Abstract
The TRIM37 (or MUL) gene is located in the 17q23 chromosomal region, which is amplified in up to ~40% of breast cancers1. TRIM37 contains a RING finger domain, a hallmark of E3 ubiquitin ligases2, but its protein substrate(s) is unknown. Here we report that TRIM37 mono-ubiquitinates histone H2A, a chromatin modification associated with transcriptional repression3. We find that in human breast cancer cell lines containing amplified 17q23, TRIM37 is upregulated and, reciprocally, the major H2A ubiquitin ligase RNF23,4 (also called RING1B) is downregulated. Genome-wide chromatin immunoprecipitation (ChIP)-chip experiments in 17q23-amplified breast cancer cells identified many genes, including multiple tumour suppressors, whose promoters were bound by TRIM37 and enriched for ubiquitinated H2A (H2A-ub). However, unlike RNF2, which is a subunit of Polycomb repressive complex 1 (PRC1)3–5, we find that TRIM37 associates with Polycomb repressive complex 2 (PRC2). TRIM37, PRC2 and PRC1 are co-bound to specific target genes resulting in their transcriptional silencing. RNA interference (RNAi)-mediated knockdown of TRIM37 results in loss of H2A-ub, dissociation of PRC1 and PRC2 from target promoters, and transcriptional reactivation of silenced genes. Knockdown of TRIM37 in human breast cancer cells containing amplified 17q23 substantially decreases tumour growth in mouse xenografts. Conversely, ectopic expression of TRIM37 renders non-transformed cells tumorigenic. Collectively, our results reveal TRIM37 as an oncogenic H2A ubiquitin ligase that is overexpressed in a subset of breast cancers and promotes transformation by facilitating silencing of tumour suppressors and other genes.
We previously identified TRIM37 as a factor required for epigenetic silencing of the Fas tumour suppressor gene, and found that TRIM37 was bound to the silenced Fas promoter in Kras-transformed NIH 3T3 cells6. Mono-ubiquitination of histone H2A at lysine 119 is associated with transcriptional repression3 and we considered the possibility that TRIM37 was a histone H2A E3 ubiquitin ligase.
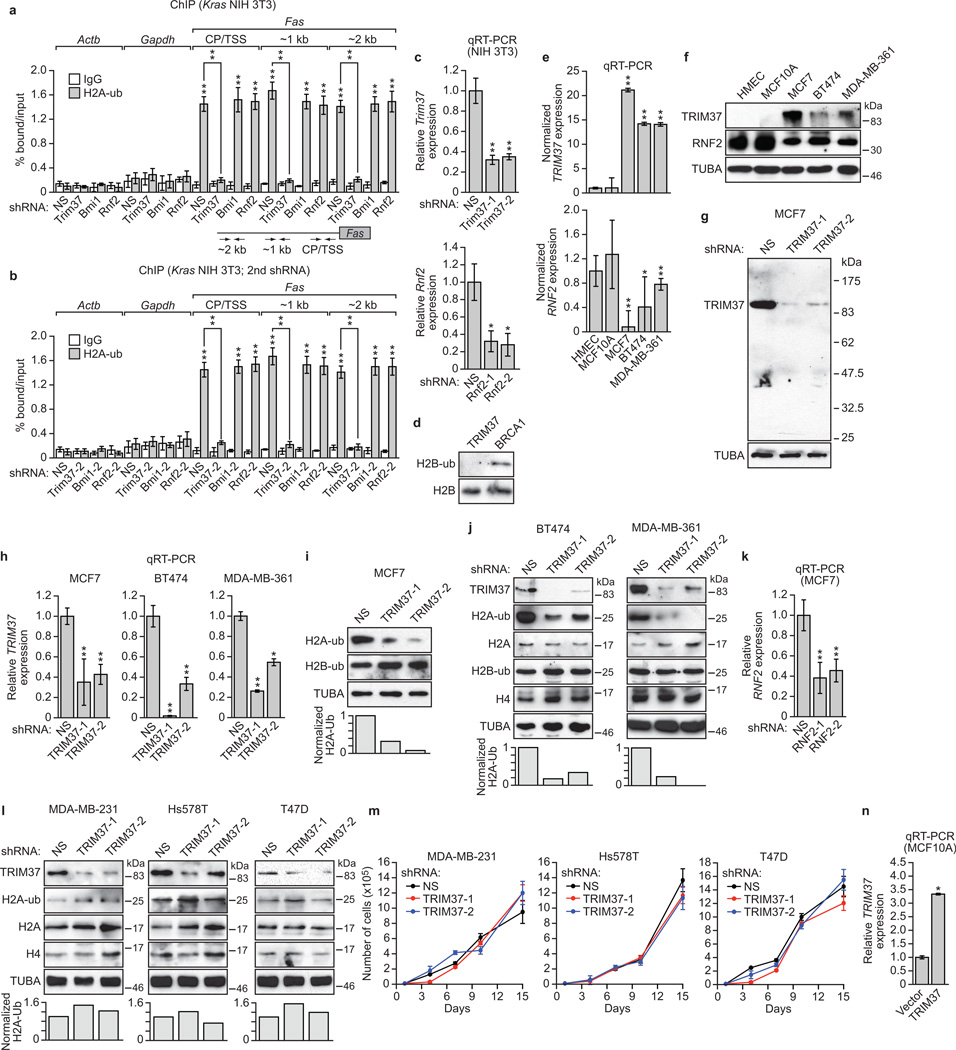
To test this idea, we first performed ChIP experiments to assess H2A-ub levels on the epigenetically repressed Fas promoter. H2A-ub was enriched at the silenced Fas promoter in Kras-transformed NIH 3T3 cells, which was lost following small hairpin RNA (shRNA)-mediated knockdown of TRIM37 (Extended Data Fig. 1a–c). By contrast, H2A-ub was retained following knockdown of RNF2, the major H2A ubiquitin ligase involved in gene regulation3,4, or BMI1, a PRC1 component7,8 that stimulates RNF2 activity7,9.
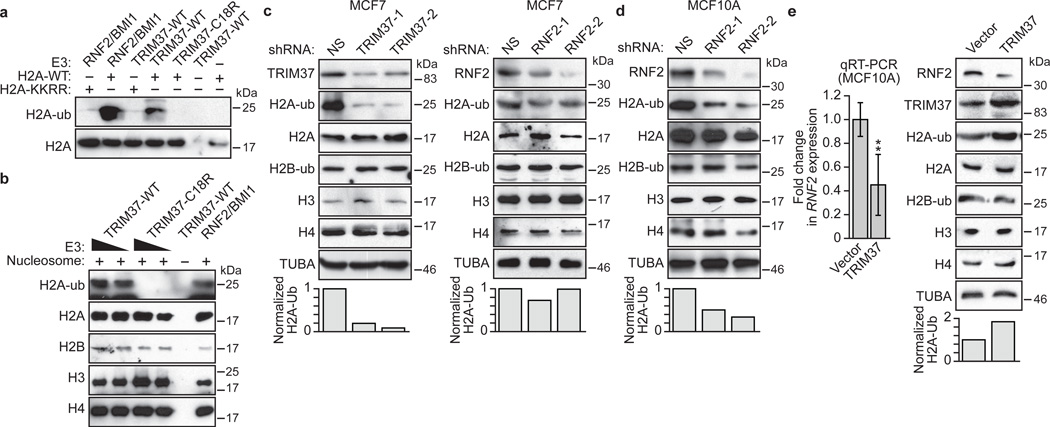
We next assessed whether TRIM37 could mono-ubiquitinate H2A at lysine 119. Purified recombinant TRIM37 or, as a positive control, the RNF2/BMI1 complex, was incubated in the presence of an E1 ubiquitin-activating enzyme, an E2 ubiquitin-conjugating enzyme, ATP and HA-tagged ubiquitin. Like RNF2/BMI1, TRIM37 could mono-ubiquitinate purified H2A, but not an H2A derivative mutated at the K118/K119 ubiquitination site (H2A-KKRR)10 or H2B (Fig. 1a and Extended Data Fig. 1d). By contrast, a TRIM37 derivative bearing a point mutation in a conserved cysteine residue in the RING finger motif (C18R), which interferes with catalytic activity2, failed to promote H2A mono-ubiquitination (Fig. 1a). Furthermore, wild-type TRIM37, but not TRIM37-C18R, could mono-ubiquitinate H2A in an assembled nucleosome (Fig. 1b).
Figure 1. TRIM37 is a histone H2A ubiquitin ligase that is over-expressed in 17q23-amplified human breast cancer cell lines.
a, b, In vitro ubiquitination assays with H2A-wild-type (WT) or H2A-KKRR (a) or reconstituted nucleosomes (b). c, (Top) Immunoblots in MCF7 cells expressing a NS, TRIM37 or RNF2 shRNA. (Bottom) Quantification of H2A-ub relative to α-tubulin (TUBA); H2A-ub in NS cells was set to 1. d, Immunoblots in MCF10A cells expressing a NS or RNF2 shRNA. e, qRT-PCR (left) and immunoblots (right) in MCF10A cells expressing TRIM37 or vector. RNF2 expression and H2A-ub levels were normalized to that obtained with vector. Error bars indicate SEM; n=3 technical replicates of a representative experiment (out of three experiments). *P<0.05; **P<0.01.
We next analysed the relationship between expression of TRIM37 and RNF2 in several human breast cancer cell lines containing amplified 17q23 (MCF7, BT474 and MDA-MB-361)11. As controls, we analysed in parallel a non-transformed human breast epithelial cell line that lacked 17q23 amplification (MCF10A) and immortalized human mammary epithelial cells (HMECs). Breast cancer cell lines containing amplified 17q23 expressed substantially higher levels of TRIM37, but lower levels of RNF2, compared to MCF10A cells and HMECs (Extended Data Fig. 1e–g).
Consistent with these expression results, knockdown of TRIM37 in MCF7, BT474 or MDA-MD-361 cells markedly decreased total H2A-ub levels (Fig. 1c, left and Extended Data Fig. 1h–j). By contrast, knockdown of RNF2 (Extended Data Fig. 1k) in MCF7 cells had only a modest effect on H2A-ub levels (Fig. 1c, right), but substantially reduced H2A-ub levels in MCF10A cells (Fig. 1d). Moreover, knockdown of TRIM37 did not have a major effect on H2A-ub levels in breast cancer cell lines lacking 17q23 amplification (Extended data Fig. 1l, m).
The expression results of Extended Data Fig. 1e, f suggested a reciprocal relationship between TRIM37 and RNF2 expression levels. Consistent with this possibility, ectopic expression of TRIM37 in MCF10A cells (Extended Data Fig. 1n) resulted in downregulation of RNF2 (Fig. 1e).
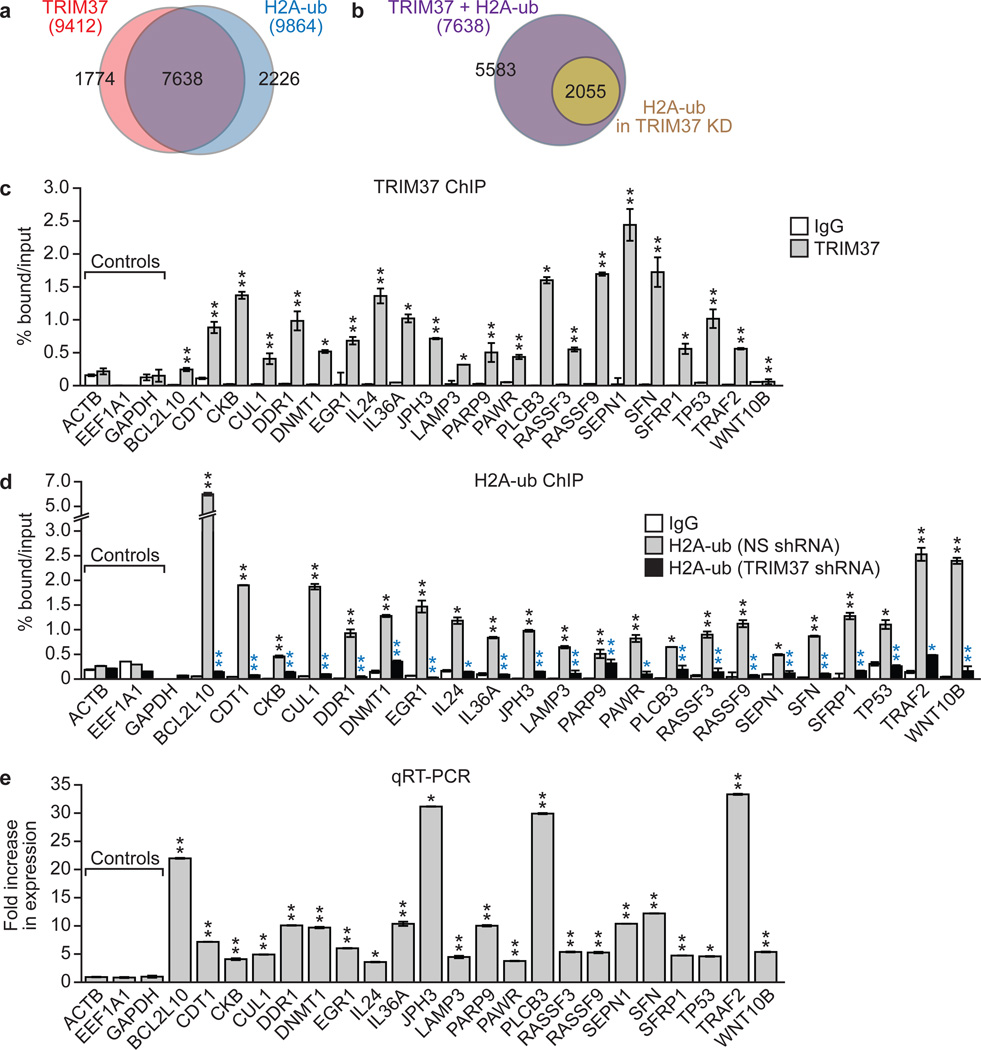
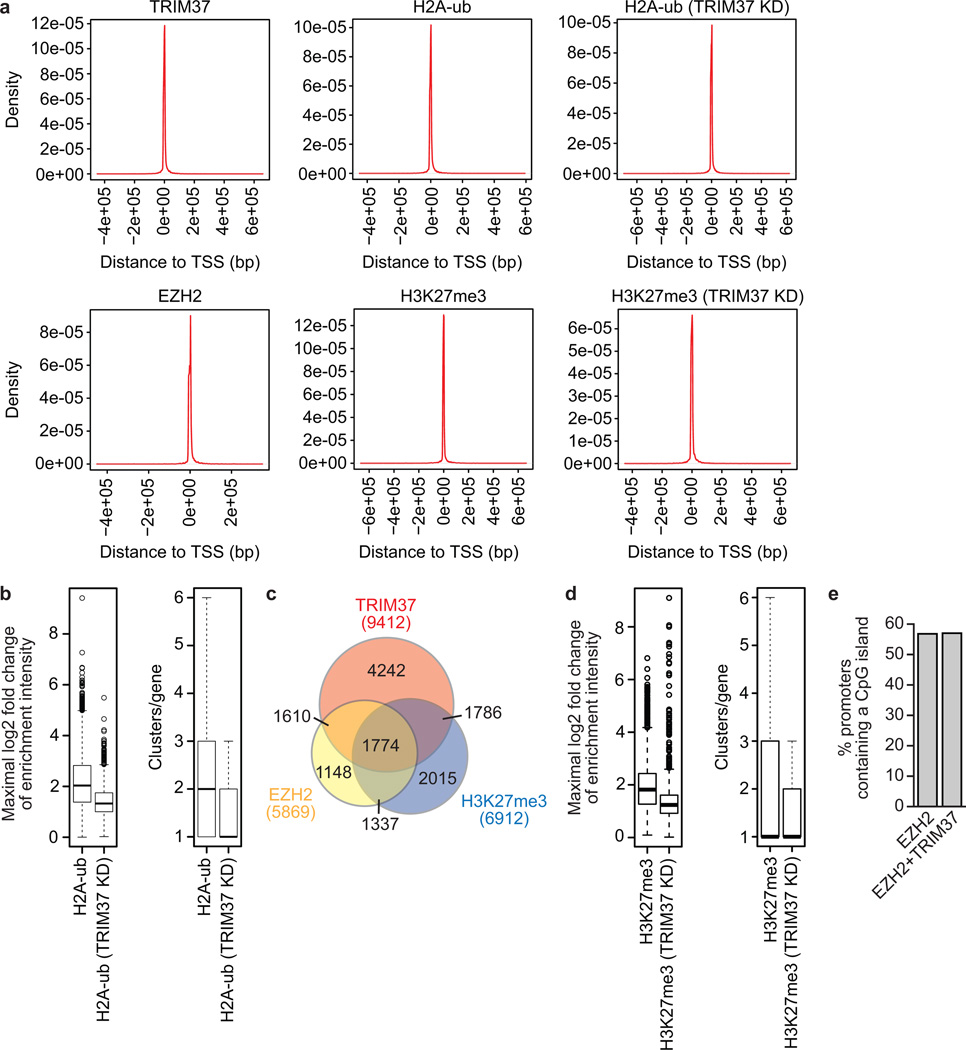
To identify TRIM37 target genes in MCF7 cells, we performed global ChIP-chip analysis and identified 9412 genes that were bound by TRIM37, 7638 of which were also enriched for H2A-ub (Fig. 2a, Extended Data Fig. 2a and Supplementary Table 1). Knockdown of TRIM37 resulted in loss or reduction of H2A-ub in ~75% of these genes (Fig. 2b, Extended Data Fig. 2a, b and Supplementary Table 1). Searches of cancer-gene databases revealed that 469 TRIM37 target genes are putative tumour suppressors (Supplementary Table 1). Notably, there was a statistically significant enrichment of putative tumour suppressors among the genes bound by TRIM37 (P = 1.99e-5).
