Abstract
Mine tailings in semiarid regions are highly susceptible to erosion and are sources of dust pollution and potential avenues of human exposure to toxic metals. One constraint to revegetation of tailings by phytostabilization is the absence of microbial communities critical for biogeochemical cycling of plant nutrients. The objective of this study was to evaluate specific genes as in situ indicators of biological soil response during phytoremediation. The abundance and activity of 16S rRNA, nifH, and amoA were monitored during a nine month phytostabilization study using buffalo grass and quailbush grown in compost-amended, metalliferous tailings. The compost amendment provided a greater than 5-log increase in bacterial abundance, and survival of this compost-inoculum was more stable in planted treatments. Despite increased abundance, the activity of the introduced community was low, and significant increases were not detected until six and nine months in quailbush, and unplanted compost and buffalo grass treatments, respectively. In addition, increased abundances of nitrogen-fixation (nifH) and ammonia-oxidizing (amoA) genes were observed in rhizospheres of buffalo grass and quailbush, respectively. Thus, plant establishment facilitated the short term stabilization of introduced bacterial biomass and supported the growth of two key nitrogen-cycling populations in compost-amended tailings.
Keywords: mine waste reclamation, bioindicator gene, qPCR
INTRODUCTION
Metal(loid)-contaminated mine tailing sites in arid and semiarid regions remain largely unvegetated for tens to hundreds of years making them highly susceptible to eolian dispersion and water erosion (reviewed in Mendez and Maier, 2008; Meza-Figueroa et al. 2009) and a potential source of human exposure to dust and toxic metals (Schaider et al. 2007). Key factors inhibiting natural revegetation of legacy tailings sites in semiarid and arid environments include low pH, high metal and salt concentrations, low nutrient and organic matter content, poor water holding capacity and severely impacted soil microbial communities (Mendez and Maier, 2008; Anawar et al. 2013). Assisted phytostabilization provides a strategy for stabilizing these sites by creating a vegetative cap that functions as a metal(loid)-containment mechanism limiting eolian and fluvial dispersion of the tailings offsite (Wong, 2003; Mendez, Glenn, and Maier, 2007; Epelde et al. 2009). The long-term success of phytostabilization is dependent upon the initiation of a progressive ecosystem reconstruction process where tailings transition from a highly disturbed matrix to a soil ecosystem supportive of self-sustaining native plant growth (Gómez-Sagasti et al. 2012).
Critical to tailings ecosystem development is the enhancement of soil biological capacity (Huang, Baumgartl, and Mulligan 2012), a process that is accomplished by establishing a resilient and diverse soil microbial community capable of promoting essential plant-microbe interactions such as nutrient cycling, phosphate solubilization, production of phytohormones and siderophores, and suppression of stress-related plant ethylene production through bacterial 1-aminocyclopropane-1-carboxylate (ACC) deaminase production (reviewed in Glick, 2004; Gómez-Sagasti et al. 2012; Sessitsch et al. 2013). Most legacy tailings sites contain depauperate microbial communities low in species richness and deficient in these critical plant-supporting activities (Moynahan, Zabinski and Gannon 2002). The microbial communities of these materials are typically dominated by autotrophic iron- and sulfur-oxidizing populations with very low levels of bacterial heterotrophs (Mendez et al. 2007; Solís-Domínguez et al. 2012). For example, a comparative analysis of tailings microbial diversity in a highly metal-contaminated, acidic, legacy mining site in southern Arizona (Mendez, Neilson and Maier, 2008) found all phylotypes identified in a pH 2.7 tailings sample to be related to chemoautotrophic iron- and sulfur-oxidizing bacteria. Phyla typically dominant in undisturbed, vegetated soils, such as Acidobacteria, Alpha- and Betaproteobacteria (Janssen, 2006) were absent from the pH 2.7 tailings, but present at moderate levels in a pH 5.7 sample (35% abundance), and at much higher levels in an offsite soil (58%). Whereas, Fe- and S-oxidizing bacterial counts and organic carbon, nitrogen and heavy metal concentrations were similar in the two acidic tailings, heterotrophic bacterial counts were 30±17, 1.5±.11 × 105, and 2.5±.52 × 106 CFU g−1 soil in the pH 2.7, 5.7, and off-site soils, respectively. Similar autotroph/heterotroph ratios were observed in acidic tailings from the Iron King Mine Humboldt Smelter Superfund (IKMHSS) site located in Dewey-Humboldt, Arizona (Solís-Domínguez et al. 2012), suggesting that acidic tailings have very low heterotrophic bacterial abundance.
Amendments such as compost, used in assisted phytostabilization, provide a microbial inoculum, essential plant nutrients, a buffer for the acidic pH, and a means of improving soil structure and water holding capacity (Mendez et al. 2007; Gómez-Sagasti et al. 2012; Solís-Dominguez et al. 2012). Short-term assisted phytostabilization greenhouse studies have repeatedly documented significant increases in neutrophilic heterotrophic plate counts (NHC) paralleled by decreases in culturable iron- and sulfur-oxidizing bacteria following compost amendment and plant establishment in acidic tailings (Mendez et al. 2007). Specifically, Solís-Dominguez et al. (2012) found a significant correlation between NHC and final plant biomass in four plant species after 60 days growth (r2 = 0.77–0.89, p=0.0001). Thus, we contend that specific transitions in microbial community development should be monitored as indicators of progress towards the goal of successful long-term phytostabilization (Mendez et al. 2008; Schippers et al. 2010). Further research is needed to characterize the sustainability and relevance of microbial communities introduced with organic matter amendments and evaluate their potential for enhancing biological capacity in severely disturbed ecosystems.
The objective of this study was to assess the abundance and activity of bacterial 16S rRNA, nifH, and amoA genes as bio-indicators of ecosystem response during a 9-month assisted phytostabilization trial in acidic, metalliferous mine tailings. The 16S rRNA gene was selected to monitor overall bacterial abundance and activity. Nitrogen-fixation (nifH gene) was targeted because nitrogen is typically limiting in mine tailings (Wong 2003; Shu et al. 2005; Solís-Dominguez et al. 2011; Solís-Dominguez et al. 2012) and nifH copy number has been shown to increase with progressing plant succession in copper mine tailings (Huang et al.2011). The ammonia-oxidizing gene (amoA) represents the rate-limiting step in nitrification and as such, is a potential indicator of community mineralization potential. In addition, bacterial amoA activity is sensitive to acidic pH (Sahrawat, 2008), soil aeration and moisture (Kowalchuk and Stephen, 2001), parameters important to successful plant establishment in tailings. Real time PCR was used to quantify the in situ genetic potential (DNA) and activity (RNA) of these genes during quailbush and buffalo grass establishment in highly-contaminated, compost-amended mine tailings during a nine month greenhouse mesocosm study.
