SUMMARY
Dyneins power microtubule motility using ring-shaped, AAA-containing motor domains. Here, we report X-ray and electron microscopy (EM) structures of yeast dynein bound to different ATP analogs, which collectively provide insight into the roles of dynein’s two major ATPase sites, AAA1 and AAA3, in the conformational change mechanism. ATP binding to AAA1 triggers a cascade of conformational changes that propagate to all six AAA domains and cause a large movement of the “linker,” dynein’s mechanical element. In contrast to the role of AAA1 in driving motility, nucleotide transitions in AAA3 gate the transmission of conformational changes between AAA1 and the linker, suggesting that AAA3 acts as a regulatory switch. Further structural and mutational studies also uncover a role for the linker in regulating the catalytic cycle of AAA1. Together, these results reveal how dynein’s two major ATP-binding sites initiate and modulate conformational changes in the motor domain during motility.
INTRODUCTION
Myosin, kinesin and dynein, ATP-driven cytoskeletal motor proteins, power various forms of biological motility including muscle contraction, ciliary beating, intracellular cargo transport, and movements during cell division. Specific transitions in the motor ATPase cycles are coupled to the binding and dissociation from a polymer track (actin for myosin and microtubules [MTs] for kinesin and dynein) and the execution of a “power stroke,” a conformational change that biases the movement of the motor in one direction along the polymer. Although they bind different cytoskeletal polymers, myosin and kinesin evolved from a common ancestor, similar to small GTPases, and share similarities in how they amplify small conformational changes in their active sites into larger structural changes that drive motility (Kull and Endow, 2013; Rayment et al., 1996; Vale and Milligan, 2000).
Dynein is a member of the AAA family (ATPases associated with diverse cellular activities) and thus evolved through an evolutionary lineage separate from that of kinesin and myosin. Each AAA domain in AAA ATPases is typically composed of a large α/β subdomain (“L” domain) and small α-helical subdomain (“s” domain). Many AAA proteins self-assemble into homo-hexameric rings and use ATP energy to translocate polypeptide chains or nucleic acids into the central pore (Baker and Sauer, 2012; Furst et al., 2003; Skordalakes and Berger, 2003; West, 2003). Dynein is unusual in the AAA protein family; its six AAA modules are contained within a single, large polypeptide chain, and each module has evolved its own unique sequence and function (Carter, 2013; Roberts et al., 2013). The first four AAA domains bind ATP, whereas AAA5 and AAA6 do not. ATP hydrolysis at AAA1 is required for dynein motility, and mutational studies indicate that it is the main hydrolytic site (Gibbons and Gibbons, 1987; Kon et al., 2004; Reck-Peterson and Vale, 2004). AAA2 lacks key residues for nucleotide hydrolysis and appears to constitutively bind nucleotide (Carter, 2013). ATP hydrolysis at AAA3 plays an important role, given that a hydrolysis mutation in AAA3 reduces dynein motility and ATPase activity by >20-fold (Cho et al., 2008; Kon et al., 2004). ATP hydrolysis at AAA4 appears to have a more subtle role, as a AAA4 hydrolysis mutation only decreases the velocity of yeast dynein by ~20% (Cho et al., 2008). Although mutational analyses suggest important roles for both AAA1 and AAA3, single-molecule studies indicate that the binding of a single ATP molecule (presumably at AAA1) is sufficient for dynein to step along the microtubule (DeWitt et al., 2012; Qiu et al., 2012). Thus, why blocking nucleotide hydrolysis at AAA3 so drastically impairs dynein motility has remained an unresolved question.
Crystal structures for cytoplasmic dyneins have been obtained recently for yeast (Saccharomyces cerevisiae) dynein in a nucleotide-free state (apo) (Carter et al., 2011; Schmidt et al., 2012) and Dictyostelium discoideum dynein in an ADP-bound state (Kon et al., 2011, 2012). These structures revealed that the motor domain consists of six AAA modules organized into an asymmetric ring and three appendages that extend from the ring. Two appendages are antiparallel coiled coils called the “stalk” and the “buttress” (or “strut”). The stalk is longer (~15 nm) and contains the microtubule-binding domain (MTBD) at its tip; the buttress interacts with the stalk near its base. The other appendage is a proposed mechanical element called the “linker,” which is a series of helical bundles that arch over one face of the ring. A transition of the linker from a bent to a straight conformation has been proposed to act as a power stroke that drives movement of an attached cargo toward the microtubule minus end (Burgess et al., 2003; Roberts et al., 2013). However, recent electron cryo-tomography studies at ~30–50 Å resolution of a dimeric axonemal dynein in intact flagella came to a different conclusion, that the power stroke may be driven by a rotational movement of the AAA ring with respect to the relatively stationary linker, rather than by a remodeling of the linker (Lin et al., 2014).
In order to understand the sequence of conformational changes that take place during dynein’s catalytic cycle, it is necessary to obtain high-resolution snapshots of dynein in several different nucleotide-bound states. In this study, we solved a crystal structure (~3.5 Å) of yeast dynein with an ATP analog (AMPPNP) and obtained 12 different electron microscopy (EM) data sets and 27 EM reconstructions of dynein in different nucleotide states. Our data show that ATP binding to AAA1 initiates a large conformational change in half of the ring (AAA1–4), whereas ATP hydrolysis in AAA3 is important for propagating this conformational change to the remainder of the ring (AAA5–6) and enabling linker bending. Together with structure-function studies performed here and information from prior structures, we generated a model for the movements of AAA domains and the linker during dynein’s ATPase cycle. Our results also suggest that the linker, in addition to its previously postulated role as a mechanical element, acts allosterically to regulate the catalytic cycle at AAA1.
RESULTS
Crystal Structure of Yeast Dynein in the AMPPNP-Bound State Reveals Closure of AAA1 and AAA3
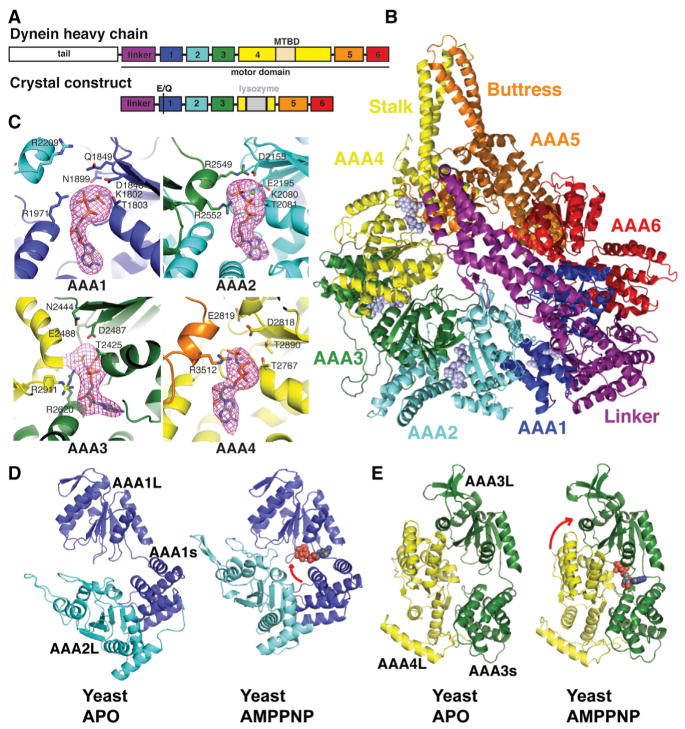
The motor domain from yeast dynein was previously crystallized in the absence of nucleotide (Carter et al., 2011). To obtain a conformation of the motor domain with a nucleotide triphosphate or triphosphate analog bound to AAA1, we blocked nucleotide hydrolysis by generating a mutation (E1849Q) in the Walker B motif of AAA1 (Babst et al., 1998; Watanabe et al., 2002). We obtained dynein crystals in the presence of AMPPNP, a nonhydrolyzable ATP analog (Lee et al., 2007), using a construct in which the MTBD and part of the stalk were replaced with lysozyme (Figures 1A and 1B). The lysozyme fusion with the motor domain, which retained basal ATPase activity (Figure S1A available online), facilitated crystallization by promoting the formation of crystal contacts (Figure S1B). The structure, which contained two monomers per asymmetric unit, was determined by molecular replacement using the individual domains of the nucleotide-free yeast motor domain as search models (see Extended Experimental Procedures) and was refined to 3.54 Å resolution with an Rwork and Rfree of 23% and 26%, respectively (Table S1). The two monomers in the asymmetric unit pack against one another via the nonlinker faces of their AAA rings (Figure S1B) and are almost identical to one another (0.45 Å root-mean-square deviation [rmsd] of the polypeptide backbone). Here, we describe the monomer structure by referring to molecule B in the asymmetric unit.
