SUMMARY
Protein-protein interactions (PPIs) play central roles in orchestrating biological processes. While some PPIs are stable, many important ones are transient and hard to detect with conventional approaches. We developed ReBiL, a recombinase enhanced bimolecular luciferase complementation platform, to enable weak PPI detection in living cells. ReBiL readily identified challenging transient interactions between an E3 ubiquitin ligase and an E2 ubiquitin-conjugating enzyme. ReBiL’s ability to rapidly interrogate PPIs in diverse conditions revealed that some stapled α-helical peptides, a new class of PPI antagonists, induce target-independent cytosolic leakage and cytotoxicity that is antagonized by serum. These results explain the requirement for serum-free conditions to detect stapled peptide activity, and define a required parameter to evaluate for peptide antagonist approaches. ReBiL’s ability to expedite PPI analysis, assess target specificity and cell permeability, and to reveal off-target effects of PPI modifiers should facilitate development of effective, cell permeable PPI therapeutics and elaboration of diverse biological mechanisms.
Keywords: P53, Mdm2, Mdm4, Ube2t, FANCL, Bimolecular luciferase complementation, BiLC, Protein-fragment complementation assay, PCA, Recombinase mediated cassette exchange, RMCE, Protein-protein interaction, PPI, Recombinase enhanced BiLC, ReBiL, Stapled peptide, Nutlin-3a
INTRODUCTION
Most cellular functions are conducted by interactions between multiple proteins comprising either static or dynamic macromolecular machines. Dynamic protein-protein interactions (PPIs) are often composed of weak and transient interactions that render their discovery and analysis in physiologically relevant environments difficult. The human protein “interactome” may involve ~130,000 to ~650,000 protein-protein interactions (Stumpf et al., 2008; Venkatesan et al., 2009). Even if a small fraction of these cause diseases through aberrant interactions (Ivanov et al., 2013), the availability of rapid PPI antagonist screens will open up vast new opportunities for developing therapeutic agents. Although fluorescent image-based assays such as the proximity ligation in situ assay (P-LISA) (Soderberg et al., 2006; Bernal et al., 2010) and two/three hybrid (F2H and F3H) assays have been used to evaluate PPI antagonists in cells (Brown et al., 2013; Herce et al., 2013), they do not enable simultaneous evaluation of on-target validation and facile high throughput analysis of target disruption in living cells. Clearly, a critical unmet need is availability of methods to enable real time PPI detection and direct measurement of their intracellular disruption by antagonists. To address this need, we integrated Cre-recombinase mediated cassette exchange (RMCE) (Wong et al., 2005) and bi-molecular luciferase complementation (BiLC) (Luker et al., 2004) in an inducible gene expression format to create a platform system for detecting and analyzing PPIs. We refer to the approach as Recombinase-enhanced BiLC, and use the acronym ReBiL for concision, and to precisely and simply describe the integration of its key components.
We applied ReBiL to two outstanding problems to assess the potential breadth of biological problems it could be used to study. First, we evaluated its ability to detect weak, transient PPIs in living cells, exemplified by the interaction of the E2 ubiquitin conjugating enzyme Ube2T and FANCL, the E3 ubiquitin ligase to which it binds. FANCL is a critical component of the Fanconi anemia DNA repair pathway. Mutations in this pathway cause hematologic abnormalities and create a predisposition to develop cancer (Moldovan and D’Andrea, 2009). The E2–E3 interactions are usually transient and typically in the low μM range (Deshaies and Joazeiro, 2009; Ye and Rape, 2009), making their interaction difficult to detect. While interaction between Ube2t and FANCL E3 ligase was first shown in a yeast two-hybrid screen (Machida et al., 2006), it has eluded detection in living mammalian cells. The Ube2t-FANCL dissociation constant was measured as 0.454 μM by isothermal titration calorimetry, but this required analysis at 8 °C (Hodson et al., 2011). Furthermore, co-crystallization required their fusion (Hodson et al., 2014), consistent with their associating with low affinity. Assessing Ube2t-FANCL association by ReBiL provides a stringent test of its ability to detect such challenging interactions.
The second system to which we applied ReBiL involves the p53 tumor suppressor. Defects in the p53 pathway occur in as many as 22 million cancer patients, with about 50% being due to inactivating mutations in p53 itself (Brown et al., 2009). Many of the remaining tumors contain alterations that lead to over-expression of either of two oncogenes, Mdm2 or Mdm4 that bind to wild type p53 and inactivate it by serving as an E3 ubiquitin ligase (Mdm2) and/or a transcriptional repressor (Mdm2 and Mdm4). Therefore, availability of drugs that interfere with p53-Mdm2 and p53-Mdm4 interactions could restore p53 function and significantly benefit patients whose cancers express wild type p53 (Brown et al., 2009; Wade et al., 2013).
Unlike classic enzyme-ligand binding pockets that can be effectively targeted by small molecules, surfaces at which protein domains interact are typically large, flat and featureless (Wells and McClendon, 2007). Nevertheless, several PPI antagonists that disrupt p53-Mdm2, p53-Mdm4 or both have been reported (Brown et al., 2009; Wade et al., 2013). Recently, stapled α-helical (SAH) peptides have been designed and suggested as the model for superior PPI antagonists because of their larger interaction interfaces, better structural stability, protease resistance, and cell permeability (Verdine and Hilinski, 2012). PPI antagonist effectiveness has conventionally been evaluated using in vitro biochemical and biophysical assays that quantify the ability of the antagonist to displace one of the interacting protein fragments. However, such assays do not reveal whether molecules that work effectively in in vitro systems can cross the cell membrane to effect target disruption in a native intracellular environment. While fluorescence-activated cell sorting (FACS) analyses have been used to indicate whether fluorophore-tagged PPI antagonists can enter cells, they do not reveal the subcellular localization (endosome versus cytoplasm) of the antagonists, nor whether they reach their targets at concentrations sufficient to disrupt the PPIs to elicit biological effects. Furthermore, assays of biologic activity such as cell death can be misleading, and do not provide direct evidence of the intracellular efficacy of a PPI antagonist. For example, since p53 can be activated by diverse cellular insults and by many different mechanisms (Beckerman and Prives, 2010), the ability of a putative PPI antagonist to activate p53 target genes or p53-dependent biological processes does not prove that these effects were directly mediated by disruption of p53-Mdm2 and/or p53-Mdm4 complexes.
Here, we report that ReBiL can detect transient and weak protein interactions such as between Ube2t and FANCL. Additionally, ReBiL enabled us to elucidate on and off-target activities of SAH peptides, and a mechanism by which serum antagonizes SAH peptide induced membrane damage. The sensitivity, specificity, and versatility of ReBiL platform should find its broad applications for elucidating biological mechanisms, and as a screen for small molecule and peptide based PPI antagonists.
RESULTS
Development of the Recombinase-enhanced BiLC (ReBiL) platform system
We use the firefly BiLC (Luker et al., 2004), one type of protein-fragment complementation assay (PCA) (Michnick et al., 2007), to study PPIs and their antagonists because its two unique advantages. First, the lack of background luminescence in mammalian cells creates an opportunity for high sensitivity. Second, while split fluorescent proteins associate irreversibly, firefly split luciferase fragments exhibit little if any interaction by themselves, enabling rapid dissociation of complementing pairs (Yang et al., 2009; Ilagan et al., 2011; Macdonald-Obermann et al., 2012). These features make BiLC ideal for analyzing PPI stability and for ascertaining the effectiveness of PPI antagonists.
Firefly luciferase PCA relies on the reconstitution of luciferase enzymatic activity from two split fragments and the interaction of the proteins to which they are genetically fused (Luker et al., 2004) (Figure 1A). BiLC experiments have typically utilized transient transfection of two plasmids encoding each split luciferase fusion partner (Luker et al., 2004; Paulmurugan and Gambhir, 2007; Cassonnet et al., 2011; Gilad et al., 2014). Consequently, the sensitivity, accuracy and reproducibility of this assay are profoundly influenced by the percentage of cells transfected and the copy numbers of each expressed plasmid. Alternatively, BiLC can also be done after selecting for stable cell clones carrying both split luciferase fusions such as the studies of EGF receptors and Notch pathway (Yang et al., 2009; Ilagan et al., 2011; Macdonald-Obermann et al., 2012). However, screening and identifying stable clones with appropriate expression levels is often time-consuming and labor-intensive, and does not enable generation of isogenic lines for comparison of activities of mutant and wild type alleles.
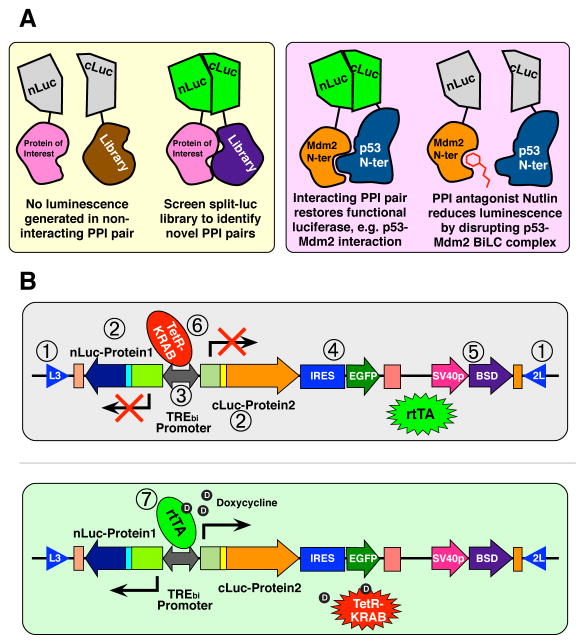
Figure 1. Description of the ReBiL platform system and expected results generated by PPIs and antagonists.
(A) Cartoon depicts BiLC strategy to detect PPIs and their disruption by antagonists. (B) The expression of a ReBiL cassette was controlled by both the TetR-KRAB in the absence of doxycycline (top panel) and rtTA2S-M2 in the presence of doxycycline (bottom panel). The numbers in open circles indicate each key component in the ReBiL platform system (see Table S1).
We overcame these drawbacks by generating the “ReBiL” platform (see Table S1 for feature descriptions). Cre recombinase is used to insert a ReBiL targeting cassette encoding both split luciferase fusion partners into a predetermined floxed acceptor locus integrated at a single chromosomal site in host cells (Wong et al., 2005; Green et al., 2013) (Figure S1). High RMCE integration efficiency and faithful transgene expression at a pre-determined genomic locus (Wong et al., 2005) circumvent the tedious and costly processes involved in screening reporter clones generated by random integration or viral infection. This greatly accelerates the production of stable reporter cell lines, and enables routine generation of 9 to 12 stable ReBiL cell lines in 4 to 6 weeks with minimum effort and could likely be scaled up using robotics.