2. MATERIALS AND METHODS
Experimental design
A nine month greenhouse study was conducted at the Controlled Environment Agriculture Center (CEAC) at The University of Arizona, Tucson, Arizona to track changes in soil quality during plant establishment in compost-amended mine tailings. Large polypropylene mesocosms 1 m in diameter and 0.5 m deep were custom designed (ProPlastics, Chandler, AZ) and fitted with tensiometers and pore water samplers at 10 cm intervals along the mesocosm profile. The following controls and treatments were established in triplicate generating a total of twelve mesocosms arranged in a spatially randomized design: unplanted controls with 1) tailings only (TO) or 2) tailings amended with 15% (w/w) compost (TC); and two planted treatments with tailings plus 15% (w/w) compost seeded with 3) buffalo grass (Buchloe dactyloides) (BG) at 8.8 g seeds m−2 (7 g per mesocosm) or 4) quailbush (Atriplex lentiformis) (QB) at 5.5 g seeds m−2 (4.3 g per mesocosm). The plants are both native species and were selected based upon their ability to grow in 15% w/w compost-amended tailings with shoot metal accumulations below the domestic animal toxicity limit (Solís-Dominguez et al. 2012).
Unamended tailings were packed to a depth of 40 cm in TO controls and to 20 cm depth in TC, BG, and QB mesocosms at a bulk density of 1.3 kg L−1. Compost-amended tailings (15% w/w) were then added to the top 20 cm of the TC, BG and QB mesocosms. The specifics of compost amendment and depth were based on preliminary greenhouse studies (Solís-Dominguez et al. 2012) and a simultaneous field trial (unpublished data). All mesocosms were irrigated immediately following the seeding of planted treatments using water from an on-site well at an initial rate of 5–10 L bi-weekly and then 5–10 L week−1 using the tensiometers to monitor soil moisture. Time zero (t0) for the experiment was defined as the first irrigation event.
Pore water was collected continuously at 5 and 15 cm depths from the mesocosms. Sub-samples removed at 0, 3, 6, and 9 months were consolidated from these two depths and analyzed for pH and EC. The pH was also analyzed from samples collected 2 to 4 months following the conclusion of the experiment. Samples from 5 and 15 cm depths were combined for analysis to represent conditions found within the 15 cm deep core that was removed and homogenized for microbial community analysis.
Mine tailings and compost analysis
Tailings used in this study were transported from the Iron King Mine Humboldt Smelter Superfund (IKMHSS) site located in Dewey-Humboldt, Arizona. The tailings mineralogy was described previously (Solís-Domínguez et al. 2012). A tailings mixture was produced that included 3 parts oxidized tailings (0–20 cm depth) from 3 locations on the tailings pile and 1 part reduced subsurface tailings (> 60 cm depth), a ratio selected to represent the heterogeneous distribution of tailings at the field site. This tailings mixture had extremely high levels of arsenic, lead and zinc ( Table 1). The compost was a mixture of composted cattle manure and green waste from a local dairy (Arizona Dairy Compost LLC, Anthem, AZ) and commercial composted steer manure (El Toro De-Odorized Steer Manure, Tempe, AZ). Tailings and compost mixtures were homogenized with a cement mixer and sieved to 0.5 cm prior to application. Select tailings biological and chemical properties are presented in Table 1. Total carbon, total inorganic carbon and total nitrogen (TN) were quantified using a Shimadzu TOC-VCSH analyzer (Columbia, MD) with a solid state module (SSM-5000A). Total organic carbon (TOC) was determined from the difference between total carbon and total inorganic carbon. Compost neutrophilic heterotrophic bacterial counts were 3.0±0.5 × 107 CFU g−1.
Table 1.
Select initial biological and chemical properties of IKMHSS mine tailings and compost amended tailings.
| Biochemical property | Unamended tailings | Tailings + 15% compost |
|---|---|---|
| NHC (CFU g−1)a | 1.7±1.3 × 102 | 1.5±1.7 × 104 |
| Feoxidizers (MPN g−1)b | 1.6 × 104 | 2.7 × 103 |
| Total organic carbon (g kg−1) | 0.14 | 42.3 |
| Total inorganic carbon (g kg−1) | 0.43 | 0.31 |
| Total nitrogen (g kg−1) | 0.013 | 1.7 |
| EC (dS m−1)c | 4.92 | 11.0 |
| pHc | 4.56 | 6.67 |
| As (mg kg−1) | 2590 | 2202d |
| Pb (mg kg−1) | 2200 | 1870d |
| Zn (mg kg−1) | 2000 | 1735d |
Neutrophilic heterotrophic bacteria plated on R2A medium at pH 7 as described previously (Mendez et al. 2007).
Values from Solís-Dominguez et al. (2011)
Measured in a DI water extraction (1:1).
Calculated from a 15 wt% dilution with compost [As] = 1.07 mg kg−1;[Pb] = 3.31 mg kg−1; [Zn] = 236 mg kg−1
Sample collection, processing and storage
Time zero (t0) analyses for all treatments were performed on triplicate samples of TO and TC materials collected immediately following homogenization and prior to seeding. TC sample data provided the t0 information for TC, BG, and QB mesocosms since the same starting material was used for the top 20 cm of these treatments. Following mesocosms establishment, samples were collected at 3, 6, and 9 months for DNA and RNA quantification as described below.
At 3, 6, and 9 months (t3, t6, t9), a sample core (3 cm diameter) was removed from each of the twelve mesocosms for DNA and RNA quantification (Figure 1). In unplanted control mesocosms, the core was removed from a random location, whereas in planted mesocosms the sampling targeted rhizosphere-influenced soil by selecting a single plant of average health. The shoot was removed at the soil surface and the core placed over the stem to sample the rhizosphere-influenced soil. The root was removed from the core and the remaining soil representing the top 15 cm of the mesocosm was removed and homogenized under sterile conditions. Samples for DNA (0.5 g) and RNA (0.25 g) extractions were collected simultaneously and stored at −80°C until extracted. Life Guard Soil Preservation Solution (MO BIO Laboratories, Inc., Carlsbad, CA) was added to the RNA sample immediately after sampling. Sample soil moisture content was determined from a sub-sample removed from each core. Sample collection and processing for all twelve mesocosms was completed within a two-day period. Final plant cover of each mesocosm was estimated from mesocosm photos using the Image J software (imagej.nih.gov/ij/). Analysis was done in triplicate for each planted mesocosm to determine the portion of mesocosms surface covered by plants. This measurement was not intended as an estimate of biomass, but rather a relative indicator of within treatment variability in plant establishment.
Figure 1.
Growth of planted treatments at 9 months during the greenhouse mesocosm experiment; A) buffalo grass mesocosms 3, 5, and 12 and B) quailbush mesocosms 4, 7, and 10. Arrows indicate positions of sample cores when visible.