Figure 2. Identification of TRIM37 target genes.
a, b, Venn diagrams showing overlap between TRIM37-bound and H2A-ub-enriched genes in parental MCF7 cells (a) or following TRIM37 knockdown (b). See also Supplementary Table 1. c, TRIM37 ChIP in MCF7 cells. ACTB, EEF1A1 and GAPDH are negative controls. Error bars indicate SEM; n=3 technical replicates of a representative experiment (out of three experiments). d, H2A-ub ChIP in MCF7 cells expressing a NS or TRIM37 shRNA. IgG from cells expressing a NS shRNA is shown. Error bars indicate SD; n=3 technical replicates of a representative experiment (out of three experiments). Black asterisks indicate significance of H2A-ub enrichment compared to IgG; blue asterisks indicate significant differences in H2A-ub enrichment in cells expressing a TRIM37 relative to NS shRNA. e, qRT-PCR monitoring target gene expression in TRIM37 knockdown MCF7 cells. Error bars indicate SEM; n=3 technical replicates of a representative experiment (out of three experiments). *P<0.05; **P<0.01.
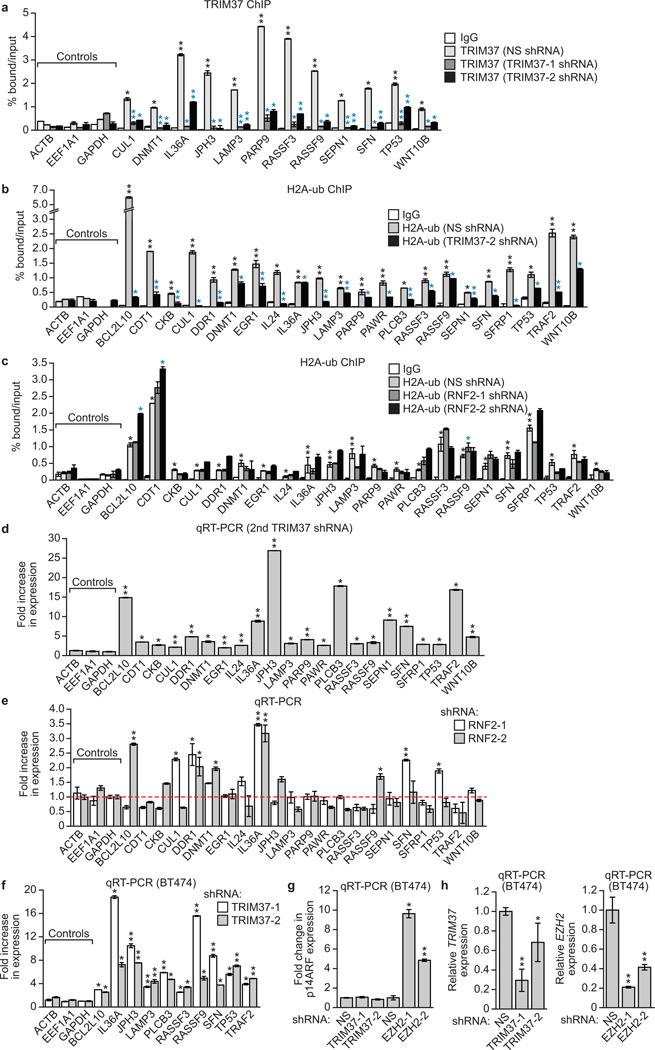
To validate the ChIP-chip results, we analysed a representative set of 22 putative tumour suppressor and other genes in directed ChIP experiments using promoter-specific primer pairs. For all 22 genes analysed, TRIM37 and H2A-ub were significantly enriched at the promoter (Fig. 2c, d and Extended Data Fig. 3a, b). For the majority of these genes, enrichment of H2A-ub substantially decreased following knockdown of TRIM37 but not RNF2 (Fig. 2d and Extended Data Fig. 3b, c). Finally, knockdown of TRIM37 but not RNF2 substantially increased expression of all TRIM37 target genes analysed (Fig. 2e and Extended Data Fig. 3d, e). Knockdown of TRIM37 in BT474 cells also resulted in increased expression of a subset of TRIM37 target genes analysed (Extended Data Fig. 3f–h).
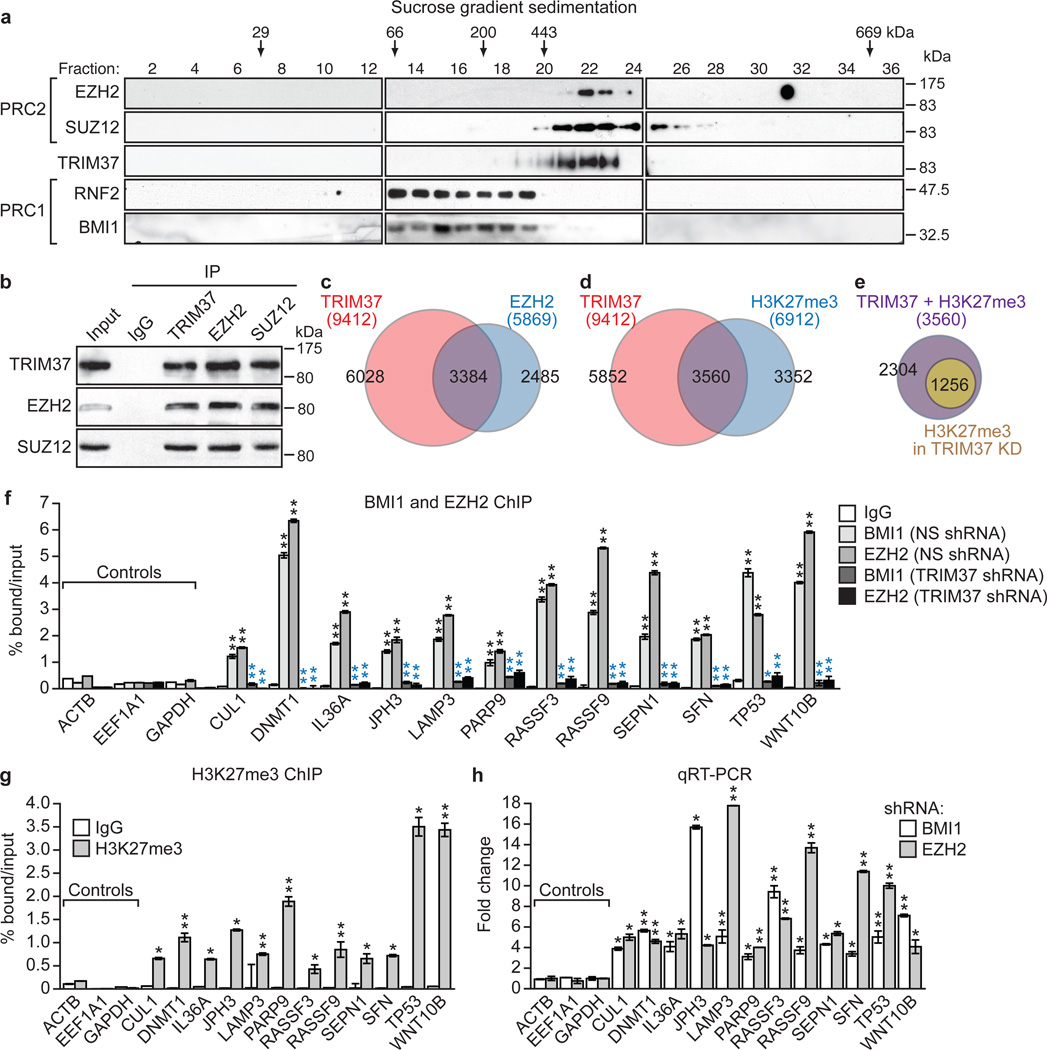
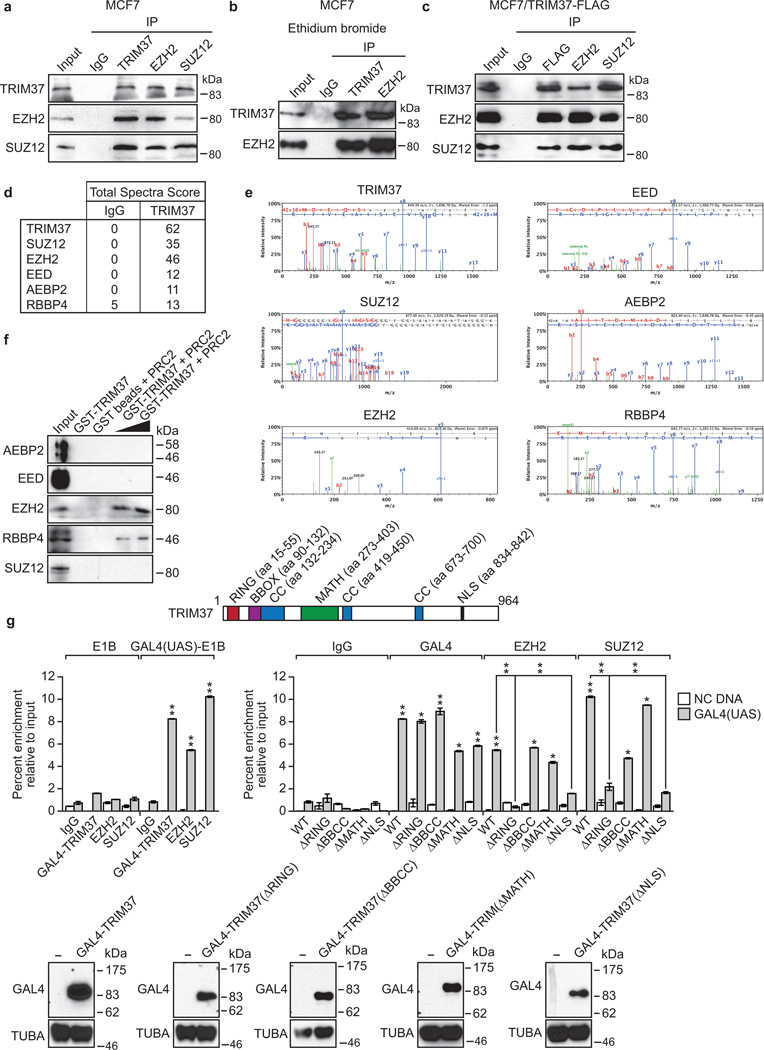
Previous studies have shown that RNF2 functions through association with PRC13–5. We next performed a series of experiments to determine whether TRIM37 also functioned through PRCs. We first asked whether TRIM37 was physically associated with PRC1 or PRC2. To test this idea, MCF7 cell nuclear extract was fractionated by sucrose gradient sedimentation and individual fractions analysed by immunoblotting for TRIM37 and PRC subunits. The results of Fig. 3a show that TRIM37 co-sedimented with PRC2 subunits EZH2 and SUZ128,12,13. Physical interactions between TRIM37 and PRC2 subunits could be demonstrated by co-immunoprecipitation (Fig. 3b, Extended Data Fig. 4a–c), liquid chromatography tandem mass spectroscopy (Extended Data Fig. 4d, e and Supplementary Table 2), an in vitro pull-down assay (Extended Data Fig. 4f), and the ability of a GAL4-TRIM37 fusion-protein to recruit PRC2 subunits to GAL4 binding sites (Extended Data Fig. 4g).
