presence of Bt; however, Bt generation time increased from 282 min/gen when grown alone to 91 min/gen when grown with C. rodentium (Fig. 4A, B).
Figure 4. C. rodentium as an EHEC infection model.
A, B, Growth curves (of 6 biological samples, experiments were repeated 2 times, each with 3 independent biological samples) of in vitro growth of A, C. rodentium or B, Bt. The error bars indicate the standard deviation of the mean. Doubling time for C. rodentium grown alone and in the presence of Bt is 69 min/gen and 66 min/gen, respectively. Doubling time for Bt grown alone and in the presence of C. rodentium is 282 min/gen and 91 min/gen, respectively. C, qRT-PCR of LEE, nleA and stx genes in C. rodentium grown alone (−) Bt or in the presence of Bt (+) Bt (n = 6; error bars, s.d.; **P < 0.01, * P < 0.05).
Transcription of key LEE (ler, espA, eae) and non-LEE encoded (nleA, stx2d) virulence genes was also increased when C. rodentium was grown in the presence of Bt (Fig. 4C). Expression of ler increased 6-fold in the presence of Bt (p = 0.0012). Expression of eae, the gene that encodes the adhesin intimin essential for AE lesion formation, increased 5-fold (p = 0.0303), and the gene encoding the T3SS filament espA increased by 7.5-fold (p = 0.0054). Bt impacted C. rodentium growth and virulence in a similar manner to EHEC, augmenting the virulence of these AE lesion pathogens in vitro. We reasoned that C. rodentium would be a suitable model to study EHEC infection in the context of an altered microflora.
Bt Mediates its Pro-Virulence Effect on C. rodentium by Enhancing Expression of the T3SS, not by a bloom in the C. rodentium population
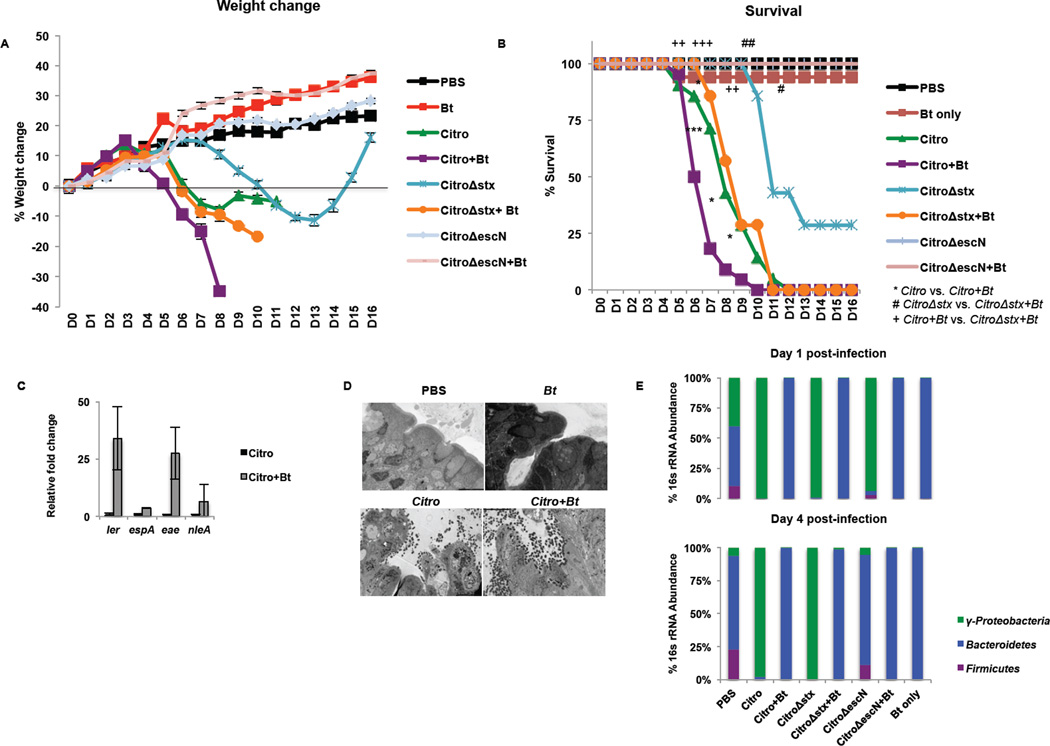
To determine the role of Bt during C. rodentium infection in mice, mice were depleted of their microflora (Kuss et al., 2011) and then either left deplete of gut microflora or reconstituted with Bt (Supplementary Fig. 2). The mice were then challenged with a wild-type C. rodentium DBS100 strain, C. rodentium strain DBS770 that is Shiga toxin (Stx)+, a DBS770 Stx- (Δstx), a T3SS-deficient (ΔescN), or a Cra-deficient (Δcra) C. rodentium (Supplementary Fig. 3 and Fig. 4). Bt-reconstituted mice lost weight and succumbed to C. rodentium infection more rapidly than microflora-deplete groups. C. rodentium -infected mice reconstituted with Bt began to lose weight on day 4 post-infection. In contrast, C. rodentium -infected mice deplete of microflora did not begin to lose weight until day 6 post-infection (Fig. 5A). By day 6 post-infection, 50% of the C. rodentium-infected mice reconstituted with Bt had succumbed to infection; whereas, only 14% of the C. rodentium-infected mice deplete of microflora had died by day 6 post-infection. Similarly, mice infected with C. rodentium Δstx and reconstituted with Bt had 100% mortality by day 11 post-infection while mice infected with C. rodentium Δstx but deplete of microflora had 43% mortality on day 11 post-infection (Fig. 5B). The presence of Stx compounded these effects, demonstrating that both the presence of Bt and the production of Stx contribute to the morbidity and mortality during C. rodentium infection. It is noteworthy that the Δstx DBS770 mutant behaves similarly to the WT DBS100 strain (that does not encode Stx) in the absence or presence of Bt (Supplementary Fig. 3; Fig. 5). However, this effect is lost if C. rodentium is unable to form a functional T3SS and colonize (Fig. 5 A , B). The cra mutant, as expected, is also attenuated for infection (Supplementary Fig. 4), highlighting the importance of fluctuations of carbon metabolites during infection.
Figure 5. Bt mediates its pro-virulence effect on C. rodentium by enhancing expression of the T3SS, not by a bloom in the C. rodentium population.