The ReBiL platform confers two other significant experimental and analytical advantages distinguishable from other stable clones. First, it facilitates structure, function, and interaction analyses because it enables BiLC fusions encoding wild type and mutant proteins to be integrated into, and expressed homogeneously from, the same chromosomal locus (Figure S1). Second, single copy integration and doxycycline-tunable regulation of transgenes (Figure 1B) generates rheostatic and uniform expression (Rossi et al., 2000; Wong et al., 2005) that can be tuned to physiologically relevant levels. Together, these implementations significantly enhance the reproducibility and signal-to-noise ratio of the firefly luciferase PCA.
ReBiL enhances detection of weak protein-protein interactions
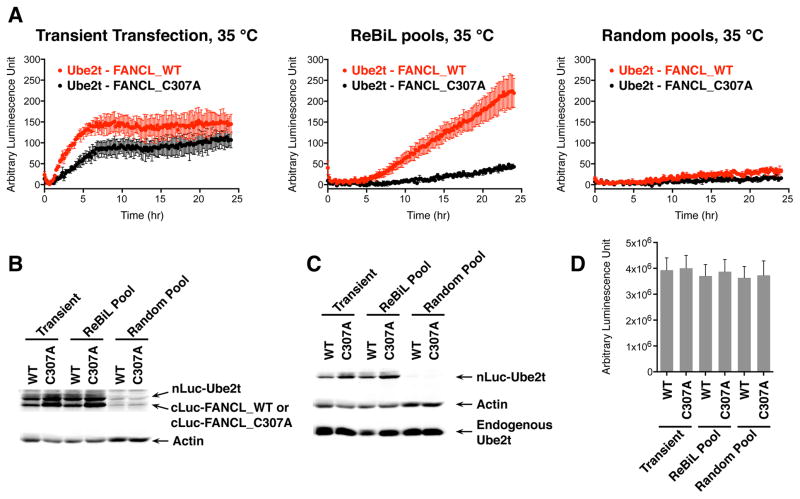
We compared BiLC signals generated by transient transfection, ReBiL, and random integration approaches. We used the identical inducible reporter constructs encoding either Ube2t and FANCL or Ube2t and the FANCL_C307A mutation (Figure S2A and Table S2). The FANCL-C307A is a RING domain mutation that prevents interaction with Ube2t (Machida et al., 2006) and serves as a negative control. The luminescent signals generated in transiently transfected cells and random integrants were barely statistically different between the specific interaction pair (Ube2t-FANCL) and the mutant pair (Ube2t-FANCL_C307A) (Figure 2A). In stark contrast, the Ube2t-FANCL ReBiL cells generated ~5 times higher luminescent signals than the Ube2t-FANCL_C307A mutant ReBiL cells (Figure 2A, middle panel) at 35 °C. The expression levels of FANCL_C307A were higher than its wild type counterpart, which corresponded to increased levels of Ube2t, regardless of whether transfection or ReBiL was used (Figure 2B). Importantly, the level of introduced Ube2t was always lower than the endogenously produced protein (Figure 2C), but due to a lack of validated FANCL antibody, we could not evaluate its endogenous level. We also note that Ube2t and FANCL expression did not affect cell viability (Figure 2D). Thus, the absence of BiLC signal in the Ube2t and FANCL_C307A cells must result from their inability to interact, and not due to a difference in protein level and cell viability. The very low BiLC signals from the random integrant pool (Figure 2A, right panel) correlates with the low expression levels of the BiLC pairs (Figure 2B), consistent with a recent report showing that PPIs depend strongly on abundance (Levy et al., 2014). It is especially significant, therefore, that ReBiL manifests a robust signal-to-noise ratio between these weakly interacting proteins even when at least one of them is expressed at lower than endogenous levels.
Figure 2. Detection of low-affinity Ube2t and FANCL interaction with ReBiL.
(A) ReBiL detected Ube2t and FANCL interaction with significantly higher signal-to-noise ratio. Luminescent signals in the transient transfection, ReBiL and randomly integrated reporter cells were compared at 35 °C. Data shown are mean ± standard error of mean (SEM) from 3 independent experiments. (B) Western blot analysis of BiLC fusion proteins. The nLuc-HA-Ube2t, cLuc-FLAG-FANCL_WT and cLuc-FLAG-FANCL_C307A were detected by anti-HA and anti-FLAG antibodies respectively. Actin was a loading control. (C) Western blot analysis of the levels of nLuc-Ube2t and endogenous Ube2t by anti-Ube2t antibody (Cell Signaling D2L7H). Actin was a loading control. (D) CellTiter-Glo assay indicated there was no growth difference between FANCL_WT and FNACL_C307A cells. Data shown are mean ± SEM from 3 independent experiments.
Intriguingly, at lower temperature (30 °C), both ReBiL and random integrated cells generated higher total BiLC signals and bigger signal-to-noise ratios (~10 fold in ReBiL cells) (Figure S2B) without much difference in the levels of expressed split luciferase fusions (Figure S2C) compared to 35 °C (Figure 2B). Similarly, the expressed split luciferase Ube2t fusion was less than the endogenous Ube2t at 30 °C (Figure S2D) and no growth differences between wild type and mutant cells were observed (data not shown). This temperature dependency is consistent with the low affinity of these interacting proteins, and may indicate that at 30 °C Ube2t-FANCL has a lower dissociation rate (higher affinity). Importantly, at both temperatures, the ReBiL platform exhibited significantly enhanced ability to detect weak and transient PPIs.
Analysis of protein interactions in the p53 pathway using ReBiL
Several small molecule and peptide-based compounds have been reported to interfere with p53-Mdm2 and p53-Mdm4 interactions in biophysical assays and to activate p53 in living cells (Brown et al., 2009; Wade et al., 2013). However, recent data raise questions about the ability of some SAH peptides to interfere with p53-Mdm2 and p53-Mdm4 interactions in cells (Brown et al., 2013). We therefore used the ReBiL system to evaluate small molecule, SAH peptide, and cyclotide based p53-Mdm2 and p53-Mdm4 PPI antagonists.
We used two strategies to avert cytotoxicity or cell cycle arrest that could be generated by wild type p53 activation. First, we used p53 null Saos-2 cells as the host for ReBiL reporter cell line. Second, we combined two p53 mutations, the R273H (Joerger and Fersht, 2008) and C312 truncation (Vassilev et al., 2004), to build a transcriptionally inactive split luciferase p53 fusion that can interact with Mdm2 and Mdm4 (Figure S3A and Table S2).
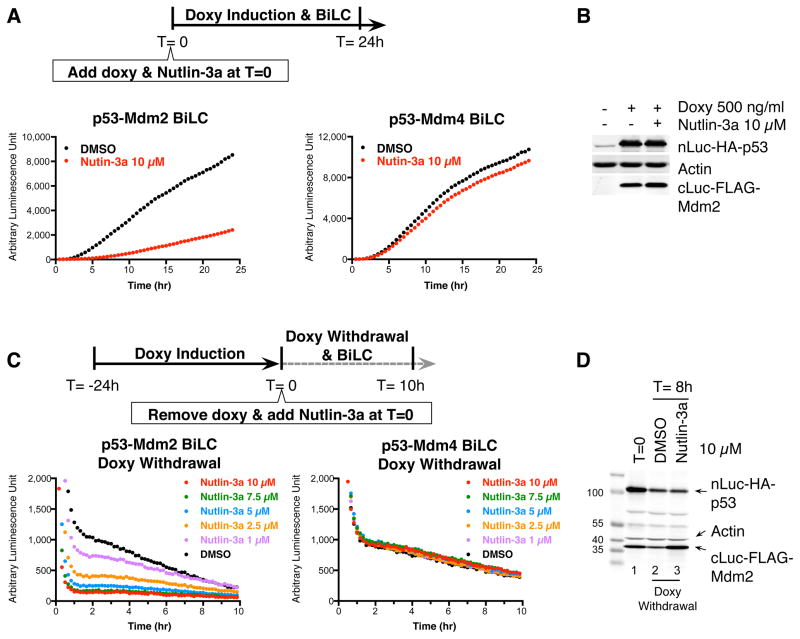
We evaluated the specificity of ReBiL for studying p53-Mdm2 interaction with their antagonist Nutlin-3a, which is known to disrupt p53-Mdm2 but not p53-Mdm4 interactions (Vassilev et al., 2004; Patton et al., 2006; Wade et al., 2008). As expected, Nutlin-3a reduced the p53-Mdm2, but not the p53-Mdm4 BiLC signal (Figure 3A). Signal reduction did not result from Nutlin-3a induced effects on protein levels (Figure 3B) and cell viability (data not shown). These data demonstrate the specificity of the ReBiL system to discriminate between a small molecule’s ability to prevent interaction between p53 and the two very similar binding sites in Mdm2 and Mdm4 in vivo.
Figure 3. ReBiL cells faithfully report PPIs and activity of p53-Mdm2 antagonist using real-time BiLC analyses.
(A) Nutlin-3a prevents newly synthesized p53-Mdm2 but not p53-Mdm4 interactions. The Saos-2 p53-Mdm2 and p53-Mdm4 ReBiL cells in 384-well plates (8,500 cells per well) were treated with 500 ng/ml doxycycline, 100 μM D-luciferin and 10 μM Nutlin-3a or DMSO at Time = 0. Luminescence was read every 30 min for 24 hr at 37 °C. Data shown are a representative experiment from more than 3 independent experiments. (B) Western blot analysis of BiLC fusion proteins showed that Nutlin-3a does not affect the expression amounts of nLuc-HA-p53 and cLuc-FLAG-Mdm2 (detected by anti-HA and anti-FLAG antibodies, respectively). Actin served as a loading control. (C) Pre-induced Saos-2 p53-Mdm2 and p53-Mdm4 ReBiL cells were re-seeded into a 384-well plate (5,000 cells per well) together with Nutlin-3a and D-luciferin at Time = 0. The p53-Mdm2 and p53-Mdm4 BiLC signals decayed over time in a biphasic fashion. The first steep decline in BiLC signal is likely due to the temperature changes of the ReBiL cells when moving from the bench (~ 24 °C) to the pre-warmed luminometer at 37 °C. The second slow decay phase of BiLC results from doxycycline withdrawal and the consequent reduction in transcription of the BiLC fusion genes. Luminescence was read every 10 min for 10 hr at 37 °C. Data shown are a representative experiment from more than 3 independent experiments. (D) Western blot analysis showed that Nutlin-3a did not promote nLuc-HA-p53 and cLuc-FLAG-Mdm2 degradation. Actin was a loading control.