Figure 3. Interaction and co-occupancy of TRIM37 and PRC2.
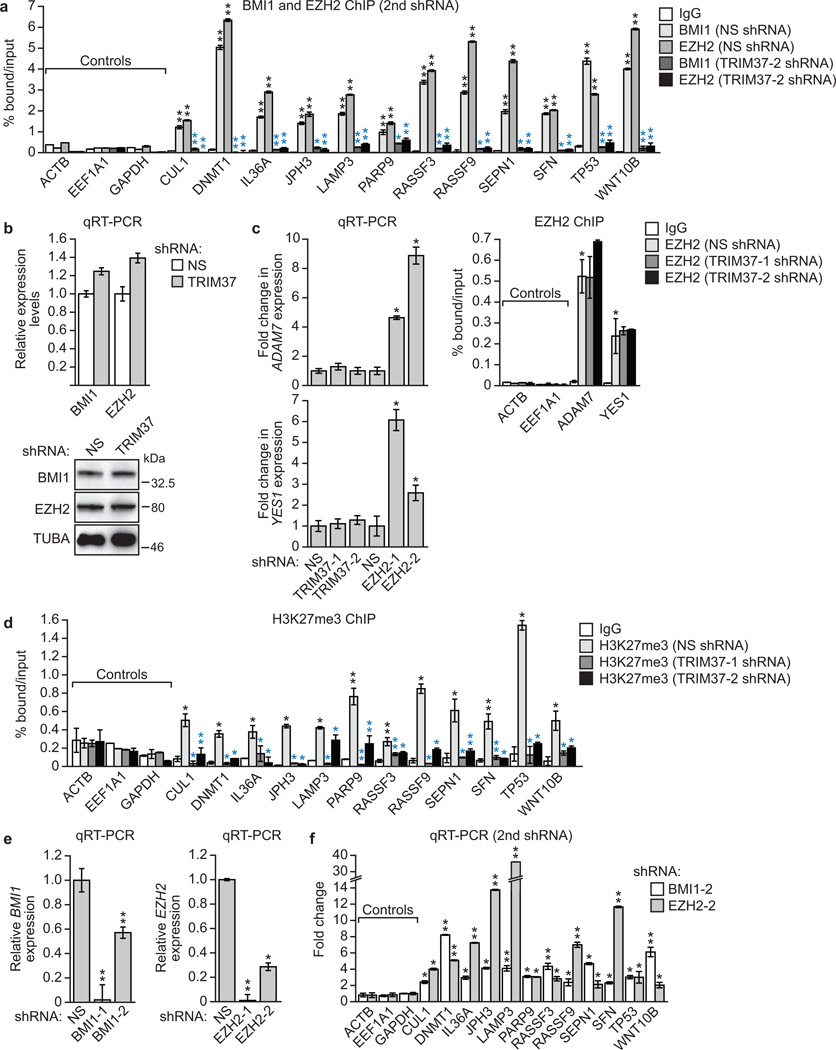
a, Immunoblots on sucrose gradient fractions. b, Sucrose gradient fractions 20–22 immunoprecipitated with a TRIM37, EZH2 or SUZ12 antibody, or an IgG control, were analysed by immunoblotting. c–e, Venn diagrams showing overlap between TRIM37- and EZH2-bound genes (c) and TRIM37-bound and H3K27me3-enriched genes in parental MCF7 cells (d) or following knockdown of TRIM37 (e). See also Supplementary Tables 3 and 4. f, BMI1 and EZH2 ChIP in MCF7 cells expressing a NS or TRIM37 shRNA. Error bars indicate SD; n=3 technical replicates of a representative experiment (out of three experiments). g, H3K27me3 ChIP in MCF7 cells. Error bars indicate SD; n=3 technical replicates of a representative experiment (out of three experiments). h, qRT-PCR monitoring TRIM37 target gene expression in BMI1 or EZH2 knockdown MCF7 cells. Error bars indicate SEM; n=3 technical replicates of a representative experiment (out of three experiments). *P<0.05; **P<0.01.
Global ChIP-chip analysis identified 3384 genes that were co-bound by TRIM37 and EZH2 (Fig. 3c, Extended Data Fig. 2a and Supplementary Table 3). In addition, 3560 genes were bound by TRIM37 and enriched for histone H3 lysine 27 trimethylation (H3K27me3) (Fig. 3d, Extended Data Fig. 2a, c and Supplementary Table 4). Knockdown of TRIM37 resulted in loss or reduction of H3K27me3 in ~65% of these genes (Fig. 3e, Extended Data Fig. 2a, d and Supplementary Table 4). Several studies have reported global correlations between PRC1/PRC2 occupancy and CpG islands14–18. We found no statistically significant difference between the percentage of promoters co-bound by TRIM37 and EZH2 or bound by EZH2 alone that contain a CpG island (Extended Data Fig. 2e).
To validate the ChIP-chip results, we analysed a representative set of TRIM37 target genes for binding of EZH2 and BMI1 by directed ChIP assays. All gene promoters analysed were enriched for BMI1 and EZH2, which was diminished following TRIM37 knockdown (Fig. 3f and Extended Data Fig. 5a–c). These gene promoters were also enriched for H3K27me3, which was reduced following TRIM37 knockdown (Fig. 3g and Extended Data Fig. 5d). As expected, knockdown of BMI1 or EZH2 resulted in increased expression of these genes (Fig. 3h and Extended Data Fig. 5e, f).
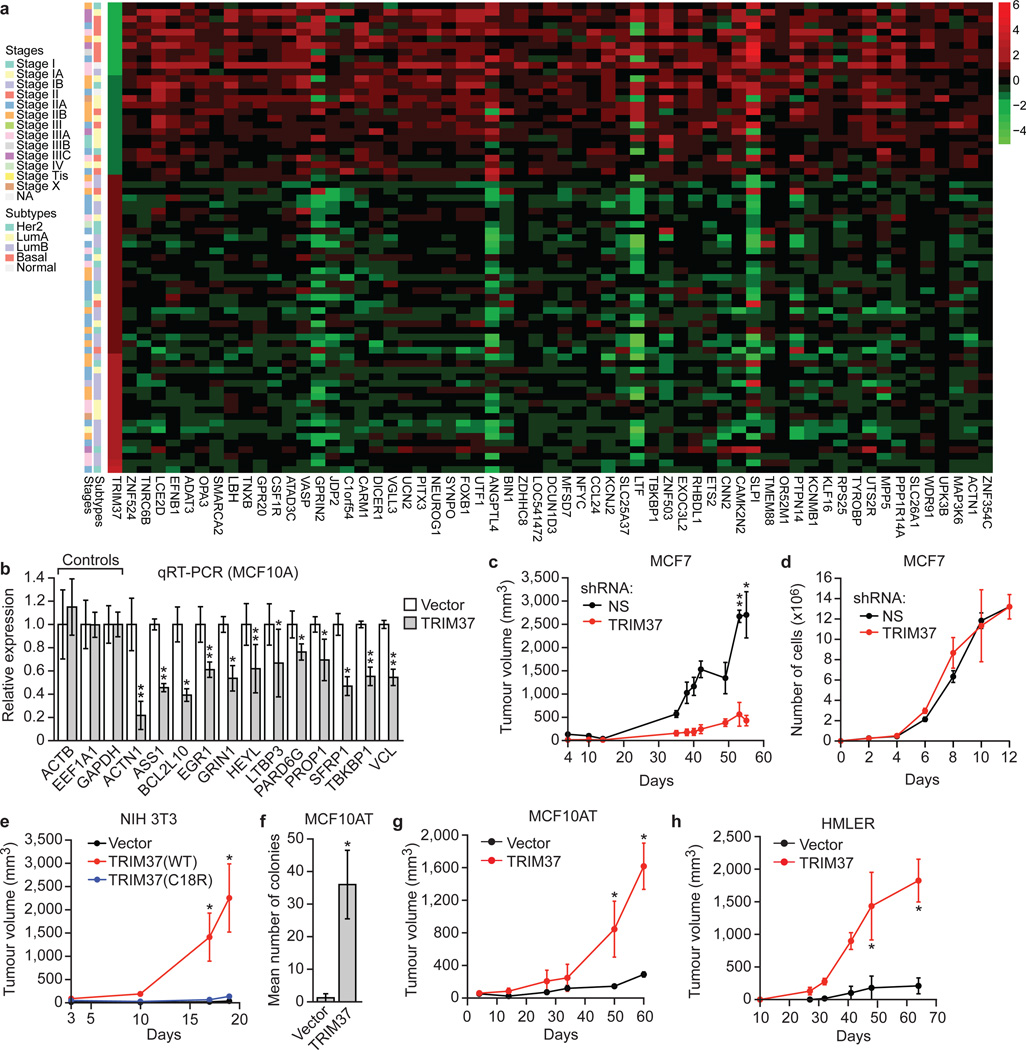
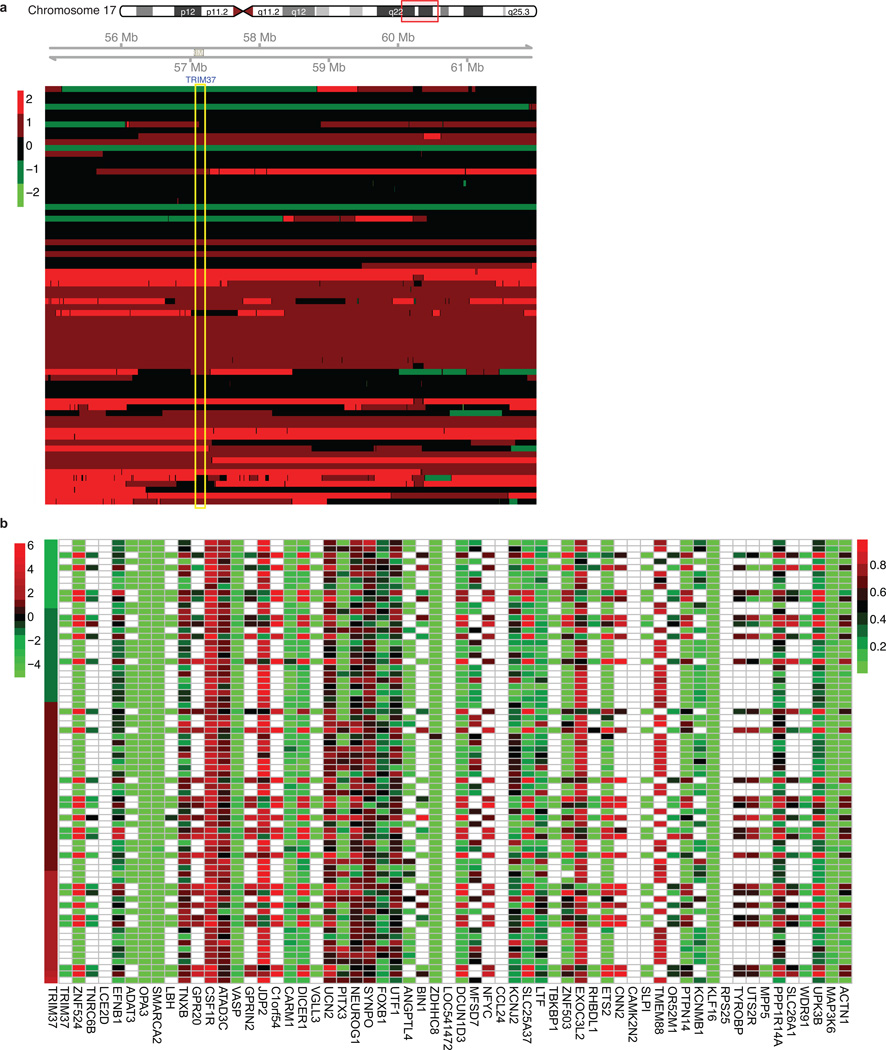
Our results raised the possibility that in breast cancers containing amplified 17q23, TRIM37 is over-expressed and contributes to transformation by silencing tumour suppressor genes. To investigate the relationship between TRIM37 levels and expression of TRIM37 target genes, we analysed a published comprehensive dataset of 466 human breast cancer samples19. Of the 9412 TRIM37 target genes we identified, there was a statistically significant correlation between increased TRIM37 levels and decreased expression of 2794 genes (Supplementary Table 5). The heat map of Fig. 4a shows the results for 60 TRIM37 target genes in a subset of breast cancer samples in which TRIM37 is either under- or over-expressed (Extended Data Fig. 6).
Figure 4. TRIM37 is an oncogene.