DNA Extraction
DNA was extracted using the FastDNA Spin Kit for Soil (MP Biomedicals, Solon, OH). All extraction reagents and materials (with the exception of MT Buffer) were UV-sterilized for 30 min to remove contaminating DNA. Soil samples were thawed on ice prior to extraction and processed following the revised manufacturer’s protocol described by Solís-Dominguez et al. (2011). In addition, a 2x rinse of the Binding Matrix-DNA complex with saturated 4M Guanidine Thiocyanate (Sigma, St. Louis, MO) was included for all compost-amended samples to remove humic materials. Kit filters were dried overnight prior to DNA elution with 50 µl of 60°C UV-sterilized Nuclease-free DEPC-treated H2O (ISC BioExpress, Kaysville, UT). Extracts were stored at −20°C for further analysis.
RNA Extraction
After testing multiple extraction kits and RNA preservation solutions, the ZR Soil/Fecal RNA MicroPrep kit (ZymoResearch Corporation) in combination with LifeGuard Soil Preservation Solution was found to generate the highest RNA yields with the lowest inhibition to downstream reactions. All equipment was pre-treated with RNaseZap Wipes (Ambion, Grand Island, NY) and reagents and tubes were UV-sterilized for 30 min with the exception of S/F RNA Lysis Buffer (ZymoResearch Corporation). Soil samples were thawed on ice and centrifuged to remove the LifeGuard Soil Preservation solution (MO BIO Laboratories, Carlsbad, CA). The ZR manufacturer’s protocol was followed using 1.0 mL of S/F RNA Lysis solution and 5 min of bead beating. The full extraction volume was processed by sequential reloading of the Zymo-Spin IIIC column followed by RNA elution in 33 µL UV-sterilized Nuclease-free DEPC-treated H2O (ISC BioExpress). Residual DNA was removed in a 35 min DNase reaction at 37°C as described previously (Neilson et al. 2010). RNA quality was confirmed by gel electrophoresis using the Alpha Imager system (Alpha Innotech, San Leandro, CA) and extracts were stored at −80°C.
Target Genes
Three target genes were quantified in this study. The V7/V8 variable region of the 16S rRNA gene was amplified using primers 1055–1070F/1392–1406R (Ferris, Muyzer and Ward 1996). The nifH gene, encoding nitrogenase reductase, the Fe-protein subunit of the nitrogenase enzyme, was amplified with degenerate primers PolF/PolR (Poly, Monrozier, and Bally 2001). The amoA gene, encoding a subunit of the bacterial ammonia monooxygenase enzyme, was amplified with amoA-1F/amoA-2R (Rotthauwe, Witzel, and Liesack 1997; Stephen et al. 1999). Limited clone libraries (25 clones) were generated for each gene to identify the dominant populations from two time points, t0-TC and t3-BG samples (Supporting Information, Table S1, Table S2). This information was used to pick representative sequences for the qPCR calibration curves.
Reverse Transcription PCR (RT-PCR)
The Quant-iT RiboGreen RNA Reagent and Kit (Molecular Probes, Eugene, OR) was used to quantify RNA concentrations with a TBS 380 fluorometer (Turner Biosystems, Sunny Valley, CA). Extracts were normalized to the lowest concentrations of t0 unamended and compost-amended RNA concentrations to minimize variability in reverse transcription (RT) reactions. TO samples were normalized to 41 pg µL−1 and compost-amended samples were normalized to 3.7 ng µL−1. All respective samples were normalized to these concentrations with the exception of isolated samples from later periods in the study with lower RNA yields.
cDNA was transcribed from RNA using iScript cDNA Synthesis kit (Bio-Rad Laboratories) with 0.33 ng RNA reaction−1 for TO samples and 30 ng RNA reaction−1 for compost-amended samples. RT-negative reactions were included with all reactions to confirm the absence of DNA. An RT reaction efficiency analysis procedure was implemented using Pseudomonas aeruginosa PAO1 RNA as a positive control according to the method proposed by Libus and Storchova (2006). This assessment strategy calculates the cDNA yield from the RT reaction using a known concentration of total RNA. This positive control was included in all RT reactions and RT efficiency was determined to be consistent across all RT reactions.
Quantification of Gene Abundance and Gene Expression
16S rRNA, amoA and nifH gene abundance and activity were quantified using quantitative PCR (qPCR) and quantitative reverse transcription PCR (qRT-PCR) of DNA and cDNA extracts, respectively. A CFX96 Real-Time Detection System (Bio-Rad Laboratories, Hercules, CA) was used for amplification of triplicate 10 µL reactions as follows: 1 µL community DNA or cDNA, 5 µL SsoFast EvaGreen Supermix and 0.4 µM (16S rRNA and nifH) or 0.45 µM (amoA) of each primer. Amplification protocols for 16S rRNA and nifH were: 95°C for 1 min, followed by 46 cycles (16S rRNA) or 50 cycles (nifH) of 95°C for 6 s and 55°C (16S rRNA) or 59°C (nifH) for 6 s. The amplification protocol for amoA was 95°C for 3 min, followed by 50 cycles of 95°C for 7 s and 56°C for 7 s. The melt curve protocol was 95°C for 10 s followed by 5 s cycles increasing from 65–95°C in 0.5°C increments and a final extension of 72°C for 5 min. Product specificity was confirmed by both melt curve analysis and gel electrophoresis. DNA controls were used to normalize for inter-plate variations in amplification efficiencies among plates being assayed for the same gene.
Calibration curve
Clones representing the most abundant taxonomic group for each gene, 16S rRNA, nifH, and amoA, from the BG-t3 libraries were selected to generate the calibration curves (Table 2, Table S1 and Table S2). Plasmids from clones containing the target gene-inserts were used as standards and were quantified using Quant-iT PicoGreen (Molecular Probes) and the TBS 380 Fluorometer. Amplification efficiencies (E%) were generated for each gene based upon Single Threshold Cq Determination Mode with Baseline Subtracted Curve Fit (Table 2). Abundance was expressed as gene copy number gram dry soil−1 for both q-PCR and qRT-PCR.
Table 2.
Clones used as standards for q-PCR/qRT-PCR of 16S rRNA, nifH, and amoA genes. Amplification efficiency and detection limits are indicated.
| Genes |
||||
|---|---|---|---|---|
| 16S rRNA | nifH | amoA | ||
| Closest GenBank identity of mesocosm clone used as standarda (% Identity) | Pseudolabrys sp. clone JF731561.1 (100%) | Uncultured DGGE band GU097349.1 (96%) | Uncultured clone S4-3 AB465017.1 (100%) | |
| Amplicon length (bp) | 350 | 359 | 491 | |
| Standard Curve Parameters | ||||
| Abundance (DNA) | Reaction Efficiency (%) | 91.9 | 76.6 | 81.2 |
| Detection threshold (Log copies g dry soil−1) | 3.6 | 6.5 | 4.0 | |
| Activity (cDNA) | Reaction Efficiency (%) | 90.9 | 75.7 | 80.5 |
| Detection threshold (Log copies g dry soil−1) | 8.3 | 4.6 | 2.9 | |
GenBank accession number listed after clone description
Minimum and maximum detection thresholds were set for qPCR and qRT-PCR of each gene based on the following criteria: 1) the highest and lowest Cq levels with highly reproducible standard curve traces for the specific target gene; 2) Cq of a lower value than the Cq of reagent contaminants from DNA and RNA extraction blanks processed in parallel with the respective soil samples; 3) Cq value lower than detection level of residual DNA in qRT-PCR from RT-negative controls; and 4) template Cq producing a product with an accurate melt curve.