Whether small molecules like Nutlin-3a disrupt pre-formed p53-Mdm2 complexes in living cells has remained an open question. We used ReBiL to investigate this by first inducing expression of the p53- and Mdm2-split luciferase fusion pairs to generate a functional BiLC complex, and then removing doxycycline to prevent their further transcription. Subsequent incubation of the “pre-loaded” cells with Nutlin-3a enabled analysis of the decay of the p53-Mdm2 BiLC complex in living cells. The p53-Mdm2 complexes decayed over time and this was accelerated significantly and dose dependently by Nutlin-3a (Figure 3C, left panel). Western blotting analysis indicates that Nutlin-3a did not promote degradation of BiLC fusion proteins (Figure 3D, compare Lane2 and Lane3). As doxycycline withdrawal only stops p53- and Mdm2-split luciferase fusion transcription but not translation, we added cycloheximide to prevent translation and then determined dissociation with and without Nutlin-3a. We determined luminescence every minute for a 40-minute time period after inhibiting transcription and translation. Similar to longer time period experiments (Figure 3C), Nutlin-3a dose dependently reduced p53-Mdm2 BiLC signals within 20 minutes, cycloheximide did not appreciably affect the kinetics (Figure S3B), and p53- and Mdm2-BiLC fusion protein levels remained unchanged during the short time period of the analysis (Figure S3C). The p53-Mdm4 complexes were not affected by Nutlin-3a, regardless of cycloheximide addition (Figure S3D). These data show that small molecule PPI antagonists such as Nutlin-3a can selectively and rapidly disrupt preformed p53-Mdm2 complexes in living cells.
Disruption of p53-Mdm4 complexes has been an important goal for reactivation of wild type p53 in cancer therapy (Wade et al., 2013). We used ReBiL to determine whether a previously reported small molecule p53-Mdm4 antagonist, SJ-172550 (Reed et al., 2010) disrupts this interaction in cells. No decrease in p53-Mdm4 BiLC signal was detected (Figure S3E). We then evaluated a second compound, RO-5963, that has been proposed to disrupt both p53-Mdm2 and p53-Mdm4 interactions by increasing Mdm2-Mdm4 association (Graves et al., 2012). Consistent with this model, RO-5963 increased the BiLC signal in the Mdm2-Mdm4 N-terminal BiLC pair (Figures S3F and S3G). However, even though the Mdm2 binding affinity of RO-5963 (17.3 nM) is similar to that of Nutlin-3a (18.7 nM) (Graves et al., 2012), RO-5963 exhibited limited ability to disrupt the p53-Mdm2 complex and no effect on the p53-Mdm4 complex in living cells (Figure S3H). Intriguingly, although Nutlin-3a binds to the same Mdm2 N-terminal domain as RO-5963 (Graves et al., 2012), it did not promote Mdm2 and Mdm4 N-terminal domain interactions (Figure S3G, right panel). These data demonstrate the exquisite specificity of the ReBiL strategy to reveal both the agonist and antagonist activities of putative PPI modifiers in vivo.
Evaluate if SAH peptides disrupt p53-Mdm2 and p53-Mdm4 interactions in living cells
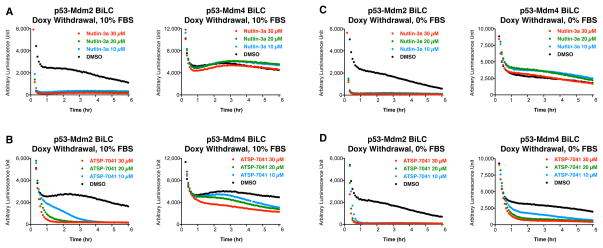
We next determined whether the ability of SAH peptide-based antagonists that have been shown to disrupt p53-Mdm2 and p53-Mdm4 interactions in vitro do so in living cells. We analyzed SAHp53-8 (Bernal et al., 2007; Bernal et al., 2010), sMTide-02 (Brown et al., 2013) and ATSP-7041 (Chang et al., 2013). The larger binding surfaces of these peptidic drugs confer far higher binding affinities than Nutlin-3a, exemplified by ATSP-7041 with a Ki = 0.9 nM for Mdm2 compared with Ki = 52 nM for Nutlin-3a (determined by (Chang et al., 2013)). Surprisingly, despite this much higher binding affinity, SAH peptides are typically used at higher concentrations (20 μM to 100 μM) to elicit cellular activities (Bernal et al., 2010; Gembarska et al., 2012; Chang et al., 2013; Brown et al., 2013). Indeed, in spite of its 57-fold higher binding affinity, ATSP-7041 (10 μM) reached full p53-Mdm2 inhibition much slower (4 hours) than Nutlin-3a (20 minutes, compare Figure 4A to 4B). ATSP-7041 exhibited only marginal activity against p53-Mdm4 complexes (Figure 4B, right panel). Surprisingly, SAHp53-8 and sMTide-02 exhibited no detectable ability to disrupt p53-Mdm2 or p53-Mdm4 complexes in living cells (Figures S4B and S4C). Paradoxically, sMTide-02 actually increased BiLC signals in a dose dependent fashion for both p53-Mdm2 and p53-Mdm4 complexes by an unclear mechanism (Figure S4C).
Figure 4. Analysis of the ability of SAH peptides to disrupt p53-Mdm2 and p53-Mdm4 complexes in living cells, and antagonism by serum.
The Saos-2 p53-Mdm2 and p53-Mdm4 ReBiL cells in 96 well plates (20,000 cells per well) were pre-induced by doxycycline (500 ng/ml for 24 hr). At Time = 0, cells were washed with DMEM and treated with new media containing different PPI antagonists with or without 10% FBS. Luminescent signals were read every 5 minutes for 6 hr by Tecan-M200 at 37 °C. The ReBiL cells were treated with (A) Nutlin-3a and 10% FBS, (B) ATSP-7041 and 10% FBS, (C) Nutlin-3a no FBS, and (D) ATSP-7041 no FBS. Data shown are a representative experiment from more than 3 independent experiments.
These results indicate that higher binding affinity in vitro does not necessarily correlate with higher intracellular PPI disruption activity, suggesting that there might be a barrier to effective entry of the SAH peptides into the cells. The increased activity of ATSP-7041 in 0% serum (Chang et al., 2013) (Figure 4D) indicates that serum itself might limit intracellular access of the SAH peptides, which would be consistent with prior studies in which the cellular activity of SAH peptides is typically measured in serum-free medium (Bernal et al., 2010; Edwards et al., 2013) (see Figure S5). Since the mechanism for serum-mediated inhibition of SAH peptides has remained elusive, we investigated it fully below.
Dual Mdm2 and Mdm4 antagonist SAH peptides exhibit p53-independent cytotoxicity
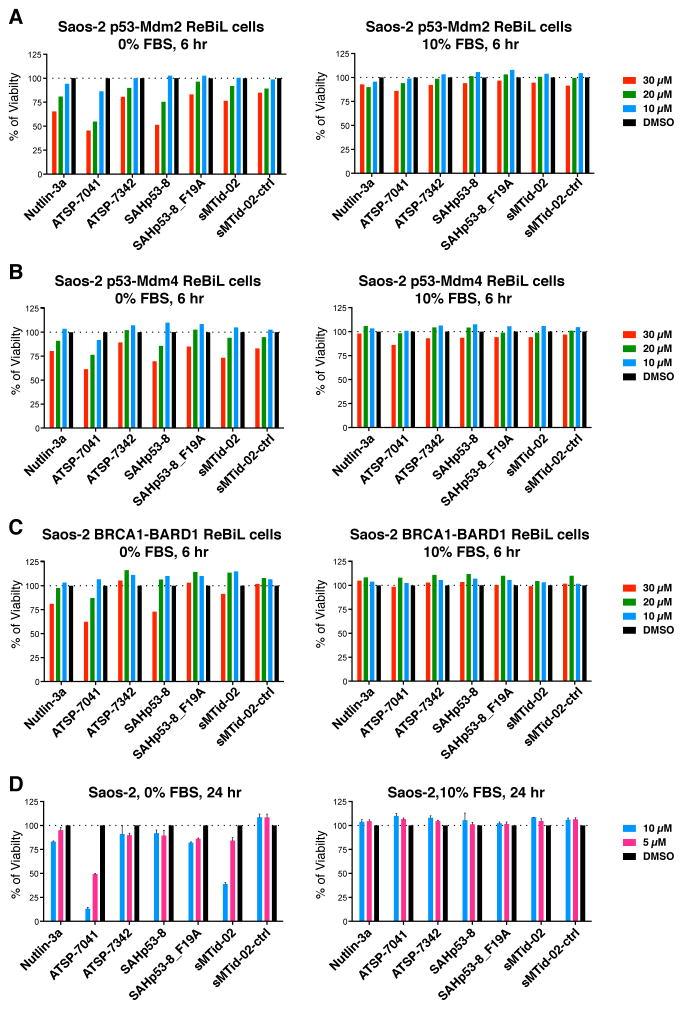
As ReBiL system enables real time analyses of the kinetics of target disruption in cells, we could determine when target disruption occurs and then correlate this with other parameters such as cell viability. We noticed that wild type SAH peptides rapidly (~6 hours) and dose dependently reduced viability of p53-null Saos-2 ReBiL cells (Figure 5). This cytotoxicity is p53 independent since the Saos-2 ReBiL cells are p53 null and they were engineered to encode a transcriptionally inactive split-luciferase p53 fusion protein. This also occurred in Saos-2 cells expressing BRCA1-BARD1 BiLC fusion proteins (Brzovic et al., 2001) (Figure 5C and Table S2). Surprisingly, the negative control mutant peptides exhibited neither PPI disruption (Figure S4) nor significantly reduced cell viability, even in serum-free media (Figure 5). Importantly, 10% serum prevented loss of cell viability induced by wild type SAH peptides (Figure 5), and also reduced the effectiveness of ATSP-7041 (Chang et al., 2013) (compare Figure 4B to 4D). Similarly, lower concentrations of ATSP-7041 and sMTide-02 peptides also reduced Saos-2 viability in 24 hours in a serum dependent manner (Figure 5D). These data imply that these p53-activating SAH peptides can elicit p53-independent cytotoxicity, which is inhibited by serum. As reported previously, high Nutlin-3a concentrations also induce p53-independent cytotoxicity (Liu et al., 2010) (Figure 5), likely by off-target effects different from those of SAH peptides (discussed in Figure 7).
Figure 5. Viability assay of Saos-2 ReBiL cells reveals p53-activating SAH peptides possess p53-independent cytotoxicity in the absence of serum.
The cell viability was measured at 6 hr after Saos-2 ReBiL cells were treated with indicated SAH peptides without and with 10% FBS by CellTiter Glo assay. (A) Saos-2 p53-Mdm2 ReBiL cells (B) Saos-2 p53-Mdm4 ReBiL cells and (C) Saos-2 BRCA1-BARD1 ReBiL cells. Data shown are a representative experiment from more than 3 independent experiments and normalized to the luminescent reading of DMSO (set to 100%). (D) Saos-2 cells were treated with indicated SAH peptides in 384-well plate (2,000 cells per well) without and with 10% FBS for 24 hr. Cell viability was detected by CellTiter Glo assay. Data shown are mean ± standard deviation from two independent experiments and normalized to the luminescent reading of DMSO (100%).