Figure 1. Crystal Structure of Yeast Dynein Motor Domain in the AMPPNP-Bound State.
(A) Cartoon of the domain organization of the yeast dynein heavy chain and the crystal construct (a stalk-truncated motor domain harboring an E1849Q mutation at AAA1).
(B) The overall structure of the motor domain-AMPPNP complex is shown in cartoon representation for the protein and in space-filling representation for AMPPNP ligands.
(C) ATP-binding sites showing the density for AMPPNP molecules. The pink mesh shows an Fo – Fc omit map for AMPPNP, contoured at 3σ level. Side chains of the Walker A motif (K1802, T1803, T2425, K2080, T2081, and T2767), Walker B motif (D1848, Q1849, D2155, D2487, E2488, D2818, and E 2819), Sensor 1 (N1899, N2444, and T2890), Sensor 2 (R1971 and R2620), and R finger (R2209, R2552, R2911, and R3512) are shown in stick representation.
(D and E) Binding to AMPPNP at AAA1 (D) and AAA3 (E) triggers closures of the AAA1–2 and AAA3–4 interfaces. The large domains are aligned. Arrows indicate a predominant rotation of the large and small domains of AAA1 and a rotation of the AAA3s-AAA4L interface. See text and Figure S1 for details. The color scheme is illustrated in (A).
In the dynein-AMPPNP structure, the binding pockets of AAA1–4 are all occupied by nucleotide (Figure 1C). Typically, in AAA proteins, residues from the Walker A (P loop), Walker B, Sensor 1 in the L domain, and Sensor 2 motifs from the s domain contact the nucleotide triphosphate. In addition, an arginine (R finger) from the L domain of the neighboring subunit accelerates nucleotide hydrolysis by contacting the γ-phosphate (Hanson and Whiteheart, 2005; Ogura et al., 2004). In our structure, the Walker A, Walker B, and Sensor 2 residues in AAA1, 3, and 4 contact the AMPPNP. The R finger from AAA5 is positioned close (3.5 Å) to the γ-phosphate of AMPPNP in AAA4. Although their electron density is less well defined, the AAA2 and AAA4 R fingers appear to be positioned much farther from the nucleotide γ-phosphate in AAA1 and AAA3 (minimum distance of ~13 Å and ~9 Å, respectively). Thus, the AAA1/AAA2 interface is not fully closed, and the R finger not optimally positioned for catalysis in this AMPPNP crystal structure. A likely reason for this will be presented later when we discuss the position of the linker domain.
A comparison of the nucleotide-binding pockets from our AMPPNP crystal structure with the yeast nucleotide-free structure (which contains a constitutively bound ATP in AAA2 and no nucleotide in AAA1, 3, 4; PDB code 4AKG) (Carter et al., 2011; Schmidt et al., 2012) showed that the gaps between AAA1L and AAA2L and between AAA3L and AAA4L are more closed in the AMPPNP versus the apo structure (Figures 1D and 1E). The other AAAL interfaces, including AAA4 and AAA5, did not change substantially (Figure S1C). As described for other AAA ATPases (Glynn et al., 2009, 2012), the AAA1–2 and AAA3–4 closures involve rotations (~28° and ~11°, respectively) of the small domain toward large domains within AAA1 and AAA3 (Figure S1D). Most homohexameric AAA ATPase proteins have rigid interfaces between small and neighboring large domains (Nyquist and Martin, 2014). However, we found that AAA2L rotates ~14° away from AAA1s (rotating its R finger away from the nucleotide), whereas AAA4L rotates 17.3° toward AAA3s (rotating the R finger toward the nucleotide) (Figure S1E). In summary, our data show that AAA1 and AAA3, upon binding of AMPPNP, close their nucleotide pockets through a rotation of their small and large domains, as is true of other AAA proteins. In addition, and somewhat unique for a AAA protein, the large domain of AAA4 rotates toward the small domain of AAA3, thus closing the pocket further.
Comparison of Dynein Motor Domain in Apo, AMPPNP, and ADP States
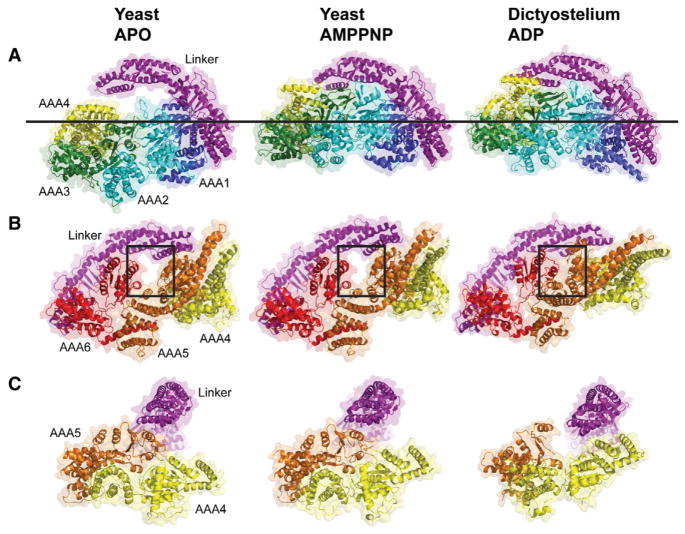
We next examined overall domain motions that occur upon nucleotide binding by comparing our yeast AMPPNP structure with a prior yeast apo structure (4AKG) (Schmidt et al., 2012), aligning the AAA1L domains as a reference point. Overall, the AAA ring with AMPPNP becomes more planar compared with the apo structure (Figure 2A). This conformational change involves a large (~28 Å), rigid-body movement of AAA2L/AAA2s/AAA3L and a smaller (~14 Å) movement of AAA3s/AAA4L toward the linker. In contrast, AAA5, AAA6, and the C-terminal helix, which extends from AAA6s to AAA5s, all superimpose well in these two states (Figure 2B), with an rmsd of 1.2 Å between main-chain atoms (excluding the coiled-coil buttress). These findings suggest that the AAA5/AAA6 side of the ring remains largely fixed in position between the apo- and AMPPNP-bound states of the motor domain, whereas the AAA1–AAA4 side of the ring undergoes pronounced conformational changes.
Figure 2. Comparison of the Motor Domain Ring between Yeast apo, Yeast AMPPNP, and Dictyostelium ADP Crystal Structures.
(A and B) Comparison of the two sides of the AAA ring in the indicated crystal structures. An upward movement of AAA2/3/4 toward the linker with the AMPPNP and ADP structures is observed (A), leading to a planar ring. In the ADP structure, AAA4 is lifted higher toward the linker. The line shows the common position of AAA1 in all structures. (B) An almost identical conformation of AAA5/6 for the apo and AMPPNP structures is observed, but the gap between AAA5 and AAA6 closes in the ADP structure (see box). Color coding of domains is the same as in Figure 1; structures are aligned on AAA1L.
(C) Movements of the large domains of AAA4 and AAA5 relative to the linker (linker subdomains 1,2 aligned in these structures). The linker is docked to AAA5L, and AAA5/6 are in similar states in the apo and AMPPNP structures. However, in the ADP structure, the linker is undocked as a result of a movement of AAA5. See Figure S2 for supporting information. PDBs: 4AKG (Schmidt et al., 2012) for yeast apo and 3VKG (Kon et al., 2012) for Dictyostelium ADP. Note: subdomain 0 of the linker, the AAA5 extension, and C sequence were removed from the Dictyostelium structure for comparison with yeast.
ATP binding has been proposed to cause a bending of the linker domain, which has been hypothesized to generate a “pre-power state” of the motor (Burgess et al., 2003; Roberts et al., 2012). Somewhat surprisingly, we found that AMPPNP binding did not substantially alter the conformation of the linker compared with the apo state of yeast dynein and produced only a subtle (~8 Å) shift of subdomains 1–2 of the linker toward AAA5 (Figure S2A). Furthermore, a similar set of contacts between linker subdomain 1 and AAA5L are observed in both the apo (Carter et al., 2011; Schmidt et al., 2012) and AMPPNP structures (Figure 2C). This result was also unexpected, as it was previously speculated that nucleotide binding might undock the linker from AAA5L (Schmidt et al., 2012).
