Abstract
Reconstruction of damaged nerves remains a significant unmet challenge in clinical medicine. To foster improvements, the control of neural stem cell (NSC) behaviors, including migration, proliferation and differentiation are critical factors to consider. Topographical and mechanical stimulation based on the control of biomaterial features is a promising approach, which are usually studied separately. The synergy between topography and mechanical rigidity could offer new insights into the control of neural cell fate if they could be utilized concurrently in studies. To achieve this need, silk fibroin self-assembled nanofibers with a beta-sheet-enriched structure are formed into hydrogels. Stiffness is tuned using different annealing processes to enable mechanical control without impacting the nanofiber topography. Compared with nonannealed nanofibers, NSCs on methanol annealed nanofibers with stiffness similar to nerve tissues differentiate into neurons with the restraint of glial differentiation, without the influence of specific differentiation biochemical factors. These results demonstrate that combining topographic and mechanical cues provides the control of nerve cell behaviors, with potential for neurogenerative repair strategies.
Keywords: silk nanofibers, hydrogels, tunable stiffness, neural stem cells, nerve regeneration
1. Introduction
Central nerve injuries generally lead to dysfunction and loss of neuronal tissue, resulting in long-term disabilities and major socio-economic costs.1,2 The multipotent property of neural stem cells (NSCs), which can differentiate into neurons and glial cells, suggests that these cells are promising candidates for the treatment of neurodegenerative diseases or neural injuries.3–6 Although incredible progress has been made using stem cell therapies in different neurological disease models,7–11 fundamental questions remain regarding the efficiency and ability of stem cells to safely and effectively repair damaged neural tissues. Sufficient control of neural stem cell fate is a major challenge to progress in the field. Recently, the critical influence of specific microenvironments or niches on the proliferation, migration and differentiation of these stem cells was reported.12–16 A multitude of biomaterials have been developed as substrates or scaffolds to regulate the behavior of NSCs by fabrication with control of microstructure, surface charge, biomechanical properties, as well as composition, to simulate the microenvironments of nerve tissues.17–20 The results suggest the feasibility of specifically enhancing the differentiation of NSCs into more neuronal phenotype than glial cells based on the biomaterial substrates.21–23 However, significant challenges remain to achieve desired neural cell fates by integrating different strategies in the biomaterials designs, including the choice of biomaterial, microstructural and mechanical features, and the incorporation of bioactive molecules.4
Silks spun by silkworms and spiders represent some of the strongest and toughest biological materials.24–26 The ability to control the release of growth factors from silk, the control of silk materials morphology, and the biocompatibility of silk make this protein a unique material platform for controlling stem cell fate and tissue regeneration.27,28 Recent studies have revealed the feasibility of silk as a supporting matrix for cells, including fibroblasts, osteoblasts, hepatocytes, nerve and stem cells, as well as a scaffolding for tissue engineering of bone, ligaments, blood vessels, skin, cartilage and other tissues.29–32 Another interesting use of silk is to treat peripheral and central nervous system injuries or diseases because of the neuro-compatibility of the material, along with regeneration using silk-based conduits that approached autografts in terms of outcomes.33–36 However, further microstructural and mechanical control during silk materials fabrication is still needed to fully exploit the protein for functional regeneration in the central nervous system (CNS).