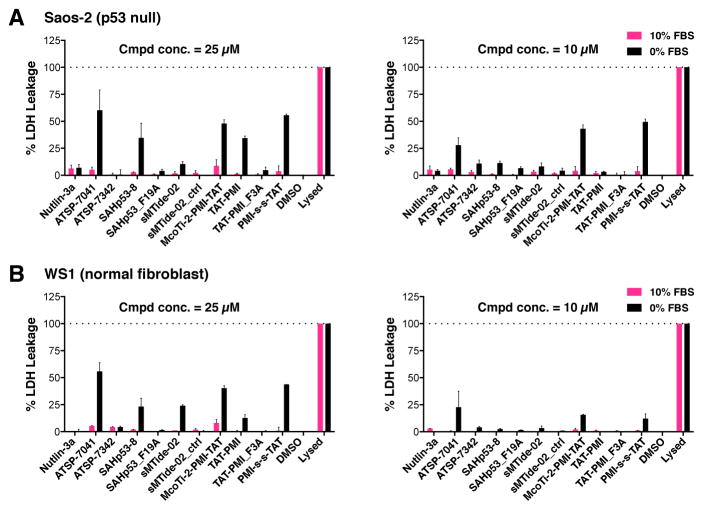
Figure 7. Stapled peptides induce membrane leakage by a p53-independent mechanism that is antagonized by serum.
(A) Saos-2 cells were treated with the indicated PPI antagonists at 25 μM and 10 μM for 6 hr. Accumulation of cytoplasmic lactate dehydrogenase (LDH) in the growth medium was used as a metric of cell membrane damage. LDH was detected by the CytoTox 96 Non-Radioactive Cytotoxicity Assay Kit (Promega). The Lysed sample represents the maximum LDH leakage in this experiment and its reading was set to 100%. DMSO treatment served as the vehicle control and its value was set to 0%. (B) Normal human fibroblasts (WS1 cells) were treated exactly as in (A). Data are shown as mean ± SEM from two independent experiments.
Dual Mdm2 and Mdm4 antagonist SAH peptides exhibit strong target disruption activity in cell lysates
We consider the following as two reasonable explanations of serum’s ability to both prevent SAH peptide-induced cytotoxicity and to reduce the efficacy of PPI disruption. First, serum may bind SAH peptides in such a way as to prevent them from disrupting their target complexes. Second, serum may prevent SAH peptide entry into cells to both reduce their efficacy and limit cytotoxicity.
We gained insights into these possibilities by developing a cell-free BiLC assay. We reasoned that if SAH peptides efficiently disrupt PPIs in cell lysates, but not in intact cells, then membrane penetration and access to their intracellular targets might be limiting. We induced expression of the BiLC complexes by doxycycline, and then prepared cell lysates using an optimized buffer (PPI lysis buffer, PLB) (Figure S6). In contrast to in vitro binding competition assays that use purified protein fragments to identify PPI antagonists, the BiLC lysate assay contains all soluble cellular proteins extracted by PLB and should therefore reveal PPI inhibitor potency in a more physiologically relevant context.
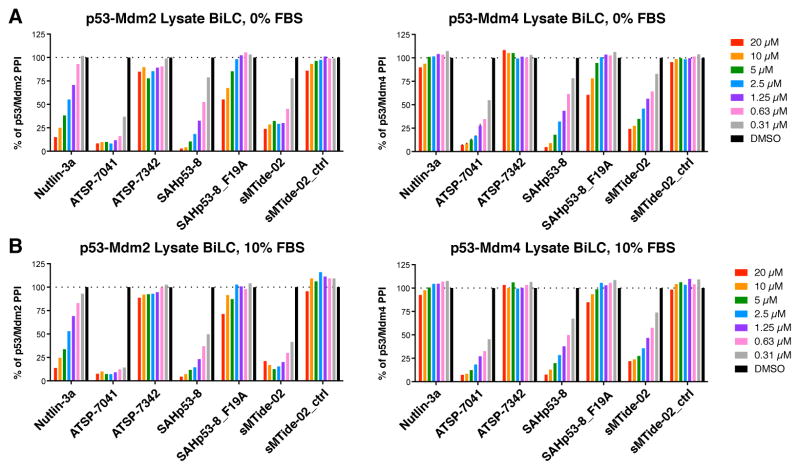
Consistent with the cell-based BiLC assay, Nutlin-3a efficiently disrupted p53-Mdm2, but not p53-Mdm4, complexes in the lysate BiLC assay (Figure 6 and S6E). SJ-172550 (Reed et al., 2010), RO-5963 (Graves et al., 2012) and pyrrolopyrimidine compound 3b (Lee et al., 2011) exhibited no activity in the lysate BiLC assays (Figure S6E), confirming their poor intracellular PPI disruption activity (Figures S3E and S3H). In contrast, the SAHp53-8, sMTide-02, and ATSP-7041 were potent p53-Mdm2 and p53-Mdm4 interaction disruptors in the lysate BiLC assays (Figure 6A); negative control peptides with mutations in amino acids (Phe, Trp and Leu) known to mediate p53 interactions with Mdm2 and Mdm4 were inactive (Figure 6A). The lack of effect of any of these compounds on BRCA1-BARD1 BiLC excludes the explanation that active SAH peptides are simply luciferase inhibitors (Figure S7). These results demonstrate that SAHp53-8 and sMTide-02 are indeed potent PPI antagonists when they can access their targets. This combination of ReBiL assays in cellular lysates and in live cells provides a simple and efficient strategy to quickly assess the specificity and potency of putative PPI antagonists, and indicates when poor activity may derive from inefficient intracellular target access.
Figure 6. The BiLC lysate assay reveals that serum does not prevent SAH peptides from disrupting p53-Mdm2 or p53-Mdm4 complexes.
(A) The cellular lysates obtained from p53-Mdm2 and p53-Mdm4 ReBiL cells were co-incubated with the indicated PPI antagonists in the absence of FBS in 384-well plates at RT for 10 minutes. Steady-Glo was added, and luminescence was detected at 26 °C. (B) The BiLC lysate assays were identical to (A) except for the inclusion of 10% FBS. Data shown are a representative experiment from more than three independent experiments and normalized to the luminescent reading of DMSO (set to 100%).
The inhibitory effect of serum on SAH peptides could result from direct binding of serum proteins to the peptide (Bird et al., 2014). We added 10% fetal bovine serum (FBS) into the lysate BiLC assays to determine directly whether serum reduces the ability of stapled peptides to disrupt p53-Mdm2 and p53-Mdm4 interactions. Serum did not reduce the potency of ATSP-7041, SAHp53-8, or sMTide-02 in cell lysates (Figure 6B). These results demonstrate that serum components or proteins do not sequester or modify SAH peptides and affect their ability to disrupt the target complexes.
Dual Mdm2 and Mdm4 antagonist SAH peptides cause p53-independent cell membrane damage
Given the data presented above, we inferred that serum might compromise the ability of SAH peptides to enter the cell. We also wondered whether there might be a mechanistic linkage between the reduced cytotoxicity and reduced PPI disruption by SAH peptides in cells exposed to serum-containing medium. We reasoned that if SAH peptides compromise membrane integrity, they would be able to gain access to the cytoplasm and their targets; by extension, the serum effect could be explained if it antagonized such membrane effects. We tested this possibility by examining membrane integrity after exposure to SAH peptides in the presence or absence of 10% serum. We quantified release of the cytosolic enzyme lactate dehydrogenase (LDH) into the culture medium as an indicator of the loss of cell membrane integrity. The data clearly show that ATSP-7041, sMTide-02, and SAHp53-8 all cause LDH leakage in the absence of serum (Figure 7). In contrast, 10% serum protects the cell membrane from the damage induced by these SAH peptides (Figure 7). This cell membrane damaging activity is p53-independent since the LDH assay was preformed in p53-null Saos-2 cells (Figure 7A). The membranes of normal human WS1 fibroblasts were also damaged by SAH peptides, indicating this is not a process restricted to cancer cells (Figure 7B). Surprisingly, all three mutant stapled peptides lacked this cell membrane damaging activity (Figure 7). Importantly, Nutlin-3a did not induce membrane leakage, excluding the possibility that the cytotoxicity observed when it is used at high concentrations results from membrane damage (Figure 5).
We also examined the activities of several PMI-based p53-Mdm2 and p53-Mdm4 antagonists, as these are high affinity Mdm2-binding peptides obtained through screening phage display libraries (Pazgier et al., 2009) (Table S3 and Supplemental Experimental Procedures). The lysate BiLC assay showed that all of them are highly potent p53-Mdm2 and p53-Mdm4 antagonists (Figure S6F). Unfortunately, all lack intracellular p53-Mdm2 and p53-Mdm4 disruption activity in 10% serum media (data not shown). In the absence of serum, all except one (McoTi-1-PMI, data not shown), damage cell membranes in a p53-independent fashion that is exacerbated by inclusion of TAT penetrating peptides (Wadia et al., 2004) (Figure 7). Taken together, our studies show that serum does not prevent SAH peptides from binding to their targets. Instead, serum protects cell membranes from being damaged by a previously unrecognized membrane disrupting activity of wild type SAH peptides.
DISCUSSION
The ReBiL vectors and cell lines described here constitute a platform system able to detect weak and transient PPIs, and expedite structure-activity relationships for PPI antagonists and agonists. It is sensitive, specific, and can be used to analyze PPIs in both living cells and cell free systems. For example, we quickly confirmed that small molecule Nutlin-3a interferes with p53-Mdm2 but not p53-Mdm4 interactions. In contrast, other published antagonists (SJ-172550 and RO-5963) showed little if any PPI disruption activity in cells. Our observations suggest that the reported p53 activating effects of these compounds may result from induction of other cellular stresses (Beckerman and Prives, 2010), and not completely from p53-Mdm2 or p53-Mdm4 disruption. Therefore, there can be a lack of concordance between in vitro biophysical and biochemical assays and in vivo biological readouts, and they emphasize the critical importance of using an assay such as ReBiL that directly analyzes target disruption in cells to deduce mechanism of action.
The ReBiL approach also led to new insights concerning conflicting results of studies using SAH peptides (Brown et al., 2013; Okamoto et al., 2013; Okamoto et al., 2014). Considerable effort and resources have been expended to develop SAH peptides as PPI antagonists. Given the difficulty of developing small molecule p53-Mdm4 disruptors, SAH peptides were developed as dual p53-Mdm2 and p53-Mdm4 antagonists since Mdm2 and Mdm4 contain similar N-terminal hydrophobic clefts that interact with p53. Consistent with expectations, SAH peptides that target this region have high binding affinities (Bernal et al., 2010; Chang et al., 2013; Brown et al., 2013) and an ability to disrupt both p53-Mdm2 and p53-Mdm4 complexes in vitro (Figure 6). However, there has been much debate about their cell permeability (Okamoto et al., 2013; Brown et al., 2013) and their ability to elicit p53-dependent biological responses (Brown et al., 2013). For example, in contrast to a previous report (Bernal et al., 2010), a recent study found that the SAHp53-8 lacked cytotoxicity and failed to activate a p53 reporter in cell-based analyses (Brown et al., 2013). Our results clearly show that the stapled peptides SAHp53-8 and sMTid-02 exhibit little to no intracellular activity in the presence or absence of serum (Figures S4B, S4C, S4F, and S4G). This likely accounts for the discordance between their nanomolar binding affinities in vitro, and their marginal ability to generate p53-dependent cytotoxic responses. ATSP-7041 did antagonize p53-Mdm2 association, but surprisingly had little effect on p53-Mdm4 complexes. However, high concentrations and long times were required for any effects to occur. We suggest that this relates to the ability of ATSP to disrupt membranes, which is partly mitigated by serum, and its strong affinity for Mdm2 enables it to accumulate to a sufficient intracellular concentration for target disruption, though less efficiently and more slowly than Nutlin-3a.
