Abstract
Nonviral gene therapy has high potential for safely promoting tissue restoration and for treating various genetic diseases. One current limitation is that conventional transfection reagents such as polyethylenimine (PEI) form electrostatically stabilized plasmid DNA (pDNA) polyplexes with poor colloidal stability. In this study, a library of poly(ethylene glycol-b-(dimethylaminoethyl methacrylate-co-butyl methacrylate)) [poly(EG-b-(DMAEMA-co-BMA))] polymers were synthesized and screened for improved colloidal stability and nucleic acid transfection following lyophilization. When added to pDNA in the appropriate pH buffer, the DMAEMA moieties initiate formation of electrostatic polyplexes that are internally stabilized by hydrophobic interactions of the core BMA blocks and sterically stabilized against aggregation by a PEG corona. The BMA content was varied from 0% to 60% in the second polymer block in order to optimally tune the balance of electrostatic and hydrophobic interactions in the polyplex core, and polymers with 40 and 50 mol% BMA achieved the highest transfection efficiency. Diblock copolymers were more stable than PEI in physiologic buffers. Consequently, diblock copolymer polyplexes aggregated more slowly and followed a reaction-limited colloidal aggregation model, while fast aggregation of PEI polyplexes was governed by a diffusion-limited model. Polymers with 40% BMA did not aggregate significantly after lyophilization and produced up to 20-fold higher transfection efficiency than PEI polyplexes both before and after lyophilization. Furthermore, poly(EG-b-(DMAEMA-co-BMA)) polyplexes exhibited pH-dependent membrane disruption in a red blood cell hemolysis assay and endosomal escape as observed by confocal microscopy.Lyophilized polyplexes made with the lead candidate diblock copolymer (40% BMA) also successfully transfected cells in vitro following incorporation into gas-foamed polymeric scaffolds. In summary, the enhanced colloidal stability, endosomal escape, and resultant high transfection efficiency of poly(EG-b-(DMAEMA-co-BMA))-pDNA polyplexes underscores their potential utility both for local delivery from scaffolds as well as systemic, intravenous delivery.
Introduction
Nonviral gene therapy has potential for use in accelerating restoration of tissue defects and treatment of a myriad of diseases. Plasmid DNA (pDNA) production is efficient and relatively inexpensive, and DNA therapy avoids the immunogenic risk associated with viral vectors. Naked pDNA uptake and utilization is very inefficient; however, synthetic polymer- and lipid-based carriers face several in vivo challenges, and there has been limited translation of efficient and nontoxic nonviral options for pDNA delivery. The endocytotic entry of pDNA is aided by condensation into stable nanoparticles. Ideally, the pDNA nanocarriers should be stabilized to minimize aggregation in physiological conditions (i.e., presence of proteins and salts) and should protect the plasmid cargo from nuclease degradation in the extracellular environment. After endocytosis, the vectors must escape the endo-lysosomal pathways to avoid degradation or exocytosis, and the plasmid must be unpackaged and trafficked to the nucleus.
Electrostatic condensation of plasmids into nanoparticles using cationic polymers or lipids is a promising approach for overcoming in vitro barriers to nonviral gene therapy. Electrostatic interactions between positively charged amine groups on polymers such as polyethylenimine (PEI) or poly(2-(dimethylamino) ethyl methacrylate) (PDMAEMA) and negatively charged phosphates on DNA result in the condensation of the pDNA into polyplexes (50 – 200 nm nanoparticles).1, 2 After entering cells through endocytosis, polyplexes made from amine-containing polymers with pKa in the range 5.0 – 7.4 are presumed to buffer the acidification of the vesicles of the endo-lysosomal trafficking pathways. This “proton sponge” behavior increases proton and counterion influx and causes osmotic swelling and rupture of endosomes, enabling pDNA cytoplasmic entry.3, 4 While direct injection of pDNA/PEI polyplexes has been used successfully in vivo for some tissue types 5-7, effective delivery of polyplexes from tissue-engineered scaffolds has been challenging due to aggregation of the polyplex nanoparticles.8, 9 Recently, lyophilization of PEI-pDNA polyplexes with excipients such as sucrose has reduced polyplex aggregation and increased transfection efficiency.10, 11 However, approaches to improve the inherent stability of the DNA/polymer polyplexes and studies on the contribution of colloidal stability to transfection efficiency have not been extensively investigated.
Block copolymers are a promising strategy to improve colloidal stability, increase transfection efficiency, and decrease cytotoxicity of nonviral carriers. Complexation of pDNA with block copolymers comprising polycations and polyethylene glycol (PEG) has been reported to enhance steric stability of the resulting polyplexes by formation of a PEG corona.12-14 Titration of hydrophobic content into the cationic, pDNA-condensing polymer block has also been investigated as a strategy for reducing charge density, increasing stability, decreasing toxicity, and enhancing endosomal escape of polycations. For example, delivery of the pro-apoptotic TRAIL gene using terpolymers synthesized by enzyme-catalyzed copolymerization of lactone with dialkyl diester and amino diol significantly inhibited tumor growth in a xenograft model.15 The high molecular weight and increased hydrophobicity were conjectured to compensate for the low charge density of the nanoparticles, resulting in low cytotoxicity and high transfection efficiency. Similarly, increasing hydrophobicity and decreasing charge density through the modification of poly(amido amines) with benzoyl groups was found to increase polyplex stability, decrease cytotoxicity, and increase transfection efficiency.16, 17 In other studies, the ratio of cationic DMAEMA to hydrophobic butyl methacrylate (BMA) or propylacrylic acid (PPAA) has been optimized to achieve pH-dependent membrane disruptive properties ideally tuned for endosomal escape.18-20 Recently, we were the first to investigate a PEG-stabilized polyplex system in which siRNA cargo was loaded into the particle core, which contained a balance of BMA and DMAEMA, and yielded enhanced performance following intravenous injection in vivo.21 Here, we extend this work and apply simple, controlled polymerization methods to synthesize polymers in a single step that can be used to form pDNA carriers that provide steric stabilization by incorporating a PEG corona as well as an optimal balance of cationic and hydrophobic content to allow improved stability, reduced toxicity, and endosomal escape. The current study also explores structure-property relationships governing the effects of poly(DMAEMA-co-BMA) composition and molecular weight on polyplex stabilization and performance.
In this study, a library of poly(ethylene glycol-b-(dimethylaminoethyl methacrylate-co-butyl methacrylate)) [poly(EG-b-(DMAEMA-co-BMA))] polymers was synthesized using a one-step reversible-addition fragmentation chain transfer (RAFT) polymerization from a PEG-based macro-chain transfer agent (macro-CTA). These polymers were designed such that DMAEMA initiates nucleic acid electrostatic interactions and triggers formation of polyplexes that are further stabilized by the hydrophobic interactions of the BMA in the polyplex core. The PEG corona was designed to shield the polyplex charge in the core and to provide enhanced steric stabilization while the poly(DMAEMA-co-BMA) block was designed to achieve pH-dependent membrane disruption tuned to promote endosomal escape. The controlled nature of RAFT22 was leveraged to synthesize a well-defined library of polymers containing a range of compositions with varied mol% BMA titrated into the cationic DMAEMA block. We also used controlled synthesis to make monodispersed polymers with varied molecular weights to enable study of the effect of molecular weight of the cationic block on performance of polymers with optimal [DMAEMA]/[BMA] ratios. The block copolymers were characterized for pDNA packaging efficiency, physicochemical properties of the polyplexes, colloidal stability, and in vitro transfection bioactivity before and after lyophilization. Finally, pDNA polyplexes were prepared from the lead-candidate block copolymer and incorporated into reactive, gas-foamed scaffolds in a proof-of-concept experiment demonstrating scaffold-mediated plasmid delivery in vitro.
