We describe our algorithm and software for determining copy number profiles from tumor genome sequencing data, and find that it compares favorably to existing algorithms for the same purpose.
Keywords: cancer genomics, copy number alterations, mutations, next-generation sequencing, software
Abstract
Background
Exome or whole-genome deep sequencing of tumor DNA along with paired normal DNA can potentially provide a detailed picture of the somatic mutations that characterize the tumor. However, analysis of such sequence data can be complicated by the presence of normal cells in the tumor specimen, by intratumor heterogeneity, and by the sheer size of the raw data. In particular, determination of copy number variations from exome sequencing data alone has proven difficult; thus, single nucleotide polymorphism (SNP) arrays have often been used for this task. Recently, algorithms to estimate absolute, but not allele-specific, copy number profiles from tumor sequencing data have been described.
Materials and methods
We developed Sequenza, a software package that uses paired tumor-normal DNA sequencing data to estimate tumor cellularity and ploidy, and to calculate allele-specific copy number profiles and mutation profiles. We applied Sequenza, as well as two previously published algorithms, to exome sequence data from 30 tumors from The Cancer Genome Atlas. We assessed the performance of these algorithms by comparing their results with those generated using matched SNP arrays and processed by the allele-specific copy number analysis of tumors (ASCAT) algorithm.
Results
Comparison between Sequenza/exome and SNP/ASCAT revealed strong correlation in cellularity (Pearson's r = 0.90) and ploidy estimates (r = 0.42, or r = 0.94 after manual inspecting alternative solutions). This performance was noticeably superior to previously published algorithms. In addition, in artificial data simulating normal-tumor admixtures, Sequenza detected the correct ploidy in samples with tumor content as low as 30%.
Conclusions
The agreement between Sequenza and SNP array-based copy number profiles suggests that exome sequencing alone is sufficient not only for identifying small scale mutations but also for estimating cellularity and inferring DNA copy number aberrations.
introduction
Cancer is a genetic disease in which specific mutations or genomic aberrations can enable tumor initiation or progression, and in certain cases can determine the effectiveness of specific anticancer therapies. Several tumor resequencing projects have collected and analyzed genetic material from large cohorts of patients in an effort to identify important somatic events that may represent drug targets or predictive biomarkers [1]. In such projects, nonsynonymous substitutions and short indels in coding regions are typically detected by analyzing exome sequencing data derived from matched pairs of tumor and normal tissues of cancer patients, whereas larger aberrations such as copy number alterations or loss of heterozygosity (LOH) are typically detected using genome-wide single nucleotide polymorphism (SNP) arrays, which remains the current state-of-the-art.
Tumor tissue specimens comprise a mixture of cancer cells and normal cells; therefore, analysis of tumor data must take the specimen cellularity into consideration [2–5]. However, it is currently not possible to make a histological estimate of tumor cellularity and extract high-quality DNA from the very same specimen; therefore, cellularity estimates based on histology are commonly made from an adjacent tumor section which often does not reflect the cellularity of the section used for DNA sequencing. Thus, using the DNA itself to make cellularity estimates is an appealing approach. Several methods have been described that estimate, and then correct for, tumor cellularity in SNP array data in order to improve copy number profiles [2–5] or in DNA sequencing data for mutation calling [6].
Copy number profiles can be inferred from sequencing data of sufficient depth and coverage, by using the relative number of reads mapped to a given genomic position (depth ratio) as an indicator of copy number. This approach has recently been demonstrated in algorithms such as VarScan 2 [7] and APOLLOH [8], wherein inferred copy number profiles from whole-exome sequencing alone (WES profiles) are largely concordant with profiles inferred from SNP array data (SNP profiles). APOLLOH estimates the tumor cellularity, whereas VarScan 2 does not. In addition, algorithms such as PurityEst [9] and PurBayes [10] are specialized to estimate tumor cellularity directly from paired tumor-normal sequence data. Only recently, newer tools including absCN-seq [11] and newer versions of ABSOLUTE [4] have provided methods to estimate cellularity and ploidy and calculate copy number profiles directly from exome sequencing data. In such algorithms, accurate cellularity and ploidy estimation is essential for the generation of correct copy number profiles.
