Abstract
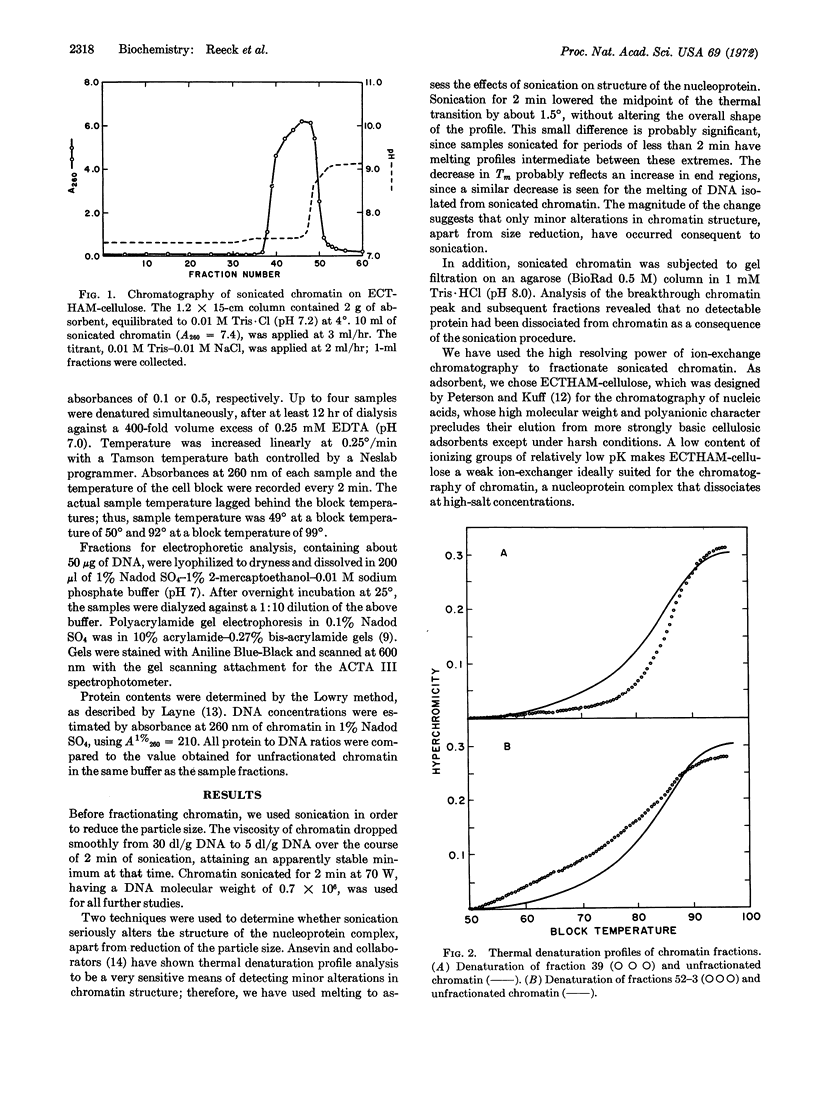
Sonicated rabbit-liver chromatin is fractionated by ion-exchange chromatography on ECTHAM-cellulose, a weakly cationic adsorbent. The chromatographic procedure provides a series of fractions having a spectrum of thermal denaturation profiles. The earliest-eluting fractions melt cooperatively at a significantly higher temperature than bulk chromatin, and totally lack any of the components of chromatin that melt at low temperatures. In contrast, the latest-eluting fractions are significantly enriched in their content of DNA sequences that melt at low temperatures. Although the protein to DNA mass ratio of all fractions is equal to that of unfractionated chromatin, polyacrylamide gel electrophoresis demonstrates that the relative amounts of individual proteins vary across the chromatographic peak. The most pronounced changes observed are the enrichment of a particular high molecular weight nonhistone protein in the later fractions, and a significant diminution of lysine-rich histone in the last fractions. The results, in conjunction with the demonstration by McConaughy, B. L. & McCarthy, B. J. (Biochemistry 11, 998-1003, 1972) that RNA transcribed in vivo in immature chick erythrocytes hybridizes to the DNA of only the portions of chromatin from these cells that melt at low temperatures, suggest that this fractionation may separate transcribable DNA sequences in chromatin from the repressed segments of a eukaryotic genome.