Materials and Methods
Materials
Luciferase reporter plasmid (pPK-CMV-R3) and PromoFluor-500 fluorescent labeling kit were purchased from Promokine (Heidelberg, Germany). Live/dead viability/cytotoxicity kit, Opti-MEM reduced serum media, Dulbecco's phosphate buffered saline (DPBS), Dulbecco's Modified Eagle Medium (DMEM), fetal bovine serum (FBS), penicillin streptomycin, and 0.4% trypan blue stain were purchased from Invitrogen (Grand Island, NY). Pierce™ BCA Protein Assay Kit was purchased from Thermo Fisher Scientific (Waltham, MA). MDA-MB-231 breast cancer cell line was obtained from ATCC (Manassas, VA). NAP-5 desalting columns were purchased from GE. Lysine triisocyanate-poly(ethylene glycol) (LTI-PEG) prepolymer was obtained from Ricerca (Concord, OH). TEGOAMIN33, a tertiary amine catalyst composed of 33 wt% triethylene diamine (TEDA) in dipropylene glycol, was received from Goldschmidt (Hopewell, VA). All other reagents were purchased from Sigma Aldrich (St. Louis, MO).
Poly(EG-b-(DMAEMA-co-BMA)) Synthesis and Characterization
RAFT polymerization was used to synthesize a library of diblock copolymers. The chain transfer agent (CTA) 4-Cyano-4-(ethylsulfanylthiocarbonyl) sulfanylvpentanoic acid was synthesized following standard procedures23 and was subsequently conjugated to 5 kDa mono-methoxy-PEG using DCC and DMAP, resulting in 91% substitution of the PEG.21, 23 The polymerization reaction was carried out at 70°C for 24 h using azobisisobutyronitrile as the initiator with a 5:1 [CTA]:[initiator] molar ratio. A series of polymerizations were carried out with monomer feed ratios of 0, 25, 40, 50, or 60 mol% BMA and 100, 75, 60, 50, or 40 mol% DMAEMA. For polymers with short block length, the degree of polymerization was 150, and the polymerization time was 6 h. For 0, 25, 40, and 60% BMA with long block length and 50% BMA with medium block length, the degree of polymerization was 150, and the polymerization time was 24 h. For 50% BMA with long block length, the degree of polymerization was 200, and the polymerization time was 24 h. All polymerizations were carried out with 40% wt/vol of monomer + CTA in dioxane. The reaction was stopped by exposing the polymerization solution to air, and the resulting diblock polymers were precipitated into an excess of pentane. The isolated polymers were vacuum-dried, re-dissolved in water, further purified using PD10 columns, and lyophilized. Polymers were characterized for composition and molecular weight by 1H nuclear magnetic resonance spectroscopy (NMR, Bruker 400Mhz Spectrometer equipped with 9.4 Tesla Oxford magnet). Absolute molecular weight of the polymers was determined using DMF mobile phase gel permeation chromatography (GPC, Agilent Technologies, Santa Clara, CA, USA) with inline Agilent refractive index and Wyatt miniDAWN TREOS light scattering detectors (Wyatt Technology Corp., Santa Barabara, CA).
Polymer-pDNA Polyplex Formation
Prior to mixing, both pDNA and poly(EG-b-(DMAEMA-co-BMA)) polymers were diluted in 100 mM citric acid/sodium citrate buffer solution (pH 4). Polyplexes were formed by mixing equal volumes of pDNA and polymer solutions by pipetting. After incubating the polyplexes 15 min at room temperature, sodium carbonate/bicarbonate buffer (pH 10.8) was added to bring the pH to 7.4. The concentration of pDNA in the final solution was 25 μg/ml, and the concentration of the polymer solution was dependent on the desired amine/phosphate (N/P) ratio (1, 2, 5, 10, 20, or 30). The N/P ratio was defined as the ratio of the total amines in the polymer to the total phosphates in pDNA.
To make control polyplexes, PEI (25,000 Da, branched) and pDNA were separately diluted in equal volumes of phosphate buffer (pH 7.4). Polyplexes were formed by mixing the PEI and pDNA solutions by pipetting. The mixture was incubated 15 min before adding to cells. The concentration of pDNA in the final solution was 25 μg/ml, and the concentration of the polymer solution was dependent on the desired amine/phosphate (N/P) ratio (1, 2, 5, 10, 20, or 30). Particle size, transfection efficiency, and cellular uptake were measured for fresh polyplexes (both poly(EG-b-(DMAEMA-co-BMA) and PEI groups) immediately after preparation without lyophilization.
Polymer-pDNA PolyplexLyophilization
Polymer-pDNA polyplexes were formed as described above. Prior to lyophilization, trehalose was added as a cryprotectant and/or excipient. For polyplex experiments, the mass ratio of trehalose:pDNA was 200:1, while for the scaffold-based transfection experiments, the weight ratio of trehalose:pDNA was varied from 50 – 200:1. Polyplexes were frozen at a rate of -1°C/min until the temperature reached -45°C. Polyplexes were lyophilized overnight with a vacuum of 0.045 mbar and a collector temperature of -45°C. For particle size measurement by DLS and cell transfection experiments, lyophilized polyplexes were reconstituted in water and incubated for 15 min prior to use.
Agarose Gel Electrophoresis
To determine the ability of the polymers to efficiently package pDNA, agarose gel electrophoresis was performed. Fresh polymer-pDNA polyplexes were formed as described above. Samples containing 300 ng pDNA were loaded onto a 0.8% agarose gel and subjected to electrophoresis at 100 V for 45 min. Ethidium bromide was then added to the gel to visualize pDNA.
In Vitro Transfection
MDA-MB-231 human breast cancer cells or immortalized murine dermal fibroblasts (IMDF) were plated in complete DMEM (10% FBS, 100 U/ml penicillin, and 100 μg/ml streptomycin) at a density of 20,000 cells/well in 96-well plates 24 hours prior to transfection. The MDA-MB-231 breast cancer cell line was used in initial experiments to study transfection efficiency of fresh and lyophilized polyplexes in the potential context of cancer therapy. Dermal fibroblasts were used in experiments involving pDNA delivery from scaffolds because the scaffolds are designed for use in skin wound healing. Immediately before transfection, media were aspirated and replaced with 100 μl Opti-MEM + 2% FBS. The serum concentration was 2% FBS for the transfection experiments because low serum conditions increases cell uptake and minimizes the introduction of experimental variability due to rapid cell proliferation 24. Fresh or reconstituted lyophilized polyplexes containing 150 ng pDNA were then added to each well. At 24 h after transfection, transfection media were aspirated and replaced with 100 μl of 1 mg/ml luciferin in complete DMEM. Luminescence was measured using a Xenogen IVIS 200 bioluminescence imaging system (Perkin Elmer, Waltham, MA) after luciferin was added. Relative luminescence in each well was quantified using Living Image™ software. To normalize luciferase signal to total protein content, cells were lysed using RIPA buffer and total protein measured using the Pierce™ BCA Protein Assay Kit according to the manufacturer's instructions.
pH-dependent Red Blood Cell Hemolysis Assay
To screen for endosomolytic activity, a red blood cell hemolysis assay was used to measure the pH-dependent membrane disruptive activity of the polyplexes.25 Fresh polyplexes with N/P ratio of 10 were prepared as described above. Human red blood cells were incubated with polyplexes for one hour in buffers with pH 7.4, 6.8, 6.2, or 5.6 to mimic different stages in the endo-lysosomal pathway. After centrifugation to remove intact cells, the absorbance of the supernatant was measured at 405 nm to determine the amount of hemoglobin released. The absorbance of supernatant from untreated cells was subtracted, and the percent red blood cell disruption was normalized to positive control samples lysed with Triton X-100.
Confocal Microscopy
Colocalization of plasmid and endosomes was analyzed using confocal microscopy. Plasmid DNA was fluorescently labeled using a PromoFluor-500 labeling kit according to the manufacturer's instructions (PromoKine). MDA-MB-231 cells were plated in complete DMEM at a density of 10,000 cells/well in 8-well chamber slides 24 h prior to transfection. Immediately before transfection, media were aspirated and replaced with Opti-MEM + 2% FBS. Fresh polyplexes containing 150 ng pDNA were then added to each well. At 24 h after transfection, media were aspirated and replaced with complete DMEM + 75 nM LysoTracker Red probe + 1 μg/ml Hoechst stain. After 30 min incubation, cells were imaged with a fluorescent confocal microscope (Zeiss LSM 710 Moieta Oberkochen, Germany) to determine distribution of plasmid and endosomes within the cells. ImageJ software was used to analyze colocalization of plasmid and endosomes.