Here we describe Sequenza, a software package that uses paired tumor-normal exome or whole-genome sequencing data to estimate tumor cellularity and ploidy and to infer allele-specific tumor copy number profiles. Using publicly available matched tumor-normal data, we compare the results of exome sequence data analyzed by Sequenza with SNP array data from the same tumors analyzed by allele-specific copy number analysis of tumors (ASCAT). For comparison, we also assess the performance of the previously described algorithms absCN-seq and ABSOLUTE.
materials and methods
algorithm
Sequenza is based on a probabilistic model applied to segmented data. The observations include the average depth ratio (tumor versus normal) and B allele frequency (the lesser of the two allelic fractions as measured at germline heterozygous positions) for each segment. The model parameters include overall tumor ploidy and cellularity, and segment-specific copy number and minor allele copy number. The location of the segments and the segment-level dispersion are taken as known constants. We estimate model parameters using a maximum a posteriori approach in which prior probabilities are defined for the copy number such that two copies (by default) are preferred over other values. Under this model, given values for cellularity and ploidy, the segment-level parameters can be quickly estimated. Thus, we solve the overall estimation problem using a grid-based search over reasonable values of cellularity and ploidy (see supplementary Methods, available at Annals of Oncology online).
implementation
The Sequenza software consists of two distinct parts: a python-based preprocessing tool, and an R package implementing the model fitting and visualization functions (supplementary Figure S1, available at Annals of Oncology online).
The python script ‘sequenza-utils’ has two roles. First, it calculates the GC content in sliding windows from a genome reference file in FASTA format. Second, it processes the sequencing data from the tumor and normal specimens, which must be in the Pileup format, as output by SAMtools [12]. For genomic positions with sufficient sequencing depth (by default, >20 reads total from tumor and normal specimens), the script extracts sequencing depth, determines homozygous and heterozygous positions in the normal specimen, and calculates the variant alleles and allelic frequency from the tumor specimen. The output is a tab-delimited text file suitable for import into R. Additionally, ‘sequenza-utils’ is compatible with the pypy python implementation [13], which performs around six times faster than the standard python implementation.
The ‘sequenza’ R package is used to perform the analysis on the output of the sequenza-utils and is implemented with three high-level functions (supplementary Figure S1B, available at Annals of Oncology online): first, sequenza.extract efficiently reads the input file into R, performs GC-content normalization of the tumor versus normal depth ratio, and performs allele-specific segmentation using the ‘copynumber’ package [14]. Second, sequenza.fit applies the model described in the supplementary Material, available at Annals of Oncology online, to infer cellularity and ploidy parameters and copy number profiles. Alternative solutions are also provided, using local maxima of the posterior probability space. Finally, sequenza.results returns the results of the estimation together with alternative solutions and visualization of the data and the model along the genome and the individual chromosomes.
Detailed methods are available in supplementary Methods, available at Annals of Oncology online. The software has a web page at http://www.cbs.dtu.dk/biotools/sequenza and is freely available from CRAN.
data and analysis
Thousands of specimens are available from the TCGA; we arbitrarily selected the first 10 ovarian serous carcinomas (OVCA) and 20 clear-cell renal cell carcinomas (KIRC) sample IDs as of May 2013, when sorted alphabetically. The SNP arrays for ovarian serous carcinomas and renal clear-cell carcinomas were obtained on 22 January 2010 and 17 November 2011, respectively. Exome sequence data, previously aligned to the human genome version hg19, was obtained in BAM format in May 2013.
The SNP array files were preprocessed using the aroma.affymetrix package [15] as described [16], and copy number variations were determined using ASCAT version 2.1 [3]; sex chromosomes were excluded from the analysis.