We next compared the AMPPNP yeast structure with the ADP crystal structure from Dictyostelium dynein (3VKG, chain A) (Kon et al., 2012), again aligning AAA1L as a reference point (Figures 2A and 2B). AAA1 and AAA2 move toward one another in both AMPPNP and ADP structures, as compared with the yeast apo structure, resulting in an upward movement of AAA2–AAA4 toward the linker (Figure S2B). The AAA5 and AAA6 large domains move toward one another in the Dictyostelium ADP structure compared with the nearly identical yeast apo and AMPPNP structures (Figure 2B). The movement of AAA5L leaves subdomain 1 of the Dictyostelium linker “undocked” from the ring and positioned closer to AAA4 (Figure 2C) (Kon et al., 2012).
In summary, yeast apo, yeast AMPPNP, and Dictyostelium ADP exhibit distinct AAA domain arrangements, particularly with respect to the two halves of the ring. Yeast apo and AMPPNP structures differ dramatically in AAA1–AAA4 but are nearly identical in their AAA5–AAA6 domains. Yeast AMPPNP and Dictyostelium ADP display a similar overall conformation of their AAA1–3 domains and differ most significantly in their positions of AAA4–6. The linker exhibits a similar extended architecture and secondary structure in the apo, AMPPNP, and ADP structures.
The Linker-AAA2 Interactions Regulate Microtubule-Stimulated ATPase Activity and Motility
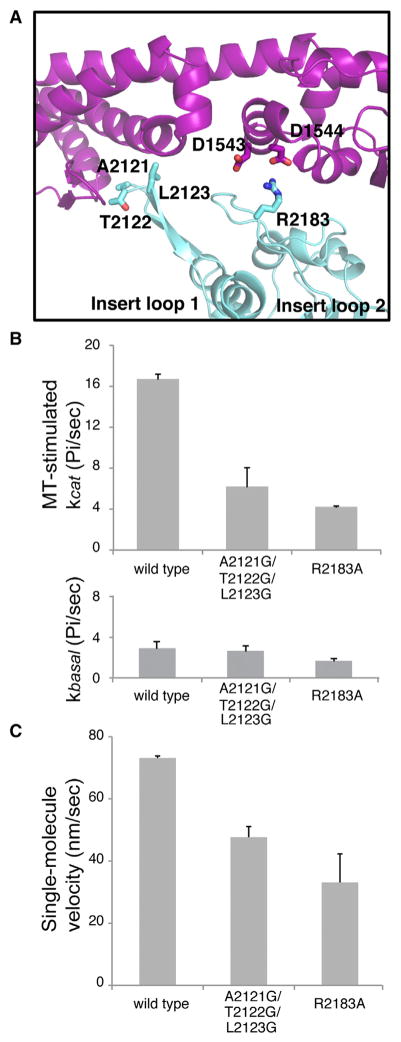
In our AMPPNP structure, two insert loops on AAA2L contact the linker (Figure 3A). These two loops (also called H2 and H3–β4 hairpin inserts) are relatively uncommon in the AAA family, only being found in dynein AAA2, NtrC/PspF, and the magnesium chelatase BchI (Cho and Vale, 2012). The AAA2 loops in our AMPPNP structure display similar, although not identical, contacts to those observed in the Dictyostelium ADP structure (Kon et al., 2012). The most notable difference is that the conserved R2384 from insert loop 2 makes an unfavorable contact with K1720 and R1723 in subdomain 3 of the linker (Kon et al., 2012), while this same arginine (R2183) in the yeast AMPPPNP structure forms favorable ionic interactions with two highly conserved aspartic acids (D1543, D1544) (Figures 3A and S3).
Figure 3. The Linker-Ring Interaction and Its Role in Dynein ATPase Activity and Motility.
(A) The linker-AAA2 contacts in the yeast AMPPNP structure.
(B) ATPase activity of dynein constructs in the absence (basal, bottom panel) or presence (MT-stimulated, top panel) of porcine MTs (see Experimental Procedures). The mean ± MT-stimulated kcat (mean ± SEM of four measurements from two independent protein preparations) is shown.
(C) TMR-labeled, GST-dimerized yeast dynein constructs were tested for velocity in a single-molecule fluorescence motility assay (see Extended Experimental Procedures). The velocity (mean ± SEM of two independent protein preparations with n > 100 moving molecules each) is shown.
See also Figure S3.
Kon et al. (Kon et al., 2012) examined the role of insert loop 1 on ATPase activity by replacing it in its entirety with a polyglycine linker and did not examine insert loop 2. Here, we created a more subtle triple mutant of the three residues at the tip of loop 1 (A2121G/T2122G/L2123G) that contact the linker and a single mutant of a residue in loop 2 (R2183A) that forms a salt bridge with the linker. We assayed the effect of these mutations on microtubule-stimulated ATPase activity and single-molecule motility. We find that a triple mutant at the tip of loop 1 (A2121G/T2122G/L2123G) exhibited a normal basal ATPase activity but 3-fold lower maximal MT-stimulated ATPase activity (Figure 3B). Mutation of the highly conserved arginine in loop 2 (R2183A) exhibited a reduced basal ATPase activity and 4-fold lower MT-stimulated ATPase activity (Figure 3B). Thus, even a single amino acid change can dramatically perturb MT stimulation of the ATPase cycle. The insert loop 1 and 2 mutants moved processively along an MT but with a lower velocity, although the reduction was less than observed for the ATPase activity (Figure 3C). These results show that residues in insert loops 1 and 2 that contact the linker influence the allosteric communication between the microtubule-binding domain and the AAA ring for controlling ATPase activity.
Cryo-EM Structures of Dynein in the ADP-AlF3 and ADP-Vanadate Bound States
Our AMPPNP crystal structure revealed a large conformational change in one half of the AAA ring but did not show a notable conformational change of the linker domain, the proposed mechanical element. Previous single-particle EM studies of Dictyostelium dynein in the presence of ADP-vanadate found that the linker density was not visible in the ~25 Å 3D reconstruction, but based upon interpreting variance maps, the linker was suggested as being bent and positioned close to AAA3 (Roberts et al., 2012). The conformation of dynein in the presence of ADP-AlF3 has not been examined previously for any dynein, and, because of its similarity to ADP-vanadate, we hypothesized that this analog also may capture dynein in the pre-power stroke state. Here, we obtained cryoelectron microscopy (cryo-EM) structures for dynein in the presence of ADP-AlF3 or ADP-vanadate and used 3D classification (Scheres, 2012a, b) to separate out conformational/compositional heterogeneity.
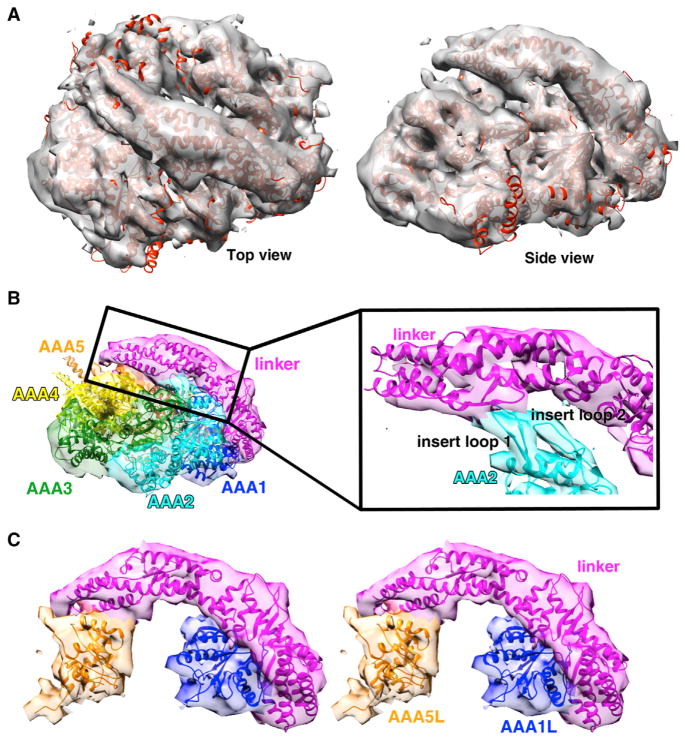
The highest resolution ADP-AlF3 dynein structure (using ~50% of particles) could be resolved to an average resolution of ~10.5 Å, as reported by Gold-standard FSC 0.143 criteria (Figure S4C) (Scheres, 2012b). Consistent with such resolution, tubular densities, indicative of helical secondary structure, could be seen in some areas of the map. However, other regions are likely at lower resolution because such tubular densities are not present. The map colored by local resolution (as determined with Resmap; Kucukelbir et al., 2014) provides information on which parts of the structure are better defined (Figure S4D). The ADP-AlF3 EM density (Figure 4A) could be fit very well with a model of the yeast dynein AMPPNP crystal structure as evidenced by the fact that (1) AAA2–4 are rotated upward and inward compared with the apo structure (Figures 4A and S4H), (2) densities corresponding to the AAA2 insert loops are clearly visible making contacts to the linker (Figure 4B), and (3) the linker is extended and docked onto AAA5 (Figure 4C).