C3H/HeJ mice were treated for 5 days with an antibiotics regimen to deplete gut microbiota. Half of the mice were reconstituted with Bt (+Bt) while the remainder of the mice were left deplete of gut microbiota. Mice were mock-infected (PBS, Bt only) or infected with C. rodentium (Citro, Stx+), C. rodentiumΔstx (CitroΔstx, Stx-), or C. rodentiumΔescN (CitroΔescN). A, Weight loss or gain from baseline (weight at day 0) over the course of infection (blue: mock-infected, reconstituted with Bt; red: Citro-infected, deplete of microbiota; green: Citro-infected, reconstituted with Bt; purple: CitroΔstx-infected, deplete of microbiota; turquoise: CitroΔstx-infected, reconstituted with Bt; orange: CitroΔescN-infected, deplete of microbiota; light blue: CitroΔescN-infected, reconstituted with Bt). B, Survival after infection (n = 7–22 mice/group; error bars, s.d.; ***P < 0.001, **P < 0.01: comparison of Citro vs. Citro+Bt; ##P < 0.01, #P < 0.05: comparison of CitroΔstx vs. CitroΔstx+Bt;+++P < 0.001, ++P < 0.01: comparison of Citro+Bt vs. CitroΔstx+Bt). C, qRT-PCR analysis of ler, espA, eae, and nleA from mRNA isolated from fecal pellets of infected animals. Significance is indicated as follows: one asterisk P≤0.05, two asterisks P≤0.01; three asterisks P≤0.001. D, Ultrastructure of the distal colon harvested five days post-infection from mock-infected (PBS, Bt) or C. rodentium-infected mice either deplete of gut microbiota or reconstituted with Bt, 2500×. Microvilli destruction and C. rodentium forming attaching and effacing (AE) lesions on the colonic epithelium can be observed. Original magnification, 2500X (TEM). E, qRT-PCR of 16s rRNA from the major phylogenetic groups (green: Proteobacteria, blue: Bacteroidetes, purple: Firmicutes) from feces collected on day 1 and day 4 post-infection.
In Bt-reconstituted mice, C. rodentium virulence gene expression was increased compared to C. rodentium from microflora-deplete mice (Fig. 5C and Supplementary Fig. 5A). Analysis of the major phylogenetic groups determined that Bacteroidetes dominated the Bt-reconstituted groups on days 1 and 4 post-infection while Proteobacteria dominated the microflora-deplete groups infected with C. rodentium. Maintenance of Proteobacteria was independent of Stx but dependent on a functional T3SS (Fig. 5E). However, the bacterial burden of C. rodentium did not significantly differ in mice reconstituted with Bt and then challenged with C. rodentium compared to those not reconstituted with Bt (Supplementary Fig. 5B). Furthermore, examination of the ultrastructure of the distal colon showed destruction to the microvilli and attachment of C. rodentium to the epithelium (Fig. 5D). The presence of Bt during C. rodentium infection does not cause a bloom in the C. rodentium population, but rather an increase in its virulence, akin to our observations in vitro (Fig. 4).
Bt Contributes to the Accelerated Loss of a Protective Mucosal Layer during C. rodentium Infection
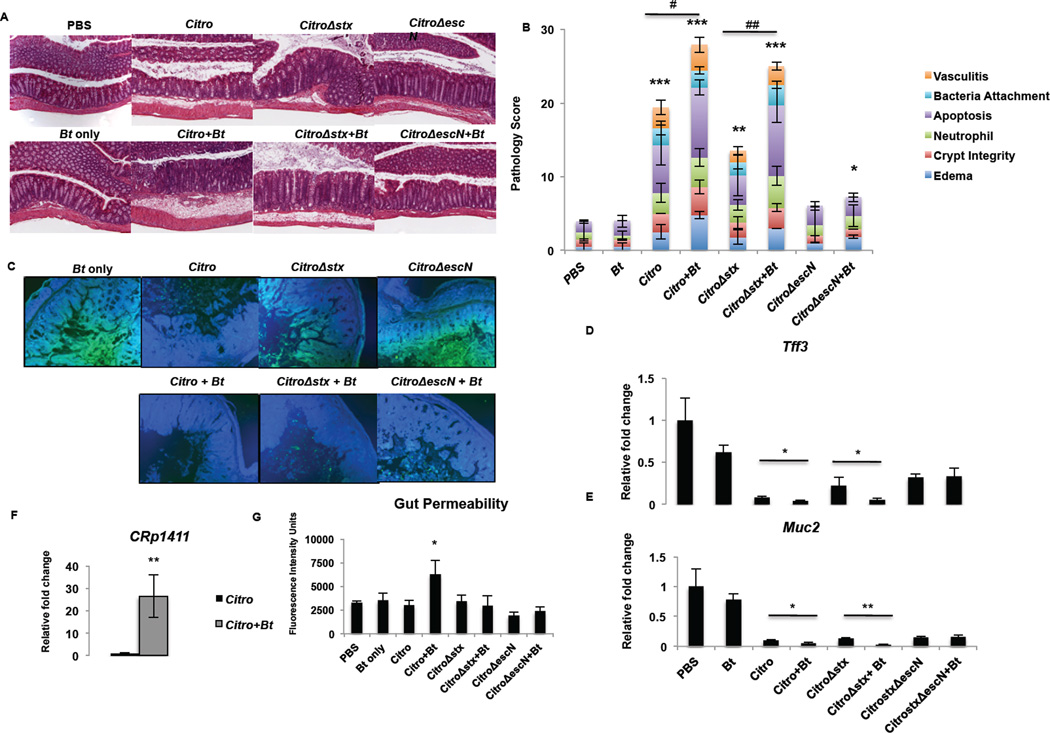
To determine if the increased morbidity and mortality that occurred during C. rodentium infection in Bt-reconstituted mice was due to increased host pathology, we evaluated the cecum and the colon for the following parameters: edema, crypt integrity, neutrophil infiltration, apoptosis, bacterial attachment, and vasculitis. The evaluation, performed in a double-blind fashion, demonstrated that host pathology was worsened in Bt-reconstituted mice, with augmented edema, vasculitis, apoptosis, and destruction of the crypts (Fig. 6A, B, Supplementary Fig. 6). We also examined expression of host genes key in epithelial repair and in innate defense. Within the gastrointestinal tract, the mucus layer is the first line of defense against pathogens. The colon consists of two mucus layers—an outer layer that serves as a home to commensal bacteria and a tight inner layer devoid of bacteria (Johansson et al., 2013). In mice deficient in Muc2, the primary component of the colonic mucus layer, infection with C. rodentium causes rapid weight loss and mortality and increased gut permeability compared to wild-type mice (Bergstrom et al., 2010). Reconstitution with Bt during C. rodentium infection augmented the loss of Muc2 (Fig. 6C). Transcription of intestinal trefoil factor 3 (Tff3), a secreted molecule important in epithelial repair and maintenance of the mucosa (Taupin and Podolsky, 2003), and Muc2 (Muc2) were significantly reduced during C. rodentium infection in Bt-reconstituted mice (Tff3: p = 0.0283; Muc2: p = 0.0148) (Fig. 6D, E). Additionally, expression of the bacterial serine protease p1411, a homologue of Pic mucinase found in Shigella spp. and enteroaggregative E. coli, increased in Bt-reconstituted animals compared to mice deplete of microflora (Fig. 6F). Consistent with the worsened damage to the crypts and mucosa, gut permeability increased in Bt-reconstituted mice during C. rodentium infection (Fig. 6G). Host pathology is worsened during C. rodentium infection in Bt-reconstituted mice and likely contributes to the increased morbidity and mortality in these mice. Conversely, transcription of the innate defense genes Reg3β and Reg3γ were increased during C. rodentium infection, and augmented when Bt was also present (Supplementary Fig. 7). Of note expression of RegIIIγ is increased in germ-free mice upon exposure to the microbiota, and this lectin kills only Gram-positive, but not Gram-negative bacteria such as Bt and C. rodentium (Cash et al., 2006).