The Sequenza results were obtained using version 2.1.0 with default parameters; the input was generated by the python script sequenza-utils.py version 2.1.0 with default binning size of 50 bases for the exome sequencing or 200 bases for the whole-genome sequencing. The absCN-seq results were obtained using version 1.0 with default parameters; the input was the same genomic segments used by Sequenza as well as high-quality somatic mutations calls detected by VarScan2 as described in the software documentation. The ABSOLUTE results were obtained using software version 1.0.6 with default parameters except that the platform was specified as ‘Illumina_WES’; the input was the same genomic segments used with Sequenza and absCN-seq.
Exome sequencing data from 31 of the NCI-60 tumor cell lines, aligned to the genome version hg19, were downloaded in May 2014 in the BAM format [17].
Whole-genome sequencing, aligned to the hg19 genome in the BAM format at ×30 of coverage, of two cell lines HCC1143 and HCC1954, matching normal blood, and simulated admixtures at tumor cellularity of 20%, 40%, 60%, and 80%, were obtained in March 2014 from the TCGA4 benchmark cohort (https://cghub.ucsc.edu/datasets/benchmark_download.html).
All BAM files were processed to remove PCR duplicates and low-quality mappings with Picard, and then converted to pileup format using SAMtools [12].
results
application of sequenza to tumor exome sequencing data
To compare Sequenza WES profiles with the current state-of-the-art, SNP profiles, we obtained paired tumor-normal exome and Affymetrix SNP6 arrays from 10 OVCA patients [18] and 20 KIRC patients [19]. We chose renal and ovarian cancer because these represent two widely different cancer types: clear-cell renal cancer has low cellularity and few copy number variations, whereas ovarian cancer typically shows extensive copy number alterations and high tumor cellularity.
The exome data were processed with Sequenza using default settings. Running on a single CPU core, this required an average per-specimen running time of 4 h for preprocessing, 30 min for segmentation, and 4 min for model fitting and parameter estimation. Results from a representative sample are shown in Figure 1. Of the 20 renal cancer copy number profiles, 17 exhibited 3p loss (supplementary Figure S5, available at Annals of Oncology online), consistent with previous observations of renal cancer [19].
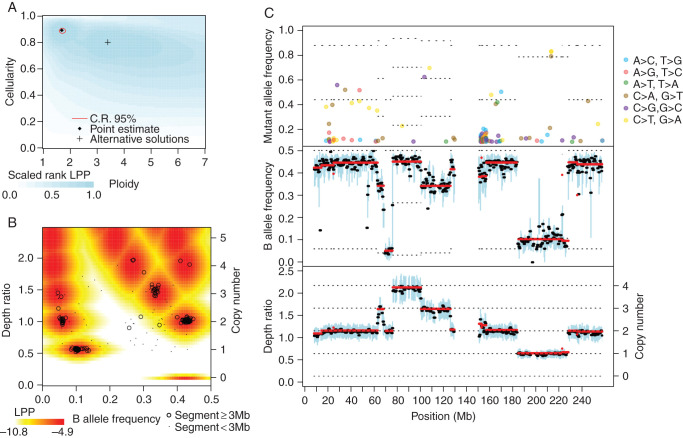
Figure 1.