Figure 4. Cryo-EM Structure of Dynein in the Presence of ADP-AlF3 at an Average Resolution of ~10.5 Å.
(A) Cryo-EM density (gray) with our AMPPNP crystal structure docked in.
(B) Side view of the cryo-EM density and the docked AMPPNP crystal structure colored by domain. Density within 5 Å of each domain in the AMPPNP X-ray structure is colored. Insert, zoomed-in view of the contact between AAA2 loops and the linker; helices and loops fit reasonably well within the EM density.
(C) Stereo view of density for linker docked to AAA5 and AAA1 is shown with the AMPPNP X-ray structure docked in. Representative data, the reconstruction colored by local resolution, other 3D classes, and negative-stain reconstructions for a construct containing the full stalk and MTBD are shown in Figure S4.
It is somewhat surprising that ADP-AlF3 did not produce a pre-power stroke conformation with a bent linker. To assess whether the lack of a linker conformational change in ADP-AlF3 was due to the truncation of the stalk in the dynein construct used, we obtained 3D reconstructions of negative-stain EM data for a monomeric wild-type dynein with its full stalk and MTBD in the presence of ADP-AlF3 (Figures S4E and S4F). These ~16–22 Å negative-stain EM maps of the full-stalk dynein revealed a similar conformation to that in the cryo-EM ADP-AlF3 maps of truncated-stalk dynein filtered to the same resolution; in both cases, clear density connecting AAA2 and the linker can be seen (Figures 4B and S4E), and this is not observed in reconstructions for apo dynein (Figure S5M). The collective EM and X-ray data indicate that binding of AMPPNP and ADP-AlF3 to yeast dynein results in a conformational change that shifts AAA2/AAA3/AAA4 upward and brings AAA2L in contact with the linker but does not cause a large conformational change in the linker.
For the ADP-vanadate state, we found multiple positions of the linker using 3D classification, both by cryo-EM (Figure 5A) and negative-stain EM (Figure S5), indicating that the linker is in an equilibrium between different conformational states. One cryo-EM 3D class could be resolved to an average resolution of ~9 Å (Figures 5, S5B, and S5E); many tubular densities representing helices are visible in this map, although not all secondary structure elements are well defined. In contrast to the situation for our ADP-AlF3 cryo-EM structure, the AMPPNP crystal structure did not dock well to the ADP-vanadate cryo-EM map (Figures 5C, 5D, 5E, and S5D). To define the ADP-vanadate state, we generated a model by simultaneously docking individual AAA large and small domains and linker subdomains as rigid bodies to the cryo-EM map (Figure 5B). As the secondary structure was not unambiguously defined in our maps, we did not modify the positions of individual helices within these domains by flexible fitting, although such movements are very likely to occur.
Figure 5. Cryo-EM Structure of Dynein in the Presence of ADP-Vanadate at an Average Resolution of ~9 Å.
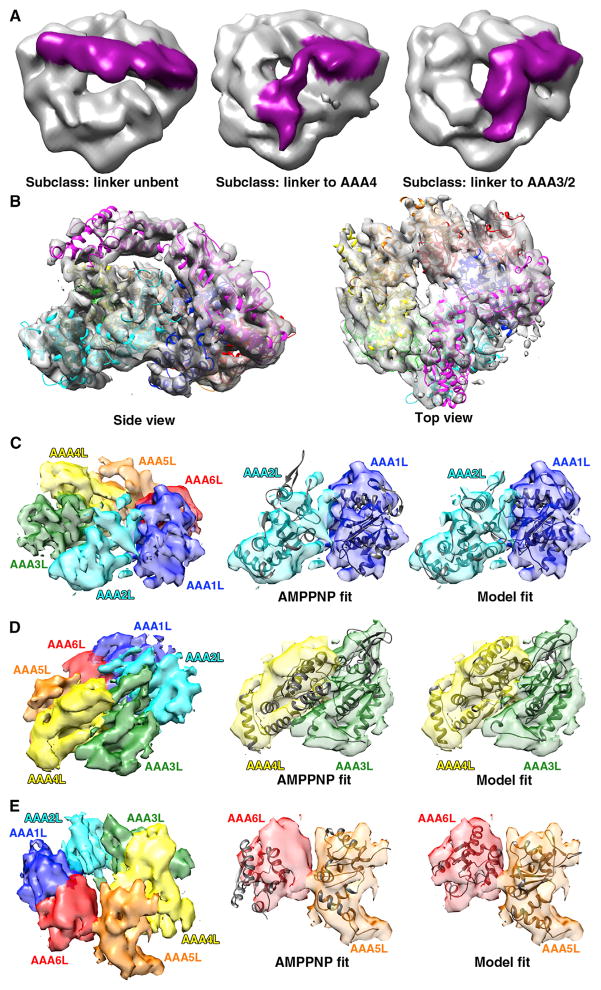
(A) 3D classes for cryo-EM data of dynein in the presence of ADP-vanadate: unbent linker (~39% particles), linker to AAA4 (~36% particles), and linker to AAA3/2 (~25% particles). The last subclass could be refined to the highest resolution, as shown in (B)–(E).
(B) ADP-vanadate cryo-EM density fit with our model, which was generated from simultaneously fitting each s and L AAA subdomain into the density as rigid bodies in UCSF Chimera.
(C–E) The large domains of the AAA ring, colored by domain, are shown on the left to provide a reference orientation for the fits of the cryo-EM electron density with the AMPPNP X-ray structure (middle) or the ADP-vanadate model (right). Domain motions of AAA2-AAA1 (C), AAA4-AAA3 (D), and AAA6-AAA5 (E) between the AMPPNP and ADP-vanadate states are shown. Representative data, the reconstruction colored by local resolution, 2D class averages, supporting 3D reconstructions from negative-stain EM data, and stereo views of Apo, AMPPNP, and the model fit in cryo-EM density as well as negative-stain data for similar complexes are shown in Figure S5.
The highest resolution ADP-vanadate model revealed significant conformational changes in the ring and the linker at the subdomain level. The linker exhibits a bent conformation, and the docked model shows that linker subdomain 1 is positioned in close proximity to the insert loop of AAA3 (~aa. 2467–2470) and likely contacts AAA2 as well (Figure 5B). This result is consistent with predictions from prior studies of this nucleotide state with Dictyostelium and axonemal dynein (Burgess and Knight, 2004; Burgess et al., 2003; Roberts et al., 2013), although prior cryo-EM studies could not directly resolve the linker density most likely due to conformational heterogeneity (Roberts et al., 2012). Indeed, we can confirm conformational heterogeneity, as other 3D classes from the same data set, but at lower resolution, show the linker docked either onto AAA4 or unbent (Figure 5A).
The AAA ring shows a number of domain rearrangements, most notably in AAA2, AAA4, and AAA6 (Figures 5C–5E), which have not been resolved before in the ADP-vanadate state. AAA2L rotates toward AAA1L as compared with the AMPPNP state, producing a more closed conformation of these two domains (Figure 5C). AAA4L moves further upward compared with the AMPPNP structure (Figure 5D), potentially creating an additional docking site for the linker seen in some 3D classes (Figures 5A and S5L). AAA6 undergoes the largest displacement. AAA6L and AAA5L move toward each other, closing a large gap found in the apo and AMPPNP crystal structures (Figure 5E). The shift of AAA5L may destabilize its docking with the linker (Figure 2C), thus allowing the linker to sample alternate conformations. In summary, conformational changes of several AAA domains could be resolved in the ADP-vanadate compared with the AMPPNP structure, which collectively produce a more closed ring (Movie S1).