Dynamic Light Scattering (DLS)
Polyplexes were formed as described above and diluted to a concentration of 1.5 μg/ml pDNA in DPBS or KCl solutions. Polyplex size was measured for fresh and lyophilized polyplexes using a Zetasizer Nano ZS (Malvern Instruments, Worchestershire, UK). DLS was performed with a wavelength of 633 nm using a 4.0 mW Helium-Neon laser at a backscattering angle of 173°. Polyplex size was determined from the average of at least 10 runs of 10 seconds each. For the aggregation study, polyplexes were incubated at room temperature and analyzed using DLS at various time points up to 72 h. To assess stability in the presence of serum, polyplexes were diluted to a concentration of 15 μg/ml pDNA in Opti-MEM + 2% or 10% FBS. Size was measured by DLS, and the peak representing serum components (∼15 nm) was omitted in the analysis. Data were fitted to diffusion-limited or reaction-limited models of colloidal aggregation. For ζ-potential measurements, polyplexes were diluted in 1 mM KCl at pH 7 and the ζ-potential calculated from the average of at least 10 runs using a universal dip cell.
Cell Viability
The cytotoxicity of lyophilized polyplexes was determined using calcein AM staining to detect live cells. In vitro transfection experiments were carried out as described above. 24 h after transfection, media were aspirated and replaced with 200 μl of a 1 μg/ml calcein AM solution in DPBS. After incubating 30 min at 37 °C, fluorescence intensity was measured using an FL600 microplate reader with an excitation wavelength of 485 nm and an emission wavelength of 530 nm (Bio-Tek Instruments, Winooski, Vermont).
In Vitro Transfection on Scaffolds
40L Polyplexes were desalted using GE NAP-5 columns according to the manufacturer's instructions (Fairfield, CT) and lyophilized with 50:1, 100:1, or 200:1 Trehalose:pDNA. Polyurethane (PUR) scaffolds were synthesized by reacting a polyester triol (900 Da) with a backbone comprising 60% caprolactone, 30% glycolide, and 10% lactide, 2.7 parts per hundred parts polyol (pphp) TEGOAMIN33 catalyst, 10 pphp water, 2 pphp calcium stearate, lyophilized polyplexes, and LTI-PEG prepolymer (index = 115). All reactants were sterilized prior to scaffold formation. After curing, scaffolds were cut into 8×2 mm pieces each containing either 2.5 or 5 μg pDNA and placed in untreated 24-well plates. 200,000 IMDF cells were seeded on each scaffold and incubated in 1 ml Opti-MEM + 2% FBS. After 24 h, media were replaced with Opti-MEM + 2% FBS + 1 mg/ml luciferin, and bioluminescence was measured using an IVIS (Perkin Elmer).
Statistical Analysis
One way analysis of variance (ANOVA) was used to evaluate the statistical significance of results. All p-values less than 0.05 were considered statistically significant.
Results
Poly(EG-b-(DMAEMA-co-BMA)) Characterization
A library of diblock copolymers with varying compositions and molecular weights were synthesized. Results of polymer characterization using GPC and 1H-NMR are shown in Table 1. The abbreviated names of the polymers indicate the mol% BMA in the DMAEMA-co-BMA block and the relative length of the DMAEMA-co-BMA block (short [S], medium [M], or long [L]). PEG block molecular weight was held constant at 5000 Da.
Table 1. Block lengths and composition of poly(EG-b-(DMAEMA-co-BMA)) polymers.
| Polymer Name | PEG (Da) | DMAEMA-co-BMA (Da) | % BMA in DMAEMA-co-BMA block | PDI |
|---|---|---|---|---|
| 0S | 5000 | 13,743 | 0.0% | 1.05 |
| 0L | 5000 | 17,035 | 0.0% | 1.09 |
| 25S | 5000 | 13,128 | 24.5% | 1.05 |
| 25L | 5000 | 18,747 | 23.8% | 1.08 |
| 40S | 5000 | 12,428 | 39.3% | 1.08 |
| 40L | 5000 | 20,765 | 39.6% | 1.12 |
| 50S | 5000 | 13,683 | 48.5% | 1.05 |
| 50M | 5000 | 18,041 | 48.3% | 1.04 |
| 50L | 5000 | 22,857 | 49.7% | 1.16 |
| 60S | 5000 | 8550 | 58.6% | 1.13 |
| 60L | 5000 | 19,939 | 58.6% | 1.08 |
Agarose Gel Electrophoresis
Agarose gel electrophoresis was performed to assess the ability of the transfection reagents to package pDNA, as shown in Figure 1.
Figure 1. Agarose gel electrophoresis of fresh polymer-pDNA polyplexes.
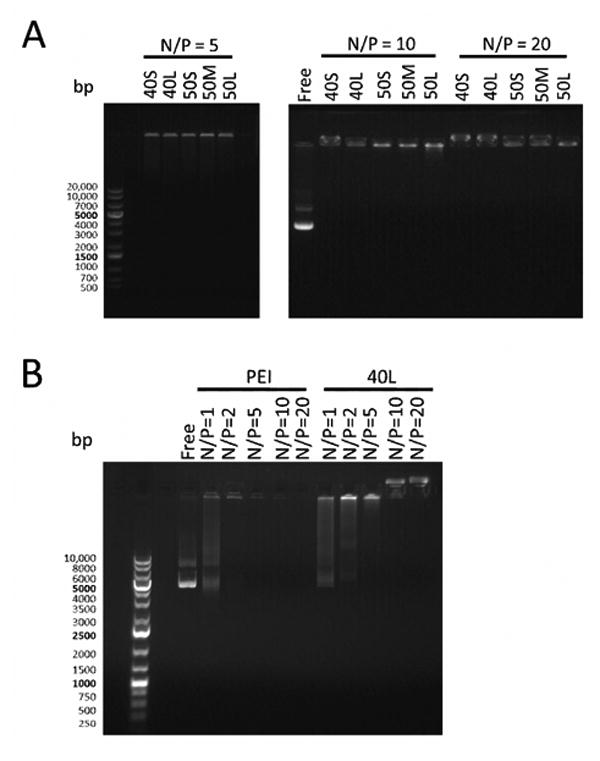
Polyplexes were electrophoresed on a 0.8% agarose gel. Free pDNA and a DNA ladder were also included on the gel.(A) Poly(EG-b-(DMAEMA-co-BMA)) polyplexes with N/P ratios of 5, 10, or 20. pDNA is visible in the loading wells in the lanes containing polyplexes, and no free pDNA is visible in these lanes. (B) 40L and PEI polyplexes with varying N/P ratios. Free pDNA is visible lanes containing 40L polyplexes with N/P ratios of 1 and 2 and PEI polyplexes with N/P ratio of 1.
Poly(EG-b-(DMAEMA-co-BMA))-pDNA polyplexes with N/P ratios of 5, 10, and 20 did not migrate out of the loading wells, and no free pDNA was detected at any of the N/P ratios investigated (Figure 1A). 40L polyplex formation was further investigated at lower N/P ratios (Figure 1B). Some free pDNA was detected at N/P ratios of 1 and 2 for 40L polyplexes and N/P ratio of 1 for PEI polyplexes. No free pDNA was visible at N/P ratios of 5 or above for 40L polyplexes and 2 or above for PEI polyplexes. These results indicate that poly(EG-b-(DMAEMA-co-BMA)) polymers efficiently encapsulate pDNA at N/P ratios of 5 or above.