Representative output of the Sequenza algorithm. Exome sequencing data from an ovarian tumor (TCGA-42-2591-01A) and matched normal (TCGA-42-2591-10A) specimen were applied to Sequenza. (A) The log posterior probability (LPP) of the observed data were calculated for a range of candidate ploidy and cellularity values. The point estimate is the ploidy and cellularity with maximum LPP. The 95% confidence region is the smallest (not necessarily contiguous) set of points with a total posterior probability >0.95. The background color indicates the rank of the LPP (blue = most likely, white = least likely), provided here to contrast other possible parameters that are very unlikely under our model but might still be of interest. Local maxima are indicated with a ‘+’ and indicate possible alternative solutions. (B) Observed depth ratio and BAF values for each genomic segment (black circles and dots) along with the representative joint LPP density (colors). The representative joint LPP density is calculated for the cellularity and ploidy estimates identified in (A), and is calculated for a hypothetical representative 10 Mb segment. The actual joint LPP density is dependent on segment size and variability and thus varies quantitatively but not qualitatively for each segment. Observed segments with highly unlikely DR and BAF values may indicate subclonality, measurement errors, or incorrect model parameters. (C) Chromosome plot indicating mutant allele frequency (top panel), B allele frequency (middle panel), and depth ratio (bottom panel) according to genomic position. Here, chromosome 1 is shown. The mutant allele frequency at a given position is the fraction of reads with a mutation, and is displayed if >0.1 for each genomic position with sufficient sequencing depth. For the sake of visualization, the B allele frequency and depth ratio are summarized within 1 Mb windows staggered every 0.5 Mb. Within each window, a thick black line indicates the median value, and a blue bar indicates the interquartile range. Red lines indicate segmented values. The thin dotted lines indicate the expectation values under the fitted model; their placement is based on the estimated cellularity, ploidy, and copy number profile. In the top panel, the dotted lines indicate the number of alleles with mutation, with the lowest line starting at one. In the middle panel, the dotted lines indicate the minor allele copy number, with the lowest line starting at zero. In the lower panel, the dotted lines indicate the copy number.
comparison between exome/Sequenza and SNP array/ASCAT profiles
There is no tumor gold standard that could be used to validate the performance of Sequenza. However, the use of SNP arrays processed by ASCAT is an established approach for determining copy number profiles; therefore, a positive agreement between these two platforms would confirm the performance of Sequenza. Hereafter, for simplicity, we use the terms ‘Sequenza’ and ‘ASCAT’ with the understanding that it is actually the combined measurement platform/software that is being considered. Sequenza and ASCAT both provide estimates of cellularity and ploidy, and we found a strong correlation for both parameters (r = 0.90 and r = 0.42, respectively, Figure 2A and B, Table 1). Interestingly, the ploidy comparison seems to be characterized by a few large outliers, many of which have low cellularity. Details about three highly discordant samples are shown in supplementary Figures S6–S8, available at Annals of Oncology online.
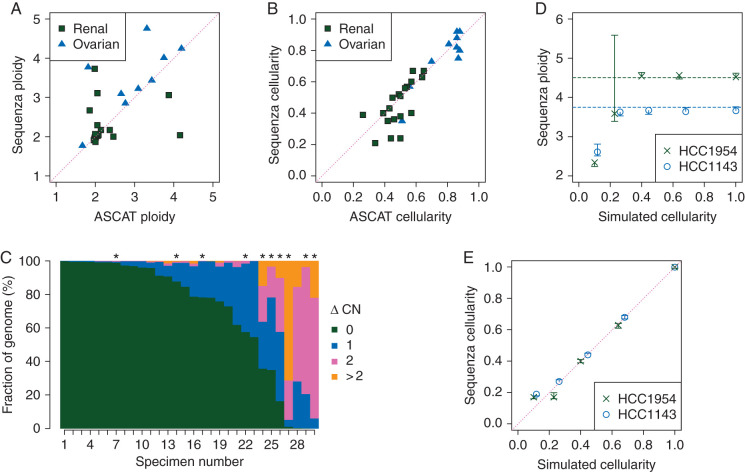
Figure 2.
Comparison of cellularity and ploidy estimates and copy number profiles derived from exome sequence to those derived from SNP array and testing on simulated data. (A–C) Matched tumor-normal exome sequencing and SNP array data from 10 ovarian cancer patients and 20 renal cell carcinoma patients were obtained from TCGA. Exome data was analyzed with Sequenza, and SNP array data were analyzed with ASCAT. (A) Ploidy and (B) cellularity estimates were compared between the two platforms. (C) Copy number profiles were compared by calculating the absolute difference in estimated copy number for each genomic position (ΔCN). The figure indicates the fraction of the covered genome with each level of ΔCN. Asterisks indicate tumors for which the Sequenza cellularity estimate is lower than 0.4. (D and E) Sequenza (D) ploidy and (E) cellularity estimates from simulated whole-genome sequencing with varying cellularity for cell lines HCC1954 and HCC1143. Vertical lines indicate 95% confidence intervals on the estimates. Dashed horizontal lines indicate ploidy estimates of the same cell lines by SNP array in an independent study [4].