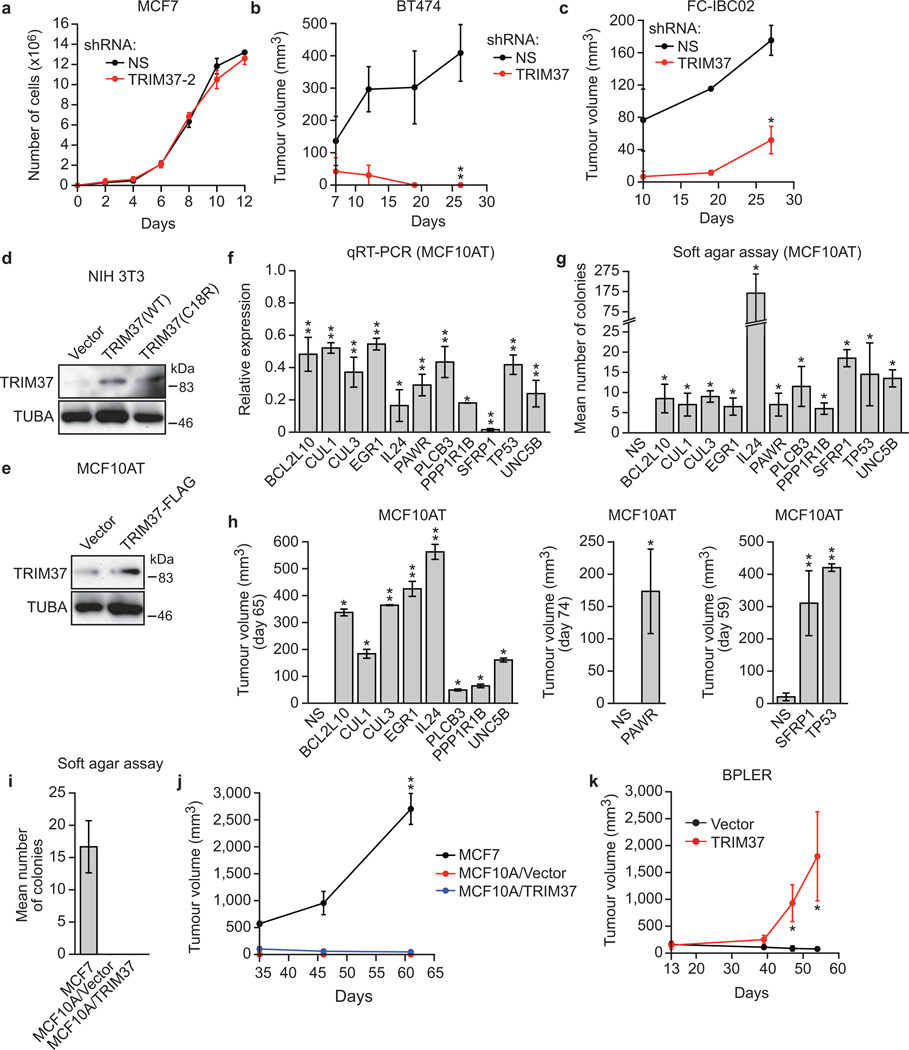
a, Heat map showing expression of TRIM37 and target genes in human breast cancer samples. See also Supplementary Table 5. b, qRT-PCR monitoring TRIM37 target gene expression in MCF10A cells expressing TRIM37 or vector. Error bars indicate SEM; n=3 technical replicates of a representative experiment (out of three experiments). c, Tumour formation in mice injected with MCF7 cells expressing a NS or TRIM37 shRNA. Error bars indicate SEM; n=3 mice per group. d, Proliferation of MCF7 cells expressing a NS or TRIM37 shRNA. Error bars indicate SD; n=3 technical replicates of a representative experiment (out of three experiments). e, Tumour formation in mice injected with NIH 3T3 cells expressing vector, TRIM37(WT) or TRIM37(C18R). Error bars indicate SEM; n=3 mice per group. f, Soft agar assay with MCF10AT cells expressing vector or TRIM37. Error bars indicate SEM; n=3 technical replicates of a representative experiment (out of three experiments). g, h, Tumour formation in mice injected with MCF10AT (g) or HMLER (h) cells expressing vector or TRIM37. Error bars indicate SEM; n=3 mice per group. *P<0.05; **P<0.01.
To confirm that elevated TRIM37 levels were responsible for decreased TRIM37 target gene expression, we analysed expression of representative TRIM37 target genes in MCF10A cells ectopically expressing TRIM37. Expression of all TRIM37 target genes analysed was significantly lower in cells ectopically expressing TRIM37 compared to empty vector (Fig. 4b).
To directly investigate the potential oncogenic activity of TRIM37, we knocked down TRIM37 in MCF7 cells and measured the effect on tumorigenicity in mouse xenografts. Knockdown of TRIM37 markedly reduced tumour growth (Fig. 4c) without affecting proliferation of cultured MCF7 cells (Fig. 4d and Extended Data Fig. 7a). Knockdown of TRIM37 also reduced tumour formation in xenografts derived from BT474 and FCIBC02 breast cancer cells (Extended Data Fig. 7b, c).
Conversely, ectopic expression of wild-type TRIM37 (Extended Data Fig. 7d), but not TRIM37-C18R, transformed NIH 3T3 cells, enabling tumour formation in mice (Fig. 4e). Moreover, ectopic expression of TRIM37 (Extended Data Fig. 7e) also transformed MCF10AT cells, a pre-malignant MCF10A derivative stably expressing activated HRAS20, enabling growth in soft agar (Fig. 4f) and tumour formation in mice (Fig. 4g). Furthermore, knockdown of a subset of TRIM37 target genes with predicted tumour suppressor activity also transformed MCF10AT cells (Extended Data Fig. 7f–h). By contrast, TRIM37 could not transform parental MCF10A cells (Extended Data Fig. 7i, j) indicating that TRIM37 can cooperate with RAS to transform human breast epithelial cells. Ectopic expression of TRIM37 also transformed HMLER and BPLER cells, immortalized human breast epithelial cell lines containing activated HRAS21 (Fig. 4h and Extended Data Fig. 7k). Notably, the RAS pathway is activated in ~50% of breast cancers22.
In this report, we have identified a new H2A ubiquitin ligase and shown how it functions in conjunction with PRCs to silence target genes. In the canonical silencing pathway, PRC2 first interacts with the promoter, followed by EZH2-catalyzed H3K27 trimethylation, which is recognized by PRC1 followed by RNF2-catalyzed H2A ubiquitination8,13. In cells containing high TRIM37 levels, TRIM37 associates with PRC2 and the TRIM37-PRC2 complex targets specific promoters. The promoter-bound PRC2-TRIM37 complex then carries out both H3K27 trimethyation and H2A mono-ubiquitination (see Extended Data Fig. 8a). Our results show that in the absence of PRC2, TRIM37 can catalyze H2A mono-ubiquitination (Fig. 1a, b) and bind to DNA (Fig. 3c). However, it remains possible that association with PRC2 could modulate, positively or negatively, the DNA-binding or H2A mono-ubiquitination activity of TRIM37. TRIM37 is also required for PRC1 occupancy, which presumably is mediated by PRC2 as in the canonical pathway. Although PRC1 does not direct H2A ubiquitination it is still required for silencing, consistent with previous studies showing that PRC1 mediates other repressive activities, such as chromatin compaction23,24.
TRIM37 alters the specificity of PRC2 and thus when TRIM37 is over-expressed there are extensive changes in gene expression that include the silencing of multiple tumour suppressor genes. Our results are consistent with previous studies showing that altered PRC2 activity, resulting, for example, from over-expression of EZH2, leads to malignancy25,26. In support of our cell culture results, we found a correlation between increased TRIM37 levels and decreased expression of TRIM37 target genes in human breast cancer samples (Fig. 4a and Supplementary Table 5). Interestingly, there is also a statistically significant correlation between high levels of TRIM37 and decreased survival in patients with estrogen-receptor positive breast cancer (Extended Data Fig. 8b). Collectively, our results reveal TRIM37 as a new H2A ubiquitin ligase that is over-expressed and promotes transformation in a subset of breast cancers.
METHODS
Cell lines and cell culture
K:Molv NIH 3T3 cells (ATCC# CRL-6361; referred to here as Kras NIH 3T3) were maintained in Dulbecco’s Modified Eagle Medium (DMEM) supplemented with 10% fetal calf serum (FCS) at 37°C and 5% CO2. HMECs (Life Technologies) were cultured in HuMec Ready Media (Life Technologies). MCF10A (ATCC) and MCF10AT27 cells were maintained in DMEM/F12 supplemented with 5% horse serum (Invitrogen), EGF (Peprotech, Inc.), hydrocortisone (Sigma), cholera toxin (Sigma), insulin (GIBCO) and Pen/Strep (Life Technologies). MCF7 cells (National Cancer Institute) were maintained in DMEM supplemented with 1× nonessential amino acids (NEAA) and 10% fetal bovine serum (FBS). BT474 cells (ATCC) were maintained in ATCC HybriCare Media supplemented with 10% FBS. MDA-MB-361 and MDA-MB-231 cells (ATCC) were cultured in ATCC-formulated Leibovitz’s L-15 medium supplemented with 20% FBS. T47D cells (NCI) were maintained in RPMI medium supplemented with 10% FBS. Hs578T cells (NCI) were maintained in DMEM medium supplemented with 0.01 mg ml−1 bovine insulin and 10% FBS. BPLER and HMLER cells21 (provided by Robert Weinberg, Whitehead Institute, Massachusetts Institute of Technology) were maintained in WIT (Stemgent) and Mammary Epithelial Cell Growth Medium (Lonza), respectively. FC-IBC02 cells28 (provided by Massimo Cristofanilli, Thomas Jefferson University) were maintained in Ham’s F12 Nutrient Mixture (Life Technologies) with 10% FBS and 0.5% insulin (Gibco) with 100 µg l−1 hydrocortisone (Sigma) and antibiotic-antimycotic (Gibco). Cells cultured at the same time were pooled together and then seeded after counting in a 6-well or 10 cm dish. Dishes/wells were then subjected, in a random order, to treatment with a control or test shRNA/vector.
RNA interference
For stable shRNA knockdowns, cells were seeded in a six-well plate to 50% confluency and subsequently transduced with 400 µl lentiviral particles expressing shRNAs (obtained from Open Biosystems/Thermo Scientific through the UMMS RNAi Core Facility, listed in Extended Data Table 1) in a total volume of 2 ml of appropriate media supplemented with 6 µg ml−1 polybrene. Media was replaced after overnight incubation to remove the polybrene and viral particles and cells were subjected to puromycin selection (2 µg ml−1) for 3 days.
In vitro ubiquitination assays
To produce purified recombinant TRIM37, the TRIM37 open reading frame was PCR amplified from a cDNA clone (Invitrogen) using high fidelity Pfu polymerase, and cloned into a derivative of pEF6/V-5HisB (Invitrogen) resulting in the addition of a protein-C epitope (MAEDQVDPRLIDGKEFT) at the N-terminus of TRIM37. The C18R mutation was introduced by overlap extension PCR using primers listed in Extended Data Table 2. All constructs were verified by sequencing.
In vitro ubiquitination reactions were set up containing purified TRIM37 or TRIM37-C18R (500 ng–2 µg), a combination of BMI1 (500 ng; Abcam) and RNF2 (500 ng; Origene) mixed together, or BRCA1 (500 ng; Abcam) plus H2A or H2B (5×, Boston Biochemicals), 10 nM UBE1 (E1; Boston Biochemicals), 10 nM UBCH5B (E2; Boston Biochemicals), and 10 nM HA-ubiquitin aldehyde (Boston Biochemicals) in 10 µl of reaction buffer (50 mM Tris-Cl, pH 7.9, 5 mM MgCl2, 2 mM NaF, 100 mM DTT and 2 mM ATP). Reactions were incubated at 32°C for 90 min. For the nucleosomal assay, in vitro ubiquitination reactions were set up similarly but contained Xenopus oocyte-derived nucleosomes; recombinant histone particles were expressed and purified as described previously29 and reconstituted using a standard salt step dialysis method30. To generate the H2A-KKRR mutant10, a pET-based plasmid expressing H2A (provided by Craig Peterson, University of Massachusetts Medical School) was used as a template in a PCR-based site-directed mutagenesis with primer extension reaction using primers listed in Extended Data Table 2. Proteins were resolved by 15% SDS-PAGE and subjected to immunoblotting with H2A-ub(K119) (Cell Signaling Technology #8240, clone D27C4), H2A (Cell Signaling Technology #2578), H2B-ub (EMD Millipore 05-1312, clone 56), H2B (Abcam ab18977), H3 (Cell Signaling Technology #9715) or H4 (Cell Signaling Technology #2592) antibodies.
Quantitative RT-PCR (qRT-PCR)
Total RNA was isolated and reverse transcribed using Superscript II Reverse Transcriptase (Invitrogen). Quantitative real-time PCR was performed as described previously6 using primers listed in Extended Data Table 2. Gene expression was normalized to GAPDH. No-RT controls were carried out in parallel to rule out the possibility of DNA contamination. Each sample was analyzed three independent times and the results from one representative experiment, with technical triplicates, are shown.
Immunoblotting
Nuclear lysates were prepared as previously described31. Nuclear extracts were separated on either 8%, 10% or 20% SDS-PAGE gels. Blots were probed with primary antibodies (H2A-Ub[K119] [Cell Signaling Technology], H2A [Cell Signaling Technology], H2B-ub [EMD Millipore], H2B [Cell Signaling Technology #12364, clone D2H6], H3 [Cell Signaling Technology], H4 [Cell Signaling Technology], HA [Cell Signaling Technology #2367, clone 6E2], TRIM37 [Abcam ab95997, or custom made by 21st Century Biochemicals against a synthetic peptide corresponding to amino acids 444–460 of the human protein followed by affinity purification] or], RNF2 [Abcam ab28629], and α-tubulin [in-house]) overnight at 4°C, washed five times in TBS plus 0.1% Tween (TBST) and then incubated with the appropriate HRP-conjugated secondary antibody for 1 h at room temperature. Membranes were washed five times in TBST and visualized on autoradiography film after incubating with ECL reagent (Supersignal West Pico or Supersignal West Femto; Thermo Scientific). Immunoblots were quantified using Image J software version 1.47v (NIH).
TRIM37 over-expression
A TRIM37 cDNA clone (Origene Technologies) was subcloned into the vector p3X-FLAG-Myc-CMV-26 (Sigma) using Not1 and Xba1 sites, and verified by full-length sequencing. The TRIM37 expression vector was transfected into cells using Effectene reagent (Qiagen) and stable clones were selected. Over-expression of TRIM37 was confirmed by immunoblotting using TRIM37 (21st Century) and anti-FLAG (Sigma M2) antibodies.