Electrospinning, a simple and versatile technique for preparing nanofibrous nonwoven mats, has been successfully developed to fabricate nanofibrous structures of silk using an all-aqueous processes.37,38 Further modification of this method allows nanofiber orientation, size control, and the addition and release of bioactive materials without loss of biological function, providing improved microenvironments for nerve regeneration.39–42 Although different studies demonstrated the potential of aligned and functionalized electrospun silk nanofibers to promote nerve growth in the central nervous system,39–42 some problems remain, such as the difficulty in designing complex three-dimensional porous structures with electrospinning. Another potential opportunity is to mimic the microstructure and elastic modulus of the extracellular environment of nerve tissues simultaneously with morphology, since evidence indicates that the differentiation of stem cells could be regulated by tuning scaffold stiffness.43–45 Neural tissues are inherently soft (elastic moduli, 0.1–1 kPa).43 Although silk fibroin has been extensively studied as potential biomaterial for neural tissue regeneration, to the best of our knowledge, the stiffness of previous silk fibroin scaffolds/hydrogels prepared for neural regeneration is still significantly higher than natural neural tissues.44 The combined control of rigidity and topography is also difficult because nanostructure fabrication usually requires higher stiffness than found in nerve tissues.4 In order to overcome the problems of electrospun silk nanofibers used in neural regeneration, silk fibroin hydrogels with tunable stiffness have been developed to facilitate neural cell growth and tissue regeneration.44 Unfortunately, ECM-mimic nanofiber fabrication is unfeasible for the silk hydrogel system. Recently, silk nanofibers with different sizes and secondary conformations were prepared in aqueous solution through controlling silk self-assembly process.46 Then silk nanofiber hydrogels were prepared after beta-sheet structure formation, making it possible to design biomimetic hydrogels for tissue regeneration in the central nervous system. The goal of the present study was to generate bioactive silk nanofibers with tunable mechanical properties by controlling the self-assembly of silk in aqueous solution, and to test the ability of these systems to regulate the differentiation of NSCs. The self-assembly strategy avoided the stiffness requirements of previous nanostructure fabrication techniques, and also overcomes the restriction of electrospinning on building complex three-dimensional structures, providing a feasible approach to achieve synergistic control of NSC fate by the dual regulation of rigidity and topography of silk materials. The self-assembled silk nanofiber hydrogels supported NSCs, facilitated their differentiation to neurons and depressed their differentiation to glial cells, by tuning the mechanical properties of the hydrogels. These results suggest options for new types of nerve guides based on bioactive silk-based materials with tunable microstructural and mechanical properties.
2. Experimental section
2.1 Preparation of aqueous silk fibroin solutions
Silk solution was prepared according to our previously described methods.46 Bombyx mori cocoons were boiled for 20 min in an aqueous solution of 0.02 M Na2CO3, and then rinsed thoroughly with distilled water to extract the sericin proteins. The extracted silk was dissolved in 9.3 M LiBr solution (Sigma-Aldrich, St. Louis, MO) at 60 °C, yielding a 20% (w/v) solution. This solution was dialyzed against distilled water, using Slide-a-Lyzer dialysis cassettes (Pierce, MWCO 3500) for 72 h to remove the salt. The solution was optically clear after dialysis and was centrifuged at 9,000 rpm for 20 min at 4 °C to remove silk aggregates. The final concentration of aqueous silk solution was about 6 wt%, determined by weighing the remaining solid after drying.
2.2 Preparation of silk nanofiber hydrogels
To prepare silk nanofiber hydrogels, the fresh silk solutions were treated by a slow concentration-dilution process. The solution (6 wt%) was slowly concentrated to about 20 wt% over 24 h at 60 °C to form metastable nanoparticles, and then diluted to below 2 wt% with distilled water. The diluted silk solution was incubated for about 24 hours at 60 °C to induce nanofiber hydrogel formation. Modulated water/methanol annealing processes were applied to further change the secondary structures for the modulation of stiffness of the hydrogels. The hydrogels were placed in desiccators filled with methanol/water blend solutions with a 25 in. Hg vacuum for 4 h. Methanol content in these blend solutions were 0%, 50% and 80%, respectively. The methanol-annealed hydrogels were kept in distilled water to replace methanol with water until further experiments.47 After dialysis, the content of methanol in silk hydrogels measured with ICO-OES (Icap 6000, Thermo Scientific, Massachusetts, USA) was < 0.03%, which confirmed that the methanol removal has almost completed. The untreated, water-annealed, 50% methanol-annealed and 80% methanol-annealed hydrogels were termed SN-H, WA-SN-H, MA50-SN-H and MA80-SN-H, respectively.