It has been observed that serum decreases the biological activity of SAH peptides (Edwards et al., 2013; Chang et al., 2013; Brown et al., 2013). Our in vitro analyses demonstrated that serum does not prevent SAH peptides from disrupting either p53-Mdm2 or p53-Mdm4 complexes. The live cell analyses showed that all wild type SAH peptides tested possessed the unexpected ability to elicit p53-independent membrane damage that correlated with cytotoxicity. Adding serum prevented both membrane damage and cytotoxicity. Surprisingly, mutations that replaced an essential phenylalanine with alanine in the α-helical region of each peptide abrogated membrane permeabilization and cytotoxicity. We infer that this derives from the ability of these mutations to alter the hydrophobicity and α-helicity of the peptides (Bernal et al., 2007). Previously, the cytotoxicity of the wild type SAH peptides was interpreted to derive from p53-dependent activity since mutant control peptides did not exert this effect (Bernal et al., 2010; Chang et al., 2013; Brown et al., 2013). However, our data show that the lack of effect in these mutant peptides is actually due to their inability to permeabilize membranes. Our data are consistent with a recent microscopic study showing that a fluorescent FAM conjugated mutant (F19A) ATSP peptide exhibited limited cellular permeability compared to the apparent cellular distribution of the wild type ATSP peptide (Fig. 3 in (Chang et al., 2013)). This agrees with our conclusion that wild type SAH peptides access the cytoplasm after first compromising membrane integrity. Taken together, the data show that the observed cytotoxicity of the wild type SAH peptide does not completely depend on functional p53, and the absence of activity in the mutant peptide is more related to its biophysical properties than to its inability to interact with Mdm2 and Mdm4.
We speculate that membrane disturbance may commonly result from positively charged cell-penetrating peptides (CPPs) appended to peptides with exposed hydrophobic residues. For example, the stapled PMI-PenArg (a lysine to arginine derivative of the CPP penetratin) elicited cell death within 3 hours in cells growing in serum free media (data not shown). Similarly, the cationic cell-penetrating D-peptide DPMI-γ-DR9 rapidly induced p53-independent cytotoxicity (Liu et al., 2010). The positively charged CPPs from N-terminal prion proteins also elicited membrane leakage in defined large unilamellar phospholipid vesicles (Magzoub et al., 2005). It is also worth noting that the unstapled DPMI-γDR9 (Liu et al., 2010), TAT-PMI, and PMI-s-s-TAT peptides induce severe membrane damage (Figure 7) and cytotoxicity, indicating that chemical stapling per se is not required for cytotoxicity (Okamoto et al., 2014).
The ReBiL strategy creates a facile platform system for PPI analyses. We have shown that this system can detect interactions between Ube2t-FANCL (Figure 2), p53-Mdm2, p53-Mdm4 (Figure 3), Mdm2-Mdm4 RING domains (Figure S6) and BRCA1-BARD1 RING domains (Figures 5C and S7), among other proteins (Y.-C. Li and G.M. Wahl, unpublished data). Furthermore, the very high signal-to-noise ratio in the lysate format enabled ReBiL to be used for high throughput drug screens in a 1,536-well format with Z’ values exceeding 0.7 (Y.-C. Li, G.M. Wahl, and K.F. Wertman, unpublished data). We expect ReBiL to have broad applications ranging from identification of noncoding RNAs that facilitate cytosolic PPIs to factors that impact plasma membrane-associated KRAS dimerization, neither of which is feasible using other strategies such as the two/three-hybrid systems with transcriptional readouts. Together, these attributes should enable ReBiL to broaden our understanding of the impact of disease relevant mutations on protein interactions, to elucidate more precisely mechanisms of drug action, to improve efficacy of PPI antagonists, and to advance our understanding of the makeup of the human protein interactome.
EXPERIMENTAL PROCEDURES
Construction of ReBiL targeting plasmids
We used standard molecular biology methods and the Gibson Assembly strategy (NEB E2611S) to construct all ReBiL targeting plasmids. See Table S2 for details.
Real-time BiLC assay in living cells
Phenol-red free DMEM/F12 (Life Technology No. 11039-021 or Sigma D2906-1L) containing 2x concentrated reagents including doxycycline (Sigma D9891) and D-luciferin (potassium salt; Biosynth L-8220) were pipetted into 384-well plate (Corning 3570); 20 μl per well. The cells were trypsinized and cell numbers were determined. The numbers of required cells were collected into 1.5 ml LoBind tubes (Eppendorf No. 022431081) and spun at 200 rcf for 5 min at RT. Cells were resuspened with DMEM/F12 (phenol-red free) and 20 μl cells were pipetted into each well. The final concentration of each component is as follows: FBS 10%, Ciprofloxacin 10 μg/ml, doxycycline 0~500 ng/ml, D-luciferin 100~200 μM, 5,000~20,000 cells per well. The plate was sealed with a MicroAmp Optical Adhesive Film (Life Technology No. 4311971), and luminescence was read in a Tecan M200: integration time 2 sec, 10~30 min per cycle for a total of 24~48 hr at indicated temperature.
Doxycycline withdrawal strategy to enable real-time BiLC analysis protein complex dissociation
(A) The following protocol was used to evaluate small molecule PPI antagonists. The ReBiL cells were cultured in 10–15 cm dishes with regular media containing doxycycline (500 ng/ml) and D-luciferin (100 μM) for 24 hr. The next day, cells were trypsinized, and cell numbers were determined. Cells were seeded into assay plates as described above except the doxycycline was eliminated from the media. (B) To evaluate SAH peptides, the cells were seeded into 96-well plate (Corning 3917) with 20,000 cells per well, and incubated in the presence of doxycycline (500 ng/ml) and D-luciferin (100 μM) for 24 hr. The next day, the media were aspirated; cells were washed once with DMEM, and 50 μl of DMEM/F12 (phenol-red free) media containing D-luciferin (100 μM) and the stapled peptides at indicated concentrations were added into each well. The plate was sealed with a MicroAmp Optical Adhesive Film, and luminescence was read in a Tecan M200: integration time 2 sec, 5~10 min per cycle for total 6 hr at 37 °C.
Cell viability assay
Luminescence-based cell viability assay was performed using CellTiter Glo (measures the amount of ATP produced by viable cells, Promega G7572) according to the manufacturer’s protocol. Luminescence was detected in a Tecan M200 with integration time 0.5 second. Trypsinized cells were treated with Trypsin inhibitor (Sigma T6522) before seeded into serum-free media for viability assay.
BiLC assay using cell lysates
The Saos-2 or U2OS ReBiL cells were cultured in regular media with doxycycline (500 ng/ml, 48~72 hr). The 4x concentrated drugs diluted in DMEM/F12 media were pipetted into 384 well plates, 10 μl per well. Cells with loaded BiLC complexes were washed and lysed with PLB buffer (100 mM Tris-HCl pH 7.5, 0.5 mM EDTA, 150 mM NaCl, 0.1% Triton X-100, 1 mM sodium orthovanadate, 50 mM sodium fluoride, and Roche Complete Mini Protease Inhibitor Cocktail). The cell lysates were transferred into 1.5 ml tubes and cleared by centrifugation (13,000 rcf, 5 min at 4 °C). The clear lysates were collected, diluted with DMEM (~300 μl DMEM added into 100 μl lysates); 10 μl of diluted lysates were immediately pipetted into each of the 384-wells containing drugs, and the plates were incubated at room temperature for 10 minutes. 20 μl of luciferin reagents (Promega Bright-Glo E2620 or Steady-Glo E2520) were added into each well and luminescence was read in a Tecan M200: integration time 0.5 sec, 3~5 min per cycle for 30 min at 26 °C.
Lactate dehydrogenase (LDH) leakage assay
LDH leakage was detected by the CytoTox 96 Non-Radioactive Cytotoxicity Assay Kit (Promega G1780) per manufacture’s protocol. For details, see Supplemental Procedures.
Supplementary Material
Figure S1. ReBiL system enables generation of isogenic wild type and mutant reporter cells, Related to Figure 1
Single copy of ReBiL targeting cassette encoding wild type or mutant reporter construct respectively can be inserted into a pre-selected chromosomal locus. The isogenic wild type and mutant ReBiL cells enhance the accuracy and reproducibility of structure, function, and interaction analyses.
Figure S2. Detection of low-affinity Ube2t and FANCL interaction with ReBiL at 30 °C, Related to Figure 2
(A) Schematic diagram of Ube2t-FANCL (pLi505) and Ube2t-FANCL_C307A (pLi506) ReBiL targeting cassettes (see Table S2 for details). (B) ReBiL system detected higher Ube2t and FANCL BiLC signals and significantly greater signal-to-noise ratio at 30 °C. Luminescent signals in ReBiL and randomly integrated reporter cells in 384-well plates were read every 10 minutes for 24 hours using a Tecan-M200 at 30 °C. Data shown are mean ± SEM from 4 independent experiments. (C) Western blot analysis of BiLC fusion proteins at 30 °C. The nLuc-Ube2t was detected by anti-HA antibody. The cLuc-FANCL_WT and cLuc-FANCL_C307A were detected by anti-FLAG antibody. Actin was used as a loading control. (D) The expressed nLuc-Ube2t in ReBiL cells was less than endogenous Ube2t at 30 °C. They were assessed by anti-Ube2t antibody (Cell Signaling D2L7H). Actin was a loading control.