Figure 6. Bt contributes to the accelerated loss of a protective mucosal layer during C. rodentium infection.
A, B, Antibiotics-treated C3H/HeJ mice either deplete of gut microbiota or reconstituted with Bt (+Bt) were mock-infected (PBS, Bt only) or infected with C. rodentium (Citro, Stx+), C. rodentiumΔstx (CitroΔstx, Stx-), or C. rodentiumΔescN (CitroΔescN). The histological changes in the colon and cecum were analyzed on day 5 post-infection based on the following scoring system: edema, 0 is no edema and 5 has the highest edema in the submucosa; crypt integrity, 1 = normal, 2 = irregular crypts, 3 = mild crypt loss, 4 = severe crypt loss, 5 = complete crypt loss; neutrophil, neutrophilic infiltration in the wall; apoptosis, number of apoptotic cells per 600× field (n = five fields); bacteria attachment, bacteria associated to the epithelial surface; vasculitis, 0 is no evidence of vasculitis and 5 is the most severe vasculitis. Scoring was performed blindly, and the scores for each parameter are an average of the cecum and distal colon, taken from two independent experiments with 3 mice/experiment. C, Cecum harvested on day 5 post-infection was stained with Muc2 (green) and DAPI (DNA, blue). Original magnification, 10X. D, E, qRTPCR of D, Tff3 (trefoil factor 3) and D, Muc2. RNA was isolated from colonic tissue harvested from mice on day 5 post-infection. (n = 6; error bars, s.d.). F, qRT-PCR of p1411 from feces collected on day 2 post-infection. G, Levels of FITC-Dextran in the serum of mice on day 5 post-infection. Mice were fasted 4 hrs prior to administration of FITC-Dextran via oral gavage. FITC-Dextran levels were determined by measuring fluorescence at excitation 490 nm, emission 525 nm (n = 6; error bars, s.d. ***P < 0.001, **P < 0.01, *P < 0.05, P > 0.05 = ns).
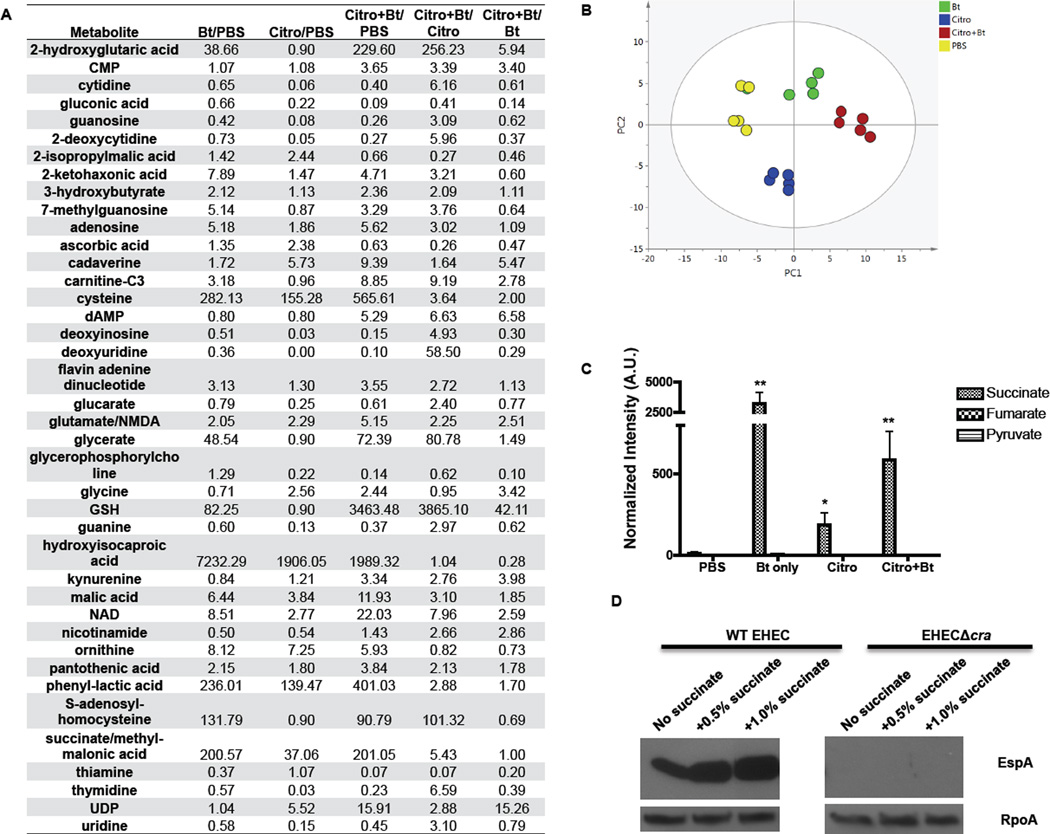
To further explore the mechanism of how Bt may be exerting its pro-virulence effect on C. rodentium, we examined metabolites present in the large intestine during infection. Our metabolomics studies showed that the metabolic landscape between mice colonized with Bt or not (PBS or C. rodentium infected animals) were quite diverse, and that the Bt alone and Bt + C. rodentium animals have a more similar metabolite profile in their intestines (Fig. 7A,B). Of note, several metabolites indicative of a gluconeogenic environment (such as lactate, succinate, glycerate, and others) were elevated in the Bt treated animals (Fig. 7A,B). Elevated concentrations of succinate, but not of fumarate or pyruvate, were measured within the cecum when Bt was present (Fig. 7C), consistent with findings identifying succinate as a major metabolic by-product of Bacteroides spp (Macy et al., 1978). Succinate produced by Bacteroides spp. has been shown previously to impair neutrophil function and inhibit phagocytic killing of E. coli (Rotstein et al., 1985; Rotstein et al., 1989). Additionally, in a recent complementary study, Sonnenburg and colleagues demonstrated that microbiota-produced succinate induced after antibiotic treatment augments expansion of C. difficile in mice and that C. difficile deficient in succinate utilization pathways displayed a competitive disadvantage (Ferreyra et al., 2014). Moreover, the addition of succinate during in vitro culture of EHEC resulted in an increase in the expression of the LEE-encoded protein EspA in wild-type EHEC; whereas, the cra mutant remains unresponsive to succinate (Fig. 7D). It is also worth noting that Enterococcus faecalis, which also increases EHEC virulence gene expression (Fig. 3), also produces and secretes copious amounts of succinate (Milstien and Goldman, 1973).