ATP Hydrolysis Mutants Reveal Unique Roles for AAA1 and AAA3
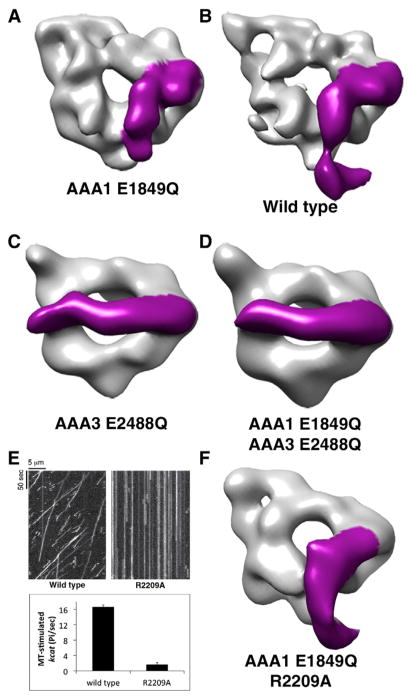
The dramatic difference in linker conformations must arise from subtle differences in the AlF3 and vanadate chemical structures, which cause them to mimic the γ-phosphate in different ways and/or potentially interact differently with AAA1 and AAA3. To explore this further, we determined the linker position in the presence of ATP, dynein’s natural substrate. We used 3D reconstructions from negative-stain EM data as an assay for linker bending, as the position of the linker can be unambiguously determined at low resolutions due to the large-scale conformational change (~75 Å movement at the N terminus of the linker). To allow ATP binding but not hydrolysis, we made Walker B mutations in either AAA1 (E1849Q) or AAA3 (E2488Q), or both. ATP is expected to be bound at the site of the Walker B E/Q mutation, whereas other sites may contain ATP, ADP, or any combinations of these nucleotides. The 3D reconstructions (15–20 Å resolution) of the AAA1 E1849Q mutant in the presence of 5 mM ATP produced conformations that were very similar to those observed by cryo-EM with ADP-vanadate, yielding a 3D class with a bent linker and similar points of contact between the linker and AAA3 (Figure 6A). Additional 3D classes were observed in which the linker contacted different AAA domains in the ring (Figure S5L), similar to Figure 5A and to the observation made for axonemal dynein in situ (Lin et al., 2014). We note that ATP addition to dynein without the Walker B mutation also resulted in a 3D class with a bent linker (Figure 6B), revealing that the bent linker conformation can occur with the natural substrate and native binding site. This result also implies that a substantial proportion of dyneins have their AAA1 binding sites occupied with ATP or ADP-Pi in the absence of MTs. Bending of the linker was not observed in yeast dynein with 5 mM ADP, as expected from the Dictyostelium ADP crystal structure (Figure S6F). Collectively, these results indicate that ATP binding to AAA1 can trigger a conformational change from a straight to a bent conformation.
Figure 6. Blocking ATP Hydrolysis in AAA3 Prevents the Linker Conformational Change.
(A–D) Negative-stain reconstructions for (A) AAA1 E1849Q, (B) wild-type, (C) AAA3 E2488Q, and (D) AAA1/AAA3 double Walker B mutant (E1849Q/E2488Q) dyneins. The dyneins were incubated with MgATP (5 mM) prior to negative staining. The electron density for the linker was clearly visible and is colored magenta.
(E) Representative kymographs for single-molecule motility assays showing no detectable motility of the AAA2 R finger mutant (R2209A) (top panel) and microtubule-stimulated ATPase activity (bottom panel) for wild-type and the R2209A mutation (mean ± SEM of two independent protein preparations).
(F) Negative-stain reconstruction for E1849Q/R2209A dynein in the presence of 5 mM ATP. Representative micrographs, additional 3D classes, and comparison with ADP instead of ATP for wild-type and the AAA1 E1849Q mutant are shown in Figure S6.
In contrast to results with wild-type dynein or the AAA1 E1849Q mutant, when ATP was incubated with the AAA3 hydrolysis mutant (E2488Q), we quite strikingly observed that the linker remained extended and positioned over AAA5 (Figure 6C); no 3D class of particles was observed with a bent linker conformation. We also tested a double Walker B mutant in both AAA1/AAA3 (E1849Q/E2488Q) and similarly found that the linker remained extended and positioned above AAA5 (Figure 6D).
Taken together, these results indicate that ATP binding to AAA1 can trigger a conformational change of the linker; however, when ATP also occupies AAA3, then this linker conformational change is essentially blocked. The model is consistent with FRET data on Dictyostelium dynein using donor and acceptor fluorescent proteins on AAA2 and the linker, which showed a large FRET signal change upon binding of ATP (consistent with a linker swing) but little FRET change with a AAA3 Walker B mutant (Kon et al., 2005). The model may seem inconsistent with the fact that linker bending is triggered by ADP-vanadate but not ADP-AlF3, even though both are often considered as mimics of an ADP-Pi or ATP transition state. However, we hypothesize that ADP- AlF3 and AMPPNP bind at both AAA1 and AAA3 and mimic an ATP-like state, whereas ADP-vanadate binds and mimics an ATP-like state at AAA1 but cannot bind effectively at AAA3, leaving ADP alone, without the vanadate, occupying this site. This hypothesis is supported by the longstanding observation that vanadate-mediated photocleavage of the dynein polypeptide chain occurs primarily at AAA1 (Gibbons and Gibbons, 1987). In addition, this is also consistent with functional differences, as assayed by single-molecule studies, which show that a dynein monomer dissociates faster from microtubules with ADP-vanadate than with ADP-AlF3 or AMPPNP, which display similar rates (Figure S5N).
Finally, we tested whether the AAA2 R finger (R2209) plays a role in the linker conformational change. Work in other AAA proteins has shown that the R finger from a neighboring subunit plays a role both in nucleotide hydrolysis as well as in inducing a conformational change (Tucker and Sallai, 2007). In dynein, a mutation in the AAA4 R finger (which interacts with AAA3) was shown to produce a similar phenotype as the AAA3 hydrolysis mutant (E2488Q), reducing motility substantially (Cho et al., 2008; Huang et al., 2012). To test the role of the AAA2 R finger, we mutated it to alanine (R2209A). The R2209A mutant statically bound but no longer moved on microtubules in the presence of ATP and showed dramatically reduced microtubule-stimulated ATPase activity (Figure 6E). When examined by negative-stain EM in the presence of ATP, the bent linker was still observed in the R2209A mutant (Figure 6F). This result suggests that the R finger is crucial for promoting AAA1 ATP hydrolysis but does not inhibit linker bending.
DISCUSSION
The large size and inherent flexibility of dynein make it challenging to address its structural mechanism using a single technique. Here, we combined X-ray crystallography and EM to gain insights into the domain motions that constitute the basis of allosteric communication in the dynein motor domain. Our AMPPNP-bound crystal structure provides the first high-resolution comparison of a dynein from the same species (yeast) bound to different nucleotides (apo and AMPPNP). Cryo-EM data processed using 3D classification methods (Scheres, 2012b; Scheres et al., 2005) enabled visualization of AAA domain and linker movements in additional nucleotide states (ADP-AlF3 and ADP-vanadate), and negative-stain EM allowed us to assay the distinct roles of AAA1 and AAA3 using several mutants. These data, in combination with biochemical studies and previous X-ray structures (Carter et al., 2011; Kon et al., 2012; Schmidt et al., 2012), allow us to generate a structural model for how nucleotide-dependent conformational changes propagate around the dynein AAA ring to produce motility.
The Linker and AAA3 as Regulators of Dynein ATPase Activity
Our structural data suggest that the linker, in addition to being a mechanical element (Roberts et al., 2009), regulates dynein ATPase activity. When the linker is docked simultaneously on AAA5 and the AAA2 insert loops (Figure 3A), the AAA1-AAA2 interface is prevented from fully closing, and in fact AAA2L is rotated away from AAA1L (Figure 1). As a result, the AAA2 R finger, which we show here is important for dynein motility and ATPase activity (Figure 6E), is likely positioned too far away to promote efficient hydrolysis of ATP at AAA1. Upon AAA5 undocking and bending of the linker, our cryo-EM data suggest that AAA2L moves toward AAA1 (Figure 5C), likely resulting in a catalytically competent position of the AAA2 R finger. This result suggests that the linker position influences the conformation of AAA2, which is critical for ATP hydrolysis to occur at AAA1. This model is consistent with the absence of a phosphate burst at AAA1 (indicative of slow hydrolysis and/or phosphate release) when nucleotide hydrolysis is blocked at AAA3 and AAA4 by Walker B mutations (Kon et al., 2012), which we show here has the consequence of inhibiting linker undocking from AAA5.