Select samples with nitrogen-cycling genes that were undetectable with qPCR due to low template concentrations or PCR inhibition were subjected to a two-step conventional and qPCR to determine the potential presence of that gene in the tailings sample. The confirmation involved a pre-qPCR conventional PCR amplification of 5–12 cycles followed by qPCR using 1 µL of the conventional PCR reaction as described in the Supporting Information.
Statistical Analysis
Treatment effects were quantified by one-way ANOVA with differences identified by the Tukey-Kramer HSD test for multiple means (p < 0.05). Significant differences in gene copy number were determined using the log of 16S rDNA or rRNA copy number g dry soil−1. Significant differences between initial and final gene copy numbers were identified by paired t-test (p<0.05). All analyses were done using JMP 8 (SAS Institute Inc., Cary, NC).
Accession numbers
Gene sequences were deposited in the GenBank database with the following accession numbers: 16S rRNA gene, KF573458-KF573479, KF573481-KF573491 and KF573493-KF573495 (Table S2); nifH gene, KF700641-KF700658; and amoA gene KF700659-KF700674.
RESULTS
Mesocosm establishment
The impact of the 15% compost amendment was revealed immediately following amendment. Increases were observed in pH, EC, TOC, TN and neutrophilic plate counts (Table 1). Germination of buffalo grass (BG) and quailbush (QB) was observed within 6 days of the first irrigation (t0), and plants were well established by 3 months. Due to higher than expected QB germination rates, plants were thinned by 50% following plant establishment. Plant stress, as defined by a qualitative decrease in leaf greenness, was observed beginning at 3 months (t3) for BG and 6 months (t6) for QB. Final plant cover was quantified at nine months (t9) as a percent coverage of the mesocosm surface. QB coverage was 100% for all three mesocosms, whereas BG coverage was 4.8±0.2%, 97.4±0.5%, and 93.9±0.8% for mesocosms 3, 5, and 12, respectively (Figure 1).
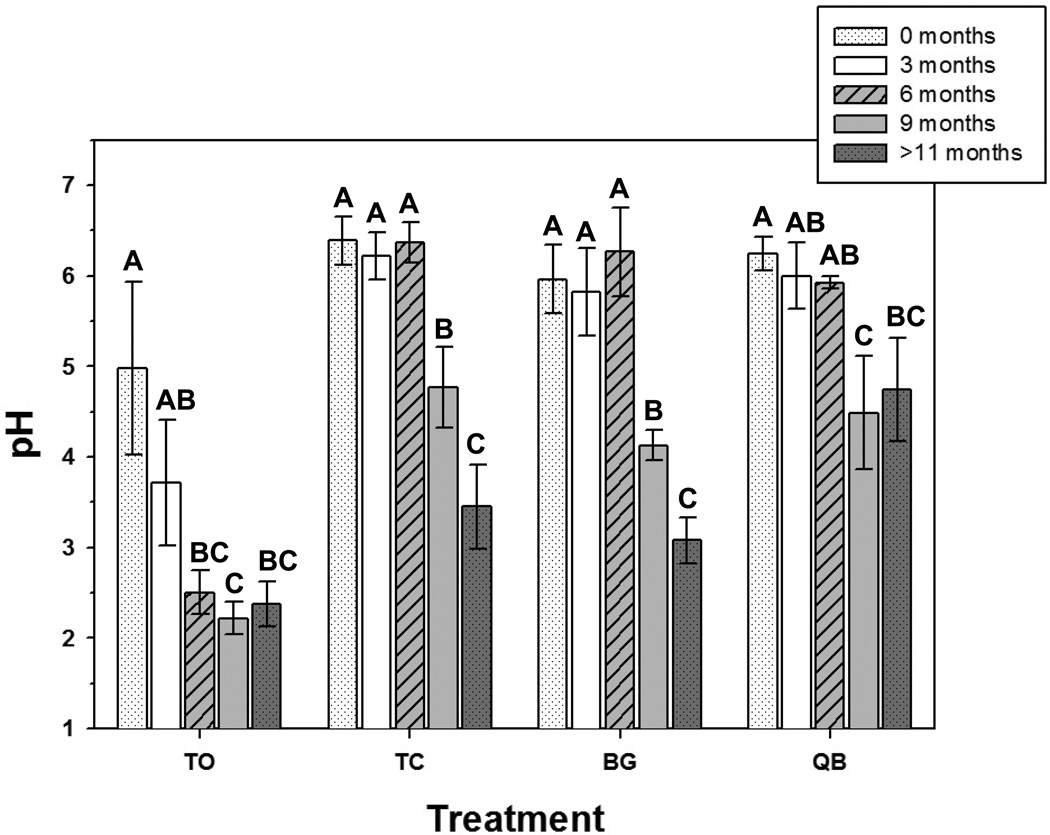
The average pore water pH for the tailings only (TO) mesocosms decreased from 5.0 ±1.0 at t0 to 2.4±0.2 at t9 (Figure 2). In contrast, the average pH of all the compost-amended mesocosms at t0 was 6.2±0.3 (TC, BG, QB) and remained stable through t6. A significant decrease in pH was then observed at t9 for TC, BG, and QB mesocosms. Analysis of pore water collected over the 3 months following the last soil sample collection (t10-t12) revealed that the pH of TC and BG mesocosms continued to decrease significantly, while the QB treatment stabilized at an average pH of 4.7±0.6 (Figure 2). Pore water EC ranged from 13.8–26.2 dS m−1 for TO and 34.6–59.8 dS m−1 for all compost amended treatments. Thus, pore water salt levels were lower in TO than in in all compost-amended treatments.
Figure 2.
Pore water pH collected from mesocosms. At each time point, water samples were collected at 5 cm and 15 cm depths and samples from the two depths from each mesocosm were consolidated prior to pH analysis. Treatment designations are TO, tailings only; TC, tailings + 15% compost (w/w); BG, tailings + 15% compost seeded with buffalo grass; and QB, tailings + 15% compost seeded with quailbush. Treatment means with different letters are significantly different (ANOVA, p<0.05; Tukey-Kramer HSD test).