Fresh Polyplex Transfection
The effects of BMA content (0 – 60% BMA) and poly(DMAEMA-co-BMA) block length (short or long) on transfection efficiency were assessed by delivering a luciferase reporter plasmid to MDA-MB-231 cells using freshly made polyplexes. As shown in Figure 2, transfection efficiency was maximized at 40% BMA, and polymers with 40 or 50 mol% BMA had significantly higher transfection than all other diblock copolymers. Furthermore, increasing the DMAEMA-co-BMA block length increased the transfection efficiency at each mol% BMA. 40L polyplexes had significantly higher transfection than PEI polyplexes. Due to their superior transfection efficiency, polymers with 40 and 50 mol% BMA were the focus of subsequent experiments.
Figure 2. Transfection efficiency of fresh polymer-pDNA polyplexes.
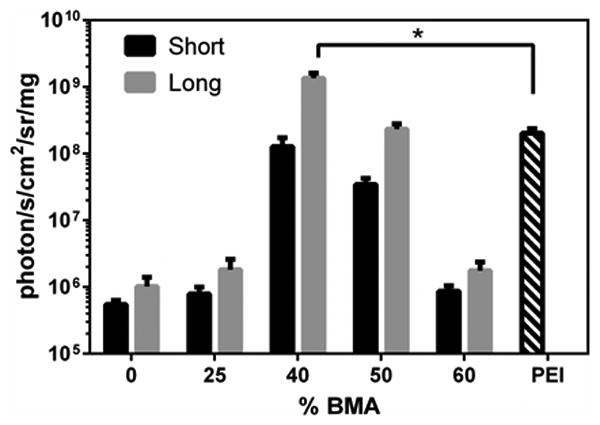
Luminescence produced by MDA-MB-231 cells transfected with fresh polyplexes containing luciferase pDNA with N/P ratio of 10 (normalized to total protein). Asterisk indicates significant difference (p < 0.05). Data are plotted as mean ± standard deviation.
pH-dependent Membrane Disruption and Endosomal Escape
A red blood cell hemolysis assay was performed to determine pH-dependent membrane disruption by fresh polyplexes with N/P ratio of 10. As shown in Figure 3A, all polyplexes had <5% hemolysis at pH 7.4 (mimicking extracellular and cytosolic pH), which is important for minimizing polyplex cytotoxicity. As pH decreased, hemolytic behavior of the polyplexes increased significantly. All polyplexes produced 100% hemolysis (statistically equivalent to Triton X detergent) at pH 5.6 (mimicking late endosomes), suggesting that the polyplexes can achieve efficient endosomal escape. These data agree with hemolysis profiles of a related family of polymers formulated to deliver siRNA 21.
Figure 3. Endosomal escape and pH-dependent membrane disruption of fresh polyplexes.
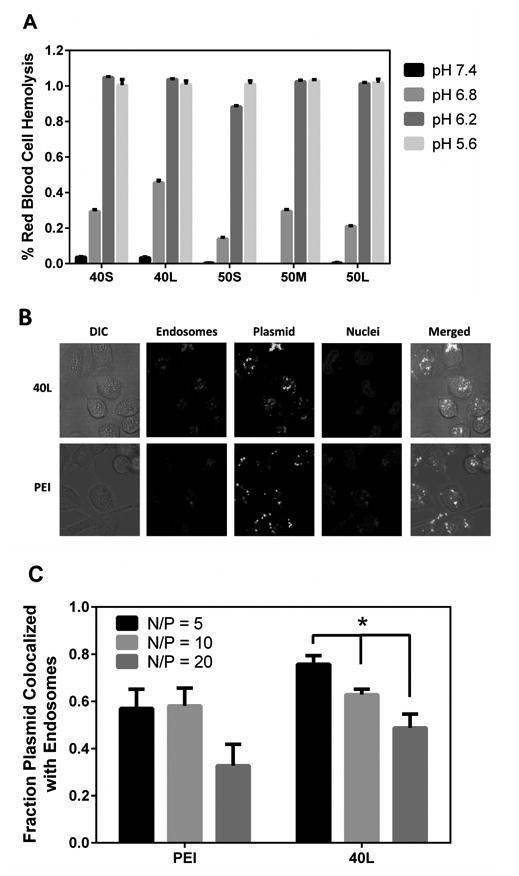
(A) pH-dependent red blood cell hemolysis assay. All polyplexes investigated exhibited pH-dependent membrane disruption, displaying minimal hemolysis at physiologic pH and switch-like transition into a membrane disruptive confirmation at endo-lysosomal pHs. Data are plotted as mean ± standard error. (B) Confocal microscopy images of MDA-MB-231 cells transfected with fresh 40L or PEI polyplexes with N/P ratio of 10 showing distribution of plasmid (green), endosomes (red), and nuclei (blue). (C) Percentage of plasmid colocalized with Lysotracker dye. Data are plotted as mean ± standard error, and the asterisk indicates statistically significant differences (p < 0.05).
The ability of the diblock copolymers to aid in endosomal escape was further investigated using confocal microscopy. Images of cells transfected with fresh 40L or PEI polyplexes (Figure 3B) were analyzed to quantify the colocalization of green (plasmid) and red (endosomes) fluorescent probes (Figure 3C). As the N/P ratio of 40L increased from 5 to 20, the fraction of green colocalized with red decreased significantly (p < 0.05). Furthermore, there were no significant differences in plasmid-endosome colocalization between 40L and PEI polyplexes. These results provide evidence that in this setting, the active, pH-dependent membrane disruptive mechanism of 40L is concentration-dependent and provides a means to escape endo-lysosomal compartments.
Polyplex Size and ζ-potential
The transfection and endosomal escape data (Figures 1 – 3) show that fresh 40L and 50L polyplexes are comparable to or more effective than PEI polyplexes. To evaluate their colloidal stability, the size of polyplexes with N/P = 10 was measured in DPBS before (fresh) and after lyophilization (Figure 4). Before lyophilization, the diameter of poly(EG-b-(DMAEMA-co-BMA)) polyplexes ranged from 130 – 180 nm. BMA content and block length did not significantly affect initial polyplex size. After lyophilization, the 40% BMA polyplexes showed a trend toward increased size, but the differences were not statistically significant (p > 0.05). The 50% BMA showed a modest but statistically significant increase in size(p<0.05). In contrast, PEI polyplexes significantly increased in size to 400% of the fresh polyplex size. In addition, the effect of N/P ratio on stability after lyophilization was investigated (data not shown). N/P ratio had no effect on polyplex size prior to lyophilization, but increasing the N/P ratio increased the stability of lyophilized polyplexes, leading to smaller increases in diameter after lyophilization.
Figure 4. Effect of lyophilization on the size of polymer-pDNA polyplexes measured by DLS.
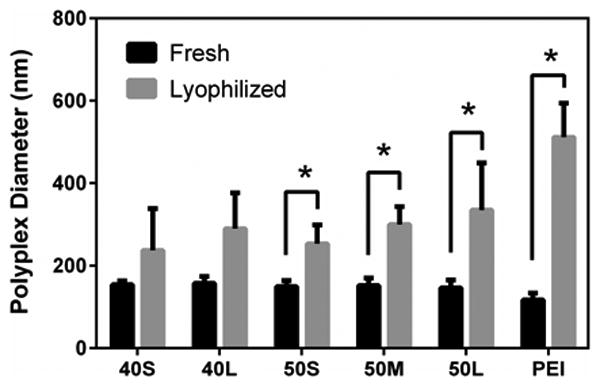
Polyplexes were diluted to a concentration of 1.5 μg/ml pDNA in DPBS. Hydrodynamic diameter of polyplexes with N/P ratio of 10 before and after lyophilization and subsequent reconstitution. Asterisks indicate significant difference (p < 0.05). Data are plotted as means ± standard deviation.
Since 40L was identified as the lead candidate for in vitro transfection and showed no significant change in polyplex size pre- versus post-lyophilization, it was the focus of subsequent investigations. As shown in Figure 5, the ζ-potential of fresh 40L and PEI polyplexes measured in 1 mM KCl at pH 7 increased with N/P ratio. At N/P ratios ≥ 5, the ζ-potential of 40L polyplexes approached 0 mV. Even at higher N/P of 20 and 30, the ζ-potential of 40L polyplexes was neutral due to shielding of the excess cationic charge in the polyplex core by the PEG corona. In contrast, the ζ-potential of PEI polyplexes increased up to an N/P ratio of 20, at which point it asymptotically approached +20 mV.