ChIP-chip and directed chIP
For ChIP-chip analysis, chromatin was sonicated and immunoprecipitation was carried out as previously described31 using a TRIM37 (Abcam), H2A-ub (Cell Signaling Technology), EZH2 (Cell Signaling Technology #5246, clone D2C9), or H3K27me3 (Cell Signaling Technology #9733, clone C36B11) antibody. Samples were prepared and hybridized to a GeneChip Human Promoter 1.0R Array (Affymetrix) according to the manufacturer’s specifications, and arrays were scanned using a GeneChip Scanner 3000 7G (Affymetrix). Each Chip-chip experiment was performed in duplicate. ChIP-chip data were analyzed using Bioconductor package Starr32,33 for data read-in, normalizing and peak calling. Quantile normalization was applied to normalize probe intensity before peak calling with default settings except that fragment length (sonication fragment length) was set to 500, minrun (minimum number of probes in a bound region) was set to 2, method (multiple test adjustment method) was set to BH (Benjamini & Hochberg)34, and alpha (false discovery rate cutoff) was set to 0.1. ChIP-chip results were annotated by R(v3.1.0)/Bioconductor(v2.14) ChIPpeakAnno(v.2.12.1) package35. Density-plots were plotted by the distance of peaks to the annotated transcription start site. Venn diagrams were drawn by R(v3.1.0) Vennerable(v3.0) package. Gene ontology analysis was done by R/Bioconductor package goProfiles36. For the statistical analysis on the global decrease in H3K27me3 and H2A-ub following TRIM37 knockdown in MCF7 cells, the Wilcoxon Signed-rank test was performed in R37. A summary table showing the overlap of proteins and epigenetic marks, along with the P values for each overlap, is shown in Supplementary Table 6. CpG island overlap analysis was performed as previously described17 except that CpG island annotation was downloaded from the UCSC Genome Browser (https://www.genome.ucsc.edu). A CpG gene list was generated for CpG islands located within 1 kb upstream of transcript start-site. The percentage of promoters bound by EZH2 alone or co-bound by TRIM37 and EZH2 and which also contain a CpG island was calculated by the intersection of the CpG gene list and the EZH2 or EZH2/TRIM37 target gene list, respectively. The ChIP-chip data have been deposited in NCBI’s Gene Expression Omnibus38 and are accessible through GEO Series accession number GSE48196.
Directed ChIP assays were performed essentially as described previously6 using cell extracts prepared 7 days post-retroviral transduction and puromycin selection, and antibodies against BMI1 (Abcam ab14389, clone 1.T.21), EZH2 (Cell Signaling Technology), H3K27me3 (Cell Signaling Technology), H2A-Ub (Cell Signaling Technology) and TRIM37 (Abcam) or, as a negative control, IgG. The sequences of primers used for amplifying ChIP products are listed in Extended Data Table 2. Based on the probe sequence used in the Affymetrix array, for each gene a 400–600 bp region was tested for enrichment of TRIM37 using at least three primer pairs. The primer pair with the consistently highest enrichment in three different experiments was chosen for subsequent enrichment analysis of BMI1, H2A-ub, H3K27me3 and EZH2. Each ChIP experiment was carried out three independent times and the results from one representative experiment, with technical triplicates, are shown.
Identification of putative tumour suppressors
Candidate TRIM37 target genes were systematically queried against three databases: DNA Tumor Suppressor and Oncogene Database (DTSOD; available at http://embryology.med.unsw.edu.au.), TSGene Database (TSGene; http://bioinfo.mc.vanderbilt.edu/TSGene/), and Tumor Associated Gene (TAG; http://www.binfo.ncku.edu.tw/TAG/GeneDoc.php). Genes listed in the TSGene database or listed as “tumor suppressor genes” in the DTSOD or TAG database were considered putative tumour suppressor genes. Statistical enrichment of putative tumour suppressor genes among TRIM37 target genes was calculated by a hypergeometric test.
Sucrose gradient fractionation
Sucrose gradient sedimentation analysis was performed as described39. Briefly, 10–40% gradients were formed by layering 500 µl NEB1 buffer containing 10%, 20%, 30%, or 40% sucrose in a 11 × 34-mm centrifuge tube (Beckman) and allowed to equilibrate at room temperature for 2 h. Gradients were chilled, loaded with 500 µg MCF7 nuclear extract (adjusted to a volume of 150 µl) or 150 µl molecular weight markers (Sigma MW-GF-1000), and centrifuged in a Beckman TLS-55 rotor at 50,000 rpm (214,000×g) for 14 h. Thirty-six fractions of •45 µl were collected. For the markers, 20 µl of each fraction was electrophoresed and Coomassie stained. For the gradient fractions, 20 µl of fractions were analysed by immunoblotting using EZH2 (Cell Signaling Technology), SUZ12 (Abcam ab12073), TRIM37 (Abcam), RNF2 (Abcam ab101273) and BMI1 (Bethyl Laboratories, A301-694A).
Co-immunprecipitation
MCF7 nuclear extract (~1000 µg) was incubated with a TRIM37 (Abcam), EZH2 (Cell Signaling Technology) or SUZ12 (Abcam) antibody at 4°C for 24 h in the presence or absence of ethidium bromide (100 µg ml−1). For immunoprecipitations from fractionated nuclear lysate, fractions 20–22 (which were enriched for TRIM37, EZH2 and SUZ12) were pooled and diluted 15-fold. Immune complexes were captured on rabbit or mouse TrueBlot IP beads (eBioScience), washed three times in NEB1 buffer, and eluted by boiling 10 min in 2× SDS sample buffer. Immunoprecipitations from MCF7 cells ectopically expressing FLAG-tagged TRIM37 were performed similarly, except anti-FLAG magnetic beads (Sigma) were used to capture the immune complexes. Immunoprecipitated proteins were analysed by immunoblotting as described above. Input lanes represent 10–25% of extract loaded in the IP lanes.
Liquid chromatography tandem mass spectrometry
Large-scale immune-precipitation was carried out as previously described40 by incubating 80 mg MCF7 nuclear extract with 50 µg of anti-TRIM37 antibody (Abcam) at 4°C for 5 h. Immune complex were captured on IgG Dynabeads (Life Technologies) by additional incubation at 4°C for 2 h. The beads were then washed with NETN [20 mM Tris-HCl (pH 8.0), 0.1 M NaCl, 1 mM EDTA, 0.5% NP-40] three times and re-suspended in Laemmli buffer. Proteins were separated on a short 10% SDS-polyacrylamide gel. A 2 cm gel slice with all the proteins was submitted for mass spectrometry at the University of Massachusetts Medical School Proteomics and Mass Spectrometry Facility.
For in gel digestion, gel slices were placed in 1.5 ml eppendorf tubes with 1 ml of water for 30 min and then replaced with 100 µl of 250 mM ammonium bicarbonate. For reduction, 20 µl of a 45 mM 1, 4 dithiothreitol (DTT) was added and the samples were incubated fat 50°C for 30 min. The alkylation was carried out by addition of 20 µl of a 100 mM iodoacetamide solution for 30 min. The gel slices were washed twice with water and then treated with 50:50 (50 mM ammonium bicarbonate: acetonitrile) for 1 h. The solution was then removed and 200 µl of acetonitrile was added. Dried gel slices were rehydrated in 50 µl of 2 ng µl−1 trypsin (Sigma) in 0.01% ProteaseMAX Surfactant (Promega): 50mM ammonium bicarbonate at 37°C for 21 h. Additional dehydration was carried out with 100 µl of 80:20 (acetonitrile: 1% formic acid). Dried samples were resuspended in 25 µl of 5% acetonitrile in 0.1% trifluroacetic acid prior to injection on LC/MS/MS.
Briefly, samples were injected onto a custom packed 2cm × 100 µm C18 Magic 5µ particle trap column. Labeled peptides were then eluted and sprayed from a custom packed emitter (75 µm × 25 cm C18 Magic 3µm particle) with a linear gradient from 95% solvent A (0.1% formic acid in water) to 35% solvent B (0.1% formic acid in acetonitrile) in 90 min at a flow rate of 300 nl per minute on a Waters Nano Acquity UPLC system. Data dependent acquisitions were performed on a Q Exactive mass spectrometer (Thermo Scientific). Raw data files were processed with Proteome Discoverer (version 1.3) prior to searching with Mascot Server (version 2.4) against the Human Index of the SwissProt database. Search parameters utilized were fully tryptic with 2 missed cleavages, parent mass tolerances of 10 ppm and fragment mass tolerances of 0.05 Da. A fixed modification of carbamidomethyl cysteine and variable modifications of acetyl (protein N-term), pyro glutamic for N-term glutamine, oxidation of methionine was considered. Search results were loaded into the Scaffold Viewer (Proteome Software, Inc.), which provided total spectra scores and interaction probabilities.
GST pull-down assay
The TRIM37 cDNA (Origene Technologies) was cloned into the vector pGEX4T-1 (Life Technologies) using EcoR1 and NotI sites and the construct was verified by full-length sequencing. Supernatant from bacteria expressing GST-TRIM37 was incubated with glutathione agarose beads (GE Healthcare) at 4°C for 3–4 h. Beads were washed thrice with PBS and 0.5% Triton X-100 before using in pull-down assays. PRC2 subunits (EZH2 [Origene Technologies], SUZ12 [Origene Technologies], AEBP2 [Open Biosystems], EED [Open Biosystems] and RBAP48 [Open Biosystems]) were in vitro translated using the TnT® T7 Quick Coupled Transcription/Translation System (Promega) according to the manufacturer’s specifications. GST-TRIM37 beads (3 µg) were incubated with in vitro translated protein (added to 1/10 volume of the reaction mixture) in Buffer A (50 mM Tris-HCl pH 7.9, 0.5 mM EDTA, 1 mM DTT, 0.2 mM PMSF, 10% glycerol, 150 mM KCl and 0.05% NP40) at 4°C for 2 h. Following incubation, beads were washed twice with wash buffer (50 mM Tris-HCl pH 7.9, 0.5 mM EDTA, 1 mM DTT, 0.2 mM PMSF, 10% glycerol, 300 mM KCl and 0.05% NP40). The interaction was detected by immune-blot using a biotin antibody (Abcam #53494).
GAL4-TRIM37 recruitment assay
The TRIM37 cDNA was cloned into the plasmid pCMX-Gal4 (Addgene), which contains amino acids 1–147 of GAL4, using KpnI and Nhe1 sites. TRIM37 deletions were generated using PCR-based site-directed deletion with primer extension using primers listed in Extended Data Table 2. All constructs were verified by full-length sequencing. 293T cells were co-transfected with the GAL4-TRIM37 plasmid and G5E1bcat-pSP7241 (containing five GAL4-binding sites) or E1bcat-pSP7242 (negative control). ChIP assays were carried out as described above.
Breast cancer biostatistical analysis
Gene expression and CpG methylation data from breast cancer samples (n=466) were obtained from The Cancer Genome Atlas Data Portal (https://tcga-data.nci.nih.gov/docs/publications/brca_2012/)19. The mRNA data was converted to relative mRNA expression levels by subtracting each gene's median level computed over 22 tumour-adjacent normal controls from its level observed in each patient sample. A subset of samples was then selected for further analysis to include those with upregulated TRIM37 (n=45 with TRIM37 value ≥1) and downregulated TRIM37 (n=26 with TRIM37 value ≤−1). Pearson correlation analysis was performed to identify TRIM37 target genes (identified in the ChIP-chip experiment) whose down-regulation significantly correlated with increased TRIM37 expression (P <0.01). Statistical analysis and plotting were done with R(v3.1.0), a system for statistical computation and graphics37. The top 60 genes with negative correlation to TRIM37 were extracted for heat map plots using R(v3.1.0) pheatmap(v0.7.7) package. Cancer subtypes were extracted from sample list information of gene expression data (level 3, PAM50). Cancer stages were extracted from TCGA clinical information.
TRIM37/17q23 copy number data were also downloaded from https://tcga-data.nci.nih.gov/docs/publications/brca_2012/, Section "SNP and Copy Number" Level3 Data Archives. Data were visualized using R/Bioconductor package Gviz and analysed as described previously19. Briefly, thresholded gene level copy number values were calculated using GISTIC 2.0 and Log2 copy number values were thresholded as followed: values <−1 are set to −2, values between −1 and −0.3 are set to −1, values between −0.3 and 0.3 are set to 0, values between −1 and −0.3 are set to 1, and values > 1 are set to 2. Tumours where all gene level thresholded copy number changes equaled 0 were excluded from the analysis.
For correlation analysis between TRIM37 and prognosis, Kaplan-Meier plotting and logrank test were done at http://kmplot.com/analysis/index.php?p=service&cancer=breast. Analysis was restricted to breast cancer patients with ER-positive subtypes. Patients were divided into two classes based on TRIM37 expression: patients with higher than upper quartile TRIM37 expression were considered as high-expressing TRIM37 group (n=352) and the other patients were considered as low-expressing TRIM37 group (n = 1061).
Animal experiments
All animal protocols were approved by the Institution Animal Care and Use Committee at UMMS (A-2016). Animal sample sizes were selected based on precedent established from previous publications and an understanding that at least n=5 is generally required to achieve statistical significance. For mouse experiments involving shRNAs, the most efficacious shRNA of multiple shRNAs tested and validated in cell culture was used, a criterion that was established prospectively. Mice were randomly allocated to each group. No blinding was done as animal groups were identified by tagging and labeling the cages with the cells injected. Animals were excluded, according to pre-established criteria, if the tumour volume reached >2000mm3; if tumour size or location affected the mobility or general health of animal, the animal was euthanized and excluded from the experiment or the complete experiment was terminated.
MCF7 (1×106), BT474 (3×106) or FC-IBC02 (3×106) cells stably expressing a non-silencing (NS) or TRIM37 shRNA were injected into the right thoracic mammary fat pad (MCF7) or flank (BT474 and FC-IBC02) of 6–8 week old BALB/c nu/nu female mice (n=3; Taconic Farms) to produce orthotopic primary tumours. Because MCF7, BT474 and FC-IBC02 cells are hormone dependent, a 60 day release tablet of 1.7 mg estradiol (Innovative Research of America) was implemented under the skin of the neck of the mouse with 11-gauge trochar. Primary tumour growth was evaluated by measurement with calipers. NIH 3T3 (5×106), MCF10AT (3×106), HMLER (3×106) or BPLER (3×106) cells expressing vector or TRIM37 or TRIM37-C18R were subcutaneously injected in the flank of the mice (n=3) and the growth of the primary tumours was monitored as previously described6. MCF10AT (5×106) cells expressing a NS shRNA or shRNA targeting a tumour suppressor gene were subcutaneously injected in the flank of the mice (n=3) and tumour growth was monitored. All mouse experiments were repeated at least twice (for a total of at least 6 mice per group).
Soft agar assay
MCF10AT cells (2.5×104) expressing vector or TRIM37, or MCF10AT cells expressing a NS shRNA or shRNA targeting a tumour suppressor gene, were re-suspended in 3 ml of top agar (MCF10AT media containing 0.3% Difco Noble agar (BD Biosciences)) warmed to 40°C. The cell suspension was layered onto 3 ml of set bottom agar (MCF10AT media containing 0.7% Noble agar) in a 6-well plate. Two milliliter of medium was added on the top agar next day and changed every other day. Visible colonies were scored after 4–5 weeks.