16S rRNA gene abundance and activity
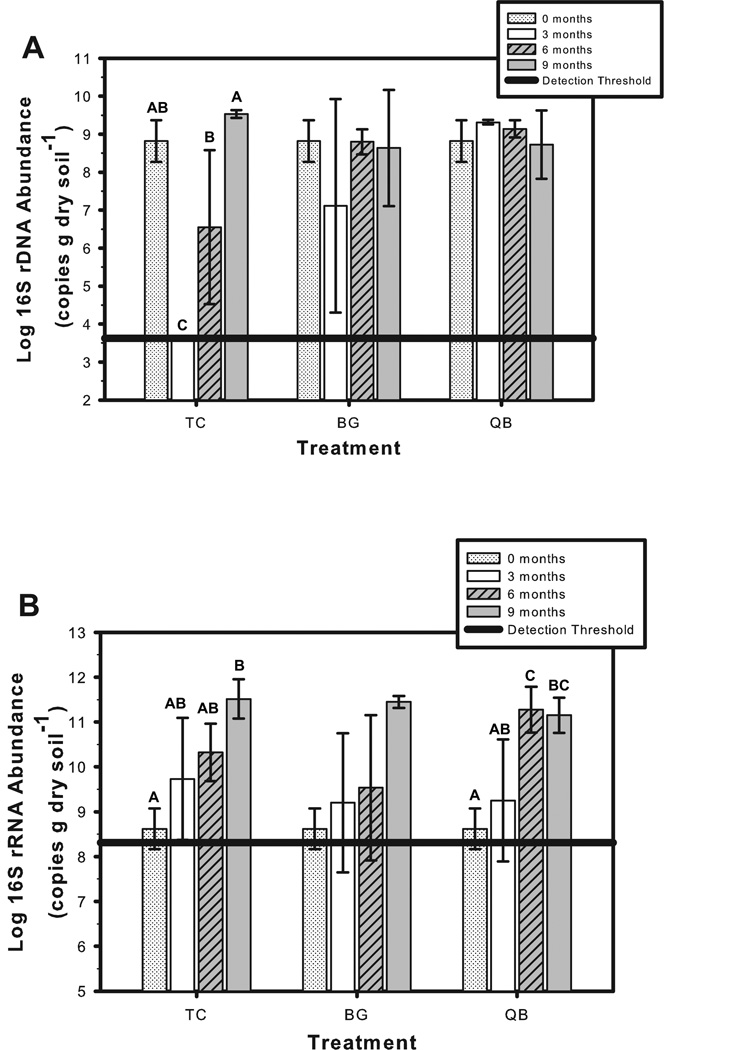
The 16S rRNA gene qPCR provided a molecular quantification of total bacterial biomass throughout the nine months of the phytostabilization trial. Bacterial abundance in unamended control mesocosms (TO) remained below the detection level throughout the experiment. A 5-log increase in 16S rDNA copy number above the detection threshold was documented immediately following compost amendment (t0 for TC, BG, and QB; Figure 3A). TC 16S rDNA abundance fell below the detection threshold at t3, but gradually returned to the t0 16S rDNA copy number by t9 (Figure 3A). In contrast, no significant change in average biomass was observed for the BG or QB treatments throughout the experiment (Figure 3A), suggesting that both buffalo grass and quailbush provided a vital buffer for stabilizing the microbial inoculum introduced by compost amendment. No significant difference in 16S rDNA abundance was observed between BG and QB at any time point during the experiment and by t9 bacterial abundance was comparable in all compost amended treatments; BG, QB, and TC (F=0.701, p=0.53).
Figure 3.
Bacterial abundance (A) measured as the log of 16S rDNA copies g dry soil−1 in mesocosm soil samples and activity (B) measured as the log of 16S rRNA copies g dry soil−1 in mesocosm soil samples. Neither 16S rRNA genes or 16S rRNA were detected in the unamended tailings (TO). Treatment designations are TC, tailings + 15% compost (w/w); BG, tailings + 15% compost seeded with buffalo grass; and QB, tailings + 15% compost seeded with quailbush. Treatment means with different letters are significantly different (ANOVA, p < 0.05; Tukey-Kramer HSD test). The absence of letters indicates no significant difference between means for that treatment.
The QB mesocosms demonstrated the greatest intra-treatment consistency in bacterial abundance throughout the experiment with a coefficient of variation (%CV) less than or equal to 10%. In contrast, the %CV for TC was 31% at t6, and for BG it was 39% and 18% at t3 and t9, respectively. Percent plant cover was also more consistent for the QB mesocosms than for BG. Notable was the high variability in 16S rDNA abundance for the BG treatment at t9 which was due to the low 16S rRNA copy number in mesocosm 3 (6.9±0.4 log copies g dry soil−1), 2.5 logs lower than for BG mesocosms 5 and 12 (9.5 and 9.6 log copies g dry soil−1, respectively). Recall that the percent plant cover was 5% in mesocosm 3 as compared to 97% and 94% coverage in mesocosms 5 and 12, respectively. These results suggest a potential association between final plant cover and 16S rDNA copy number in planted treatments.
16S rRNA qRT-PCR was used as a proxy for the relative activity of the bacterial community in the four treatments during the experiment. No activity was detected in the TO treatment throughout the experiment, whereas a low level of activity could be detected in all compost-amended treatments immediately following amendment (Figure 3B). This low level of activity corresponds to the pattern observed for cultured NHC at t0. A calculation based on the theoretical heterotrophic bacterial inoculum provided by the compost compared with actual plate counts recovered from the tailings-compost mixture (Table 1) shows that just 0.3% of the expected NHC were recovered from the compost-amended tailings. This data suggests that a major portion of the compost bacterial inoculum is compromised when exposed to the tailings, a finding that explains the low 16S rRNA gene activity at t0. A significant increase in bacterial activity was observed by t6 for the QB treatment and t9 for TC (p<0.05; Figure 3B). Bacterial activity also increased in BG mesocosms, but the change was less significant (F=3.58, p=0.07), due to the high intra-treatment variability at t3 and t6. However, a paired t-test showed that BG activity at t9 was significantly higher than at t0 (p=0.0006). Whereas plant establishment was associated with sustained bacterial abundance levels at t3 in QB and BG treatments, no plant associated enhancement of 16S rRNA gene activity was observed in BG, and QB mesocosms at any given time point (data not shown).