Figure S3. ReBiL analysis of Nutlin-3a, SJ-172550 and RO-5963 in living cells, Related to Figure 3
(A) Schematic diagram of the p53-Mdm2 (pLi385) and p53-Mdm4 (pLi354) ReBiL targeting cassettes (Table S2). (B) Nutlin-3a disrupts preformed p53-Mdm2 BiLC complexes. Saos-2 p53-Mdm2 ReBiL cells were seeded in 96-well plate (20,000 cells per well). The p53-Mdm2 BiLC fusion proteins were induced by doxycycline (500 ng/ml for 24 hours). Doxycycline media were aspirated. Cells were washed with DMEM/F12 and treated with new media containing Nutlin-3a and D-luciferin with or without cycloheximide (50 μg/ml) at Time = 0. The p53-Mdm2 BiLC signals were detected using a Tecan luminometer M200 at 37 °C. Integration time 0.8 sec, 1 minute per cycle for 40 minutes. Data shown are mean ± SEM from 3 independent experiments. (C) Western blot analysis showed that Nutlin-3a did not promote nLuc-HA-p53 (detected by anti-HA antibody) and cLuc-FLAG-Mdm2 (detected by anti-FLAG antibody) degradation at 30 min. Actin was used as a loading control. (D) Nutlin-3a has no effect on preformed p53-Mdm4 BiLC complexes. Saos-2 p53-Mdm4 ReBiL cells were treated exactly as in (B). (E) SJ-172550 lacks p53-Mdm4 disruption activity in living cells. Saos-2 p53-Mdm4 ReBiL cells were treated with SJ-172550 in a 384-well plate (12,000 cells per well) for 48 hours at 37 °C with 500 ng/ml doxycycline and 100 μM D-luciferin. (F) The schematic diagram shows the Mdm4-Mdm2 N-terminal ReBiL targeting cassette in plasmid pLi544 (Table S2). (G) RO-5963 enhances Mdm2-Mdm4 association. U2OS Mdm4_111 - Mdm2_108 ReBiL cells (U2OS 134–544) in 384-well plate (5,000 cells per well) were treated with 500 ng/ml doxycycline and 100 μM D-luciferin plus RO-5963 or Nutlin-3a for 24 hours at 37 °C. (H) RO-5963 moderately reduces p53-Mdm2 interaction but not p53-Mdm4 interaction. A doxycycline withdrawal ReBiL experiment was performed exactly as in Figure 3C.
Figure S4. Effect of SAH peptides on the stability of intracellular p53-Mdm2 and p53-Mdm4 complexes, Related to Figure 4
The assay conditions are described in the Figure 4. Saos-2 ReBiL reporter cells in 10% FBS media were treated with (A) ATSP-7342 (F19A mutant stapled peptide), (B) SAHp53-8, (C) sMTid-02, and (D) sMTid-02-ctrl. Saos-2 ReBiL reporter cells in 0% FBS media were treated with (E) ATSP-7342, (F) SAHp53-8, (G) sMTid-02, and (H) sMTid-02-ctrl.
Figure S5. Serum inhibits cytotoxicity induced by SAHp53-8, Related to Figure 5
SJSA-1 cells in a 384-well plate were treated with SAHp53-8 and SAHp53-8_F19A in the presence or absence of 10% FBS and incubated for 24 hours. Trypsinized SJSA-1 cells were treated with Trypsin inhibitor (Sigma T6522, per manufacture’s recommendation) before seeded into serum-free media in a 384-well plate. Cell viability was measured using CellTiter Glo.
Figure S6. Derivation of optimal lysis buffers for using BiLC in cell free extracts and analyses of compounds reported to disrupt p53-Mdm2 and/or p53-Mdm4 interactions using the BiLC lysate assay, Related to Figure 6
We identified an optimized lysis buffer for the BiLC lysate assay by measuring the signal-to-noise ratio of a set of validated positive and negative BiLC PPI pairs. The interaction between the Mdm4_RING and Mdm2_RING (Tanimura et al., 1999) serves as the positive signal and any detectable interactions between Mdm4_RING domain and Mdm2_RING_C464A are the negative control or background noise because this cysteine to alanine mutation collapses the RING domain structure and prevents interaction with the Mdm4_RING domain (Kostic et al., 2006). The U2OS ReBiL cells carrying (Mdm4_RING)-(Mdm2_RING) (U2OS 134-283) and (Mdm4_RING)-(Mdm2_RING_C464A) (U2OS 134-285) were induced by 500 ng/ml doxycycline for 24 hours. Cells were washed with PBS- and lysed with following three lysis buffers: (1) CA-630 Lysis Buffer (CLB): 50 mM Tris-HCl pH8.0, 5 mM EDTA, 150 mM NaCl, 0.5% CA-630, 1 mM sodium orthovanadate, 50 mM sodium fluoride, and Complete Mini Protease Inhibitor Cocktail (Roche). (2) PPI Lysis Buffer (PLB): 100 mM Tris-HCl pH 7.5, 0.5 mM EDTA, 150 mM NaCl, 0.1% Triton X-100, 1 mM sodium orthovanadate, 50 mM sodium fluoride, and Complete Mini Protease Inhibitor Cocktail (Roche). (3) Proprietary Promega Glo Lysis Buffer E2661 (GLB). Cell lysates were transferred to microcentrifuge tubes and cleared by centrifugation (13,000 rcf for 3 ~ 5 minutes at 4 °C). The cleared lysates (20 μl) and luciferin reagent (20 μl) (Promega Bright-Glo E2620 or Steady-Glo E2520) were pipetted into each well of a 384-well plate (Corning 3570). The plate was incubated at 26 °C for 15 min and luminesces were read in a Tecan M200 luminometer with 0.5 second integration time at 26 °C. (A) Schematic diagram of (Mdm4_RING)-(Mdm2_RING) (pLi283) and (Mdm4_RING)-(Mdm2_RING_C464A) (pLi285) ReBiL targeting cassettes (see Table S2). (B) Specific BiLC signals (Mdm4_RING)-(Mdm2_RING) and background BiLC noise (Mdm4_RING)-(Mdm2_RING_C464A) were detected in three different lysis buffers. (C) The signal to noise ratios (indicated on the top of the bar graph) in three different lysis buffers were calculated as signal luminescence divided by noise luminescence. (D) The western blot showed that the amounts of nLuc-HA-Mdm4_RING, cLuc-HA-Mdm2_RING and cLuc-HA-Mdm2_RING-C464A BiLC fusion proteins (detected by an anti-HA antibody) were very similar across all three lysis buffers. Actin was used as a loading control. (E) The p53-Mdm2 and p53-Mdm4 BiLC lysate assays were performed exactly like Figure 6. Small molecule PPI antagonists were tested in p53-Mdm2 and p53-Mdm4 BiLC lysate assays without FBS. (F) The potency of PMI-derived PPI antagonists (summarized in Table S3) were validated in p53-Mdm2 and p53-Mdm4 BiLC lysate assays in 0% FBS (top panel) and 10% FBS (bottom panel). Data are normalized to the luminescent reading of DMSO (set to 100%).
Figure S7. The BRCA1-BARD1 BiLC pair served as a counter assay for non-specific luciferase inhibition, Related to Figures 5, 6, and S6
The interaction surfaces of BRCA1-BARD1 RING domain (Brzovic et al., 2001) are structurally different from the interaction surfaces of the p53-Mdm2 (Kussie et al., 1996) and p53-Mdm4 (Popowicz et al., 2007). Thus, the only common component among these three BiLC pairs is the split luciferase portions. If a PPI antagonist is able to reduce luminescent signals in all three BiLC pairs, it must be a split luciferase inhibitor that targets the split luciferase portion. (A) The schematic diagram of BRCA1-BARD1 RING domain ReBiL targeting cassette in plasmid pLi367 (Table S2). (B) The BRCA1-BARD1 lysate BiLC assay in the absence (top panel) or presence (bottom panel) of 10% FBS. Data are normalized to the luminescent reading of DMSO (set to 100%).
Table S1. Composition and features of RMCE cassette components and reporter cell lines, Related to Figure 1
Table S2. The detailed descriptions of ReBiL targeting plasmids, Related to Experimental Procedures
Table S3. The p53-Mdm2 and p53-Mdm4 antagonists derived from PMI peptide, Related to Figures 7, S6 and S7
Acknowledgments
This work was supported by the US National Institutes of Health (R01- CA61449 and R03- MH089489- 01), Cancer Center Support Grant CA014195, Salk Innovation Grant, a Sanofi sponsored research grant, and the Leona M. and Harry B. Helmsley Charitable Trust grant #2012-PG-MED002 and Susan G. Komen for the Cure grant SAC110036. We thank M. Dyer for providing p53_R273H construct, R. Klevit for providing BRCA1, BARD1 and Ube2t constructs, A. Dutta for providing FANCL_WT and FANCL_C307A constructs, C. Brown and D. Lane for providing sMTide-02 and sMTide-02_ctrl, F. Bernal for providing SAHp53-8 and SAHp53_F19A, S. Dowdy for providing disulfide-linked PMI-s-s-TAT peptide and insightful discussions, and D. Piwnica-Worms for discussions of the BiLC assay. We thank A. Saghatelian, T. Hunter, B. Eisenman, R. Sobol, M. Ridinger, B. Spike and M. Wade for comments on the manuscript. We also want to thank L. DiTacchio, P. Rychestsky, members in Sanofi Tucson Innovation Center and members in the Wahl lab for discussions and technical assistances.