Keywords: histones, nonhistone proteins, ECTHAM-cellulose, thermal denaturation, chromatography
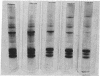
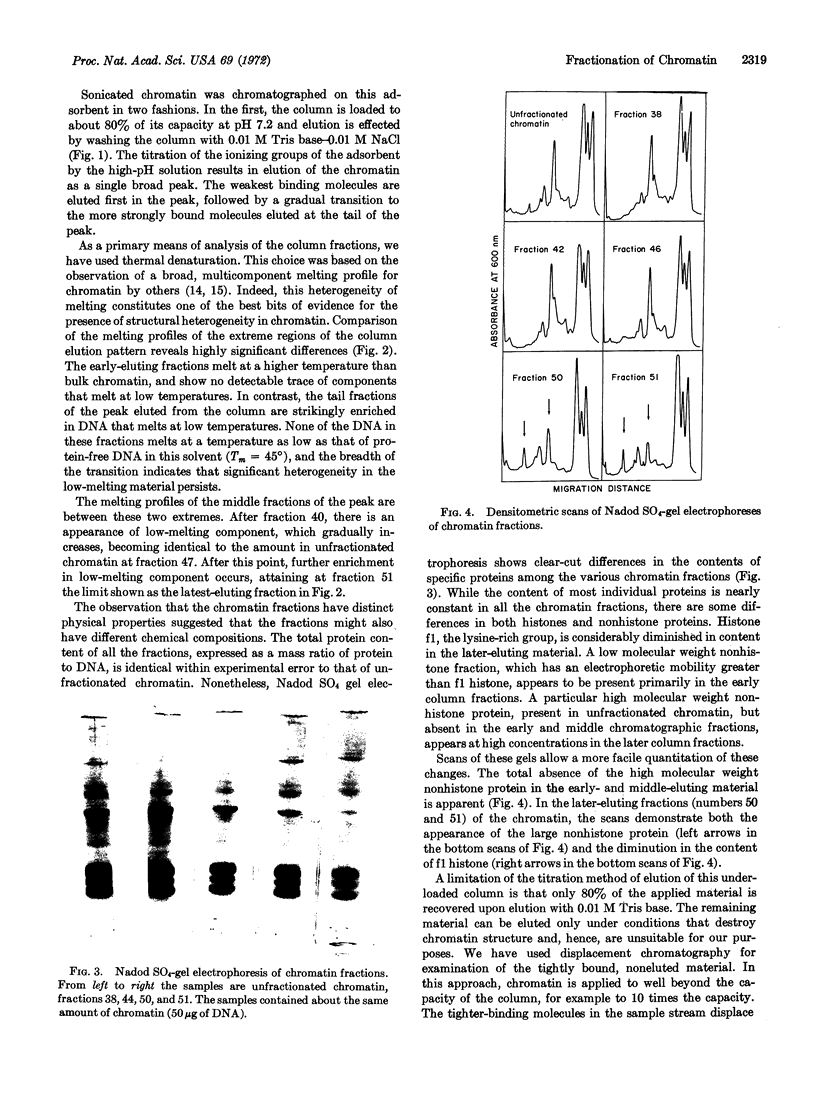
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Alberts B. M., Amodio F. J., Jenkins M., Gutmann E. D., Ferris F. L. Studies with DNA-cellulose chromatography. I. DNA-binding proteins from Escherichia coli. Cold Spring Harb Symp Quant Biol. 1968;33:289–305. doi: 10.1101/sqb.1968.033.01.033. [DOI] [PubMed] [Google Scholar]
- Alberts B. M. Function of gene 32-protein, a new protein essential for the genetic recombination and replication of T4 bacteriophage DNA. Fed Proc. 1970 May-Jun;29(3):1154–1163. [PubMed] [Google Scholar]
- Ansevin A. T., Hnilica L. S., Spelsberg T. C., Kehm S. L. Structure studies on chromatin and nucleohistones. Thermal denaturation profiles recorded in the presence of urea. Biochemistry. 1971 Dec 7;10(25):4793–4803. doi: 10.1021/bi00801a030. [DOI] [PubMed] [Google Scholar]
- Chalkley R., Jensen R. H. A study of the structure of isolated chromatin. Biochemistry. 1968 Dec;7(12):4380–4388. doi: 10.1021/bi00852a034. [DOI] [PubMed] [Google Scholar]
- Clark R. J., Felsenfeld G. Structure of chromatin. Nat New Biol. 1971 Jan 27;229(4):101–106. doi: 10.1038/newbio229101a0. [DOI] [PubMed] [Google Scholar]
- Duerksen J. D., McCarthy B. J. Distribution of deoxyribonucleic acid sequences in fractionated chromatin. Biochemistry. 1971 Apr 13;10(8):1471–1478. doi: 10.1021/bi00784a031. [DOI] [PubMed] [Google Scholar]
- EIGNER J., SCHILDKRAUT C., DOTY P. Concentration effects in the hydrodynamic properties of deoxyribonucleic acid. Biochim Biophys Acta. 1962 Jan 22;55:13–21. doi: 10.1016/0006-3002(62)90926-5. [DOI] [PubMed] [Google Scholar]
- FRENSTER J. H., ALLFREY V. G., MIRSKY A. E. REPRESSED AND ACTIVE CHROMATIN ISOLATED FROM INTERPHASE LYMPHOCYTES. Proc Natl Acad Sci U S A. 1963 Dec;50:1026–1032. doi: 10.1073/pnas.50.6.1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HYMER W. C., KUFF E. L. ISOLATION OF NUCLEI FROM MAMMALIAN TISSUES THROUGH THE USE OF TRITON X-100. J Histochem Cytochem. 1964 May;12:359–363. doi: 10.1177/12.5.359. [DOI] [PubMed] [Google Scholar]