Functional gene abundance and activity
The abundance of the nifH and amoA genes was quantified to evaluate the bacterial nitrogen fixation and nitrification potential of the tailings during the assisted phytostabilization experiment. The nifH gene was first quantified at t9 in the BG treatment. As observed with 16S rRNA gene abundance, mesocosms 5 and 12 (8.33±0.02 and 7.94±0.07 log copies g−1 dry soil, respectively) had higher nifH abundances than mesocosm 3 (6.54±0.04 log copies g−1 dry soil). Quantifiable levels of the amoA gene were observed at t6 and t9 in the QB treatment (Table 3). Sensitive detection of these functional genes was complicated by low template concentrations and inhibition from materials co-extracted from the mine tailings. Thus, a 2-step conventional and qPCR protocol (described in the Methods) was applied as a qualitative presence/absence assessment for samples found to be below the qPCR detection limit. The 2-step method confirmed that both of these functional genes were present at other time points during the study in the TC, BG and QB mesocosms (Table 3), but at abundances below qPCR detection levels. Taken together, the results reveal that nifH and amoA genes were most abundant in the planted treatments. Of particular interest was the fact that nifH was most abundant in the BG rhizosphere, whereas amoA was more abundant in QB rhizosphere. These results emphasize the importance of monitoring multiple functional genes to detect plant-induced changes in rhizosphere microbial communities.
Table 3.
qPCR and qRT-PCR quantification of nifH and amoA genes representing nitrogen fixation and ammonia oxidation potential and activity (gene abundance and expression).
| Gene Quantification (log copies g dry soil−1)1 |
|||||
|---|---|---|---|---|---|
| Time (Months) | |||||
| Gene | Treatment2 | 0 | 3 | 6 | 9 |
| nifH Abundance | TC | * | * | * | * |
| BG | * | * | * | 7.4 ± 1.0 | |
| QB | * | * | * | * | |
| nifH Expression | TC | ND | ND | ND | ND |
| BG | ND | ND | ND | ND | |
| QB | ND | ND | ND | ND | |
| amoA Abundance | TC | * | ND | ND | * |
| BG | * | ND | ND | * | |
| QB | * | * | 5.6 ± 0.1 | 4.9 ± 0.4 | |
| amoA Expression | TC | ND | ND | ND | ND |
| BG | ND | ND | ND | ND | |
| QB | ND | ND | ND | ND | |
Non-numerical labels represent classifications of gene presence based upon a minimum occurrence in 2 of 3 mesocosm replicates.
Only compost-amended treatments are represented since gene abundance and expression were below detection limits in the TO treatment for the duration of the experiment.
Gene was not detected using real time PCR, but could be detected using a double conventional qPCR as explained in the Materials and Methods
ND: Non-detectable
No nifH or amoA gene activity was detected by qRT-PCR in any of the mesocosms throughout the experiment (Table 3). The 16S rRNA qRT-PCR provides an important control for this analysis because all RNA genes were quantified from the same pool of cDNA. Thus, the successful quantification of 16S rRNA confirms that RNA was successfully extracted from the samples. However, mRNA represents less than 10% of total RNA, thus it is possible that amoA and nifH transcripts may have been present in some samples at levels below the qPCR detection limit.
DISCUSSION
Successful plant establishment was observed for BG and QB treatments by t3. Beginning at 6 months, a decline in plant health was observed as evidenced by a decrease in plant greenness for both BG and QB and a loss of plant cover in BG mesocosms. This decline was accompanied by a significant decrease in pH as the buffering capacity of the compost was exhausted. Of interest was the fact that in the QB treatment pH stabilized at 4.7±0.6, whereas the TC and BG treatments continued to decrease to 3.5±0.5 and 3.1±0.3, respectively. These results contrast with our previous short-term greenhouse pot-study in compost-amended IKMSS tailings, where pH remained stable in the compost amended treatment for 60 days and actually increased significantly in planted treatments (Solís-Domínguez et al., 2012). Surprisingly, after 60 days, BG treatments maintained a higher pH in the more acidic bulk and rhizosphere influenced tailings than QB. Plants are known to be capable of controlling rhizosphere pH by exuding protons or anions (reviewed in Hinsinger et al. 2003), and lab experiments demonstrated that buffalo grass and mesquite seedling roots increased agarose pH from 5.5 to greater than 6.8 (Solís-Domínguez et al., 2012). We hypothesize that the higher pH observed in QB mesocosms at t9, is a result of the more robust plant growth observed for this plant species at this time point allowing the plant to continue to alkanize the rhizosphere. QB is best known for growth in nutrient poor alkaline and saline soils, but it can also grow in acidic soils and thus may be more resilient than BG under extended adverse conditions. The results of these greenhouse studies highlight the importance of long-term monitoring of phytostabilization to ensure that initial conditions are sustainable.
The target gene analyses presented in this study facilitated an in situ evaluation of different bacterial responses to compost amendment and plant establishment in acidic metalliferous mine tailings. The combined use of 16S rRNA gene qPCR and qRT-PCR provided a general temporal quantification of bacterial biomass and activity during the phytostabilization trial. 16S rRNA genes and their activities were detected in all compost-amended treatments, but not in the unamended tailings, thus confirming that compost provided a significant microbial inoculum to the system. The cultured estimates for Fe-oxidizers and heterotrophs in unamended tailings (Tables 1 and Table 2) indicate that viable microbes were present in the unamended tailings, but at levels below the qPCR detection limit. As explained previously, we have shown that sensitive detection by qPCR in mine tailings extracts is limited by inhibitory substances, thus the analysis presented here is intended to identify relative in situ differences between treatments. Compost amendment led to a greater than 5-log increase in gene copy number g dry soil−1 and the assay revealed that plants sustained bacterial abundance at initial inoculum levels and prevented the sharp decline in biomass observed at t3 for the TC treatment. In addition, the %CV in 16S rRNA copy number was lowest for QB, the plant with the most consistent plant cover, again suggesting that successful plant establishment was associated with stable microbial biomass.
The differential patterns observed for gene abundance and activity in TC mesocosms suggest that the introduced compost bacterial community was functioning at a low metabolic activity level following amendment. Looking specifically at t3, the significant decrease in 16S rRNA gene abundance in the TC treatment indicates that many of the introduced compost populations did not survive in the tailings in the absence of plants. However, despite the decrease in abundance, gene activity in the TC mesocosms increased slightly from t0 to t3 at levels similar to the planted treatments suggesting that the populations that did survive were adapting. Taken together, these results indicate that either 1) the microbial activity of all bacteria was too low to reflect the significant differences in abundance or 2) that only a small fraction of the total community was metabolically active. The latter hypothesis is supported by the fact that the t0 NHC assay recovered just 0.3% of the heterotrophic bacteria theoretically introduced with the compost amendment, suggesting that the majority became nonviable or viable but non-culturable due to the stress of the tailings environment. A significant increase in activity was observed in all treatments by t9 with no significant difference observed between planted and unplanted compost-amended treatments at any time point. Thus, the increase in activity was not specifically associated with plant establishment.