Figure 7. Comparison of metabolic profiles of Bt-reconstituted and microflora-deplete animals.
A, The expression profiling of metabolites present in the cecum on day 2 post-infection (n = 5). The data are presented as ratios to show fold-changes of the specified metabolite in the individual groups. B, The normalized data were analyzed by Principal Component Analysis (PCA) with unit variance scaling in order to obtain an unbiased overview of the data and to visualize clustering, trends, and outlier metabolites among all groups on the score plots. PCA score plots showed clear separation among the four groups in different colors (yellow: PBS, red: Citro+Bt, blue: Citro, green: Bt), with no outliers detected. The amount of variance in the X matrix explained by PC1 (R2) was 0.732, and estimate of the predictive ability of the model (Q2) (cumulative) was 0.446. C, Absolute levels of succinate, fumarate, and pyruvate present in the cecum on day 2 post-infection. (A.U. Absolute Units; n = 5; error bars, s.d. **P < 0.01, *P < 0.05). D, Protein level of the T3SS translocon protein EspA, encoded within LEE4, secreted into the supernatant of wild-type or Δcra EHEC cultures grown in the presence of 0.1% glucose, plus increasing levels of succinate. RpoA protein levels in the cell lysate serve as the loading control.
Interestingly, Bt colonization increases the levels of γ-L- glutamyl-L-cysteinyl-glycine, or glutathione (GSH), and its precursors cysteine and S-adenosyl-homocysteine (Fig. 7A). GSH plays a role in nutrient metabolism, antioxidant defense, and a number of key cellular metabolic functions (Aquilano et al., 2014). Additionally, Bt colonization alone increases the gluconeogenic metabolites succinate, glycerate, and lactate, creating a different metabolic landscape for C. rodentium than what the pathogen encounters in the microflora-deplete animals. In the C. rodentium infected animals deplete of a microflora, the levels of nucleosides (cytidine, guanosine, thymidine, uridine) are decreased compared to mock-infected animals. The presence of Bt in C. rodentium-infected animals rescues this decrease in nucleosides and nucleic acid precursors (Fig. 7A). Our findings begin to delve into the specific metabolites that a key constituent of the microbiota, Bt, contributes to the intestinal environment, and we link this metabolic environment to an increase in virulence of an enteric pathogen.
DISCUSSION
The interaction between a host and its gastrointestinal microflora is a delicate balance, and if disturbed, can result in dysbiosis (Spor et al., 2011). Host balance with the microbiota is maintained in part through the C-type lectins RegIIIβ and RegIIIγ that limit direct interaction of the microbiota with the intestinal microbiota (Cash et al., 2006; Vaishnava et al., 2011) and in part through maintenance of the integrity of the intestinal epithelium. Intestinal trefoil factor (TFF3) secreted by goblet cells plays an essential role in epithelial repair and restoration of the mucosa (Taupin and Podolsky, 2003). Some pathogens, however, evade these host defenses and can even benefit from them. Salmonella typhimurium thrives in an inflammatory environment due to the production of RegIIIβ that eliminates competing microbiota (Stelter et al., 2011). In hosts with a high-fiber diet, butyrate—a beneficial metabolite in colonic health—enhances the expression of the Stx receptor, globotriaosylceramide (Gb3), and increases susceptibility to EHEC O157:H7 infection (Zumbrun et al., 2013). Our studies indicate that a normally beneficial member of the microbiota has the capacity to enhance virulence and disease progression of an enteric pathogen, altering the metabolic landscape towards a gluconeogenic environment and secreting large amounts of the succinate metabolite within the intestine that is interpreted by the pathogen, through the transcription factor Cra, as a nutritional cue to activate virulence gene expression. Recent findings corroborate the idea that the microbiota can provide a competitive advantage to enteric pathogens (Ng et al., 2013). Our findings have fundamental implications into how the composition of the microbiota impacts pathogen-mediated disease susceptibility and progression, and show that certain enteric pathogens learned to exploit the microbiota to enhance their virulence.
EXPERIMENTAL PROCEDURES
Strains and culture conditions
Strains used in this study are listed in Table S1. B. thetaiotaomicron VPI-5482 was grown anaerobically overnight at 37°C in TYG medium (Betian et al., 1977) plus 200 µg/ml gentamicin. Wild-type EHEC O157:H7 strain 86–24 (Griffin et al., 1988) and its isogenic mutants (ΔqseC, ΔqseE, Δcra, ΔkdpE, ΔcraΔkdpE, ΔfusK) were grown anaerobically overnight at 37°C in LB plus 50 µg/ml streptomycin. C. rodentium strains DBS770 (λstx2dact) and its isogenic mutant MMC01 (CRΔescN) were grown anaerobically overnight in LB plus 25 µg/ml chloramphenicol. The lysogenized, Stx-deficient C. rodentium strain DBS771 was grown in LB plus 50 µg/ml kanamycin (Mallick et al., 2012). Bt overnight culture was pelleted and concentrated 10-fold in LB. Bt was plated in a 10-fold excess over EHEC or C. rodentium to represent the composition of the intestinal microbiota. E. faecalis V583 (Paulsen et al., 2003) was grown anaerobically in LB overnight, and the overnight culture was plated at a 1:1 with EHEC. The bacteria were grown anaerobically in Petri dishes in 25 ml of pre-reduced low-glucose Dulbecco’s modified Eagle’s medium (DMEM) (Invitrogen) for 6 h, unless otherwise noted, in a GasPak EZ anaerobe container (Becton Dickinson). RNA was extracted using a RiboPure Bacteria RNA isolation kit (Ambion) according to manufacturer’s guidelines.