Statistics
To achieve statistical significance, all qRT-PCR and ChIP data were collected from experiments performed in technical triplicate; each experiment was repeated at least twice and statistically significant results were obtained in independent biological replicates. Differences between groups were assayed using a two-tailed student t-test using Microsoft Excel. In cases where the assumption of the t-test was not valid a non-parametric statistical method was used (e.g., Wilcoxon signed-rank test). Significant differences were considered when P<0.05; *P≤0.05 and **P≤0.01. The error bars indicate the standard deviation or standard error of the mean for the technical replicates, as indicated in the legend.
Extended Data
Extended Data Fig. 1. Control experiments for Fig. 1.
a, ChIP monitoring H2A-ub enrichment on Fas in Kras NIH 3T3 cells expressing a non-silencing (NS) shRNA or a Trim37, Rnf2 or Bmi1 shRNA. Three regions of the Fas promoter were analysed: the core promoter/transcription start site (CP/TSS), and 1 and 2 kb upstream of the TSS. Actb and Gapdh are shown as negative controls. Error bars indicate SEM; n=9 (three biological replicates with three technical replicates per sample). b, H2A-ub ChIP as described in (a) using a second Trim37, Rnf2 or Bmi1 shRNA unrelated to that used in (a), c, qRT-PCR analysis monitoring knockdown efficiencies of Trim37 and Rnf2 shRNAs in NIH 3T3 cells. The results are given relative to expression following treatment with a NS shRNA, which was set to 1. For knockdown efficiencies of the Bmi1 shRNAs see Gazin et al., 2007 (ref. 6). Error bars indicate SEM; n=3 technical replicates of a representative experiment (out of three experiments). d, In vitro ubiquitination assay. Purified H2B was incubated with E1 (UBE1), E2 (UBCH5B), E3 (TRIM37 or BRCA1), ATP and HA-ub. Blots were probed with antibodies against H2B-ub or H2B. The results show that TRIM37 does not ubiquitinate H2B. BRCA1, which is known to ubiquitinate H2B at K12039,40, was used as a positive control. e, f, qRT-PCR (e) and immunoblots (f) monitoring TRIM37 and RNF2 in various cell lines. Expression of TRIM37 and RNF2 expression were was normalized to that obtained in HMECs, which was set to 1. Error bars indicate SD; n=3 technical replicates of a representative experiment (out of three experiments). g, Immunoblot monitoring TRIM37 levels in MCF7 cells expressing a NS or one of two unrelated TRIM37 shRNAs. α-tubulin (TUBA) was monitored as a loading control. The results indicate that the TRIM37 antibody is highly specific. h, qRT-PCR analysis monitoring knockdown efficiencies of TRIM37 shRNAs in MCF7, BT474 and MDA-MD-361 cells. Error bars indicate SEM; n=3 technical replicates of a representative experiment (out of three experiments). i, (Top) Immunoblots monitoring levels of H2A-ub and H2B-ub in MCF7 cells expressing a NS or one of two unrelated TRIM37 shRNAs. (Bottom) Quantification of the H2A-ub immunoblots relative to TUBA. The relative level of H2A-ub in NS cells was set to 1. In this experiment, histones were acid extracted. j, (Top) Immunoblots monitoring TRIM37, H2A-ub, H2A, H2B-ub and H4 in BT474 and MDA-MB-361 cells expressing a NS or one of two unrelated TRIM37 shRNAs. (Bottom) H2A-ub quantification, as described in (g). k, qRT-PCR analysis monitoring knockdown efficiencies of RNF2 shRNAs in MCF7 cells. Error bars indicate SEM; n=3 technical replicates of a representative experiment (out of three experiments). l, (Top) Immunoblots monitoring TRIM37, H2A-ub, H2A and H4 in MDA-MB-231, Hs578T and T47D cells expressing a NS or one of two unrelated TRIM37 shRNAs. (Bottom) H2A-ub quantification, as described in (g). m, Proliferation of cultured MDA-MB-231, Hs578T and T47D cells expressing a NS or TRIM37 shRNA. Error bars indicate SD; n=3 technical replicates of a representative experiment (out of three experiments). The results show that knockdown of TRIM37 has no effect on proliferation of breast cancer cell lines lacking 17q23 amplification. n, qRT-PCR analysis monitoring TRIM37 expression in an MCF10A cell line ectopically expressing TRIM37 or, as a control, empty vector. The results were normalized to TRIM37 expression in MCF10A cells expressing empty vector, which was set to 1. Error bars indicate SEM; n=3 technical replicates of a representative experiment (out of three experiments). *P<0.05; **P<0.01.
Extended Data Fig. 2. Extra details related to ChIP-chip analysis.
a, Histograms showing enrichment of TRIM37, H2A-ub (in parental MCF7 cells or following TRIM37 knockdown [KD]), EZH2, and H3K27me3 (in parental MCF7 cells or following TRIM37 KD) as a function of distance to the nearest transcription start site (TSS). b, (Left) Box plot of maximal gene-level log2 fold change of enrichment intensity comparing input for H2A-ub in parental MCF7 cells and following TRIM37 KD. (Right) Box plot of number of enriched regions per gene for H2A-ub in parental MCF7 cells or following TRIM37 KD. The results show that there was a significant difference in H2A-ub enrichment at TRIM37 target genes following TRIM37 knockdown with respect to both enrichment intensity (P < 1e-22) and number of enriched regions (P = 2.787008e-222). A region with a fold change ≥ 2 and a false discovery rate < 0.1 was considered a differentially enriched site. c, Venn diagram showing overlap between TRIM37- and EZH2-bound genes and H3K27me3-enriched genes in MCF7 cells. d, (Left) Box plot of maximal gene-level log2 fold change of enrichment intensity comparing input for H3K27me3 in parental MCF7 cells or following TRIM37 KD. (Right) Box plot of number of enriched regions per gene for H3K27me3 in parental MCF7 cells or following TRIM37 KD. The results show that there was a significant difference in H3K27me3 enrichment at TRIM37 target genes following TRIM37 knockdown with respect to both enrichment intensity (P = 5.872777e-109) and number of enriched regions (P = 1.178392e-52). A region with a fold change ≥ 2 and a false discovery rate < 0.1 was considered a differentially enriched site. e, Percentage of promoters (defined as 1 kb upstream of the transcription start-site) bound by EZH2 alone that contain a CpG island (3332/5869; 56.77%) or co-bound by EZH2 and TRIM37 that contain a CpG island (1929/3384; 57.00%). A two-sample test for equality of proportions with continuity correction showed that there was no statistically significant difference in CpG island content between the EZH2-bound and EZH2, TRIM37 co-bound promoters.
Extended Data Fig. 3. Control experiments for Fig. 2.
a, ChIP monitoring binding of TRIM37 at promoters of TRIM37 target genes in MCF7 cells expressing a NS or one of two unrelated TRIM37 shRNAs. As a negative control, TRIM37 binding at three non-TRIM37 target genes, ACTB, EEF1A1 and GAPDH, is shown. Black asterisks indicate significance of TRIM37 enrichment compared to the IgG control (from cells expressing a NS shRNA); blue asterisks indicate significant differences in TRIM37 enrichment in cells expressing a TRIM37 shRNA relative to NS shRNA. Error bars indicate SD; n=3 technical replicates of a representative experiment (out of three experiments). The results confirm that TRIM37 occupancy is reduced upon TRIM37 knockdown, demonstrating the specificity of the TRIM37 antibody in ChIP experiments. b, ChIP monitoring enrichment of H2A-ub at promoters of TRIM37 target genes in MCF7 cells expressing a second TRIM37 shRNA unrelated to that used in Fig. 2d. The IgG control and H2A-ub signal in cells expressing a NS shRNA are the same as those shown in Fig. 2d. Error bars indicate SD; n=3 technical replicates of a representative experiment (out of three experiments). c, ChIP monitoring enrichment of H2A-ub at the promoters of TRIM37 target genes in MCF7 cells expressing a NS or one of two unrelated RNF2 shRNAs. Error bars indicate SD; n=3 technical replicates of a representative experiment (out of three experiments). d, qRT-PCR analysis monitoring expression of TRIM37 target genes in MCF7 cells expressing a second TRIM37 shRNA unrelated to that used in Fig. 2e. Expression of each gene was normalized to that obtained with a NS shRNA, which was set to 1. Error bars indicate SEM; n=3 technical replicates of a representative experiment (out of three experiments). e, qRT-PCR analysis monitoring expression of TRIM37 target genes following RNF2 knockdown in MCF7 cells. Expression of each gene was normalized to that obtained with a NS shRNA, which was set to 1 (indicated by the dotted red line). Error bars indicate SEM; n=3 technical replicates of a representative experiment (out of three experiments). f, qRT-PCR analysis monitoring expression of TRIM37 target genes following TRIM37 knockdown in BT474 cells. Error bars indicate SEM; n=3 technical replicates of a representative experiment (out of three experiments). g, qRT-PCR analysis monitoring expression of p14ARF following knockdown of TRIM37 or EZH2 in BT474 cells. Error bars indicate SEM; n=3 technical replicates of a representative experiment (out of three experiments). The results indicate that knockdown of EZH2 but not TRIM37 de-represses p14ARF expression in BT474 cells. h, qRT-PCR analysis monitoring knockdown efficiencies of TRIM37 (left) and EZH2 (right) shRNAs in BT474 cells. Error bars indicate SEM; n=3 technical replicates of a representative experiment (out of three experiments). *P<0.05; **P<0.01.
Extended Data Fig. 4. Additional experiments showing interaction between TRIM37 and PRC2 subunits.
a, Co-immunoprecipitation analysis. MCF7 nuclear extracts were immunoprecipitated with a TRIM37, EZH2 or SUZ12 antibody, or an IgG control, and the immunoprecipitates were analyzed for TRIM37, EZH2 or SUZ12 by immunoblotting. b, Co-immunoprecipitation analysis. MCF7 nuclear extracts were immunoprecipitated with a TRIM37 or EZH2 antibody, or an IgG control, in the presence of ethidium bromide, and the immunoprecipitates were analyzed for TRIM37 or EZH2 by immunoblotting. The results show that interaction between TRIM37 and EZH2 occurs in the presence of ethidium bromide and is thus not mediated by DNA. c, Co-immunoprecipitation analysis. Nuclear extracts from MCF7 cells ectopically expressing FLAG-TRIM37 were immunoprecipitated with anti-FLAG magnetic beads, a EZH2 or SUZ12 antibody, or an IgG control, and the immunoprecipitates were analyzed for TRIM37, EZH2 or SUZ12 by immunoblotting. d, Mass spectroscopy. Selected results from the liquid chromatography tandem mass spectroscopy analysis listing TRIM37 or PRC2 subnunits, and their total spectra score in samples immunoprecipitated using either IgG or a TRIM37 antibody. The probability of interaction, derived from the Scaffold Viewer software, indicates a ≥95% probability of interaction in all cases. See also Supplementary Table 2. e, Tandem mass spectra of representative peptides of proteins identified in the TRIM37 immunoprecipitate are shown with corresponding spectral counts for each protein shown in the table. f, In vitro interaction pull-down assay. Purified glutathione-S-transferase (GST)-TRIM37 was incubated with an in vitro translated biotinylated PRC2 subunit (indicated on left). GST-TRIM37 was purified using GST-agarose beads, and the presence of the PRC2 subunit was analyzed by immunoblotting with an anti-biotin antibody. The results indicate that TRIM37 interacts strongly with EZH2 and weakly with RBBP4, and does not detectably interact with AEBP2, EED or SUZ12. g, GAL4-TRIM37 fusion experiment. (Top) Schematic diagram of TRIM37 showing the location of the RING, BBOX, coiled coil (CC), MATH and nuclear localization sequence (NLS) motifs. Locations of the motifs were obtained from UniProtKB (www.uniprot.org/uniprot/O94972#section_attribute). The ΔBBCC deletion comprises the BBOX and first CC motif (Δ aa 89–235). (Middle, left) ChIP monitoring binding of GAL4-TRIM37, EZH2 or SUZ12 to the adenovirus E1B promoter containing (GAL4(UAS)-E1B) or lacking (E1B) GAL4-binding sites or to an irrelevant, negative control (NC) DNA region. The indicated GAL4-TRIM37 fusion protein was co-expressed with a plasmid containing or lacking five GAL4-binding sites upstream of the adenovirus E1B gene in 293T cells followed by ChIP analysis. Error bars indicate SD; n=3 technical replicates of a representative experiment (out of three experiments). The results indicate that the GAL4-TRIM37 fusion protein was able to recruit both EZH2 and SUZ212 to the GAL4-binding sites. (Middle, right) ChIP monitoring binding of EZH2 and SUZ12 in the presence of GAL4-TRIM37 deletion mutants. The GAL4-TRIM37 wild-type (WT) samples are the same as those shown in the left panel. The results indicate that the ability of GAL4-TRIM37 to recruit EZH2 and SUZ212 requires the TRIM37 RING domain and NLS (presumably for nuclear entry), but not the BBCC or MATH domains. (Bottom) Immunoblot analysis monitoring expression of GAL4-TRIM37 fusion proteins using a GAL4 antibody. α-tubulin (TUBA) was monitored as a loading control.