2.3 Nanostructure of silk hydrogels
To study nanostructure, the hydrogels were fixed in liquid nitrogen for 30 min firstly, placed at −20 °C for about 12 h, and then lyophilized for about 48 h to achieve freeze-dried samples. The morphology of samples was observed using scanning electron microscopy (SEM, S-4800, Hitachi, Tokyo, Japan) at 3 kV. Before SEM examination, the dried samples were coated with platinum. The nanostructures of the hydrogels were also assessed with atomic force microscopy (AFM). Silk hydrogels were diluted to below 0.1 wt% to avoid masking the original morphology by multilayers of silk. Two microliters of the diluted samples was dropped onto freshly cleaved 4 × 4 mm2 mica surfaces and spin-coated using a Spin Processor (WS-400, Laurell Technologies, PA, USA). The morphology of silk was observed by AFM (Nanoscope V, Veeco, NY, USA) in air. A 225 μm long silicon cantilever with a spring constant of 3 N m−1 was used in tapping mode at 0.5–1 Hz scan rate.
2.4 Secondary structures of silk hydrogels
The secondary structures of the silk hydrogels were measured with a CD spectrophotometer (JASCO-815, Japan). The hydrogels were diluted before the CD measurement. CD spectra were recorded from 250 to 190 nm wavelengths with an accumulation of five scans at a scanning rate of 100 nm min−1 at 25 °C. The results were averaged from three repeated experiments. The secondary structures were further analyzed by fourier transform infrared spectroscopy (FTIR) and X-ray diffraction (XRD) based on the freeze-dried samples. The hydrogels were placed at −20 °C for about 12 h, and then lyophilized for about 48 h to achieve freeze-dried samples.48 FTIR was conducted on a Nicolet FTIR 5700 spectrometer (Thermo Scientific, FL, USA). For each measurement, 64 scans were coded with a resolution of 4 cm−1, with the wavenumber ranging from 400 to 4000 cm−1. Fourier self-deconvolution (FSD) of the infrared spectra covering the amide I region (1595–1705 cm−1) was performed by Opus 5.0 software to identify silk secondary structures. FSD spectra were curve-fitted to measure the relative areas of the amide I region components. The XRD experiments were conducted with an X-ray diffractometer (X’Pert-Pro MPD, PANalytical, Almelo, Holland) with Cu Kα radiation at 40 kV and 30 mA and scanning rate of 0.6 °/min. Before examination, the dried samples were pressed into sheets with a hydraulic compressor.
2.5 Dynamic oscillatory rheology
The mechanical properties of the hydrogels were measured on a Rheometer (AR2000, TA Instruments, New Castle, USA) fitted with a 20 mm cone plate (Ti, 20/1°). Prior to each experimental day the rheometer underwent a torque map with a 10 Pa s calibration oil. Frequency sweeps were performed in the linear viscoelastic regime to determine values of the elastic (G') modulus. All samples (2%) were allowed to equilibrate for 20 min before the measurement, which were taken under ambient conditions at 25 °C.
2.6 Zeta potential
Surface charges of silk samples were determined via zeta potential measurement.46 One milliliter of the sample was loaded to a Zetasizer (Nano ZS, Malvern, Worcesteshire, UK) for the zeta potential measurement at 25 °C.
2.7 Cell culture
The neural stem cells were isolated from the lateral ventricle walls of E14 C57/BL6J mice as described.49 All mice used in this study were handled according to the protocols approved by the Institutional Animal Care and Use Committee of Soochow University. Neurospheres were growing in DMEM/F12 culture medium (Gibco) containing B27 (Gibco), 20 ng/ml basic fibroblast growth factor (bFGF) (Peprotech) and 20 ng/ml epidermal growth factor (EGF) (Peprotech). For cell culture in vitro, 1 ml of silk nanofiber hydrogel was spin-coated onto glass three times to form coatings using a Spin Processor (WS-400, Laurell Technologies, PA, USA) and sterilized with 60 Co γ-irradiation at the dose of 50 kGy. For proliferation in vitro, NSCs were cultured for 4~5 h in NSC culture medium containing 10μM BrdU. For migration assays, NSCs were cultured for 24 h in DMEM/F12 culture medium containing N2 supplement and 0.5% fetal calf serum (FCS). For differentiation, dissociated cells from primary neurospheres were seeded into 24-well dishes and were induced in DMEM/F12 culture medium containing N2 supplement and 0.5% FCS (Gibco) for 3~5 days.