Microarray preparation and analyses
Affymetrix 2.0 E. coli gene arrays were used to compare gene expression in strain 86–24 when cultured alone to that in strain 86–24 when grown in the presence of Bt, grown anaerobically in pre-reduced DMEM at 37°C for 6 h to late-log phase in a GasPak EZ anaerobe container. The relative abundance of EHEC to Bt at the time of RNA harvest was 70% EHEC, 30% Bt. Bt grown alone was included as a control to ensure that Bt cDNA did not hybridize to the E. coli gene array. The RNA processing, labeling, hybridization, and slide-scanning procedures were performed as described in the Affymetrix Gene Expression technical manual. The array data analyses were performed as described previously (Kendall et al., 2011). Based on array data analyses, the biological processes of the genes significantly increased ≥ 4-fold (406 genes, 297 with known functions, 109 with unknown function) or significantly decreased ≥ 2-fold (72 genes, 58 with known functions, 14 with unknown function) were characterized using the Universal Protein Resource (UniProt) Knowledgebase (UniProtKB/Swiss-Prot). (http://www.uniprot.org).
Growth curves
Bacteria were cultured as described above for 9 h. Bacteria were serially diluted in phosphate-buffered saline (PBS) each hour and then plated on the following agar plates: LB agar plus 50 µg/ml streptomycin (isolation of EHEC O157:H7), LB agar plus 25 µg/ml chloramphenicol (isolation of C. rodentium), or Brain Heart Infusion agar (BHI) (VWR) plus 10% calf blood (Colorado Serum Company) plus 200 µg/ml gentamicin (isolation of Bt). EHEC O157:H7 and C. rodentium plates were grown aerobically at 37°C for 24 h. Bt plates were grown anaerobically at 37°C for 48 h. In each experiment, bacteria were grown on both selection plates to confirm that growth was selective. To determine the growth rate, two points from the exponential phase were selected (t = 2 h, t = 5 h) and calculated using the formula: where Xt = higher CFU/ml, X0 = lower CFU/ml, and t = time interval (in hours) between the points. Generation time was then calculated as tgen = 1/k.
Real time qPCR
For in vitro experiments, cultures were grown to late-log phase in pre-reduced DMEM under anaerobic conditions (6 h). For in vivo experiments, fecal pellets were collected from infected mice on days 0, 1 and 4 after C. rodentium infection. RNA was extracted using the RiboPure Bacteria isolation kit according to the manufacturer’s protocols (Ambion). To assess host gene expression, tissue from the distal colon of infected mice was harvested on day 5 post-infection and homogenized in 1 ml TRIzol® (Life Technologies) per 100 mg tissue. RNA was isolated using standard molecular biological procedures. The primers used for quantitative reverse transcription-PCR (qRT-PCR) (Table S2) were validated for amplification efficiency and template specificity. qRT-PCR was performed as previously described (Hughes et al., 2009) in a one-step reaction using an ABI 7500 sequence detection system (Applied Biosystems). Data were collected using the ABI Sequence Detection 1.2 software (Applied Biosystems).
All data were normalized to an endogenous control (rpoA for virulence gene expression in EHEC and C. rodentium, Eubacteria 16S rRNA for total bacteria present in feces, or GAPDH for murine host gene expression in colonic tissue) and analyzed using the comparative critical threshold (CT) method. Virulence gene expression was presented as fold changes over the expression level of WT EHEC or C. rodentium cultured alone in vitro or C. rodentium (DBS770) infected alone in vivo. Host gene expression was presented as fold changes over the expression level present in mock-infected (PBS) mice.
The relative abundances of Bacteroidetes, Firmicutes, and Proteobacteria (family Enterobacteriaceae) were measured by qPCR with taxon-specific or universal 16S rRNA gene primers. The primers used were as follows: Eubacteria (universal bacteria), Eub338F, 5’-actcctacgggaggcagcagt-3’, and Eub338R, 5’- attaccgcggctgctggc-3’; Firmicutes, 928F–Firm, 5’- tgaaactyaaaggaattgacg-3’, and 1040FirmR, 5’-accatgcaccacctgtc-3’ (Bacchetti De Gregoris et al., 2011); Bacteroidetes, 798cfbF, 5’- craacaggattagataccct-3’, and cfb967R, 5’-ggtaaggttcctcgcgtat-3’ (Guo et al., 2008); and γ-Proteobacteria, 1080gF, 5’- tcgtcagctcgtgtygtga-3’, and g1202R, 5’- cgtaagggccatgatg-3’ (Bacchetti De Gregoris et al., 2011). Expression of each taxon was normalized to Eub388 and then compared to the expression level present in mock-infected (PBS) fecal pellets on D0. Percentage of taxa was determined by dividing the expression of the taxon-specific 16S rRNA over the combined expression of Firmicutes, Bacteroidetes, and γ-Proteobacteria. The Student unpaired t test was used to determine statistical significance.
Fluorescein actin staining (FAS)
Fluorescein actin staining assays were performed as described by Knutton et al (Knutton et al., 1989). Briefly, HeLa cells were grown on coverslips in 12-well culture plates with DMEM supplemented with 10% fetal bovine serum (FBS) and 1% penicillin/streptomycin/gentamicin (PSG) antibiotic mix at 37°C, 5% CO2, overnight to 80% confluency. The wells were washed with PBS and replaced with low-glucose DMEM supplemented with 10% FBS. Bacterial cultures were grown anaerobically overnight as described above at 37°C, and overnight Bt culture was concentrated 10-fold in LB. Overnight bacterial cultures were diluted 1:100 to infect confluent monolayers of HeLa cells for 6 h at 37°C, 5% CO2. After a 6 h infection, the coverslips were washed, fixed, permeabilized, and then treated with fluorescein isothiocyanate (FITC)-labeled phalloidin to visualize actin accumulation and propidium iodide to visualize host nuclei and bacteria. The coverslips were mounted on slides and visualized with a Zeiss Axiovert microscope. To determine percentage infected cells, 5–7 fields from three separate coverslips (triplicate) were counted, and calculated as (Cells with Pedestals/Total cells)*100. Statistical analyses were performed using the Student unpaired t test.
Mice
C3H/HeJ mice were purchased from The Jackson Laboratory and housed in a specific pathogen-free facility at UT Southwestern Medical Center. All experiments were performed under IACUC approved protocols.