Figure 5. ζ-potential of fresh polymer-pDNA complexes measured by DLS.
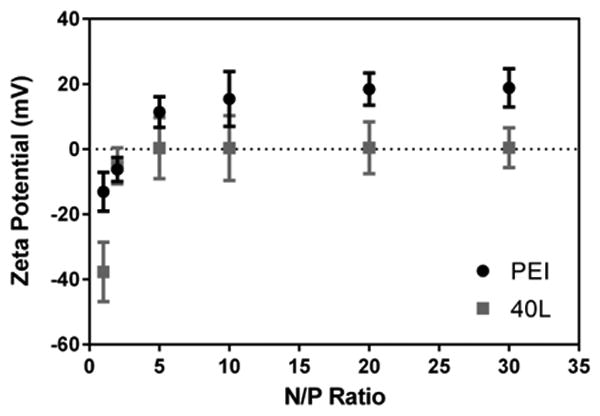
ζ-potential was measured in 1 mM KCl pH 7.0. ζ-potential of 40L and PEI polyplexes increased with N/P ratio. ζ-potential of 40L-pDNA polyplexes approached ∼0 mV for N/P of 5 or greater while ζ-potential of PEI-pDNA polyplexes approached ∼20 mV for N/P of 20 or greater. Data are plotted as mean ± standard deviation.
Aggregation Studies and Modeling Analysis
The increase in particle size observed after lyophilization is indicative of inadequate colloidal stability, prompting us to perform aggregation studies. Due to the presence of the PEG corona and a combination of electrostatic and hydrophobic interactions to stabilize the core, the poly(EG-b-(DMAEMA-co-BMA)) polyplexes were expected to show increased collodal stability compared to PEI polyplexes. The interaction potential between PEI polyplexes was calculated using Derjaguin, Landau, Verwey, and Overbeek (DLVO) theory on the stability of colloidal suspensions. Summing the short-range van der Waals attraction and the long-range electrostatic repulsion forces (shown in the Supplemental Material) revealed no potential maximum for PEI polyplexes, which points to the absence of an energy barrier and consequent rapid (i.e., diffusion-controlled) flocculation (stability ratio W ≈ 1). In contrast, poly(EG-b-(DMAEMA-co-BMA)) polyplexes were stabilized by the PEG corona, resulting in an interfacial layer ∼6 nm thick (calculations shown in the Supplemental Material). Thus, the relatively short-range van der Waals forces are screened by the long-range steric repulsion, resulting in a high stability ratio (W≫ 1) and consequent slow (i.e., reaction-limited) flocculation.
To quantify fresh poly(EG-b-(DMAEMA-co-BMA)) polyplex stability, the aggregation kinetics were assessed in pH 7.4 DPBS at N/P ratios of 5 (Figure 6A) and 10 (Figure 6B). The aggregation kinetics of fresh PEI polyplexes are shown in Figure 6C. PEI polyplexes exhibited faster aggregation than poly(EG-b-(DMAEMA-co-BMA)) polyplexes, forming aggregates >900 nm in less than 5 hours. For all polymers, polyplexes with N/P ratio of 5 were less stable than those with N/P ratio of 10. 40L polyplexes with N/P ratio of 10 exhibited the greatest stability, remaining <200 nm after more than 60 hours in DPBS. These data suggest that the PEI polyplexes are stabilized by electrostatic repulsive forces, which are screened at high electrolyte concentrations, while the 40L polyplexes are sterically stabilized.
Figure 6. Aggregation kinetics of polyplexes in the presence of buffer salts, KCl, and serum.
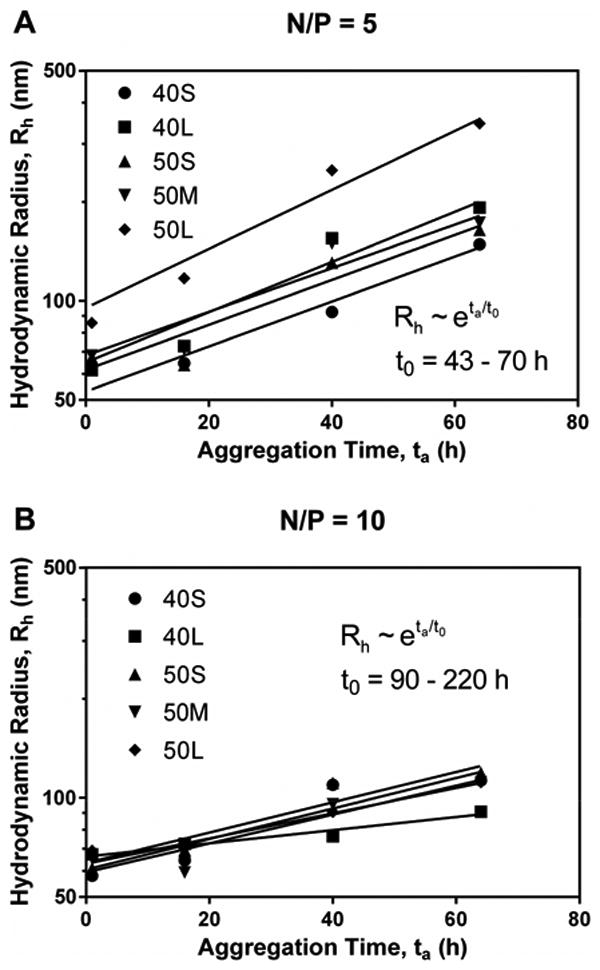
Panels A and B illustrate aggregation of fresh polyplexes with N/P ratio of 5 (A) and 10 (B) in DPBS over time. Data for diblock copolymers were fit to the reaction-limited colloidal aggregation model. (C) Data for PEI in DPBS were fit to the diffusion-limited colloidal aggregation model. Panels D and E show effects of ionic strength of KCl on aggregation of fresh 40L (D) and PEI (E) polyplexes. PEI polyplexes aggregated more quickly in solutions with higher ionic strength, while the size of 40L polyplexes was not affected by ionic strength. Data are plotted as mean ± standard deviation. (F) Data for PEI in KCl were fit to the diffusion-limited colloidal aggregation model. Panels G and H show the effects of serum concentration on aggregation of fresh 40L and PEI polyplexes with N/P ratio of 10. (G) Size of 40L and PEI polyplexes incubated in Opti-MEM + 2% or 10% FBS. (H) Data for PEI were fit to the diffusion-limited colloidal aggregation model. All data are plotted as mean ± standard deviation.