Footnotes
Supplemental information includes seven figures, three tables, and Supplemental Experimental Procedures can be found with this article online at http://
AUTHOR CONTRIBUTIONS
Y.-C.L. and G.M.W. conceived the study and designed experiments. Y-C.L. and L.W.R performed experiments. C.H., E.T.W., S.L., P.S., M.P., L.W., and K.F.W. contributed reagents. Y.-C.L. and G.M.W. analyzed data and wrote the paper with input from all authors.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Beckerman R, Prives C. Transcriptional regulation by p53. Cold Spring Harb Perspect Biol. 2010;2:a000935. doi: 10.1101/cshperspect.a000935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernal F, Tyler AF, Korsmeyer SJ, Walensky LD, Verdine GL. Reactivation of the p53 tumor suppressor pathway by a stapled p53 peptide. J Am Chem Soc. 2007;129:2456–2457. doi: 10.1021/ja0693587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernal F, Wade M, Godes M, Davis TN, Whitehead DG, Kung AL, Wahl GM, Walensky LD. A stapled p53 helix overcomes HDMX-mediated suppression of p53. Cancer Cell. 2010;18:411–422. doi: 10.1016/j.ccr.2010.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird GH, Gavathiotis E, Labelle JL, Katz SG, Walensky LD. Distinct BimBH3 (BimSAHB) stapled peptides for structural and cellular studies. ACS Chem Biol. 2014;9:831–837. doi: 10.1021/cb4003305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown CJ, Lain S, Verma CS, Fersht AR, Lane DP. Awakening guardian angels: drugging the p53 pathway. Nat Rev Cancer. 2009;9:862–873. doi: 10.1038/nrc2763. [DOI] [PubMed] [Google Scholar]
- Brown CJ, Quah ST, Jong J, Goh AM, Chiam PC, Khoo KH, Choong ML, Lee MA, Yurlova L, Zolghadr K, et al. Stapled peptides with improved potency and specificity that activate p53. ACS Chem Biol. 2013;8:506–512. doi: 10.1021/cb3005148. [DOI] [PubMed] [Google Scholar]
- Brzovic PS, Rajagopal P, Hoyt DW, King MC, Klevit RE. Structure of a BRCA1-BARD1 heterodimeric RING-RING complex. Nat Struct Biol. 2001;8:833–837. doi: 10.1038/nsb1001-833. [DOI] [PubMed] [Google Scholar]
- Cassonnet P, Rolloy C, Neveu G, Vidalain PO, Chantier T, Pellet J, Jones L, Muller M, Demeret C, Gaud G, et al. Benchmarking a luciferase complementation assay for detecting protein complexes. Nat Methods. 2011;8:990–992. doi: 10.1038/nmeth.1773. [DOI] [PubMed] [Google Scholar]
- Chang YS, Graves B, Guerlavais V, Tovar C, Packman K, To KH, Olson KA, Kesavan K, Gangurde P, Mukherjee A, et al. Stapled alpha-helical peptide drug development: A potent dual inhibitor of MDM2 and MDMX for p53-dependent cancer therapy. Proc Natl Acad Sci USA. 2013;110:E3445–E3454. doi: 10.1073/pnas.1303002110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshaies RJ, Joazeiro CA. RING domain E3 ubiquitin ligases. Annu Rev Biochem. 2009;78:399–434. doi: 10.1146/annurev.biochem.78.101807.093809. [DOI] [PubMed] [Google Scholar]
- Edwards AL, Gavathiotis E, LaBelle JL, Braun CR, Opoku-Nsiah KA, Bird GH, Walensky LD. Multimodal interaction with BCL-2 family proteins underlies the proapoptotic activity of PUMA BH3. Chem Biol. 2013;20:888–902. doi: 10.1016/j.chembiol.2013.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gembarska A, Luciani F, Fedele C, Russell EA, Dewaele M, Villar S, Zwolinska A, Haupt S, de Lange J, Yip D, et al. MDM4 is a key therapeutic target in cutaneous melanoma. Nat Med. 2012;18:1239–1247. doi: 10.1038/nm.2863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilad Y, Shiloh R, Ber Y, Bialik S, Kimchi A. Discovering protein-protein interactions within the programmed cell death network using a protein-fragment complementation screen. Cell Rep. 2014;8:909–921. doi: 10.1016/j.celrep.2014.06.049. [DOI] [PubMed] [Google Scholar]
- Graves B, Thompson T, Xia M, Janson C, Lukacs C, Deo D, Di Lello P, Fry D, Garvie C, Huang KS, et al. Activation of the p53 pathway by small-molecule-induced MDM2 and MDMX dimerization. Proc Natl Acad Sci USA. 2012;109:11788–11793. doi: 10.1073/pnas.1203789109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green JL, Bauer M, Yum KW, Li YC, Cox ML, Willert K, Wahl GM. Use of a molecular genetic platform technology to produce human Wnt proteins reveals distinct local and distal signaling abilities. PLoS ONE. 2013;8:e58395. doi: 10.1371/journal.pone.0058395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herce HD, Deng W, Helma J, Leonhardt H, Cardoso MC. Visualization and targeted disruption of protein interactions in living cells. Nat Commun. 2013;4:2660. doi: 10.1038/ncomms3660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodson C, Cole AR, Lewis LP, Miles JA, Purkiss A, Walden H. Structural analysis of human FANCL, the E3 ligase in the Fanconi anemia pathway. J Biol Chem. 2011;286:32628–32637. doi: 10.1074/jbc.M111.244632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodson C, Purkiss A, Miles JA, Walden H. Structure of the human FANCL RING-Ube2T complex reveals determinants of cognate E3-E2 selection. Structure. 2014;22:337–344. doi: 10.1016/j.str.2013.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ilagan MX, Lim S, Fulbright M, Piwnica-Worms D, Kopan R. Real-time imaging of notch activation with a luciferase complementation-based reporter. Sci Signal. 2011;4:rs7. doi: 10.1126/scisignal.2001656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov AA, Khuri FR, Fu H. Targeting protein-protein interactions as an anticancer strategy. Trends Pharmacol Sci. 2013;34:393–400. doi: 10.1016/j.tips.2013.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joerger AC, Fersht AR. Structural biology of the tumor suppressor p53. Annu Rev Biochem. 2008;77:557–582. doi: 10.1146/annurev.biochem.77.060806.091238. [DOI] [PubMed] [Google Scholar]
- Lee JH, Zhang Q, Jo S, Chai SC, Oh M, Im W, Lu H, Lim HS. Novel pyrrolopyrimidine-based alpha-helix mimetics: cell-permeable inhibitors of protein-protein interactions. J Am Chem Soc. 2011;133:676–679. doi: 10.1021/ja108230s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy ED, Kowarzyk J, Michnick SW. High-resolution mapping of protein concentration reveals principles of proteome architecture and adaptation. Cell Rep. 2014;7:1333–1340. doi: 10.1016/j.celrep.2014.04.009. [DOI] [PubMed] [Google Scholar]
- Liu M, Li C, Pazgier M, Li C, Mao Y, Lv Y, Gu B, Wei G, Yuan W, Zhan C, et al. D-peptide inhibitors of the p53-MDM2 interaction for targeted molecular therapy of malignant neoplasms. Proc Natl Acad Sci USA. 2010;107:14321–14326. doi: 10.1073/pnas.1008930107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luker KE, Smith MC, Luker GD, Gammon ST, Piwnica-Worms H, Piwnica-Worms D. Kinetics of regulated protein-protein interactions revealed with firefly luciferase complementation imaging in cells and living animals. Proc Natl Acad Sci USA. 2004;101:12288–12293. doi: 10.1073/pnas.0404041101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macdonald-Obermann JL, Piwnica-Worms D, Pike LJ. Mechanics of EGF receptor/ErbB2 kinase activation revealed by luciferase fragment complementation imaging. Proc Natl Acad Sci USA. 2012;109:137–142. doi: 10.1073/pnas.1111316109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machida YJ, Machida Y, Chen Y, Gurtan AM, Kupfer GM, D’Andrea AD, Dutta A. UBE2T is the E2 in the Fanconi anemia pathway and undergoes negative autoregulation. Mol Cell. 2006;23:589–596. doi: 10.1016/j.molcel.2006.06.024. [DOI] [PubMed] [Google Scholar]
- Magzoub M, Oglecka K, Pramanik A, Goran Eriksson LE, Graslund A. Membrane perturbation effects of peptides derived from the N-termini of unprocessed prion proteins. Biochim Biophys Acta. 2005;1716:126–136. doi: 10.1016/j.bbamem.2005.09.009. [DOI] [PubMed] [Google Scholar]
- Michnick SW, Ear PH, Manderson EN, Remy I, Stefan E. Universal strategies in research and drug discovery based on protein-fragment complementation assays. Nat Rev Drug Discov. 2007;6:569–582. doi: 10.1038/nrd2311. [DOI] [PubMed] [Google Scholar]
- Moldovan GL, D’Andrea AD. How the Fanconi anemia pathway guards the genome. Annu Rev Genet. 2009;43:223–249. doi: 10.1146/annurev-genet-102108-134222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto T, Segal D, Zobel K, Fedorova A, Yang H, Fairbrother WJ, Huang DC, Smith BJ, Deshayes K, Czabotar PE. Further insights into the effects of pre-organizing the BimBH3 helix. ACS Chem Biol. 2014;9:838–839. doi: 10.1021/cb400638p. [DOI] [PubMed] [Google Scholar]
- Okamoto T, Zobel K, Fedorova A, Quan C, Yang H, Fairbrother WJ, Huang DC, Smith BJ, Deshayes K, Czabotar PE. Stabilizing the pro-apoptotic BimBH3 helix (BimSAHB) does not necessarily enhance affinity or biological activity. ACS Chem Biol. 2013;8:297–302. doi: 10.1021/cb3005403. [DOI] [PubMed] [Google Scholar]
- Patton JT, Mayo LD, Singhi AD, Gudkov AV, Stark GR, Jackson MW. Levels of HdmX expression dictate the sensitivity of normal and transformed cells to Nutlin-3. Cancer Res. 2006;66:3169–3176. doi: 10.1158/0008-5472.CAN-05-3832. [DOI] [PubMed] [Google Scholar]
- Paulmurugan R, Gambhir SS. Combinatorial library screening for developing an improved split-firefly luciferase fragment-assisted complementation system for studying protein-protein interactions. Anal Chem. 2007;79:2346–2353. doi: 10.1021/ac062053q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pazgier M, Liu M, Zou G, Yuan W, Li C, Li C, Li J, Monbo J, Zella D, Tarasov SG, et al. Structural basis for high-affinity peptide inhibition of p53 interactions with MDM2 and MDMX. Proc Natl Acad Sci USA. 2009;106:4665–4670. doi: 10.1073/pnas.0900947106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed D, Shen Y, Shelat AA, Arnold LA, Ferreira AM, Zhu F, Mills N, Smithson DC, Regni CA, Bashford D, et al. Identification and characterization of the first small molecule inhibitor of MDMX. J Biol Chem. 2010;285:10786–10796. doi: 10.1074/jbc.M109.056747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi FM, Kringstein AM, Spicher A, Guicherit OM, Blau HM. Transcriptional control: rheostat converted to on/off switch. Mol Cell. 2000;6:723–728. doi: 10.1016/s1097-2765(00)00070-8. [DOI] [PubMed] [Google Scholar]
- Soderberg O, Gullberg M, Jarvius M, Ridderstrale K, Leuchowius KJ, Jarvius J, Wester K, Hydbring P, Bahram F, Larsson LG, et al. Direct observation of individual endogenous protein complexes in situ by proximity ligation. Nat Methods. 2006;3:995–1000. doi: 10.1038/nmeth947. [DOI] [PubMed] [Google Scholar]
- Stumpf MP, Thorne T, de Silva E, Stewart R, An HJ, Lappe M, Wiuf C. Estimating the size of the human interactome. Proc Natl Acad Sci USA. 2008;105:6959–6964. doi: 10.1073/pnas.0708078105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vassilev LT, Vu BT, Graves B, Carvajal D, Podlaski F, Filipovic Z, Kong N, Kammlott U, Lukacs C, Klein C, et al. In vivo activation of the p53 pathway by small-molecule antagonists of MDM2. Science. 2004;303:844–848. doi: 10.1126/science.1092472. [DOI] [PubMed] [Google Scholar]
- Venkatesan K, Rual JF, Vazquez A, Stelzl U, Lemmens I, Hirozane-Kishikawa T, Hao T, Zenkner M, Xin X, Goh KI, et al. An empirical framework for binary interactome mapping. Nat Methods. 2009;6:83–90. doi: 10.1038/nmeth.1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdine GL, Hilinski GJ. Stapled peptides for intracellular drug targets. Methods Enzymol. 2012;503:3–33. doi: 10.1016/B978-0-12-396962-0.00001-X. [DOI] [PubMed] [Google Scholar]
- Wade M, Li YC, Wahl GM. MDM2, MDMX and p53 in oncogenesis and cancer therapy. Nat Rev Cancer. 2013;13:83–96. doi: 10.1038/nrc3430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wade M, Rodewald LW, Espinosa JM, Wahl GM. BH3 activation blocks Hdmx suppression of apoptosis and cooperates with Nutlin to induce cell death. Cell Cycle. 2008;7:1973–1982. doi: 10.4161/cc.7.13.6072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wadia JS, Stan RV, Dowdy SF. Transducible TAT-HA fusogenic peptide enhances escape of TAT-fusion proteins after lipid raft macropinocytosis. Nat Med. 2004;10:310–315. doi: 10.1038/nm996. [DOI] [PubMed] [Google Scholar]
- Wells JA, McClendon CL. Reaching for high-hanging fruit in drug discovery at protein-protein interfaces. Nature. 2007;450:1001–1009. doi: 10.1038/nature06526. [DOI] [PubMed] [Google Scholar]
- Wong ET, Kolman JL, Li YC, Mesner LD, Hillen W, Berens C, Wahl GM. Reproducible doxycycline-inducible transgene expression at specific loci generated by Cre-recombinase mediated cassette exchange. Nucleic Acids Res. 2005;33:e147. doi: 10.1093/nar/gni145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang KS, Ilagan MX, Piwnica-Worms D, Pike LJ. Luciferase fragment complementation imaging of conformational changes in the epidermal growth factor receptor. J Biol Chem. 2009;284:7474–7482. doi: 10.1074/jbc.M808041200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Y, Rape M. Building ubiquitin chains: E2 enzymes at work. Nat Rev Mol Cell Biol. 2009;10:755–764. doi: 10.1038/nrm2780. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. ReBiL system enables generation of isogenic wild type and mutant reporter cells, Related to Figure 1
Single copy of ReBiL targeting cassette encoding wild type or mutant reporter construct respectively can be inserted into a pre-selected chromosomal locus. The isogenic wild type and mutant ReBiL cells enhance the accuracy and reproducibility of structure, function, and interaction analyses.