At 3–4 weeks of age, female C3H/HeJ mice were orally administered a combination of four antibiotics: ampicillin, neomycin, metronidazole, and vancomycin (Sigma-Aldrich) via oral gavage for 5 or 6 days to deplete gut microbiota (5 mg of each antibiotic per mouse per day) (Rakoff-Nahoum et al., 2004). Fecal pellets were collected before and after antibiotics treatment to confirm depletion of the gut microbiota. Feces were resuspended in PBS at 1 g/ml, serially diluted, and plated on BHI-Blood agar plates containing no antibiotics. Colony counts were performed after 48 h incubation at 37°C under both aerobic and anaerobic conditions. Following antibiotics treatment, half of the mice were reconstituted with 3×109 colony-forming units (CFU) Bt via oral gavage while the remainder of the mice were left deplete of gut microbiota. Twenty hours later, mice were mock-infected with PBS or orally infected with 1×109 CFU C. rodentium (DBS770, Stx+), C. rodentiumΔstx (DBS771, Stx-), C. rodentiumΔescN (MMC01, CitroΔescN), or C. rodentiumΔcra (MMC02, CitroΔcra).
Baseline weight of mice was measured on the initial day of C. rodentium infection (day 0, D0). Mice were weighed daily for the course of infection. Percentage weight was calculated in the following way: ((WeightDayn – Baseline weight)/Baseline weight)*100. Change in weight reflects weight gain or weight loss over the course of infection. Mice were monitored daily for survival. The experiments were performed at least twice with a total of 7–22 mice per group. Weight change and survival curves reflect the average of experiments plus the standard deviation. The Student’s unpaired t test was used to determine the statistical significance of each day on the survival curve.
To measure gut permeability, mice were fasted for 4 h prior to administration of 20 ml/kg of FITC-dextran (Sigma) via oral gavage on day 5 post-infection with C. rodentium. Fasting conditions were maintained for an additional 3 h, at which point blood was collected and centrifuged to collect plasma. Serum was diluted 1:2 in PBS and fluorescence at excitation 490 nm, emission 520 nm was measured using a fluorimeter. A standard curve was prepared by serially diluting FITC-dextran in a consistent PBS:mouse serum with the samples.
Histopathology
Portions of the distal colon and cecum were harvested five days post-infection with C. rodentium. The tissues were washed in PBS and then fixed in Bouin’s fixative for 48h. The tissues were embedded in paraffin, cut into 5-µm sections and stained with hematoxylin and eosin (H&E) in the UT Southwestern Pathology Core. Histological changes were analyzed in a double-blind fashion. The severity of intestinal pathology was analyzed based on the following scoring system: edema, 0 is no edema and 5 has the highest edema in the submucosa; crypt integrity, 1 = normal, 2 = irregular crypts, 3 = mild crypt loss, 4 = severe crypt loss, 5 = complete crypt loss; neutrophil, neutrophilic infiltration in the wall; apoptosis, number of apoptotic cells per 600× field (n = five fields); bacteria attachment, bacteria associated to the epithelial surface; goblet cell, average number of goblet cell in each crypt (n > 10 crypts); vasculitis, 0 is no evidence of vasculitis and 5 is the most severe vasculitis. The scores for each parameter are an average of the cecum and distal colon, taken from two independent experiments with 3 mice/experiment.
Supplementary Material
Acknowledgements
Thank you to the Microarray Core, the Pathology Core, and the Electron Microscopy Core at UT Southwestern for their help in processing samples. This work was supported by the National Institute of Health (NIH) Grants AI053067, AI77853 and AI077613, and the Burroughs Wellcome Fund (V.S.). M.M.C. was supported through NIH Training Grant 5 T32 AI7520–14. The contents are solely the responsibility of the authors and do not represent the official views of the NIH NIAID.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Supplemental Methods
Additional methods are included in the Supplementary Materials.
Author Contributions M.M.C. led the project, performed and designed experiments, and wrote the paper. Z.H., C.K., and R.J.D. performed the metabolomics experiments. S.N. performed the double-blind histopathology analysis. V.S. designed experiments and wrote the paper.
Microarray data are deposited in the Gene Expression Omnibus under accession number GSE47418. Reprints and permissions information is available at www.nature.com/reprints. The authors declare no competing financial interests.
References
- Aquilano K, Baldelli S, Ciriolo MR. Glutathione: new roles in redox signaling for an old antioxidant. Front Pharmacol. 2014;5:196. doi: 10.3389/fphar.2014.00196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bacchetti De Gregoris T, Aldred N, Clare AS, Burgess JG. Improvement of phylum- and class-specific primers for real-time PCR quantification of bacterial taxa. J Microbiol Methods. 2011;86:351–356. doi: 10.1016/j.mimet.2011.06.010. [DOI] [PubMed] [Google Scholar]
- Bergstrom KS, Kissoon-Singh V, Gibson DL, Ma C, Montero M, Sham HP, Ryz N, Huang T, Velcich A, Finlay BB, et al. Muc2 protects against lethal infectious colitis by disassociating pathogenic and commensal bacteria from the colonic mucosa. PLoS Pathog. 2010;6:e1000902. doi: 10.1371/journal.ppat.1000902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betian HG, Linehan BA, Bryant MP, Holdeman LV. Isolation of a cellulotytic Bacteroides sp. from human feces. Appl Environ Microbiol. 1977;33:1009–1010. doi: 10.1128/aem.33.4.1009-1010.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borody TJ, Khoruts A. Fecal microbiota transplantation and emerging applications. Nat Rev Gastroenterol Hepatol. 2012;9:88–96. doi: 10.1038/nrgastro.2011.244. [DOI] [PubMed] [Google Scholar]