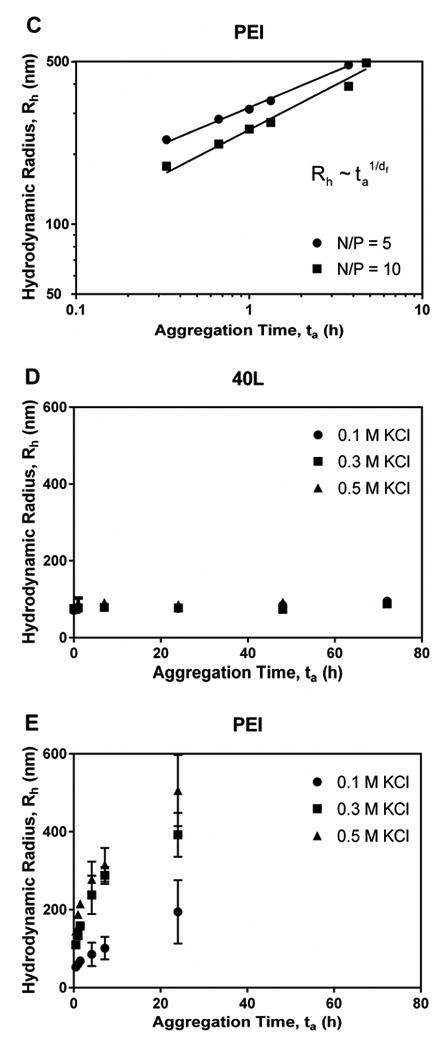
To confirm these observations, the stability of fresh 40L (Figure 6D) and PEI (Figure 6E) polyplexes was also investigated more systematically as a function of ionic strength by varying KCl concentration. Previous studies have shown that for slow flocculation, the floc diameter grows exponentially with time. Weitz described a reaction-limited colloidal aggregation model for systems with W » 1:26
| (1) |
where Rh is a scaling parameter approximating the average hydrodynamic radius, ta is aggregation time, and t0 is an aggregation time constant that varies with initial particle concentration and single particle sticking probability. For systems with W ≈ 1, a power law model describing diffusion-limited aggregation can be applied:
| (2) |
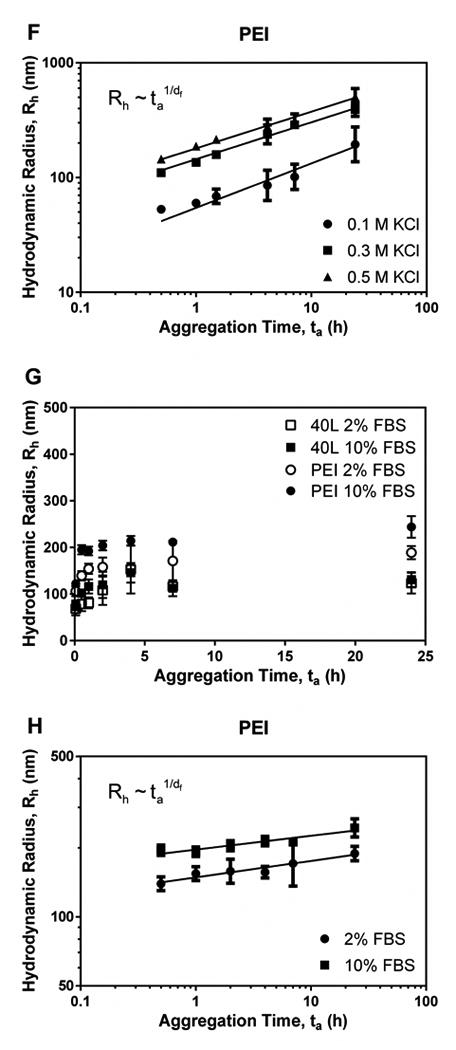
As [KCl] increased, PEI polyplexes aggregated more rapidly, a behavior that was consistent with the notion that PEI polyplexes are stabilized by electrostatic repulsion. In contrast, [KCl] had no effect on aggregation rate of 40L polyplexes. 40L polyplexes did not aggregate substantially, remaining <200 nm in diameter after 72 h at all KCl concentrations investigated. These observations indicate that 40L polyplexes are stabilized by steric interactions and not by electrostatic repulsion; hence, ionic strength does not affect their aggregation rate. In contrast, the diffusion-limited aggregation model fit the data for PEI polyplexes well (Figure 6F; R2 = 0.95 – 0.99), and PEI polyplex stability decreased with increasing [KCl].
Aggregation of fresh polyplexes in the presence of Opti-MEM + 2% or 10% FBS serum was also investigated (Figure 6G-H). While 40L polyplexes showed no increase in aggregation rate in response to serum dose, the PEI polyplexes aggregated in the presence of serum at a rate that correlated with serum concentration.
In Vitro Transfection and Cytotoxicity of Lyophilized Polyplexes
Improved colloidal stability of the 40L polyplexes resulted in less aggregation during lyophilization (Figure 6) and was thus anticipated to yield better transfection performance post-lyophilization. This is relevant both for the purpose of long-term storage (i.e., for clinical use) and also for incorporation of a polyplex powder into tissue engineered scaffolds during fabrication. The ability of the lyophilized polyplexes to transfect MDA-MB-231 cells in vitro was assessed by delivering a luciferase reporter plasmid (Figure 7A). At N/P ratio of 5, 40L polyplexes had significantly higher transfection than PEI. At N/P ratio of 10, all poly(EG-b-(DMAEMA-co-BMA)) polyplexes investigated had significantly higher transfection efficiency than PEI. At N/P ratio of 20, 40S, 40L, 50S, and 50M polyplexes produced significantly higher transfection than PEI. To determine cytotoxicity of lyophilized polyplexes, calcein AM staining was used to quantify number of live cells after transfection (Figure 7B). All wells treated with poly(EG-b-(DMAEMA-co-BMA)) polyplexes had >70% viability relative to untreated wells. The viability of cells treated with poly(EG-b-(DMAEMA-co-BMA)) polyplexes was comparable to or higher than cells treated with PEI polyplexes.
Figure 7. Transfection efficiency and cytotoxicity of lyophilized polymer-pDNA polyplexes.
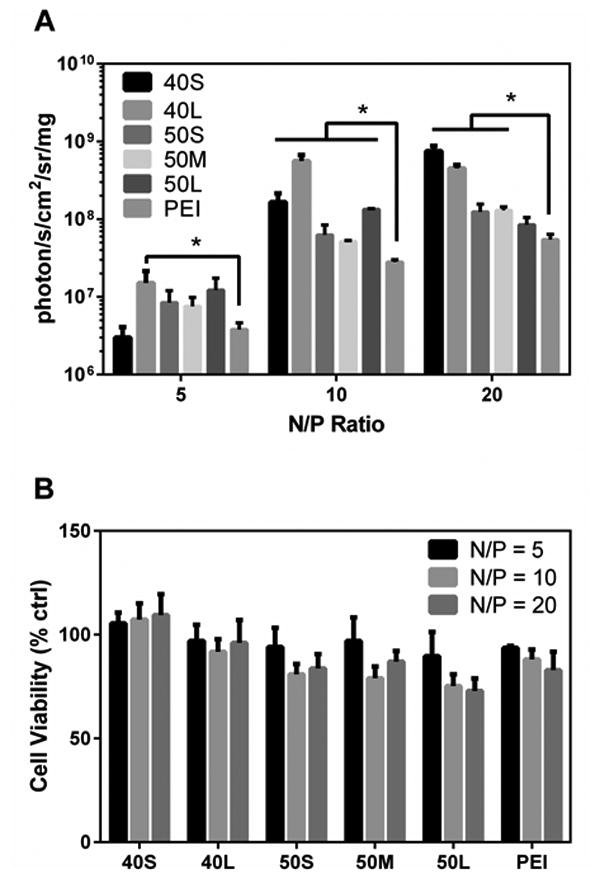
MDA-MB-231 cells were transfected with lyophilized polyplexes containing luciferase pDNA. (A) Luminescence produced by cells transfected with lyophilized polyplexes normalized to total protein. Asterisks indicate significant increase in transfection compared to PEI (p < 0.05). Data are plotted as mean ± standard deviation. (B) Cell viability after transfection with lyophilized polyplexes relative to untreated cells was assessed using calcein AM staining. Data are plotted as mean ± standard deviation.
In Vitro Transfection of Lyophilized 40L Polyplexes Incorporated in PUR Scaffolds
Desalted, lyophilized polyplexes were incorporated in PUR tissue-engineered scaffolds. After desalination and prior to lyophilization, trehalose was added at a weight ratio of 50, 100, or 200:1 trehalose:pDNA. Varied quantities of excipient were added to accelerate the release of the nanoparticles from the scaffolds 27, which facilitated transfection measurements at 24 h post-seeding in vitro. As shown in Figure 8, transfection increased with the trehalose:pDNA ratio, which is consistent with previous studies reporting that similar nanoparticles were not released on this timeframe from PUR scaffolds in vitro without addition of trehalose 27. Furthermore, the quantity of trehalose added correlates to the rate of nanoparticle release 27. This result provides evidence that incorporation of stabilized 40L-pDNA polyplexes in PUR scaffolds is a promising approach for local, scaffold-based delivery of plasmids that encode therapeutic proteins.
Figure 8. Transfection of fibroblasts seeded on PUR scaffolds loaded with lyophilized 40L polyplexes.