Figure S2. Detection of low-affinity Ube2t and FANCL interaction with ReBiL at 30 °C, Related to Figure 2
(A) Schematic diagram of Ube2t-FANCL (pLi505) and Ube2t-FANCL_C307A (pLi506) ReBiL targeting cassettes (see Table S2 for details). (B) ReBiL system detected higher Ube2t and FANCL BiLC signals and significantly greater signal-to-noise ratio at 30 °C. Luminescent signals in ReBiL and randomly integrated reporter cells in 384-well plates were read every 10 minutes for 24 hours using a Tecan-M200 at 30 °C. Data shown are mean ± SEM from 4 independent experiments. (C) Western blot analysis of BiLC fusion proteins at 30 °C. The nLuc-Ube2t was detected by anti-HA antibody. The cLuc-FANCL_WT and cLuc-FANCL_C307A were detected by anti-FLAG antibody. Actin was used as a loading control. (D) The expressed nLuc-Ube2t in ReBiL cells was less than endogenous Ube2t at 30 °C. They were assessed by anti-Ube2t antibody (Cell Signaling D2L7H). Actin was a loading control.
Figure S3. ReBiL analysis of Nutlin-3a, SJ-172550 and RO-5963 in living cells, Related to Figure 3
(A) Schematic diagram of the p53-Mdm2 (pLi385) and p53-Mdm4 (pLi354) ReBiL targeting cassettes (Table S2). (B) Nutlin-3a disrupts preformed p53-Mdm2 BiLC complexes. Saos-2 p53-Mdm2 ReBiL cells were seeded in 96-well plate (20,000 cells per well). The p53-Mdm2 BiLC fusion proteins were induced by doxycycline (500 ng/ml for 24 hours). Doxycycline media were aspirated. Cells were washed with DMEM/F12 and treated with new media containing Nutlin-3a and D-luciferin with or without cycloheximide (50 μg/ml) at Time = 0. The p53-Mdm2 BiLC signals were detected using a Tecan luminometer M200 at 37 °C. Integration time 0.8 sec, 1 minute per cycle for 40 minutes. Data shown are mean ± SEM from 3 independent experiments. (C) Western blot analysis showed that Nutlin-3a did not promote nLuc-HA-p53 (detected by anti-HA antibody) and cLuc-FLAG-Mdm2 (detected by anti-FLAG antibody) degradation at 30 min. Actin was used as a loading control. (D) Nutlin-3a has no effect on preformed p53-Mdm4 BiLC complexes. Saos-2 p53-Mdm4 ReBiL cells were treated exactly as in (B). (E) SJ-172550 lacks p53-Mdm4 disruption activity in living cells. Saos-2 p53-Mdm4 ReBiL cells were treated with SJ-172550 in a 384-well plate (12,000 cells per well) for 48 hours at 37 °C with 500 ng/ml doxycycline and 100 μM D-luciferin. (F) The schematic diagram shows the Mdm4-Mdm2 N-terminal ReBiL targeting cassette in plasmid pLi544 (Table S2). (G) RO-5963 enhances Mdm2-Mdm4 association. U2OS Mdm4_111 - Mdm2_108 ReBiL cells (U2OS 134–544) in 384-well plate (5,000 cells per well) were treated with 500 ng/ml doxycycline and 100 μM D-luciferin plus RO-5963 or Nutlin-3a for 24 hours at 37 °C. (H) RO-5963 moderately reduces p53-Mdm2 interaction but not p53-Mdm4 interaction. A doxycycline withdrawal ReBiL experiment was performed exactly as in Figure 3C.
Figure S4. Effect of SAH peptides on the stability of intracellular p53-Mdm2 and p53-Mdm4 complexes, Related to Figure 4
The assay conditions are described in the Figure 4. Saos-2 ReBiL reporter cells in 10% FBS media were treated with (A) ATSP-7342 (F19A mutant stapled peptide), (B) SAHp53-8, (C) sMTid-02, and (D) sMTid-02-ctrl. Saos-2 ReBiL reporter cells in 0% FBS media were treated with (E) ATSP-7342, (F) SAHp53-8, (G) sMTid-02, and (H) sMTid-02-ctrl.
Figure S5. Serum inhibits cytotoxicity induced by SAHp53-8, Related to Figure 5
SJSA-1 cells in a 384-well plate were treated with SAHp53-8 and SAHp53-8_F19A in the presence or absence of 10% FBS and incubated for 24 hours. Trypsinized SJSA-1 cells were treated with Trypsin inhibitor (Sigma T6522, per manufacture’s recommendation) before seeded into serum-free media in a 384-well plate. Cell viability was measured using CellTiter Glo.
Figure S6. Derivation of optimal lysis buffers for using BiLC in cell free extracts and analyses of compounds reported to disrupt p53-Mdm2 and/or p53-Mdm4 interactions using the BiLC lysate assay, Related to Figure 6
We identified an optimized lysis buffer for the BiLC lysate assay by measuring the signal-to-noise ratio of a set of validated positive and negative BiLC PPI pairs. The interaction between the Mdm4_RING and Mdm2_RING (Tanimura et al., 1999) serves as the positive signal and any detectable interactions between Mdm4_RING domain and Mdm2_RING_C464A are the negative control or background noise because this cysteine to alanine mutation collapses the RING domain structure and prevents interaction with the Mdm4_RING domain (Kostic et al., 2006). The U2OS ReBiL cells carrying (Mdm4_RING)-(Mdm2_RING) (U2OS 134-283) and (Mdm4_RING)-(Mdm2_RING_C464A) (U2OS 134-285) were induced by 500 ng/ml doxycycline for 24 hours. Cells were washed with PBS- and lysed with following three lysis buffers: (1) CA-630 Lysis Buffer (CLB): 50 mM Tris-HCl pH8.0, 5 mM EDTA, 150 mM NaCl, 0.5% CA-630, 1 mM sodium orthovanadate, 50 mM sodium fluoride, and Complete Mini Protease Inhibitor Cocktail (Roche). (2) PPI Lysis Buffer (PLB): 100 mM Tris-HCl pH 7.5, 0.5 mM EDTA, 150 mM NaCl, 0.1% Triton X-100, 1 mM sodium orthovanadate, 50 mM sodium fluoride, and Complete Mini Protease Inhibitor Cocktail (Roche). (3) Proprietary Promega Glo Lysis Buffer E2661 (GLB). Cell lysates were transferred to microcentrifuge tubes and cleared by centrifugation (13,000 rcf for 3 ~ 5 minutes at 4 °C). The cleared lysates (20 μl) and luciferin reagent (20 μl) (Promega Bright-Glo E2620 or Steady-Glo E2520) were pipetted into each well of a 384-well plate (Corning 3570). The plate was incubated at 26 °C for 15 min and luminesces were read in a Tecan M200 luminometer with 0.5 second integration time at 26 °C. (A) Schematic diagram of (Mdm4_RING)-(Mdm2_RING) (pLi283) and (Mdm4_RING)-(Mdm2_RING_C464A) (pLi285) ReBiL targeting cassettes (see Table S2). (B) Specific BiLC signals (Mdm4_RING)-(Mdm2_RING) and background BiLC noise (Mdm4_RING)-(Mdm2_RING_C464A) were detected in three different lysis buffers. (C) The signal to noise ratios (indicated on the top of the bar graph) in three different lysis buffers were calculated as signal luminescence divided by noise luminescence. (D) The western blot showed that the amounts of nLuc-HA-Mdm4_RING, cLuc-HA-Mdm2_RING and cLuc-HA-Mdm2_RING-C464A BiLC fusion proteins (detected by an anti-HA antibody) were very similar across all three lysis buffers. Actin was used as a loading control. (E) The p53-Mdm2 and p53-Mdm4 BiLC lysate assays were performed exactly like Figure 6. Small molecule PPI antagonists were tested in p53-Mdm2 and p53-Mdm4 BiLC lysate assays without FBS. (F) The potency of PMI-derived PPI antagonists (summarized in Table S3) were validated in p53-Mdm2 and p53-Mdm4 BiLC lysate assays in 0% FBS (top panel) and 10% FBS (bottom panel). Data are normalized to the luminescent reading of DMSO (set to 100%).
Figure S7. The BRCA1-BARD1 BiLC pair served as a counter assay for non-specific luciferase inhibition, Related to Figures 5, 6, and S6
The interaction surfaces of BRCA1-BARD1 RING domain (Brzovic et al., 2001) are structurally different from the interaction surfaces of the p53-Mdm2 (Kussie et al., 1996) and p53-Mdm4 (Popowicz et al., 2007). Thus, the only common component among these three BiLC pairs is the split luciferase portions. If a PPI antagonist is able to reduce luminescent signals in all three BiLC pairs, it must be a split luciferase inhibitor that targets the split luciferase portion. (A) The schematic diagram of BRCA1-BARD1 RING domain ReBiL targeting cassette in plasmid pLi367 (Table S2). (B) The BRCA1-BARD1 lysate BiLC assay in the absence (top panel) or presence (bottom panel) of 10% FBS. Data are normalized to the luminescent reading of DMSO (set to 100%).
Table S1. Composition and features of RMCE cassette components and reporter cell lines, Related to Figure 1
Table S2. The detailed descriptions of ReBiL targeting plasmids, Related to Experimental Procedures
Table S3. The p53-Mdm2 and p53-Mdm4 antagonists derived from PMI peptide, Related to Figures 7, S6 and S7