- Cash HL, Whitham CV, Behrendt CL, Hooper LV. Symbiotic bacteria direct expression of an intestinal bactericidal lectin. Science. 2006;313:1126–1130. doi: 10.1126/science.1127119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin AM, Feldheim DA, Saier MH., Jr Altered transcriptional patterns affecting several metabolic pathways in strains of Salmonella typhimurium which overexpress the fructose regulon. J Bacteriol. 1989;171:2424–2434. doi: 10.1128/jb.171.5.2424-2434.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dicksved J, Ellstrom P, Engstrand L, Rautelin H. Susceptibility to campylobacter infection is associated with the species composition of the human fecal microbiota. MBio. 2014;5 doi: 10.1128/mBio.01212-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldheim DA, Chin AM, Nierva CT, Feucht BU, Cao YW, Xu YF, Sutrina SL, Saier MH., Jr Physiological consequences of the complete loss of phosphoryl-transfer proteins HPr and FPr of the phosphoenolpyruvate:sugar phosphotransferase system and analysis of fructose (fru) operon expression in Salmonella typhimurium. J Bacteriol. 1990;172:5459–5469. doi: 10.1128/jb.172.9.5459-5469.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreyra JA, Wu KJ, Hryckowian AJ, Bouley DM, Weimer BC, Sonnenburg JL. Gut microbiota-produced succinate promotes C. difficile infection after antibiotic treatment or motility disturbance. Cell Host Microbe. doi: 10.1016/j.chom.2014.11.003. (in press). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gevers D, Kugathasan S, Denson LA, Vazquez-Baeza Y, Van Treuren W, Ren B, Schwager E, Knights D, Song SJ, Yassour M, et al. The treatment-naive microbiome in new-onset Crohn's disease. Cell Host Microbe. 2014;15:382–392. doi: 10.1016/j.chom.2014.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffin PM, Ostroff SM, Tauxe RV, Greene KD, Wells JG, Lewis JH, Blake PA. Illnesses associated with Escherichia coli O157:H7 infections. A broad clinical spectrum. Ann Intern Med. 1988;109:705–712. doi: 10.7326/0003-4819-109-9-705. [DOI] [PubMed] [Google Scholar]
- Guo X, Xia X, Tang R, Zhou J, Zhao H, Wang K. Development of a real-time PCR method for Firmicutes and Bacteroidetes in faeces and its application to quantify intestinal population of obese and lean pigs. Lett Appl Microbiol. 2008;47:367–373. doi: 10.1111/j.1472-765X.2008.02408.x. [DOI] [PubMed] [Google Scholar]
- Honda K, Littman DR. The microbiome in infectious disease and inflammation. Annu Rev Immunol. 2012;30:759–795. doi: 10.1146/annurev-immunol-020711-074937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooper LV, Midtvedt T, Gordon JI. How host-microbial interactions shape the nutrient environment of the mammalian intestine. Annu Rev Nutr. 2002;22:283–307. doi: 10.1146/annurev.nutr.22.011602.092259. [DOI] [PubMed] [Google Scholar]
- Hughes DT, Clarke MB, Yamamoto K, Rasko DA, Sperandio V. The QseC adrenergic signaling cascade in Enterohemorrhagic E. coli (EHEC) PLoS Pathog. 2009;5:e1000553. doi: 10.1371/journal.ppat.1000553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson ME, Sjovall H, Hansson GC. The gastrointestinal mucus system in health and disease. Nat Rev Gastroenterol Hepatol. 2013;10:352–361. doi: 10.1038/nrgastro.2013.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaper JB, Nataro JP, Mobley HL. Pathogenic Escherichia coli. Nat Rev Microbiol. 2004;2:123–140. doi: 10.1038/nrmicro818. [DOI] [PubMed] [Google Scholar]
- Kelly CR, Ihunnah C, Fischer M, Khoruts A, Surawicz C, Afzali A, Aroniadis O, Barto A, Borody T, Giovanelli A, et al. Fecal microbiota transplant for treatment of Clostridium difficile infection in immunocompromised patients. Am J Gastroenterol. 2014;109:1065–1071. doi: 10.1038/ajg.2014.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kendall MM, Gruber CC, Rasko DA, Hughes DT, Sperandio V. Hfq virulence regulation in enterohemorrhagic Escherichia coli O157:H7 strain 86–24. J Bacteriol. 2011;193:6843–6851. doi: 10.1128/JB.06141-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knutton S, Baldwin T, Williams PH, McNeish AS. Actin accumulation at sites of bacterial adhesion to tissue culture cells: basis of a new diagnostic test for enteropathogenic and enterohemorrhagic Escherichia coli. Infect Immun. 1989;57:1290–1298. doi: 10.1128/iai.57.4.1290-1298.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuss SK, Best GT, Etheredge CA, Pruijssers AJ, Frierson JM, Hooper LV, Dermody TS, Pfeiffer JK. Intestinal microbiota promote enteric virus replication and systemic pathogenesis. Science. 2011;334:249–252. doi: 10.1126/science.1211057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen N, Vogensen FK, van den Berg FW, Nielsen DS, Andreasen AS, Pedersen BK, Al-Soud WA, Sorensen SJ, Hansen LH, Jakobsen M. Gut microbiota in human adults with type 2 diabetes differs from non-diabetic adults. PLoS One. 2010;5:e9085. doi: 10.1371/journal.pone.0009085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macy JM, Ljungdahl LG, Gottschalk G. Pathway of succinate and propionate formation in Bacteroides fragilis. J Bacteriol. 1978;134:84–91. doi: 10.1128/jb.134.1.84-91.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallick EM, McBee ME, Vanguri VK, Melton-Celsa AR, Schlieper K, Karalius BJ, O'Brien AD, Butterton JR, Leong JM, Schauer DB. A novel murine infection model for Shiga toxin-producing Escherichia coli. J Clin Invest. 2012;122:4012–4024. doi: 10.1172/JCI62746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milstien S, Goldman P. Role of intestinal microflora in the metabolism of guanidinosuccinic acid. Journal of bacteriology. 1973;114:641–644. doi: 10.1128/jb.114.2.641-644.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mundy R, MacDonald TT, Dougan G, Frankel G, Wiles S. Citrobacter rodentium of mice and man. Cell Microbiol. 2005;7:1697–1706. doi: 10.1111/j.1462-5822.2005.00625.x. [DOI] [PubMed] [Google Scholar]