The nitrogen cycling genes, nifH and amoA, were only detectable by qPCR in BG and QB plant rhizospheres, respectively, and only in the latter half of the experiment. This pattern is of interest because the timing for detection of these functional genes corresponded to the acidification of tailings pore water and the decline in plant health. Thus, plant establishment enhanced the abundance of these bacterial populations despite adverse substrate conditions. Of particular interest, was the 1.6-log lower nifH gene copy number in BG mesocosm 3 as compared to the average of mesocosms 5 and 12. Recall that the plant cover in mesocosm 3 was 5% compared with 97% and 94% for 5 and 12, respectively. This observation suggests that plant cover influences the abundance of these two key nitrogen cycling genes. These results follow a similar pattern observed in a recent survey evaluating successional plant establishment in nonacidic copper mine tailings. In this study nifH genes were detectable in bare tailings, but the copy number abundance was two logs higher in vegetated areas (Huang et al. 2011). In addition, research has shown amoA gene abundance to be strongly influenced by changes in land use practices and AOB have been proposed as indicators of biological soil ecosystem response to perturbations such as metal contamination (Wessén and Hallin, 2011; Zeglin et al. 2011). Unlike the gene abundances, nifH and amoA gene activity was not detected in any treatment during the 9 month phytostabilization trial. Recent studies suggest that a major portion of microorganisms are dormant in soil ecosystems and that this dormancy may range from occasionally active to nonviable (Yarwood et al. 2012). For example, in a study of forest soils, Yarwood et al. (2012) could not detect amoA bacterial activity despite the presence of amoA gene copy numbers at levels comparable to those detected in the QB rhizosphere (Yarwood et al. 2012). Despite the fact that amoA expression was not detectable, studies have found that the size of AOB communities is positively correlated with nitrification activity (reviewed in Wessén and Hallin, 2011). Thus, the relative abundance of amoA and nifH genes may be more suitable indicators of ecosystem potential than their respective transcripts.
The molecular analysis used in this study identified key patterns in bacterial community development during the assisted phytostabilization mesocosm study. The16S rRNA qPCR confirmed that compost introduced a significant bacterial inoculum, but that the introduced community was largely inactive immediately following inoculation. By the conclusion of the experiment a significant increase in activity was observed in all treatments. In addition, established BG and QB plants provided a protective buffer for the initial bacterial inoculum introduced with compost amendment and supported increased abundances of nifH and amoA genes. The observed increase in 16S rRNA activity combined with the absence of detectable amoA and nifH activity and the significant decrease in pH suggests that Fe- and S-oxidizing populations may be the more active members of the communities by the end of the experiment. Current community analysis using iTag sequencing and Geochip functional gene microarrays is characterizing the taxonomic and functional profiles from this mesocosm experiment to explain the quantitative patterns profiled in this study. The most important observation from this work was that whereas plant health was compromised by tailings acidification (Ginocchio et al. 2009; Soils-Dominguez et al. 2012), the functional gene analysis revealed increased abundances of two key nitrogen cycling genes in plant rhizosphere communities.
To our knowledge this is the first study tracking specific functional gene abundances during an extended phytostabilization trial. The target gene results of this study suggest that plant establishment can sustain microbial inocula in severely disturbed ecosystems such as mine tailings and specifically enhance the survival of plant growth promoting populations as represented by the two key nitrogen cycling genes. Further, the presence of detectable levels of nifH genes in the BG rhizosphere despite the poor health of BG suggests that initial plants used in assisted phytostabilization may support important plant growth promoting bacteria even if plant health is poor. The end-goal of phytostabilization is to develop a complex, species rich and self-regenerating plant community, but the achievement of this goal in acid-generating tailings may be an incremental process requiring successive plantings to accomplish ecosystem development. (reviewed in Bradshaw and Hüttl, 2001; Wiegleb and Felinks, 2001). The long-term success of phytostabilization requires tools to monitor progress in soil ecosystem development. The results of this study suggest that amoA and nifH are two candidate genes with potential for documenting incremental improvements in the microbial community of the tailings material during the phytostabilization process.
Supplementary Material
Acknowledgements
This research was supported by Grants P42 ES04940 and R01 ES017079 from the National Institute of Environmental Health Sciences Superfund Research Program, NIH (USA). We wish to thank Steven Schuchardt, president of North American Industries, for providing access to the IKMHSS site and for supporting the research presented in this study.
Footnotes
Supporting Information
The supporting information includes methods and results for the clone libraries generated to identify representative gene sequences to be used for qPCR calibration curves. Tables are included characterizing all clones in the respective target gene libraries.
References
- Anawar HM, Canha N, Santa-Regina I, Freitas MC. Adaptation, tolerance, and evolution of plant species in a pyrite mine in response to contamination level and properties of mine tailings: sustainable rehabilitation. J Soils Sediments. 2013;13:730–741. [Google Scholar]
- Bradshaw AD, Hüttl RF. Future minesite restoration involves a broader approach. Ecol Eng. 2001;17:87–90. [Google Scholar]
- Epelde L, Becerril JM, Mijangos I, Garbisu C. Evaluation of the efficiency of a phytostabilization process with biological indicators of soil health. J Environ Qual. 2009;38:2041–2049. doi: 10.2134/jeq2009.0006. [DOI] [PubMed] [Google Scholar]
- Ferris MJ, Muyzer G, Ward DM. Denaturing gradient gel electrophoresis profiles of 16S rRNA-defined populations inhabiting a hot spring microbial mat community. App Environ Microbiol. 1996;62:340–346. doi: 10.1128/aem.62.2.340-346.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginocchio R, de la Fuente LM, Sanchez P, Bustamante E, Silva Y, Urrestarazu P, Rodriguez PH. Soil acidification as a confounding factor on metal phytotoxicity in soils spiked with copper-rich mine wastes. Environ Toxicol Chem. 2009;28:2069–2081. doi: 10.1897/08-617.1. [DOI] [PubMed] [Google Scholar]
- Glick BR. Bacterial ACC deaminase and the alleviation of plant stress. Adv Appl Microbiol. 2004;56:291–312. doi: 10.1016/S0065-2164(04)56009-4. [DOI] [PubMed] [Google Scholar]