Table 1.
Performance of various algorithms on TCGA exome data
| Algorithm | |||||
|---|---|---|---|---|---|
| Sequenza | 0.90 (0.91) | 0.42 (0.94) | 0.69 | 0.095 (0.087) | 0.95 (0.25) |
| ABSOLUTE | 0.19 (0.61) | 0.13 (0.50) | 0.08 | 0.35 (0.19) | 1.81 (1.08) |
| absCN-seq | 0.46 (0.65) | −0.26 (0.46) | 0.02 | 0.16 (0.13) | 1.91 (0.76) |
, = Pearson correlation of cellularity or ploidy estimates (respectively) with those of ASCAT. = median (over all samples) fraction of the genome with copy number estimate equal to that of ASCAT. = median (over all samples) Pearson correlation of copy number profile with that of ASCAT. The numbers in parentheses indicate the result when the set of alternative solutions is visually inspected.
Both Sequenza and ASCAT return a list of genomic segments, each with an estimated copy number state. However, the breakpoints between segments are different, and the genomic coverage of the two platforms is not the same. We compared only the positions covered in the segmentation for both platforms (Figure 2C). Aside from samples where the Sequenza and ASCAT ploidy estimates disagree, the genome fraction with perfect agreement ( equal to zero) was generally high, with a median value of 69%. However, as expected, samples with ploidy disagreement were also discordant in their copy number profiles.
To assess copy number profile agreement between the two platforms in a ploidy-independent manner, we carried out hierarchical clustering of the sample profiles using a Pearson correlation distance metric. In all but one case, sample profiles from the same patient derived from different platforms clustered together (data not shown), and are thus more similar to each other than to other profiles, even when the ploidy call is substantially different between the two algorithms.
comparison with other methods
We are aware of two previously published methods to generate copy number profiles from exome sequencing data in a way that accounts for tumor cellularity: ABSOLUTE [4] and absCN-seq [11]. We assessed the performance of these methods using the same criteria as we used to evaluate Sequenza. Similarly to above, we use the terms ‘ABSOLUTE’ and ‘absCN-seq’ to indicate the results derived from exome sequencing applied to each specific method, and we compared the results of each algorithm with the results from ASCAT applied to SNP array data. To focus the comparison on ploidy and copy number estimation algorithms rather than segmentation algorithms, we used the same segmented input (processed by copynumber [14]) as input to each algorithm. The comparison results for each algorithm are summarized in Table 1.
First, we compared cellularity and ploidy estimates. The ABSOLUTE estimates of cellularity and ploidy were weakly correlated with ASCAT estimates (r = 0.19 and r = 0.13, supplementary Figure S2A and B). The absCN-seq estimates were moderately correlated with the ASCAT cellularity estimate (r = 0.46), but had a negative correlation with the ASCAT ploidy estimate (r = −0.26, supplementary Figure S3A and B, available at Annals of Oncology online). Next, we compared segment-wise copy number estimates. As expected from the low agreement in ploidy estimates, the majority of samples showed substantial disagreement with ASCAT copy number estimates (supplementary Figure S2C and S3C, available at Annals of Oncology online).
Previous publications of copy number inference algorithms have stated the performance obtained after manual selection of a single solution from a set of multiple solutions proposed by the algorithm. Thus, we manually inspected the list of possible solutions from the three algorithms and selected the solution with best agreement to the SNP array solution. As expected, this resulted in increased accuracy for all three algorithms, with Sequenza obtaining the highest agreement (Table 1).