- Ng KM, Ferreyra JA, Higginbottom SK, Lynch JB, Kashyap PC, Gopinath S, Naidu N, Choudhury B, Weimer BC, Monack DM, et al. Microbiota-liberated host sugars facilitate post-antibiotic expansion of enteric pathogens. Nature. 2013;502:96–99. doi: 10.1038/nature12503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Njoroge J, Sperandio V. Enterohemorrhagic Escherichia coli virulence regulation by two bacterial adrenergic kinases, QseC and QseE. Infect Immun. 2012;80:688–703. doi: 10.1128/IAI.05921-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Njoroge JW, Gruber C, Sperandio V. The interacting Cra and KdpE regulators are involved in the expression of multiple virulence factors in enterohemorrhagic Escherichia coli. J Bacteriol. 2013;195:2499–2508. doi: 10.1128/JB.02252-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Njoroge JW, Nguyen Y, Curtis MM, Moreira CG, Sperandio V. Virulence meets metabolism: Cra and KdpE gene regulation in enterohemorrhagic Escherichia coli. MBio. 2012;3:e00280–e00212. doi: 10.1128/mBio.00280-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pacheco AR, Curtis MM, Ritchie JM, Munera D, Waldor MK, Moreira CG, Sperandio V. Fucose sensing regulates bacterial intestinal colonization. Nature. 2012;492:113–117. doi: 10.1038/nature11623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulsen IT, Banerjei L, Myers GS, Nelson KE, Seshadri R, Read TD, Fouts DE, Eisen JA, Gill SR, Heidelberg JF, et al. Role of mobile DNA in the evolution of vancomycin-resistant Enterococcus faecalis. Science. 2003;299:2071–2074. doi: 10.1126/science.1080613. [DOI] [PubMed] [Google Scholar]
- Peterson DA, Frank DN, Pace NR, Gordon JI. Metagenomic approaches for defining the pathogenesis of inflammatory bowel diseases. Cell Host Microbe. 2008;3:417–427. doi: 10.1016/j.chom.2008.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakoff-Nahoum S, Paglino J, Eslami-Varzaneh F, Edberg S, Medzhitov R. Recognition of commensal microflora by toll-like receptors is required for intestinal homeostasis. Cell. 2004;118:229–241. doi: 10.1016/j.cell.2004.07.002. [DOI] [PubMed] [Google Scholar]
- Reading NC, Torres AG, Kendall MM, Hughes DT, Yamamoto K, Sperandio V. A novel two-component signaling system that activates transcription of an enterohemorrhagic Escherichia coli effector involved in remodeling of host actin. J Bacteriol. 2007;189:2468–2476. doi: 10.1128/JB.01848-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rotstein OD, Pruett TL, Fiegel VD, Nelson RD, Simmons RL. Succinic acid, a metabolic by-product of Bacteroides species, inhibits polymorphonuclear leukocyte function. Infect Immun. 1985;48:402–408. doi: 10.1128/iai.48.2.402-408.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rotstein OD, Vittorini T, Kao J, McBurney MI, Nasmith PE, Grinstein S. A soluble Bacteroides by-product impairs phagocytic killing of Escherichia coli by neutrophils. Infect Immun. 1989;57:745–753. doi: 10.1128/iai.57.3.745-753.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saier MH, Jr, Ramseier TM. The catabolite repressor/activator (Cra) protein of enteric bacteria. J Bacteriol. 1996;178:3411–3417. doi: 10.1128/jb.178.12.3411-3417.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommer F, Backhed F. The gut microbiota--masters of host development and physiology. Nat Rev Microbiol. 2013;11:227–238. doi: 10.1038/nrmicro2974. [DOI] [PubMed] [Google Scholar]
- Sonnenburg JL, Xu J, Leip DD, Chen CH, Westover BP, Weatherford J, Buhler JD, Gordon JI. Glycan foraging in vivo by an intestine-adapted bacterial symbiont. Science. 2005;307:1955–1959. doi: 10.1126/science.1109051. [DOI] [PubMed] [Google Scholar]
- Sperandio V, Torres AG, Kaper JB. Quorum sensing Escherichia coli regulators B and C (QseBC): a novel two-component regulatory system involved in the regulation of flagella and motility by quorum sensing in E. coli. Mol Microbiol. 2002;43:809–821. doi: 10.1046/j.1365-2958.2002.02803.x. [DOI] [PubMed] [Google Scholar]
- Spor A, Koren O, Ley R. Unravelling the effects of the environment and host genotype on the gut microbiome. Nat Rev Microbiol. 2011;9:279–290. doi: 10.1038/nrmicro2540. [DOI] [PubMed] [Google Scholar]
- Stelter C, Kappeli R, Konig C, Krah A, Hardt WD, Stecher B, Bumann D. Salmonella-induced mucosal lectin RegIIIbeta kills competing gut microbiota. PLoS One. 2011;6:e20749. doi: 10.1371/journal.pone.0020749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taupin D, Podolsky DK. Trefoil factors: initiators of mucosal healing. Nat Rev Mol Cell Biol. 2003;4:721–732. doi: 10.1038/nrm1203. [DOI] [PubMed] [Google Scholar]
- Turnbaugh PJ, Hamady M, Yatsunenko T, Cantarel BL, Duncan A, Ley RE, Sogin ML, Jones WJ, Roe BA, Affourtit JP, et al. A core gut microbiome in obese and lean twins. Nature. 2009a;457:480–484. doi: 10.1038/nature07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbaugh PJ, Ridaura VK, Faith JJ, Rey FE, Knight R, Gordon JI. The effect of diet on the human gut microbiome: a metagenomic analysis in humanized gnotobiotic mice. Sci Transl Med. 2009b;1:6ra14. doi: 10.1126/scitranslmed.3000322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaishnava S, Yamamoto M, Severson KM, Ruhn KA, Yu X, Koren O, Ley R, Wakeland EK, Hooper LV. The antibacterial lectin RegIIIgamma promotes the spatial segregation of microbiota and host in the intestine. Science. 2011;334:255–258. doi: 10.1126/science.1209791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter J, Ley R. The human gut microbiome: ecology and recent evolutionary changes. Annu Rev Microbiol. 2011;65:411–429. doi: 10.1146/annurev-micro-090110-102830. [DOI] [PubMed] [Google Scholar]
- Wang Y, Hoenig JD, Malin KJ, Qamar S, Petrof EO, Sun J, Antonopoulos DA, Chang EB, Claud EC. 16S rRNA gene-based analysis of fecal microbiota from preterm infants with and without necrotizing enterocolitis. ISME J. 2009;3:944–954. doi: 10.1038/ismej.2009.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willing BP, Vacharaksa A, Croxen M, Thanachayanont T, Finlay BB. Altering host resistance to infections through microbial transplantation. PLoS One. 2011;6:e26988. doi: 10.1371/journal.pone.0026988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Bjursell MK, Himrod J, Deng S, Carmichael LK, Chiang HC, Hooper LV, Gordon JI. A genomic view of the human-Bacteroides thetaiotaomicron symbiosis. Science. 2003;299:2074–2076. doi: 10.1126/science.1080029. [DOI] [PubMed] [Google Scholar]
- Zumbrun SD, Melton-Celsa AR, Smith MA, Gilbreath JJ, Merrell DS, O'Brien AD. Dietary choice affects Shiga toxin-producing Escherichia coli (STEC) O157:H7 colonization and disease. Proc Natl Acad Sci U S A. 2013;110:E2126–E2133. doi: 10.1073/pnas.1222014110. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.