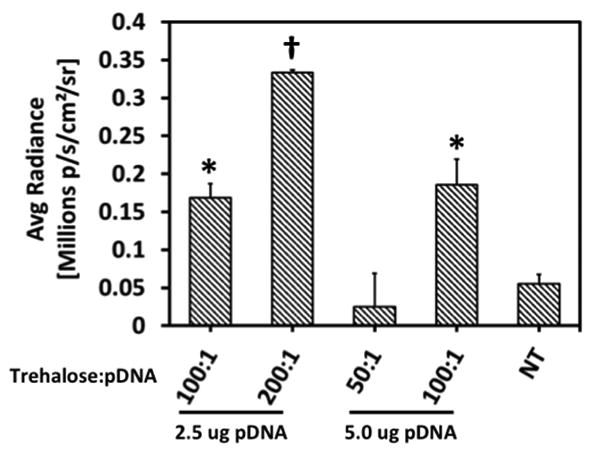
Transfection of IMDF cells seeded on PUR scaffolds containing 2.5 or 5 μg pDNA for 24 h and varied quantities of trehalose:pDNA. Data are plotted as mean ± standard error. Asterisks indicate significant difference from all other treatments (p < 0.05). Transfection increased with increasing trehalose/pDNA ratio.
Discussion
Local gene therapy has applications for tissue restoration, wound healing, and treatment of disease. Previous research on delivery of naked or nanoparticulate pDNA incorporated into hydrogels and biomaterial scaffolds has produced transgene expression and tissue responses in vivo. However, these systems often suffer from low transfection efficiency, burst release of pDNA, or nanoparticle aggregation in physiologic conditions, necessitating delivery of high doses of pDNA. In this study, we designed pDNA polyplexes with high colloidal stability and enhanced cellular uptake and transfection efficiency compared to conventional PEI-pDNA polyplexes. The new polyplexes were composed of diblock polymers containing DMAEMA to electrostatically bind DNA, BMA to provide hydrophobic interactions and enhance polyplex core stability, and a PEG corona to shield charge and enhance steric stability. These polyplexes exhibited significantly less aggregation than PEI polyplexes after lyophilization or in the presence of high salt concentrations. In addition to being optimized to overcome extracellular barriers to polyplex stability, the poly(EG-b-(DMAEMA-co-BMA)) polyplexes were also designed to overcome intracellular endo-lysosomal delivery barriers through finely-tuned pH-dependent membrane disruptive activity. Several of the new polyplex formulations tested had higher transfection efficiency than PEI complexes both before and after lyophilization. Furthermore, cells seeded on PUR scaffolds incorporating lyophilized polyplexes were efficiently transfected. These results point to the potential broad utility of diblock copolymer-pDNA polyplexes for tissue, intravenous, or local gene delivery when incorporated into tissue engineering scaffolds.
Polymer-pDNA nanoparticles used for local gene delivery from tissue engineering scaffolds must not only be effectively stabilized against colloidal aggregation but also support efficient endosomal escape in order to achieve high levels of transfection. PEGylation and titration of hydrophobic content into polycations are known to have several beneficial effects on nonviral gene therapy.17 Triblock copolymers composed of PEG, hydrophobic poly(n-butyl acrylate) (PnBA), and cationic DMAEMA have been reported to form polyplexes with DNA that exhibit improved stability against aggregation and reduced interactions with negatively charged serum components.28 While NMR spectroscopy measurements revealed that the PEG corona shielded the charged DMAEMA-DNA polyplex, incorporation of the hydrophobic PnBA homopolymer block decreased transfection efficiency, which was conjectured to occur due to decreased intracellular release of pDNA from the polyplexes.
Poly(EG-b-(DMAEMA-co-BMA)) polymers have been shown to be effective for packaging of siRNA for intravenous delivery21 but had not been tested yet for formation and stabilization of pDNA polyplexes that mediate endosomal escape through an active, pH-dependent membrane disruption mechanism. Previous studies have reported that active membrane disruption mechanisms can be used to enhance endosome escape and transfection efficiency relative to relying solely on proton sponge osmotic disruption of endosomes (i.e., as achieved using PEI or homopolymers of DMAEMA). For example, the membrane-porating peptide melittin has been shown to aid in endosome escape and increase transfection efficiency of PEI- and lysine-based polyplexes.29, 30In the present study, the composition and molecular weight of the DMAEMA-co-BMA block were tuned to achieve an optimal balance of pH-responsiveness and hydrophobic interactions in order to form colloidally stable nanoparticles that increase endosomal escape and transfection efficiency.
Despite being considered a gold standard for nonviral gene delivery, PEI has several disadvantageous properties. PEI polyplexes have been shown to have poor stability at high concentrations, in the presence of salt and serum, and during lyophilization.10, 11, 31 Excipients can reduce aggregation through increased viscosity 10, but sucrose and agarose excipients add to the overall complexity of the formulation. Furthermore, they may have adverse biological effects or alter the curing or mechanical properties of the delivery vehicle when used in the context of tissue engineering hydrogels or scaffolds. It has been reported that plasma proteins adsorb to PEI-pDNA polyplexes, resulting in rapid aggregation that can be reduced by polyplex PEGylation.32 In the present study, DLS measurements revealed that reconstituted PEI polyplexes were in an aggregated state post-lyophilization. Consistent with the notion that PEI polyplexes are electrostatically stabilized, their aggregation rate increased with increasing salt concentration due to screening of the electrostatic repulsion forces. While PEI polyplexes aggregated slowly in 0.1 M KCl, they aggregated rapidly in 0.3 M and 0.5 M KCl, in DPBS (a buffer that mimics physiologic conditions), and in serum.
Inadequate colloidal stability has been previously reported for PEI 8, 9, but the aggregation kinetics of PEI polyplexes has not been extensively investigated. In the present study, we fit the particle size time-course data (Figure 6) to established colloidal aggregation models.26 In contrast to PEI, the best-performing diblock copolymer-pDNApolyplex (40L with N/P = 10) was stable after lyophilization and at high salt and serum concentrations. Diblock copolymer polyplex aggregation was reaction-limited while PEI polyplex aggregation was diffusion-controlled, providing evidence that diblock copolymers have greater stability than PEI. The diblock copolymer polyplexes are stabilized sterically by the PEG corona as well as by DMAEMA-pDNA electrostatic interactions and BMA hydrophobicity in the core. Thus, they can be readily tuned by varying the BMA content to optimize polyplex stability.
Previous studies suggest that PEGylation increases stability but decreases transfection efficiency. In the present study, several poly(EG-b-(DMAEMA-co-BMA)) polyplexes had better transfection efficiency in serum than PEI before and after lyophilization despite having neutral ζ-potential. The best performing diblock copolymer-pDNApolyplex, 40L with N/P = 10, had greater luminescence production than PEI both before and after lyophilization. This observation is consistent with a previous study reporting that dextran-PEI complexes had moderately lower transfection efficiency than PEI in serum-free medium at 4h, but in the presence of serum at longer time points (48 h), significantly higher transfection efficiencies were observed for dextran-PEI complexes compared to PEI.33 Another study has reported that PEI transfection decreases 2-20 fold in the presence of 10% serum compared to serum-free media.34 Taken together with these previous studies, our data suggest that sterically stabilized polyplexes (i.e., 40L and dextran-PEI) have a slower initial rate of uptake in vitro due to lower ζ-potential, but their increased steric stability in the presence of salts and serum provides them with superior long-term performance under physiologically relevant conditions.
Conclusions
Poly(EG-b-(DMAEMA-co-BMA) diblock copolymers were synthesized and tested for stability, endosomal escape, and transfection efficiency of pDNA. The polymers formed pDNA polyplexes that exhibited increased colloidal stability after lyophilization and in the presence of salt relative to PEI-pDNA polyplexes, with polymer 40L exhibiting optimal performance. Poly(EG-b-(DMAEMA-co-BMA))-pDNA polyplexes had greater transfection efficiency than PEI-pDNA polyplexes and achieved pH-dependent membrane disruption leading to improved endosomal escape. The lyophilized 40L polyplexes incorporated in and delivered from PUR scaffolds were also shown to transfect cells in vitro. The enhanced colloidal stability and transfection efficiency of the lyophilized diblock copolymer-pDNA polyplexes underscores their potential utility for numerous applications, including local nonviral gene delivery from 3D scaffolds and storage for clinical use for local or systemic delivery applications.