- Huang R. C., Huang P. C. Effect of protein-bound RNA associated with chick embryo chromatin on template specificity of the chromatin. J Mol Biol. 1969 Jan;39(2):365–378. doi: 10.1016/0022-2836(69)90323-4. [DOI] [PubMed] [Google Scholar]
- Li H. J., Bonner J. Interaction of histone half-molecules with deoxyribonucleic acid. Biochemistry. 1971 Apr 13;10(8):1461–1470. doi: 10.1021/bi00784a030. [DOI] [PubMed] [Google Scholar]
- Marushige K., Bonner J. Fractionation of liver chromatin. Proc Natl Acad Sci U S A. 1971 Dec;68(12):2941–2944. doi: 10.1073/pnas.68.12.2941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuyama A., Tagashira Y., Nagata C. A circular dichroism study on the conformation of DNA in rat liver chromatin. Biochim Biophys Acta. 1971 Jun 30;240(2):184–190. doi: 10.1016/0005-2787(71)90656-3. [DOI] [PubMed] [Google Scholar]
- McConaughy B. L., McCarthy B. J. Fractionation of chromatin by thermal chromatography. Biochemistry. 1972 Mar 14;11(6):998–1003. doi: 10.1021/bi00756a008. [DOI] [PubMed] [Google Scholar]
- Peterson E. A., Kuff E. L. Chromatographic isolation of 80S ribosomes from rat liver and mouse plasma cell tumor. Biochemistry. 1969 Jul;8(7):2916–2923. doi: 10.1021/bi00835a033. [DOI] [PubMed] [Google Scholar]
- Simpson R. T. Modification of chromatin by trypsin. The role of proteins in maintainance of deoxyribonucleic acid conformation. Biochemistry. 1972 May 23;11(11):2003–2008. doi: 10.1021/bi00761a002. [DOI] [PubMed] [Google Scholar]
- Walker I. O. Electrometric and spectrophotometric titration of histone and deoxyribonucleohistone. J Mol Biol. 1965 Dec;14(2):381–398. doi: 10.1016/s0022-2836(65)80189-9. [DOI] [PubMed] [Google Scholar]
- Weaver R. F., Blatti S. P., Rutter W. J. Molecular structures of DNA-dependent RNA polymerases (II) from calf thymus and rat liver. Proc Natl Acad Sci U S A. 1971 Dec;68(12):2994–2999. doi: 10.1073/pnas.68.12.2994. [DOI] [PMC free article] [PubMed] [Google Scholar]