Extended Data Fig. 5. Confirmation of the results of Fig. 3f, h using second, unrelated shRNAs.
a, ChIP monitoring binding of BMI1 and EZH2 to the promoters of TRIM37 target genes in MCF7 cells expressing a NS shRNA or a second TRIM37 shRNA unrelated to that used in Fig. 3f. The IgG control and BMI1 and EZH2 signal in cells expressing a NS shRNA are the same as those shown in Fig. 3f. Black asterisks indicate significance of BMI1 or EZH2 enrichment compared to the IgG control (from cells expressing a NS shRNA); blue asterisks indicate significant differences in BMI1 or EZH2 enrichment in cells expressing a TRIM37 shRNA relative to NS shRNA. Error bars indicate SD; n=3 technical replicates of a representative experiment (out of three experiments). b, qRT-PCR (top) and immunoblot (bottom) monitoring BMI1 and EZH2 levels in MCF7 cells expressing a NS or TRIM37 shRNA. Error bars indicate SEM; n=3 technical replicates of a representative experiment (out of three experiments). The results indicate that BMI1 and EZH2 levels are unaffected by TRIM37 knockdown in MCF7 cells. c, (Left) qRT-PCR analysis monitoring expression of two genes that are bound by EZH2 but not TRIM37, ADAM7 (top) and YES1 (bottom), following knockdown of TRIM37 or EZH2 in MCF7 cells. Error bars indicate SEM; n=3 technical replicates of a representative experiment (out of three experiments). The results indicate that knockdown of EZH2 but not TRIM37 de-represses ADAM7 and YES1 expression. (Right) ChIP monitoring EZH2 enrichment at the promoters of ADAM7 and YES1 in MCF7 cells expressing a NS or TRIM37 shRNA. Error bars indicate SD; n=3 technical replicates of a representative experiment (out of three experiments). The results show that knockdown of TRIM37 has no effect on EZH2 binding at ADAM7 and YES1 promoters. Thus, loss of EZH2 binding and de-repression following TRIM37 knockdown is not general to PRC2-bound genes but rather is selective for TRIM37 target genes. d, ChIP monitoring H3K27me3 enrichment at promoters of TRIM37 target genes in MCF7 cells expressing a NS or TRIM37 shRNA. Error bars indicate SD; n=3 technical replicates of a representative experiment (out of three experiments). e, qRT-PCR analysis monitoring BMI1 (left) and EZH2 (right) knockdown efficiency in MCF7 cells following shRNA-mediated knockdown using two unrelated shRNAs against each gene. Expression of each gene was normalized to that obtained with a NS shRNA, which was set to 1. Error bars indicate SEM; n=3 technical replicates of a representative experiment (out of three experiments). f, qRT-PCR monitoring TRIM37 target gene expression in MCF7 cells following knockdown of BMI1 or EZH2 using a second shRNA unrelated to that used in Fig. 3h. Error bars indicate SEM; n=3 technical replicates of a representative experiment (out of three experiments). *P<0.05; **P<0.01.
Extended Data Fig. 6. TRIM37/17q23 copy number and promoter methylation analysis for TRIM37 target genes in human breast cancer samples.
a, Analysis of 17q23 copy number in a panel of 71 human breast cancer samples; the region corresponding to the TRIM37 gene is highlighted in yellow. The breast cancer samples shown here are the same as those shown in Fig. 4a. Red indicates increased copy number and green indicates decreased copy number (see Methods). b, Promoter methylation analysis. Each of the 60 TRIM37 target genes shown in Fig. 4a were analyzed for promoter CpG methylation in human breast cancer samples. The level of DNA methylation is shown ranging from low (green) to high (red); white indicates data were not available. The first column is a heat map showing expression of TRIM37. The results indicate no significant correlation between TRIM37 expression and promoter methylation of TRIM37 target genes.
Extended Data Fig. 7. Control experiments related to Fig. 4.
a, Proliferation of cultured MCF7 cells expressing an NS shRNA or a second TRIM37 shRNA unrelated to that used in Fig. 4d. Error bars indicate SD; n=3 technical replicates of a representative experiment (out of three experiments). b, c, Tumour formation in mice subcutaneously injected with BT474 (b) or FC-IBC02 (c) cells expressing an NS or TRIM37 shRNA. Error bars indicate SEM; n=3 mice per group. d, Immunoblot monitoring TRIM37 levels in NIH 3T3 cells expressing vector, wild-type (WT) TRIM37 or TRIM37-C18R. α-tubulin (TUBA) was monitored as a loading control. e, Immunoblot monitoring TRIM37 levels in MCF10AT cells expressing vector or FLAG-tagged TRIM37. f, qRT-PCR analysis monitoring knockdown efficiency of a tumor suppression gene in MCF10AT cells expressing the indicated shRNA. The results are given relative to expression following treatment with a NS shRNA, which was set to 1. Error bars indicate SD; n=3 technical replicates of a representative experiment (out of three experiments). g, Soft agar assay monitoring colony formation of MCF10AT cells expressing a NS or the indicated shRNA. Error bars indicate SEM; n=3 technical replicates of a representative experiment (out of three experiments). h, Tumour formation in mice subcutaneously injected with MCF10AT cells expressing a NS or the indicated shRNA. Error bars indicate SEM; n=3 mice per group. i, Soft agar assay monitoring colony formation of MCF10A cells expressing vector or TRIM37. Error bars indicate SEM; n=3 technical replicates of a representative experiment (out of three experiments). j. Tumour formation in mice subcutaneously injected with MCF10A cells expressing vector or TRIM37. Error bars indicate SEM; n=3 mice per group. k, Tumour formation in mice subcutaneously injected with BPLER cells expressing vector or TRIM37. Error bars indicate SEM; n=3 mice per group. *P<0.05; **P<0.01.
Extended Data Fig. 8. Schematic model of TRIM37-directed transcriptional repression and Kaplan-Meier analysis comparing survival of breast cancer patients expressing high or low TRIM37 expression.
a, Schematic models for canonical (left) and TRIM37 (right) pathways for target gene silencing. b, Kaplan-Meier analysis of survival of patients with low (black) or high (red) TRIM37 expression. The number of surviving patients at 0, 5, 10 and 15 year time-points is indicated below the graph. High expression of TRIM37 was significantly correlated with lower survival rate (p value = 0.00046, hazard ratio (HR) = 1.46 with 95% confidence interval 1.18–1.81). The analysis was performed using an online survival analysis tool to analyze the effect of gene expression on breast cancer prognosis using microarray data of 1809 patients41.
Extended Data Table 1.
List of catalog numbers for shRNAs obtained from Open Biosystems/GE Dharmacon or The RNAi Consortium (TRC)/Broad Institute.
| Gene | Catalog number for lst shRNA | Catalog number for 2nd shRNA |
|---|---|---|
| BCL2L10 | TRCN0000033595 | TRCN0000033596 |
| BMI1 | RHS3979-201751271 | RHS3979-201751275 |
| CUL1 | TRCN0000003391 | TRCN0000003392 |
| CUL3 | TRCN0000073343 | TRCN0000073344 |
| EGR1 | TRCN0000013073 | TRCN0000013074 |
| EZH2 | TRCN0000040074, | TRCN0000040076, |
| IL24 | RHS3979-9607463 | RHS3979-9607465 |
| PAWR | TRCN0000058443 | TRCN0000058444 |
| PLCB3 | TRCN0000020304 | TRCN0000020305 |
| PPP1R1B | TRCN0000000434 | TRCN0000000433 |
| Rnf2 (mouse) | TRCN0000052879 | TRCN0000052880 |
| RNF2 | TRCN0000040579 | TRCN0000040580 |
| SFRP1 | RMM3981-201769168 | RHS3979-9601103 |
| TP53 | TRCN0000062172 | TRCN0000062172 |
| Trim37 (mouse)* | TRCN0000018866 | TRCN0000018869 |
| TGCTGTTGACAGTGAGCGACCGGGATCAGCACTG- TATATTAGTGAAGCCACAGATGTAATA- TACCAGTGCTGATCCCGGCTGCCTACTGCCTCGGA |
TGCTGTTGACAGTGAGCGAGGGATCAGCACTGG- TATATTATAGTGAAGCCACAGATGTATA- ATATACCAGTGCTGATCCCGTGCCTACTGCCTCGGA |
|
| TRIM37 (human) | RHS4430-200178645 | RHS4430-200209374 |
| UNC5B | TRCN0000061816 | TRCN0000061813 |
These shRNAs are no longer commercially available from Open Biosystems/GE Dharmacon, and thus shRNA sequences are provided in lieu of catalog numbers.
Extended Data Table 2.
List of primers used for quantitative real-time RT-PCR, ChIP and vector construction.
| Gene | Forward primer (5’ → 3’) | Reverse primer (5’ → 3’) | |
|---|---|---|---|
| qRT-PCR | ACTB | ACGTTGACATCCGTAAAGACCT | GCAGTAATCTCCTTCTGCATCC |
| ACTN1 | TCCATCGGAGCCGAAGAAATC | GTGTCGGTGGATCAAAGCACA | |
| ADAM7 | GGTGCCGTATGGTGCCAATTA | CCCCAAATTCGGTTCCTTAGTT | |
| ASS1 | TCCGTGGTTCTGGCCTACA | GGCTTCCTCGAAGTCTTCCTT | |
| BCL2L10 | CGGAAGAAGCGCCATCTCC | AGGCGTCCTTCTTCACTTCAG | |
| BMI1 | TTCTGCTGATGCTGCCAATG | TCCGATCCAATCTGTTCTGGTC | |
| CDT1 | ATGTCAAGGAGCACCACAG | ACTTCATCCACGTTGAAGCG | |
| CKB | ACAACAACCACATGGCCAAG | AGCCCACGGTCATGATGTAC | |
| CUL1 | AACAGTGCAAGTGACGATGC | TTGCTCACGCCAATGTCTTG | |
| CUL3 | TAAACCAACACAGCGGGTTC | TCACCTTGTTTGGCAGCAAC | |
| DDR1 | TTCCCCCTTAATGTGCGTAAGG | AGCAGCCGAATGATGTTTGG | |
| DNMT1 | GAGCTACCACGCAGACATCA | CGAGGAAGTAGAAGCGGTTG | |
| EEF1A1 | GAAGACCCACATCAACATCGT | CTCCGCATTTGTAGATGAGGTG | |
| EGR1 | CAGCACCTTCAACCCTCAGG | GAGATGTCAGGAAAAGACTCTGC | |
| EZH2 | AGTTCGTGCCCTTGTGTGATAG | TGGTGTTGCACTGTGCTTTG | |
| GAPDH | TGCACCACCAACTGCTTAGC | GGCATGGACTGTGGTCATGAG | |
| GRIN1 | ACGCCATCCTAGTTAGCCATC | GCACGGGTATGCGGTAGAAG | |
| HEYL | GGCTGCTTACGTGGCTGTT | GACCCAGGAGTGGTAGAGCAT | |
| IL24 | TTCTGGGCTGTGAAAGACAC | TTTCTTGCGAGACGTTCTGC | |
| IL36A | CCAGACGCTCATAGCAGTCC | AGATGGGGTTCCCTCTGTCTT | |
| JPH3 | AGAATGGGGATGAGCTCAAGTC | TTTTGCTACCGCGACATCTC | |
| LAMP3 | GCGTCCCTGGCCGTAATTT | TGCTTGCTTAGCTGGTTGCT | |
| LTBP3 | TCCCCAGGGCTACAAGAGG | AGACACAGCGATAGGAGCCA | |
| PARD6G | GTCTCAGACCTTGCGATTCTAC | GGTCCAGAGAGAACCTTCGGA | |
| PARP9 | TGGTTGTGGCCATTTCTGTG | ACCATGTTCACCAGCACAAC | |
| PAWR | TGATGAAGCAGGGCAGAAAG | AAACTGTTCTAGGTGGCTCCTG | |
| PLCB3 | TTCATTACCCACGGCTTCAC | TCTTTGCCTGTTGCTTTGCC | |
| PPP1R1B | CAAGTCGAAGAGACCCAACCC | GCCTGGTTCTCATTCAAATTGCT | |
| PROP1 | GCCCAGTGCAGTTGGAACA | TGCTTCCGTTGCTTAGCTCTG | |
| P14ARF | CCCTCGTGCTGATGCTACTG | ACCTGGTCTTCTAGGAAGCGG | |
| RASSF3 | GCCTGCAAGCTCTCAGACC | TAGGCTGTGTAGCGCCTCTT | |
| RASSF9 | GAGGACCTGAGCGAAAGTGAT | TCTGGATGCCACTCAAATGAGA | |
| RNF2 | GTGCAGACAAACGGAACTCA | TTGCCACTTCTAAGGGCTGT | |
| SEPN1 | CCTGACCCTAGCGAGGAGAC | GGCTGTCCAGTTTCGGAGG | |
| SFN | TGACGACAAGAAGCGCATCAT | GTAGTGGAAGACGGAAAAGTTCA | |
| SFRP1 | TCATGCAGTTCTTCGGCTTC | TTGTCACAGGGAGGACACAC | |
| TBKBP1 | GGCGGCTAGAAGAGGCTTTG | CTCCGTCAGCTCGGTGTAG | |
| TP53 | ACTTGTCGCTCTTGAAGCTAC | GATGCGGAGAATCTTTGGAACA | |
| TRAF2 | TGCCCCAAGTTCCCCTTAAC | TTGCCACAAGTCTTGACGTG | |
| TRIM37 | AACAGAGCGTGGAGAGCATT | CTTCTGCCCAACGACAATTT | |
| UNC5B | GTCGGACACTGCCAACTATAC | CCGCCATTCACGTAGACGAT | |
| VCL | AAACTGAACCAGGCCAAAGG | TTTGCCTGCACAGAGTTCAC | |
| WNT10B | GTGAGCGAGACCCCACTATG | CACTCTGTAACCTTGCACTCATC | |
| YES1 | GCCTGTCAGTACAAGTGTGAG | AAAGGCGTTACCCCTGAGGAT | |
| ChIP | Actb (mouse) | CGAAGCCGGTGAGTGAGC | ACCGCCGAGACCGCGTCC |
| ACTB | CCGTTCCGAAAGTTGCCTT | CGCCGCCGGGTTTTATA | |
| ADAM7 | AACTGGGAGCAAGTGAACTG | CTGCTCCAAGGACGACTTTTTC | |
| BCL2L10 | AGGAGGTTGTGGTTCAGACC | ACTTAGCAGCCAGAGTGATCC | |
| CDT1 | GGGATTACAGGCGTGAACCA | ATCCGCTCTCTTGGAACTGC | |
| CKB | CAGGTGTCCCTTAAGCCTGG | CCCTTCAGGTTTTGTGGGGT | |
| CUL1 | CGAGGGAGGCTGTAAAACCTG | CGTGGGGAAGCTTCTACCTG | |
| DDR1 | ACCTGCTTCTTCCTCTGTGC | CCCCAAACGAGAACCTCCAA | |
| DNMT1 | CCACGCCTGGCTAATTTTTG | ATCACGAGGTCAGGAGTTCG | |
| EEF1A | AGCTCGGGATCAAGAATCAC | TGGCCTCTTTACGGGTTATG | |
| EGR1 | CTGGGAGTGGAGAGGGAAC | AATCCGAMGAGGGGCTTCC | |
| Fas (CP/TSS) | GCCGCCTGTGCAGTGGTGA | CTGTGTGTGGGCAGCCTGCGGC | |
| Fas (1 kb) | GGCTATAGATCACCTTCATGTA | GCAGTTAACTCAGGGACCAAG | |
| Fas (2 kb) | GCGTTGCCATAGCATGAACT | GAGTTAGGGGACCATAGTCA | |
| Gapdh (mouse) | CACGCTCGGTGCGTGCCC | GCTCAGCCAGTCCCAGCC | |
| GAPDH | TCCGGGTGATGCTTTTCCTAG | TTTGCGGTGGAAATGTCCTTTTC | |
| IL24 | TTCTTCGCTGTGGACTACTGTC | AACAACACAGAGGTGCTTGG | |