2.8 Immunochemistry and quantification
The cells were fixed in 4% paraformaldehyde for 15 min at room temperature and then permeabilized with PBS containing 0.1% TritonX-100 for 10 min and washed with PBS twice, each for 10 min. The cells were then incubated with primary antibodies for 60 min at room temperature. The cells were incubated with corresponding fluorescence labeled secondary antibody for 60 min at room temperature after being washed three times with PBS. For BrdU staining, the cells were treated with 2N HCL for 10 min before being blocked with PBS containing 10% FCS. Antibodies: Goat anti-Sox2 (1: 80; Santa Cruz Biotechnology; SC-17320); anti-Brdu (1: 100; Covance; MMS-139S), Mouse anti-TUJ1 (1: 800; 6Sigma; T5076), Rabbit anti-Caspase3 (1: 100; R&D; AF835); The secondary antibodies used were species-specific antibody conjugated with Alexa fluorophore 488 or 555 (1: 500; Invitrogen).
The quantification of immunostained cultured cells was performed as previously described.49 In brief, images of fields of cultured cells were captured by digital photomicrography under a 20X objective systematically from top-to-bottom and left-to-right across the entirety of each coverslip. All labeled cells were then counted in each photomicrograph. The percentage of neurons/astrocytes and proliferative cells was quantified as the numbers of TUJ1+/GFAP+ and BrdU+ cells divided by the total number of DAPI+ cells in the same fields, respectively. All data was collected from at least three independent experiments. Two-tailed Student’s t-test or one way ANOVA (LSD) was performed using SPSS software. Values were presented as mean+SEM. Significance was accepted at p<0.05 (*: p<0.05, **: p<0.01, ***: p<0.001).
3. Results
3.1 Morphology and Structure of Self-Assembled Nanofibers
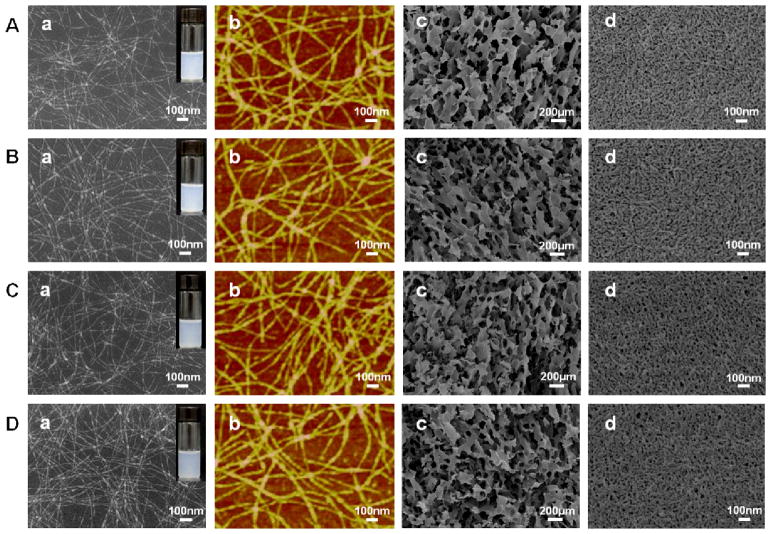
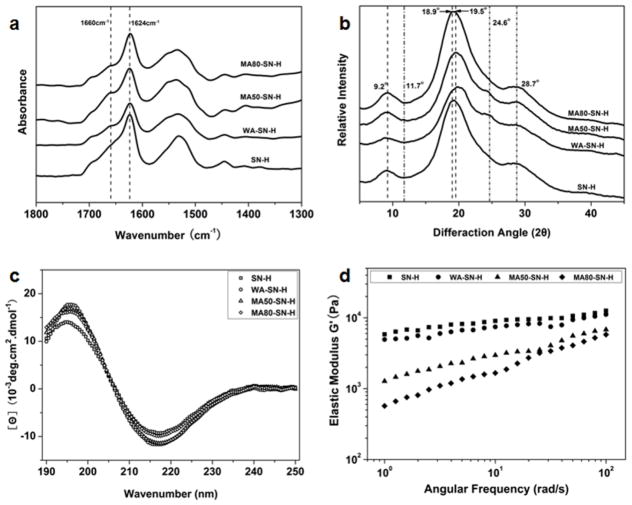
In order to facilitate cell growth and tissue repair, water stability is a prerequisite for neural tissue matrix. Different water-insoluble silk scaffolds and hydrogels have been used in various tissue engineerings including neural tissue regeneration. These scaffolds and hydrogels are generally prepared through methanol/water annealing, or ultrasonic treatment processes that could induce β-sheet formation and finally endow the silk materials enough hydrophobic property and stability in water.48,50 In our present study, different nanostructures of silk could be self-assembled in water by adjusting temperature, time and concentration as we have previously reported.46,51 Freshly prepared silk solution (6 wt%) was slowly concentrated to about 20% at 60 °C for more than 24 hours to form metastable nanoparticles. These particles were then diluted to 2 wt% with distilled water to induce disassembly.46 After further incubation for about 24 hours at 60 °C, the diluted silk solution (2%) transformed into nanofiber-based hydrogels. SEM and AFM images indicated nanofiber formation with lengths of about 1 μm (Fig. 1). The nanofibers were mainly composed of β-sheet structure (Fig. 2), endowing silk enough