- Gómez-Sagasti MT, Alkorta I, Becerril JM, Epelde L, Anza M, Garbisu C. Microbial monitoring of the recovery of soil quality during heavy metal phytoremediation. Water, Air, Soil Pollut. 2012;223:3249–3262. [Google Scholar]
- Hinsinger P, Plassard C, Tang C, Jaillard B. Origins of root-mediated pH changes in the rhizosphere and their responces to environmental constraints: a review. Plant Soil. 2003;248:43–59. [Google Scholar]
- Huang L, Baumgartl T, Mulligan D. Is rhizosphere remediation sufficient for sustainable revegetation of mine tailings? Ann Bot. 2012;110:223–238. doi: 10.1093/aob/mcs115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang L, Tang F, Song Y, Wan C, Wang S, Liu W, Shu WS. Biodiversity, abundance, and activity of nitrogen-fixing bacteria during primary succession on a copper mine tailings. FEMS Microbiol Ecol. 2011;78:439–450. doi: 10.1111/j.1574-6941.2011.01178.x. [DOI] [PubMed] [Google Scholar]
- Janssen PH. Identifying the dominant soil bacterial taxa in libraries of 16S rRNA and 16S rRNA genes. Appl Environ Microbiol. 2006;72:1719–1728. doi: 10.1128/AEM.72.3.1719-1728.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowalchuk GA, Stephen JR. Ammonia-oxidizing bacteria: a model for molecular microbial ecology. Annu Rev Microbiol. 2001;55:485–529. doi: 10.1146/annurev.micro.55.1.485. [DOI] [PubMed] [Google Scholar]
- Libus J, Storchova H. Quantification of cDNA generated by reverse transcription of total RNA provides a simple alternative tool for quantitative RT-PCR normalization. Biotechniques. 2006;41:156–162. doi: 10.2144/000112232. [DOI] [PubMed] [Google Scholar]
- Mendez MO, Glenn EP, Maier RM. Phytostabilization potential of quailbush for mine tailings: growth, metal accumulation, and microbial community changes. J Environ Qual. 2007;36:245–253. doi: 10.2134/jeq2006.0197. [DOI] [PubMed] [Google Scholar]
- Mendez MO, Maier RM. Phytostabilization of mine tailings in arid and semiarid environments-an emerging remediation technology. Environ Health Perspectives. 2008;116:278–283. doi: 10.1289/ehp.10608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendez MO, Neilson JW, Maier RM. Characterization of a bacterial community in an abandoned semiarid lead-zinc mine tailing site. Appl Environ Microbiol. 2008;74:3899–3907. doi: 10.1128/AEM.02883-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meza-Figueroa D, Maier RM, de la O-Villanueva M, Gómez-Alvarez A, Moreno-Zazueta A, Rivera J, Campillo A, Grandlic CJ, Anaya R, Palafox-Reyes J. The impact of unconfined mine tailings in residential areas from a mining town in a semi-arid environment: Nacozari, Sonora, Mexico. Chemosphere. 2009;77:140–147. doi: 10.1016/j.chemosphere.2009.04.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moynahan OS, Zabinski CA, Gannon JE. Microbial community structure and carbon-utilization diversity in mine tailings revegetation study. Restoration Ecol. 2002;10:77–87. [Google Scholar]
- Neilson JW, Zhang L, Veres TA, Chandler KB, Neilson CH, Crispin JD, Pemberton JE, Maier RM. Cadmium effects on transcriptional expression of rhlB/rhlC genes and congener distribution of monorhamnolipid and dirhamnolipid in Pseudomonas aeruginosa IGB83. Appl Microbiol Biotechnol. 2010;88:953–963. doi: 10.1007/s00253-010-2808-8. [DOI] [PubMed] [Google Scholar]
- Poly F, Monrozier LJ, Bally R. Improvement in the RFLP procedure for studying the diversity of nifH genes in communities of nitrogen fixers in soil. Res Microbiol. 2001;152:95–103. doi: 10.1016/s0923-2508(00)01172-4. [DOI] [PubMed] [Google Scholar]
- Rotthauwe JH, Witzel KP, Liesack W. The ammonia monooxygenase structural gene amoA as a functional marker: molecular fine-scale analysis of natural ammonia-oxidizing populations. Appl Environ Microbiol. 1997;63:4704–4712. doi: 10.1128/aem.63.12.4704-4712.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahrawat KL. Factors affecting nitrification in soils. Commun Soil Sci Plant Anal. 2008;39:1436–1446. [Google Scholar]
- Schaider LA, Senn DB, Brabander DJ, McCarthy KD, Shine JP. Characterization of zinc, lead and cadmium in mine waste: Implications for transport, exposure, and bioavailability. Environ Sci Technol. 2007;41:4164–4171. doi: 10.1021/es0626943. [DOI] [PubMed] [Google Scholar]
- Schippers A, Breuker A, Blazejak A, Bosecker K, Kock D, Wright TL. The biogeochemistry and microbiology of sulfidic mine waste and bioleaching dumps and heaps, and novel Fe(II)-oxidizing bacteria. Hydrometallurgy. 2010;104:342–350. [Google Scholar]
- Sessitsch A, Kuffner M, Kidd P, Vangronsveld J, Wenzel WW, Fallmann K, Puschenreiter M. The role of plant-associated bacteria in the mobilization and phytoextraction of trace elements in contaminated soils. Soil Biol Biochem. 2013;60:182–194. doi: 10.1016/j.soilbio.2013.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu WS, Ye ZH, Zhang ZQ, Lan CY, Wong MH. Natural colonization of plants on five lead/zinc mine tailings in southern China. Restoration Ecol. 2005;13:49–60. [Google Scholar]
- Solís-Dominguez FA, Valentin-Vargas A, Chorover J, Maier RM. Effect of arbuscular mycorrhizal fungi on plant biomass and the rhizosphere microbial community structure of mesquite grown in acidic lead/zinc mine tailings. Sci Total Environ. 2011;409:1009–1016. doi: 10.1016/j.scitotenv.2010.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solís-Dominguez FA, White SA, Borillo-Hutter T, Amistadi MK, Root RA, Chorover J, Maier RM. Response of key soil parameters during compost-assisted phytostabilization in extremely acidic tailings: effect of plant species. Environ Sci Technol. 2012;46:1019–1027. doi: 10.1021/es202846n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephen JR, Chang YJ, Macnaughton SJ, Kowalchuk GA, Leung KT, Flemming CA, White DC. Effect of toxic metals on indigenous soil p-subgroup proteobacterium ammonia oxidizer community structure and protection against toxicity by inoculated metal-resistant bacteria. Appl Environ Microbiol. 1999;65:95–101. doi: 10.1128/aem.65.1.95-101.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiegleb G, Felinks B. Predictability of early stages of primary succession in post-mining landscapes of Lower Lusatia, Germany. Appl Veg Sci. 2001;4:5–18. [Google Scholar]
- Wessén E, Hallin S. Abundance of archaeal and bacterial ammonia oxidizers - possible bioindicator for soil monitoring. Ecol Indic. 2011;11:1696–1698. [Google Scholar]
- Wong M. Ecological restoration of mine degraded soils, with emphasis on metal contaminated soils. Chemosphere. 2003;50:775–780. doi: 10.1016/s0045-6535(02)00232-1. [DOI] [PubMed] [Google Scholar]
- Yarwood S, Brewer E, Yarwood R, Lajtha K, Myrold D. Soil microbe active community composition and capacity of responding to litter addition after 12 years of no inputs. Appl Environ Microbiol. 2012;79:1385–1392. doi: 10.1128/AEM.03181-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeglin LH, Taylor AE, Myrold DM, Bottomley PJ. Bacterial and archaeal amoA gene distribution covaries with soil nitrification properties across a range of land uses. Environ Microbiol Rep. 2011;3:717–726. doi: 10.1111/j.1758-2229.2011.00290.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.