application to cell line data
We applied Sequenza to exome sequencing data from 31 cell lines from the NCI-60 panel [17], and compared the estimated ploidy with previously published modal chromosome numbers derived from spectral karyotyping [20]. These particular samples were selected to compare Sequenza performance with previously published results [11]. To accommodate the lack of matched normals in this dataset, we modified our algorithm to calculate the depth ratio and identify the heterozygous positions from two different sources: we used the near-diploid hematopoietic cell line SR as the normal genome for depth ratio calculation, and the selected cell line itself to determine heterozygous positions. However, with this approach, any LOH regions in the cell line would result in the absence of identified heterozygous positions; thus, we adjusted to zero the B allele frequency of segments with fewer than three heterozygous positions per megabase. Despite the suboptimal input data, we obtained a root mean square error (RMSE) between the karyotype-derived ploidy and Sequenza-estimated ploidy of 1.2 (supplementary Figure S4A, available at Annals of Oncology online), comparable with results of absCN-seq applied to the same data (0.55) [11].
For comparison to previously published results in which manual inspection of solutions was carried out, we carried out a similar analysis in which we visually inspected two to four alternative solutions, and for eight of the samples selected a solution different from the point estimate, resulting in an RMSE of 0.44 (supplementary Figure S4B, available at Annals of Oncology online). This can be compared with previously published results in which absCN-seq obtained an RMSE of 0.34 using the same data [11], or to results obtained with SNP array of the NCI-60 cohort with an RMSE of 0.54 using ABSOLUTE and 0.85 using ASCAT [4].
To assess how ploidy estimation accuracy is affected by low cellularity, we analyzed simulated tumor-normal admixtures at proportions of 100%, 80%, 60%, 40%, and 20% provided by the ‘TCGA benchmark 4’ whole-genome sequencing of the HCC1143 and HCC1954 cell lines [21]. Transformations from the normal-tumor reads admixture percentage to tumor content have to consider the tetraploid genomes of the cell line. Result from the simulations shows that the algorithm estimates the correct ploidy until the cellularity values decrease to below 0.3 (Figure 2D and E).
discussion
We have described a simple model to infer accurate copy number profiles from next-generation sequencing data and its implementation in the software package Sequenza. For the majority of specimens we analyzed, we observed a strong agreement between the output of Sequenza and the output from ASCAT using matched SNP array data. The few cases with substantial disagreement in copy number profile seem to stem from disagreement in the ploidy and were more common in specimens with low cellularity. It is possible to determine ploidy experimentally using flow cytometry [22], but this was not carried out on the TCGA specimens. In cases where experimentally derived ploidy data are available, it is possible with Sequenza to explicitly specify the ploidy rather than determine it by model fitting.
One advantage of SNP arrays over exome sequencing is the genomic coverage. SNP arrays are often designed to both determine SNP genotypes and detect copy number changes. In particular, the Affymetrix SNP6.0 platform used for the samples tested in this manuscript covers more than 900 000 positions evenly distributed in the genome for copy number detection, and another 900 000 SNP positions, of which on average ∼26% are heterozygous in a given individual. This design allows for highly accurate allele-specific determination of copy number profiles. In contrast, exome enrichment kits are generally not designed for the purpose of determining copy number states. Covered genomic regions are based on known exons, which are on average ∼150 bases in size, and are not evenly distributed throughout the genome. Inference of allele-specific copy numbers requires heterozygous positions and thus can only be achieved for those exons that include SNPs. When working with the exome sequencing data, we recorded an average of 45 000 heterozygous positions for each patient, corresponding to ∼1/5 the number identified by the SNP arrays.
However, it seems likely that whole-genome sequencing will eventually become more cost efficient and widely used than exome sequencing. Sequenza is compatible with whole-genome data, and we expect this to result in increased accuracy due to better genomic coverage and increased number of heterozygous positions. In fact, when processing available whole-genome sequencing data (data not shown), we identified an average of 1.7 × 106 heterozygous SNPs, and genotyping and depth information for 2.6 × 109 positions.