AAA1 is the primary catalytic site that drives dynein motility (Kon et al., 2004; Reck-Peterson and Vale, 2004), and single-molecule studies indicate that a single ATP molecule (presumably binding at AAA1) can trigger a dynein step (DeWitt et al., 2012; Qiu et al., 2012). However, blocking ATP hydrolysis at AAA3 severely impairs dynein motility (Cho et al., 2008; Kon et al., 2004). These data may seem conflicting: ATP hydrolysis at AAA3 is required for motility, but the single-molecule results imply that ATP turnover at AAA1 suffices for dynein stepping. This conflict can be reconciled if AAA3 serves a regulatory function rather than being integrally involved in the chemomechanical cycle. Our data suggest that ATP, or an ATP analog, bound at AAA3 blocks the conformational change initiated by ATP binding at AAA1 from propagating around the ring, rendering dynein in a “repressed” state (Figure 7A, state II). This result provides a structural explanation for the low ATPase activity of the AAA3 E2488Q mutant. We propose that once AAA3 is in an ADP conformation, dynein is then in an “active” state that can execute multiple rounds of ATP binding/hydrolysis at AAA1 (Figure 7A, III–V).
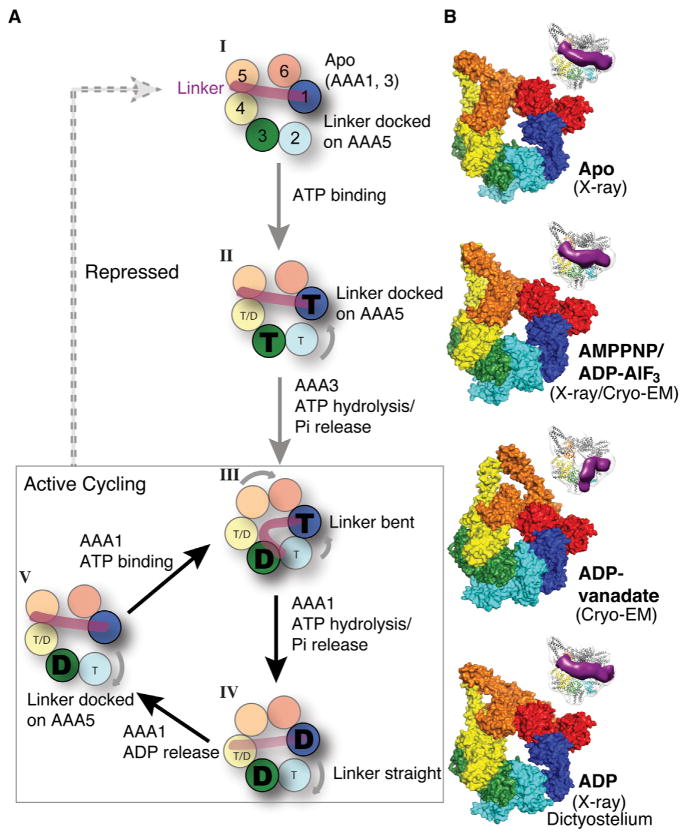
Figure 7. A Model for Structural Changes during Dynein’s ATPase Cycle.
(A) The actively cycling states of the dynein motor are boxed (III, IV, and V), and repressed states are shown outside the box (I and II). Beginning with state III, ATP (“T”) binding to AAA1 results in the closures AAA1–2, which triggers a series of domain movements around the ring and closure of the AAA5–6 interface; movement of AAA5 results in linker detachment from AAA5 and a bent conformation of the linker. After phosphate release from AAA1, the linker straightens (the proposed power stroke) but remains undocked (IV). Linker docking to AAA5 promotes the further opening of AAA1–2 and ADP (“D”) release from AAA1, returning it to an apo state at AAA1 to begin a new cycle (V). If ADP is released (broken line from V) and ATP rebinds at AAA3 (II), the motor returns to the repressed state. See Discussion for more details. We denote the nucleotide state of AAA4 as “T/D” because our present model dos not incorporate a nucleotide-specific role at this site. A subtle modulatory role is possible, as a mutation blocking nucleotide hydrolysis at AAA4 produces a modest decrease in velocity and increase in processivity (Cho et al., 2008).
(B) Surface representation of the AAA ring in yeast apo (PDB code 4AKG; Schmidt et al., 2012), yeast AMPPNP/ADP-AlF3 (our data), yeast ADP-vanadate (our data), and Dictyostelium ADP (PDB code 3VKG; Kon et al., 2012) used to synthesize the model presented in (A). We illustrate a model based on the Dictyostelium ADP X-ray structure, as a crystal structure for yeast ADP has not been obtained. Although the yeast ADP structure may differ it some details from Dictyostelium, the yeast ADP EM structure also clearly exhibits a “post-power-stroke” extended linker conformation (Figure S6F). Insets highlight the linker position in each state based on our EM data. See also Figure S7. The structural transitions in the dynein cycle can be viewed in Movie S1.
An important question arises from this study: what is the function of the “repressed” state with ATP loaded in AAA3? We speculate that this state serves as a switch for turning dynein off. If ATP hydrolysis at AAA3 is blocked for an extended period of time, dynein will be immotile but tightly bound to an MT (the E2488Q mutant has a strong affinity for MTs; Cho et al., 2008). In a cellular context, a “repressed” dynein might tenaciously hold on to a microtubule at the cortex or a kinetochore. Alternatively, the ATP turnover at AAA3 could occur at a slow rate (slower than AAA1 turnover) and thereby tune dynein’s speed and its affinity for MTs. Potentially dynein-associated proteins or posttranslational modifications could regulate the rate of ATP hydrolysis at AAA3. An example of AAA ATPase regulation by associated proteins has been documented for torsin, whose ATPase activity is strongly regulated by two protein cofactors (Zhao et al., 2013). Interestingly, the nucleotide-binding pocket of the AAA3 site in cytoplasmic dynein 2, which is involved in intraflagellar transport, is substantially divergent and thus cytoplasmic dynein 2 may not use AAA3 to regulate its motility in the same way as cytoplasmic dynein 1. Further work will be needed to resolve how cytoplasmic dynein 1 uses AAA3 for its cell biological functions, but the present structural study, combined with previous functional studies, suggests that its nucleotide cycle might be used to regulate rather than drive dynein motility.
Model for Conformational Changes during the ATPase Cycle
X-ray structures (apo, AMPPNP, and ADP) and EM reconstructions (ADP-AlF3 and ADP-vanadate) now provide information on the positions of the AAA domains and the linker, which are collectively summarized in Figure 7 and Movie S1. These structures also allow us to formulate a model for the sequence of domain motions within the dynein motor during the ATPase cycle (Figure 7A). We start the cycle with ADP at AAA3 (Figure 7A, state III) because, as discussed above, our results show that this is a prerequisite state for a dynein ATPase cycle to occur at AAA1.
ATP bound at AAA1 triggers a series of domain movements, which can be appreciated by examining the positions of the AAA domains around the ring (Figure 7B). ATP binding to AAA1 closes the gap between AAA1L and AAA1s, triggering the movement of AAA2–4 toward the linker (Figure 2). With AAA3 in an ADP state, the ATP-induced conformational change at AAA1 propagates fully around the ring, resulting in the upward movement of AAA4L and the movements of AAA5L and AAA6L toward one another (Figure 5). These combined movements may be responsible for detaching linker subdomain 1 from its docking site on AAA5. With the linker no longer attached to AAA5, we postulate that the AAA2 insert loops can break their relatively few contacts with the linker (Figure 3A); no longer restrained by the linker, AAA2 can rotate further toward the nucleotide-bound AAA1. An upward displacement of the linker and partial closure of AAA1–2 is captured in one of the four Dictyostelium ADP structures (PDB 3VKH; Kon et al., 2012; chain A; Figure S3B). However, a more complete closure of AAA1–2 accompanies the bending of the linker (Figure 5C).