Supplementary Material
Acknowledgments
Research reported in this publication was supported by the National Institutes of Health through National Institute of Arthritis and Musculoskeletal and Skin Diseases Award Number 5R01AR056138-02 and the National Institute of Biomedical Imaging and Bioengineering 1R21EB012750. Elizabeth Adolph acknowledges support from the Department of Education for a Graduate Assistance in Areas of National Need Fellowship under grant number P200A090323. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
References
- 1.Boussif O, Lezoualc'h F, Zanta MA, Mergny MD, Scherman D, Demeneix B, Behr JP. Proc Natl Acad Sci U S A. 1995;92:7297–7301. doi: 10.1073/pnas.92.16.7297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cherng JY, van de Wetering P, Talsma H, Crommelin DA, Hennink W. Pharmaceutical Research. 1996;13:1038–1042. doi: 10.1023/a:1016054623543. [DOI] [PubMed] [Google Scholar]
- 3.Behr JP. M S-Med Sci. 1996;12:56–58. [Google Scholar]
- 4.Behr JP. Chimia. 1997;51:34–36. [Google Scholar]
- 5.Wang S, Ma N, Gao SJ, Yu H, Leong KW. Molecular therapy : the journal of the American Society of Gene Therapy. 2001;3:658–664. doi: 10.1006/mthe.2001.0324. [DOI] [PubMed] [Google Scholar]
- 6.Kichler A, Chillon M, Leborgne C, Danos O, Frisch B. J Control Release. 2002;81:379–388. doi: 10.1016/s0168-3659(02)00080-9. [DOI] [PubMed] [Google Scholar]
- 7.Aoki K, Furuhata S, Hatanaka K, Maeda M, Remy JS, Behr JP, Terada M, Yoshida T. Gene Ther. 2001;8:508–514. doi: 10.1038/sj.gt.3301435. [DOI] [PubMed] [Google Scholar]
- 8.Lei P, Padmashali RM, Andreadis ST. Biomaterials. 2009;30:3790–3799. doi: 10.1016/j.biomaterials.2009.03.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Trentin D, Hubbell J, Hall H. J Control Release. 2005;102:263–275. doi: 10.1016/j.jconrel.2004.09.029. [DOI] [PubMed] [Google Scholar]
- 10.Lei Y, Huang S, Sharif-Kashani P, Chen Y, Kavehpour P, Segura T. Biomaterials. 2010;31:9106–9116. doi: 10.1016/j.biomaterials.2010.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lei Y, Rahim M, Ng Q, Segura T. J Control Release. 2011;153:255–261. doi: 10.1016/j.jconrel.2011.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Itaka K, Yamauchi K, Harada A, Nakamura K, Kawaguchi H, Kataoka K. Biomaterials. 2003;24:4495–4506. doi: 10.1016/s0142-9612(03)00347-8. [DOI] [PubMed] [Google Scholar]
- 13.Verbaan FJ, Oussoren C, Snel CJ, Crommelin DJ, Hennink WE, Storm G. The journal of gene medicine. 2004;6:64–75. doi: 10.1002/jgm.475. [DOI] [PubMed] [Google Scholar]
- 14.Vader P, van der Aa LJ, Engbersen JF, Storm G, Schiffelers RM. Pharmaceutical research. 2012;29:352–361. doi: 10.1007/s11095-011-0545-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhou J, Liu J, Cheng CJ, Patel TR, Weller CE, Piepmeier JM, Jiang Z, Saltzman WM. Nature materials. 2012;11:82–90. doi: 10.1038/nmat3187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Piest M, Engbersen JFJ. Journal of Controlled Release. 2010;148:83–90. doi: 10.1016/j.jconrel.2010.07.109. [DOI] [PubMed] [Google Scholar]
- 17.Liu Z, Zhang Z, Zhou C, Jiao Y. Progress in Polymer Science. 2010;35:1144–1162. [Google Scholar]
- 18.Manganiello MJ, Cheng C, Convertine AJ, Bryers JD, Stayton PS. Biomaterials. 2012;33:2301–2309. doi: 10.1016/j.biomaterials.2011.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Convertine AJ, Benoit DSW, Duvall CL, Hoffman AS, Stayton PS. Journal of Controlled Release. 2009;133:221–229. doi: 10.1016/j.jconrel.2008.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Duvall CL, Convertine AJ, Benoit DSW, Hoffman AS, Stayton PS. Molecular Pharmaceutics. 2009;7:468–476. doi: 10.1021/mp9002267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nelson CE, Kintzing JR, Hanna A, Shannon JM, Gupta MK, Duvall CL. ACS Nano. 2013;7:8870–8880. doi: 10.1021/nn403325f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chiefari J, Chong YK, Ercole F, Krstina J, Jeffery J, Le TPT, Mayadunne RTA, Meijs GF, Moad CL, Moad G, Rizzardo E, Thang SH. Macromolecules. 1998;31:5559–5562. [Google Scholar]
- 23.Convertine AJ, Benoit DS, Duvall CL, Hoffman AS, Stayton PS. J Control Release. 2009;133:221–229. doi: 10.1016/j.jconrel.2008.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Smith PJ, Giroud M, Wiggins HL, Gower F, Thorley JA, Stolpe B, Mazzolini J, Dyson RJ, Rappoport JZ. International journal of nanomedicine. 2012;7:2045–2055. doi: 10.2147/IJN.S29334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Evans BC, Nelson CE, Yu SS, Beavers KR, Kim AJ, Li H, Nelson HM, Giorgio TD, Duvall CL. Journal of visualized experiments : JoVE. 2013:e50166. doi: 10.3791/50166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lin MY, Lindsay HM, Weitz DA, Ball RC, Klein R, Meakin P. Phys Rev A. 1990;41:41. doi: 10.1103/physreva.41.2005. [DOI] [PubMed] [Google Scholar]
- 27.Nelson CE, Kim AJ, Adolph EJ, Gupta MK, Yu F, Hocking KM, Davidson JM, Guelcher SA, Duvall CL. Adv Mater. 2014;26:607–614. 506. doi: 10.1002/adma.201303520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sharma R, Lee JS, Bettencourt RC, Xiao C, Konieczny SF, Won YY. Biomacromolecules. 2008;9:3294–3307. doi: 10.1021/bm800876v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Boeckle S, Fahrmeir J, Roedl W, Ogris M, Wagner E. Journal of Controlled Release. 2006;112:240–248. doi: 10.1016/j.jconrel.2006.02.002. [DOI] [PubMed] [Google Scholar]
- 30.Schellinger JG, Pahang JA, Johnson RN, Chu DSH, Sellers DL, Maris DO, Convertine AJ, Stayton PS, Horner PJ, Pun SH. Biomaterials. 2013;34:2318–2326. doi: 10.1016/j.biomaterials.2012.09.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Oster CG, Wittmar M, Unger F, Barbu-Tudoran L, Schaper AK, Kissel T. Pharm Res. 2004;21:927–931. doi: 10.1023/b:pham.0000029279.50733.55. [DOI] [PubMed] [Google Scholar]
- 32.Ogris M, Brunner S, Schuller S, Kircheis R, Wagner E. Gene Ther. 1999;6:595–605. doi: 10.1038/sj.gt.3300900. [DOI] [PubMed] [Google Scholar]
- 33.Jiang D, Salem AK. Int J Pharm. 2012;427:71–79. doi: 10.1016/j.ijpharm.2011.10.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dong X, Lin L, Chen J, Guo Z, Tian H, Li Y, Wei Y, Chen X. Macromol Biosci. 2013;13:512–522. doi: 10.1002/mabi.201200346. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