We are aware of four other methods also designed to estimate copy number profiles in tumor samples of unknown cellularity, but only two of these are designed to work on exome sequencing data. The three algorithms have many common elements in their models, but several important differences. AbsCN-seq uses a least squares method to estimate the most likely model, providing a fast running time; whereas ABSOLUTE and Sequenza use likelihood or posterior probability to estimate the best solution. ABSOLUTE incorporates prior probabilities from previous karyotype analyses, whereas Sequenza uses much simpler prior probabilities on copy numbers that are the same on each segment to estimate the best solution. AbsCN-seq does not incorporate prior probabilities. Additionally, Sequenza and ABSOLUTE provide graphical reports to further inspect the alternative solutions, whereas absCN-seq reports only the numerical alternative cellularity and ploidy values. One possible advantage of Sequenza over the other two algorithms is the use of the B allele frequency, which not only provides additional information beyond the depth ratio, but also enables calculation of allele-specific copy number, whereas the other algorithms provide only absolute copy number profiles. However, the requirement for the B allele frequency is a drawback in cases where it is not possible to accurately determine the heterozygous positions, for example in cell lines where the normal sample is not available.
In our comparison with previously published methods ABSOLUTE and absCN-seq, we found that Sequenza shows substantially stronger agreement with SNP array-based cellularity, ploidy, and copy number estimates. However, in the analysis of cell line exome data, absCN-seq performed better than Sequenza, likely because Sequenza relies on identification of heterozygous positions from a matched normal sample that was not available for these cell lines.
One limitation of Sequenza as well as its competing algorithms is that the segmentation is taken as a given; a more sophisticated analysis would consider uncertainty in the assignment of segment boundaries. Also, Sequenza does not account for possible heterogeneity of mutations within a tumor specimen, which has important consequences for patient diagnosis and for identification of driver mutations [23, 24]. However, it is possible to use the variant allele frequency and corresponding copy number states from Sequenza as input for external software such as PyClone [25] in order to resolve subclonal structures.
funding
This work was supported by the European Commission 7th Framework Programme (HEALTH-2010-F2-259303); the Danish Council for Independent Research (09-073053/FSS); and the Breast Cancer Research Foundation (to ZS). Funding for open access charge: the Danish Council for Independent Research (09-073053/FSS).
disclosure
The authors have declared no conflicts of interest.
Supplementary Material
acknowledgement
We thank Anders Gorm Petersen for helpful discussions. The results published here are based on data generated by the Cancer Genome Atlas pilot project established by the NCI and NHGRI. Information about TCGA and the investigators and institutions who constitute the TCGA research network can be found at http://cancergenome.nih.gov/. The data were retrieved through dbGaP authorization (Accession No. phs000178.v8.p7).
references
- 1.Hudson TJ, Anderson W, Artez A, et al. International network of cancer genome projects. Nature. 2010;464(7291):993–998. doi: 10.1038/nature08987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Popova T, Manié E, Stoppa-Lyonnet D, et al. Genome Alteration Print (GAP): a tool to visualize and mine complex cancer genomic profiles obtained by SNP arrays. Genome Biol. 2009;10(11):R128. doi: 10.1186/gb-2009-10-11-r128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Van Loo P, Nordgard SH, Lingjærde OC, et al. Allele-specific copy number analysis of tumors. Proc Natl Acad Sci U S A. 2010;107(39):16910–5. doi: 10.1073/pnas.1009843107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carter SL, Cibulskis K, Helman E, et al. Absolute quantification of somatic DNA alterations in human cancer. Nat Biotechnol. 2012;30(5):413–421. doi: 10.1038/nbt.2203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lamy P, Andersen CL, Dyrskjot L, et al. A Hidden Markov Model to estimate population mixture and allelic copy-numbers in cancers using Affymetrix SNP arrays. BMC Bioinformatics. 