The bent linker is thought to constitute a pre-power stroke state. After nucleotide hydrolysis and phosphate release at AAA1, yielding an ADP state, the linker returns to its extended conformation (Figure 7A, state IV). Linker straightening might act as power stroke to move the partner head of the dynein dimer toward the minus end of an MT (Burgess et al., 2003). However, high-resolution stepping data show that the front dynein head can move forward without detaching the rear head (DeWitt et al., 2012; Qiu et al., 2012), suggesting that there might be other mechanisms for biasing movement toward the MT minus end. Interestingly, a recent in situ EM study of a dimeric axonemal dynein at 30–50 Å resolution suggested that a power stroke is facilitated by rotation of the ring relative to a straight linker and stalk. We also observed a small angular shift of the stalk (~9°) relative to the ring between the apo and AMPPNP states, which would be predicted to produce an ~4 nm displacement of the distal MTBD toward the MT minus end in a molecule with a full-length stalk (Figure S7). Thus, it is possible that more than one type of conformational change helps to bias the movement of dynein toward the microtubule minus end.
Finally, after the power stroke of the linker, the motor must reset itself for another ATP hydrolysis cycle by releasing ADP from AAA1. This nucleotide-release step might require the re-docking of the linker to AAA5, as suggested by Schmidt et al. (Schmidt et al., 2012), which could potentially provide the binding energy needed to pry AAA2 further apart from AAA1. The state in which ADP is released from AAA1 but bound to AAA3 (Figure 7A, state V) has not been captured by EM or X-ray, but we model it speculatively in Figure 7 as being similar to the yeast apo structure. From this state, dynein can rebind ATP at AAA1 and begin another chemomechanical cycle (Figure 7A, state III).
This model raises many questions that remain to be answered in future studies. The bending of the linker is a significant structural change of this helical domain, and an X-ray structure of this state will be required to understand the details of how and where this bending occurs. In addition, the model described above does not take into account how MT binding affects this cycle. Previous studies have suggested that a half-heptad shift in the elongated antiparallel coiled-coil stalk, which emerges from AAA4s and interacts with the buttress in AAA5s, controls the affinity of MT binding (Gibbons et al., 2005; Kon et al., 2009). Conversely, conformational changes in the stalk buttress driven by MT binding may regulate rates of ATP hydrolysis or ADP release at AAA1 by controlling linker undocking/docking at AAA5 (see Figure 2C). Obtaining higher-resolution structures of the stalk in different conformational states, particularly the low-affinity MT-binding state, will be important for understanding how the dynein ring and the MTBD allosterically regulate one another.
EXPERIMENTAL PROCEDURES
Detailed experimental procedures are outlined in the Extended Experimental Procedures.
Cloning, Protein Purification, and Activity Assays
A DNA fragment encoding dynein motor domain was integrated in the yeast genome DNA. All constructs were made by homologous recombination in yeast. Constructs are listed in Table S2. Proteins were purified by IgG affinity and subsequent size-exclusion chromatography. ATPase assays were conducted using the EnzChek phosphate assay kit (Life Technologies). GST-Halo-tagged dyneins were labeled with TMR, as previously described (Cho et al., 2008). Single-molecule motility on taxol-stabilized MTs was measured by total internal fluorescence microscopy (Cho et al., 2008).
Crystallization and Structure Determination
Diffraction-quality crystals grew with ~6 mg/ml of lysozyme-fused motor domain in the presence of ~4 mM AMPPNP in 4%–10% PEG 3350 and 200–300 mM NaAc at 22°C by the hanging drop vapor diffusion method. Diffraction data were collected at beamline 8.3.1 at Advanced Light Source in the Lawrence Berkeley National Laboratory and merged from multiple data sets from multiple crystals to improve data quality and resolution. The initial structural solution was obtained by molecular replacement using separated fragments of yeast apo structure as search models. Iterative model building and refinement were conducted using the programs Coot and Phenix (Adams et al., 2002; Emsley and Cowtan, 2004).
Electron Microscopy and Image Processing
Cryo-EM data were collected on a TF20 microscope using a phosphor scintillator based TemF816 8K × 8K CMOS camera (TVIPS GmbH) or Polara microscope using a K2 Summit direct electron detector. Negative-stain EM data were collected on a TF20 microscope using a Tietz TemF416 4k × 4k CMOS camera (UltraScan 4000, Gatan). Technical details of image processing and particle-picking procedures are described in the Extended Experimental Procedures. Three-dimensional classification was done using RELION, as described in the Extended Experimental Procedures. Rigid body fitting of subdomains was done in UCSF Chimera (Goddard et al., 2007; Pettersen et al., 2004), and figures were prepared using UCSF Chimera or PyMol (Delano Scientific).
Supplementary Material
Acknowledgments
We gratefully acknowledge Bridget Carragher, Clint Potter, and their team (NRAMM, Scripps) and staff at ALS beamline 8.3.1 at the Lawrence Berkeley Laboratory for assistance with data collection. We thank Carol Cho (Stanford), Luke Rice (UT Southwestern Medical Center), and all members in the Vale and Cheng labs for helpful discussions. Funding was provided by NIH (R01GM09731, R.D.V.), NIH (S10RR026814; Y.C.), UCSF (Breakthrough Biomedical Research Award, Y.C.), the American Heart Association (11POST7110022, H.-C.C.), the Merck fellowship of the Damon Runyon Cancer Research Foundation (DRG-2136-12, G.B.), and NIH (9 P41 GM103310, to the National Resource for Automated Molecular Microscopy).
Footnotes
ACCESSION NUMBERS
Coordinates and structure factors of the yeast dynein:AMPPNP complex have been deposited in the Protein Data Bank (PDB code 4W8F), and all EM maps have been deposited in the EMDB (accession codes listed in Table S2).
Supplemental Information includes Extended Experimental Procedures, seven figures, two tables, and one movie and can be found with this article online at http://dx.doi.org/10.1016/j.cell.2014.10.018.
AUTHOR CONTRIBUTIONS
G.B. and H.-C.C. contributed equally to this work. G.B., R.D.V., and H.-C.C. conceived the research; H.-C.C., G.B., N.Z., M.L., A.M., and J.A.S. collected data; H.-C.C., G.B., R.D.V., A.M., and Y.C. analyzed data; H.-C.C., G.B., and R.V. wrote the manuscript. All authors commented on the manuscript.