| IL36A | TCAACTATGTGTGGCTTGTGC | GCTAAACTCATTCCACAAGAAGC | |
| JPH3 | GTTTGCAAAGGGCCTCAGTC | AGGACTGCGTCATGGGAAG | |
| LAMP3 | AGCACCTGGACAGCAATTAC | AGGTCAGGTTGTGAGGTGAATG | |
| PARP9 | GTGTGGTTCTGGCTAAATGCAC | TGCGGCACATTTGTACAAGG | |
| PAWR | TGTTGTGGGAAACGTATGGC | TTAGGTAAACAGCCGCTCAGC | |
| PLCB3 | TGTGAAGGTCTCCAGCCATG | ATGGGAACTGGACAGGAATCTG | |
| RASSF3 | TAATCCCAGCTACTTGGGAAGC | TTTTGCTCTGTCACCCAGAC | |
| RASSF9 | ATCATCGCCACCTGGTATCAC | GCAAACAGAGGGAACAACATGG | |
| SEPN1 | AGGCGCAGTGTATGTGTCAC | TCGGAGGAAGGAAATGACAGAC | |
| SFN | ACAGGCCACCAACTTGAAAG | TAACAAGCAGCCTCTGAAGC | |
| SFRP1 | CCCTCCACCTCAAGCCAAAT | ACAAGTCCTCCTGCCATTCG | |
| TP53 | TTGCCAGGAAAGATGCACAG | ATGTGGCAGTTGTGTTGCTC | |
| TRAF2 | GCGCGCTCTTTGAGGAAAAA | CAGCCCTCAGGAAGCTGTAG | |
| WNT10B | TTCTTTCTGCCTCCACACTCTC | AGCACTTTCTCGGAAACCTG | |
| YES1 | TGATGAGGGTGTGAGTAACGC | TGGTTGGCTGTTTGCTTCTC | |
| GAL4 recruitment | GAL4-UAS | GACACTATAGAATCGAGCAGCTG | ATGAATTCGAGCTCGGTACCC |
| TRIM37 | AACAGAGCGTGGAGAGCATT | CTTCTGCCCAACGACAATTT | |
| Vector construction | TRIM37-C18R | CATACGTATGAAACATCGGAAAACCTCAGC | CATACGTATGGAGAAATTGCGGGATG |
| H2A-KKRR (N terminus) | ATGTCTGGTCGTGGCAAACA | ATGCGCGTCCTCCTGTTGTC | |
| H2A-KKRR (C terminus) | (P)-CATCCCTCGTCACCTCCAG | TGGGTGGCTCTAAAAAGAGCC | |
| Cloning TRIM37 into pGEX4T-1 | GAATTCATGGATGAACAGAGCGTGGA | GCGGCCGCTCTTCCACTATTTT | |
| Cloning TRIM37 into pCMX-Ga14 | GGTACCAAATGGATGAACAGAGCGTGGA | GCTAGCTCTTCCACTATTTT | |
| TRIM37ΔRING | GAATTCATGGATGAACAGAGCGTGGAACTCCAGC- TACGAGAACTAGTAAA |
CTCCAGCTACGAGAACTAGTAAA | |
| TRIM37ΔBBOX | (P)-ATGTTTGGTGAGACTGCAGAG | ATTTATGAGCAACACGTCACTAAA | |
| TRIM37ΔCC | (P)-AGTGACGTGTTGCTCATAAATTTCT | GAGTTGATATCTAAGAGCTCAGAGA | |
| TRIM37ΔMATH | (P)-AGTAGCTGAATCGTAAGATGGCA | GCAAATGAAGGATACTTGAATCCACA | |
| TRIM37ΔNLS | (P)-CAAGCCACTGAAAACTGCAACC | ATGGTCACCTTGGGGGCTAAT |
Supplementary Material
Acknowledgments
We thank C. Peterson for providing Xenopus nucleosomes; R. Weinberg and M. Cristofanilli for providing cell lines; A. Virbasius and the UMMS RNAi Core Facility for providing shRNAs; P. Spatrick at the UMMS Genomics Core Facility; the UMMS Proteomics and Mass Spectroscopy Facility for mass spectrometry analysis; and S. Deibler for providing editorial assistance. N.W. is a Sidney Kimmel Scholar for Cancer Research and is supported by young investigator awards from National Lung Cancer Partnership/Uniting Against Lung Cancer, Melanoma Research Alliance and International Association for the Study of Lung Cancer. This work was also supported by grants from CEA-DSV and ATIGE Genopole to C.G., the NIH (R01GM033977) to M.R.G. M.R.G. is an investigator of the Howard Hughes Medical Institute.
Footnotes
Online Content Any additional Methods, Extended Data display items and Source Data are available in the online version of the paper; references unique to these sections appear only in the online paper.
Author contributions C.G. made the initial observation that TRIM37 has an H2A mono-ubiqutination activity. S.B., J.S.T. and M.R.G. designed the experiments. S.B., L.C., X.Z., L.L. and N.W. performed the research. C-M.V. provided critical reagents. J.O. and L.J.Z. performed biostatistical analysis for ChIP-chip experiments and database mining. S.B. and M.R.G. analysed and interpreted the data and wrote the paper. All authors reviewed the paper and provided comments.
The authors declare no competing financial interests.
References
- 1.Sinclair CS, Rowley M, Naderi A, Couch FJ. The 17q23 amplicon and breast cancer. Breast Cancer Res. Treat. 2003;78:313–322. doi: 10.1023/a:1023081624133. [DOI] [PubMed] [Google Scholar]
- 2.Budhidarmo R, Nakatani Y, Day CL. RINGs hold the key to ubiquitin transfer. Trends Biochem. Sci. 2012;37:58–65. doi: 10.1016/j.tibs.2011.11.001. [DOI] [PubMed] [Google Scholar]
- 3.Weake VM, Workman JL. Histone ubiquitination: triggering gene activity. Mol. Cell. 2008;29:653–663. doi: 10.1016/j.molcel.2008.02.014. [DOI] [PubMed] [Google Scholar]
- 4.Wang H, et al. Role of histone H2A ubiquitination in Polycomb silencing. Nature. 2004;431:873–878. doi: 10.1038/nature02985. [DOI] [PubMed] [Google Scholar]
- 5.de Napoles M, et al. Polycomb group proteins Ring1A/B link ubiquitylation of histone H2A to heritable gene silencing and X inactivation. Dev. Cell. 2004;7:663–676. doi: 10.1016/j.devcel.2004.10.005. [DOI] [PubMed] [Google Scholar]
- 6.Gazin C, Wajapeyee N, Gobeil S, Virbasius CM, Green MR. An elaborate pathway required for Ras-mediated epigenetic silencing. Nature. 2007;449:1073–1077. doi: 10.1038/nature06251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cao R, Tsukada Y, Zhang Y. Role of Bmi-1 and Ring1A in H2A ubiquitylation and Hox gene silencing. Mol. Cell. 2005;20:845–854. doi: 10.1016/j.molcel.2005.12.002. [DOI] [PubMed] [Google Scholar]
- 8.Lanzuolo C, Orlando V. Memories from the polycomb group proteins. Annu. Rev. Genet. 2012;46:561–589. doi: 10.1146/annurev-genet-110711-155603. [DOI] [PubMed] [Google Scholar]
- 9.Buchwald G, et al. Structure and E3-ligase activity of the Ring-Ring complex of polycomb proteins Bmi1 and Ring1b. EMBO J. 2006;25:2465–2474. doi: 10.1038/sj.emboj.7601144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Richly H, et al. Transcriptional activation of polycomb-repressed genes by ZRF1. Nature. 2010;468:1124–1128. doi: 10.1038/nature09574. [DOI] [PubMed] [Google Scholar]
- 11.Monni O, et al. Comprehensive copy number and gene expression profiling of the 17q23 amplicon in human breast cancer. Proc. Natl. Acad. Sci. USA. 2001;98:5711–5716. doi: 10.1073/pnas.091582298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kirmizis A, et al. Silencing of human polycomb target genes is associated with methylation of histone H3 Lys 27. Genes Dev. 2004;18:1592–1605. doi: 10.1101/gad.1200204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Margueron R, Reinberg D. The Polycomb complex PRC2 and its mark in life. Nature. 2011;469:343–349. doi: 10.1038/nature09784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ku M, et al. Genomewide analysis of PRC1 and PRC2 occupancy identifies two classes of bivalent domains. PLoS Genet. 2008;4:e1000242. doi: 10.1371/journal.pgen.1000242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mohn F, et al. Lineage-specific polycomb targets and de novo DNA methylation define restriction and potential of neuronal progenitors. Mol. Cell. 2008;30:755–766. doi: 10.1016/j.molcel.2008.05.007. [DOI] [PubMed] [Google Scholar]
- 16.Velichutina I, et al. EZH2-mediated epigenetic silencing in germinal center B cells contributes to proliferation and lymphomagenesis. Blood. 2010;116:5247–5255. doi: 10.1182/blood-2010-04-280149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Orlando DA, Guenther MG, Frampton GM, Young RA. CpG island structure and trithorax/polycomb chromatin domains in human cells. Genomics. 2012;100:320–326. doi: 10.1016/j.ygeno.2012.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Riising EM, et al. Gene silencing triggers polycomb repressive complex 2 recruitment to CpG islands genome wide. Mol. Cell. 2014;55:347–360. doi: 10.1016/j.molcel.2014.06.005. [DOI] [PubMed] [Google Scholar]
- 19.Network CGA. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miller FR, et al. Xenograft model of progressive human proliferative breast disease. J. Natl. Cancer Inst. 1993;85:1725–1732. doi: 10.1093/jnci/85.21.1725. [DOI] [PubMed] [Google Scholar]
- 21.Ince TA, et al. Transformation of different human breast epithelial cell types leads to distinct tumor phenotypes. Cancer Cell. 2007;12:160–170. doi: 10.1016/j.ccr.2007.06.013. [DOI] [PubMed] [Google Scholar]
- 22.von Lintig FC, et al. Ras activation in human breast cancer. Breast Cancer Res. Treat. 2000;62:51–62. doi: 10.1023/a:1006491619920. [DOI] [PubMed] [Google Scholar]
- 23.Grau DJ, et al. Compaction of chromatin by diverse Polycomb group proteins requires localized regions of high charge. Genes Dev. 2011;25:2210–2221. doi: 10.1101/gad.17288211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Eskeland R, et al. Ring1B compacts chromatin structure and represses gene expression independent of histone ubiquitination. Mol. Cell. 2010;38:452–464. doi: 10.1016/j.molcel.2010.02.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chang CJ, Hung MC. The role of EZH2 in tumour progression. Br. J. Cancer. 2012;106:243–247. doi: 10.1038/bjc.2011.551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Deb G, Thakur VS, Gupta S. Multifaceted role of EZH2 in breast and prostate tumorigenesis: epigenetics and beyond. Epigenetics. 2013;8:464–476. doi: 10.4161/epi.24532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dawson PJ, Wolman SR, Tait L, Heppner GH, Miller FR. MCF10AT: a model for the evolution of cancer from proliferative breast disease. Am. J. Pathol. 1996;148:313–319. [PMC free article] [PubMed] [Google Scholar]
- 28.Fernandez SV, et al. Inflammatory breast cancer (IBC): clues for targeted therapies. Breast Cancer Res. Treat. 2013;140:23–33. doi: 10.1007/s10549-013-2600-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Luger K, Rechsteiner TJ, Richmond TJ. Expression and purification of recombinant histones and nucleosome reconstitution. Methods Mol. Biol. 1999;119:1–16. doi: 10.1385/1-59259-681-9:1. [DOI] [PubMed] [Google Scholar]
- 30.Hansen JC, Ausio J, Stanik VH, van Holde KE. Homogeneous reconstituted oligonucleosomes, evidence for salt-dependent folding in the absence of histone H1. Biochemistry. 1989;28:9129–9136. doi: 10.1021/bi00449a026. [DOI] [PubMed] [Google Scholar]
- 31.Maston GA, et al. Non-canonical TAF complexes regulate active promoters in human embryonic stem cells. Elife. 2012;1:e00068. doi: 10.7554/eLife.00068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gentleman RC, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zacher B, Kuan PF, Tresch A. Starr: Simple Tiling ARRay analysis of Affymetrix ChIP-chip data. BMC Bioinformatics. 2010;11:194. doi: 10.1186/1471-2105-11-194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Benjamini Y, Hochberg Y. Controlling the false discovery rate:a practical and powerful approach to multiple testing. J. R. Statis. Soc. B. 1995;57:289–300. [Google Scholar]
- 35.Zhu LJ, et al. ChIPpeakAnno: a Bioconductor package to annotate ChIP-seq and ChIP-chip data. BMC Bioinformatics. 2010;11:237. doi: 10.1186/1471-2105-11-237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Salicru M, Ocana J, Sanchez-Pla A. Comparison of lists of genes based on functional profiles. BMC Bioinformatics. 2011;12:401. doi: 10.1186/1471-2105-12-401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ihaka R, Gentleman R. R: A language for data analysis and graphics. J. Comput. Graph. Stat. 1996;5:299–314. [Google Scholar]
- 38.Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30:207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tanese N. Small-scale density gradient sedimentation to separate and analyze multiprotein complexes. Methods. 1997;12:224–234. doi: 10.1006/meth.1997.0475. [DOI] [PubMed] [Google Scholar]
- 40.Wang Y, et al. BASC, a super complex of BRCA1-associated proteins involved in the recognition and repair of aberrant DNA structures. Genes Dev. 2000;14:927–939. [PMC free article] [PubMed] [Google Scholar]
- 41.Liu F, Green MR. A specific member of the ATF transcription factor family can mediate transcription activation by the adenovirus E1a protein. Cell. 1990;61:1217–1224. doi: 10.1016/0092-8674(90)90686-9. [DOI] [PubMed] [Google Scholar]
- 42.Lillie JW, Green MR. Transcription activation by the adenovirus E1a protein. Nature. 1989;338:39–44. doi: 10.1038/338039a0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.