2007;8:434. doi: 10.1186/1471-2105-8-434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cibulskis K, Lawrence MS, Carter SL, et al. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat Biotechnol. 2013;31(3):213–219. doi: 10.1038/nbt.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Koboldt DC, Zhang Q, Larson DE, et al. VarScan 2: somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res. 2012;22(3):568–576. doi: 10.1101/gr.129684.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ha G, Roth A, Lai D, et al. Integrative analysis of genome-wide loss of heterozygosity and monoallelic expression at nucleotide resolution reveals disrupted pathways in triple-negative breast cancer. Genome Res. 2012;22(10):1995–2007. doi: 10.1101/gr.137570.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Su X, Zhang L, Zhang J, et al. PurityEst: estimating purity of human tumor samples using next-generation sequencing data. Bioinformatics. 2012;28(17):2265–2266. doi: 10.1093/bioinformatics/bts365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Larson NB, Fridley BL. PurBayes: estimating tumor cellularity and subclonality in next-generation sequencing data. Bioinformatics. 2013;29(15):1888–1889. doi: 10.1093/bioinformatics/btt293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bao L, Pu M, Messer K. AbsCN-seq: a statistical method to estimate tumor purity, ploidy and absolute copy numbers from next-generation sequencing data. Bioinformatics. 2014;30(8):1056–1063. doi: 10.1093/bioinformatics/btt759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li H, Handsaker B, Wysoker A, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25(16):2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Rigo A, Pedroni S. PyPy's approach to virtual machine construction. Companion to 21st ACM SIGPLAN symposium on Object-oriented programming systems, languages, and applications. In OOPSLA ′06. New York: ACM Press 2006; 944–953. [Google Scholar]
- 14.Nilsen G, Liestøl K, Van Loo P, et al. Copynumber: efficient algorithms for single- and multi-track copy number segmentation. BMC Genomics. 2012;13:591. doi: 10.1186/1471-2164-13-591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bengtsson H, Wirapati P, Speed TP. A single-array preprocessing method for estimating full-resolution raw copy numbers from all Affymetrix genotyping arrays including GenomeWideSNP 5 & 6. Bioinformatics. 2009;25(17):2149–2156. doi: 10.1093/bioinformatics/btp371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Martinez P, Birkbak NJ, Gerlinger M, et al. Parallel evolution of tumour subclones mimics diversity between tumours. J Pathol. 2013;230(4):356–364. doi: 10.1002/path.4214. [DOI] [PubMed] [Google Scholar]
- 17.Abaan OD, Polley EC, Davis SR, et al. The exomes of the NCI-60 panel: a genomic resource for cancer biology and systems pharmacology. Cancer Res. 2013;73(14):4372–4382. doi: 10.1158/0008-5472.CAN-12-3342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.The Cancer Genome Atlas Research Network. Integrated genomic analyses of ovarian carcinoma. Nature. 2011;474(7353):609–615. doi: 10.1038/nature10166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.The Cancer Genome Atlas Research Network. Comprehensive molecular characterization of clear cell renal cell carcinoma. Nature. 2013;499(7456):43–49. doi: 10.1038/nature12222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Roschke AV, Tonon G, Gehlhaus KS, et al. Karyotypic complexity of the NCI-60 drug-screening panel. Cancer Res. 2003;63(24):8634–8647. [PubMed] [Google Scholar]
- 21.TCGA Mutation Calling Benchmark 4 Datasets. https://cghub.ucsc.edu/datasets/benchmark_download.html. (29 October 2014, date last accessed)
- 22.Gerlinger M, Rowan AJ, Horswell S, et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med. 2012;366(10):883–892. doi: 10.1056/NEJMoa1113205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ding L, Raphael BJ, Chen F, Wendl MC. Advances for studying clonal evolution in cancer. Cancer Lett. 2013;340(2):212–219. doi: 10.1016/j.canlet.2012.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Horswell S, Matthews N, Swanton C. Cancer heterogeneity and “The Struggle for Existence”: diagnostic and analytical challenges. Cancer Lett. 2012;340(2):220–226. doi: 10.1016/j.canlet.2012.10.031. [DOI] [PubMed] [Google Scholar]
- 25.Shah SP, Roth A, Goya R, et al. The clonal and mutational evolution spectrum of primary triple-negative breast cancers. Nature. 2012;486(7403):395–399. doi: 10.1038/nature10933. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.