References
- Adams PD, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Terwilliger TC. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr D Biol Crystallogr. 2002;58:1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- Babst M, Wendland B, Estepa EJ, Emr SD. The Vps4p AAA ATPase regulates membrane association of a Vps protein complex required for normal endosome function. EMBO J. 1998;17:2982–2993. doi: 10.1093/emboj/17.11.2982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker TA, Sauer RT. ClpXP, an ATP-powered unfolding and protein-degradation machine. Biochim Biophys Acta. 2012;1823:15–28. doi: 10.1016/j.bbamcr.2011.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgess SA, Knight PJ. Is the dynein motor a winch? Curr Opin Struct Biol. 2004;14:138–146. doi: 10.1016/j.sbi.2004.03.013. [DOI] [PubMed] [Google Scholar]
- Burgess SA, Walker ML, Sakakibara H, Knight PJ, Oiwa K. Dynein structure and power stroke. Nature. 2003;421:715–718. doi: 10.1038/nature01377. [DOI] [PubMed] [Google Scholar]
- Carter AP. Crystal clear insights into how the dynein motor moves. J Cell Sci. 2013;126:705–713. doi: 10.1242/jcs.120725. [DOI] [PubMed] [Google Scholar]
- Carter AP, Cho C, Jin L, Vale RD. Crystal structure of the dynein motor domain. Science. 2011;331:1159–1165. doi: 10.1126/science.1202393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho C, Reck-Peterson SL, Vale RD. Regulatory ATPase sites of cytoplasmic dynein affect processivity and force generation. J Biol Chem. 2008;283:25839–25845. doi: 10.1074/jbc.M802951200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho C, Vale RD. The mechanism of dynein motility: insight from crystal structures of the motor domain. Biochim Biophys Acta. 2012;1823:182–191. doi: 10.1016/j.bbamcr.2011.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeWitt MA, Chang AY, Combs PA, Yildiz A. Cytoplasmic dynein moves through uncoordinated stepping of the AAA+ ring domains. Science. 2012;335:221–225. doi: 10.1126/science.1215804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Furst J, Sutton RB, Chen J, Brunger AT, Grigorieff N. Electron cryomicroscopy structure of N-ethyl maleimide sensitive factor at 11 A resolution. EMBO J. 2003;22:4365–4374. doi: 10.1093/emboj/cdg420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbons BH, Gibbons IR. Vanadate-sensitized cleavage of dynein heavy chains by 365-nm irradiation of demembranated sperm flagella and its effect on the flagellar motility. J Biol Chem. 1987;262:8354–8359. [PubMed] [Google Scholar]
- Gibbons IR, Garbarino JE, Tan CE, Reck-Peterson SL, Vale RD, Carter AP. The affinity of the dynein microtubule-binding domain is modulated by the conformation of its coiled-coil stalk. J Biol Chem. 2005;280:23960–23965. doi: 10.1074/jbc.M501636200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glynn SE, Martin A, Nager AR, Baker TA, Sauer RT. Structures of asymmetric ClpX hexamers reveal nucleotide-dependent motions in a AAA+ protein-unfolding machine. Cell. 2009;139:744–756. doi: 10.1016/j.cell.2009.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glynn SE, Nager AR, Baker TA, Sauer RT. Dynamic and static components power unfolding in topologically closed rings of a AAA+ proteolytic machine. Nat Struct Mol Biol. 2012;19:616–622. doi: 10.1038/nsmb.2288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goddard TD, Huang CC, Ferrin TE. Visualizing density maps with UCSF Chimera. J Struct Biol. 2007;157:281–287. doi: 10.1016/j.jsb.2006.06.010. [DOI] [PubMed] [Google Scholar]
- Hanson PI, Whiteheart SW. AAA+ proteins: have engine, will work. Nat Rev Mol Cell Biol. 2005;6:519–529. doi: 10.1038/nrm1684. [DOI] [PubMed] [Google Scholar]
- Huang J, Roberts AJ, Leschziner AE, Reck-Peterson SL. Lis1 acts as a “clutch” between the ATPase and microtubule-binding domains of the dynein motor. Cell. 2012;150:975–986. doi: 10.1016/j.cell.2012.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kon T, Nishiura M, Ohkura R, Toyoshima YY, Sutoh K. Distinct functions of nucleotide-binding/hydrolysis sites in the four AAA modules of cytoplasmic dynein. Biochemistry. 2004;43:11266–11274. doi: 10.1021/bi048985a. [DOI] [PubMed] [Google Scholar]
- Kon T, Mogami T, Ohkura R, Nishiura M, Sutoh K. ATP hydrolysis cycle-dependent tail motions in cytoplasmic dynein. Nat Struct Mol Biol. 2005;12:513–519. doi: 10.1038/nsmb930. [DOI] [PubMed] [Google Scholar]
- Kon T, Imamula K, Roberts AJ, Ohkura R, Knight PJ, Gibbons IR, Burgess SA, Sutoh K. Helix sliding in the stalk coiled coil of dynein couples ATPase and microtubule binding. Nat Struct Mol Biol. 2009;16:325–333. doi: 10.1038/nsmb.1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kon T, Sutoh K, Kurisu G. X-ray structure of a functional full-length dynein motor domain. Nat Struct Mol Biol. 2011;18:638–642. doi: 10.1038/nsmb.2074. [DOI] [PubMed] [Google Scholar]
- Kon T, Oyama T, Shimo-Kon R, Imamula K, Shima T, Sutoh K, Kurisu G. The 2.8 Å crystal structure of the dynein motor domain. Nature. 2012;484:345–350. doi: 10.1038/nature10955. [DOI] [PubMed] [Google Scholar]
- Kucukelbir A, Sigworth FJ, Tagare HD. Quantifying the local resolution of cryo-EM density maps. Nat Methods. 2014;11:63–65. doi: 10.1038/nmeth.2727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kull FJ, Endow SA. Force generation by kinesin and myosin cytoskeletal motor proteins. J Cell Sci. 2013;126:9–19. doi: 10.1242/jcs.103911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Choi JM, Tsai FT. Visualizing the ATPase cycle in a protein disaggregating machine: structural basis for substrate binding by ClpB. Mol Cell. 2007;25:261–271. doi: 10.1016/j.molcel.2007.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J, Okada K, Raytchev M, Smith MC, Nicastro D. Structural mechanism of the dynein power stroke. Nat Cell Biol. 2014;16:479–485. doi: 10.1038/ncb2939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nyquist K, Martin A. Marching to the beat of the ring: polypeptide translocation by AAA+ proteases. Trends Biochem Sci. 2014;39:53–60. doi: 10.1016/j.tibs.2013.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogura T, Whiteheart SW, Wilkinson AJ. Conserved arginine residues implicated in ATP hydrolysis, nucleotide-sensing, and inter-subunit interactions in AAA and AAA+ ATPases. J Struct Biol. 2004;146:106–112. doi: 10.1016/j.jsb.2003.11.008. [DOI] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera—a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Qiu W, Derr ND, Goodman BS, Villa E, Wu D, Shih W, Reck-Peterson SL. Dynein achieves processive motion using both stochastic and coordinated stepping. Nat Struct Mol Biol. 2012;19:193–200. doi: 10.1038/nsmb.2205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rayment I, Smith C, Yount RG. The active site of myosin. Annu Rev Physiol. 1996;58:671–702. doi: 10.1146/annurev.ph.58.030196.003323. [DOI] [PubMed] [Google Scholar]
- Reck-Peterson SL, Vale RD. Molecular dissection of the roles of nucleotide binding and hydrolysis in dynein’s AAA domains in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 2004;101:1491–1495. doi: 10.1073/pnas.2637011100. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Roberts AJ, Numata N, Walker ML, Kato YS, Malkova B, Kon T, Ohkura R, Arisaka F, Knight PJ, Sutoh K, Burgess SA. AAA+ Ring and linker swing mechanism in the dynein motor. Cell. 2009;136:485–495. doi: 10.1016/j.cell.2008.11.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts AJ, Malkova B, Walker ML, Sakakibara H, Numata N, Kon T, Ohkura R, Edwards TA, Knight PJ, Sutoh K, et al. ATP-driven remodeling of the linker domain in the dynein motor. Structure. 2012;20:1670–1680. doi: 10.1016/j.str.2012.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts AJ, Kon T, Knight PJ, Sutoh K, Burgess SA. Functions and mechanics of dynein motor proteins. Nat Rev Mol Cell Biol. 2013;14:713–726. doi: 10.1038/nrm3667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheres SH. A Bayesian view on cryo-EM structure determination. J Mol Biol. 2012a;415:406–418. doi: 10.1016/j.jmb.2011.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheres SH. RELION: implementation of a Bayesian approach to cryo-EM structure determination. J Struct Biol. 2012b;180:519–530. doi: 10.1016/j.jsb.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheres SH, Valle M, Nuñez R, Sorzano CO, Marabini R, Herman GT, Carazo JM. Maximum-likelihood multi-reference refinement for electron microscopy images. J Mol Biol. 2005;348:139–149. doi: 10.1016/j.jmb.2005.02.031. [DOI] [PubMed] [Google Scholar]
- Schmidt H, Gleave ES, Carter AP. Insights into dynein motor domain function from a 3.3-A crystal structure. Nat Struct Mol Biol. 2012;19:492–497. S491. doi: 10.1038/nsmb.2272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skordalakes E, Berger JM. Structure of the Rho transcription terminator: mechanism of mRNA recognition and helicase loading. Cell. 2003;114:135–146. doi: 10.1016/s0092-8674(03)00512-9. [DOI] [PubMed] [Google Scholar]
- Tucker PA, Sallai L. The AAA+ superfamily—a myriad of motions. Curr Opin Struct Biol. 2007;17:641–652. doi: 10.1016/j.sbi.2007.09.012. [DOI] [PubMed] [Google Scholar]
- Vale RD, Milligan RA. The way things move: looking under the hood of molecular motor proteins. Science. 2000;288:88–95. doi: 10.1126/science.288.5463.88. [DOI] [PubMed] [Google Scholar]
- Watanabe YH, Motohashi K, Yoshida M. Roles of the two ATP binding sites of ClpB from Thermus thermophilus. J Biol Chem. 2002;277:5804–5809. doi: 10.1074/jbc.M109349200. [DOI] [PubMed] [Google Scholar]
- West SC. Molecular views of recombination proteins and their control. Nat Rev Mol Cell Biol. 2003;4:435–445. doi: 10.1038/nrm1127. [DOI] [PubMed] [Google Scholar]
- Zhao C, Brown RS, Chase AR, Eisele MR, Schlieker C. Regulation of Torsin ATPases by LAP1 and LULL1. Proc Natl Acad Sci USA. 2013;110:E1545–E1554. doi: 10.1073/pnas.1300676110. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.