hydrophobic properties to maintain the hydrogel state. Since achieving water stability for silk fibroin generally needs β-sheet formation, which further results in the increase of stiffness,50 the preparation of water insoluble silk materials with stiffness similar to natural neural tissues once remained a challenge. In recent studies, water insoluble silk fibroin films and scaffolds with reduced β-sheet contents have been fabricated through water annealing process, slow drying process or glycerol treatment.52,53 These silk materials have softer mechanical properties than previous silk materials with higher β-sheet contents, implying the possibility of designing further softer silk hydrogels by changing their secondary structure composition. On the other hand, our recent studies revealed that the mechanical properties of silk fibroin also depended on hydrophilic interaction in random domains.54 Silk fibroin films with high β-sheet content but lower mechanical strength were prepared through destroying hydrophilic interaction between silk fibroin molecules. All these studies indicated that the mechanical properties of silk-based materials can be regulated by their secondary structure composition. Therefore methanol/water annealing processes with different methanol ratios were used to further tune the secondary structures of the hydrogels. After the treatments, the morphology of silk nanofibers showed no significant difference when compared to the untreated fibers. All the samples maintained nanofiber structures of about 10–20 nm in diameter (Fig. 1, Fig. S1). A positive ellipticity at 195 nm and a negative ellipticity at 217 nm appeared in all the CD curves, suggestive of a typical silk II conformation of the samples with and without the treatments (Fig. 2c).46 FTIR and XRD results of the freeze-dried samples further confirmed the predominance of silk II, but also revealed some changes in secondary conformations after the different treatments (Fig. 2a and b). Based on previous studies,48,55 the typical silk I peaks in XRD curves appear at 11.7°, 19.5°, 24.6° and 28.7° while the silk II peaks appear at 9.2°, 18.9° and 20.7°, respectively. Compared to silk nanofiber hydrogels without treatments, the intensity of the peak at 19.5° increased in the water-annealed and 50% methanol-annealed hydrogels and decreased in 80% methanol-annealed hydrogels, accompanied by an opposite changes in the peak at 18.9°. At the same time, the peak at 24.6° (silk I structure) appeared in the water-annealed and 50% methanol-annealed hydrogels, but disappeared in the 80% methanol annealed hydrogels. All the results indicated that β-turn content increased in the water-annealed and 50% methanol-annealed hydrogels while β-sheet content increased in the 80% methanol annealed hydrogels, which was further confirmed by FSD results of the infrared spectra in the amide I region (1595–1705 cm−1) (Table 1). Therefore, after water-annealed and 50% methanol annealed treatments, random structures of the hydrogels mainly transformed into intermediate conformations while more silk II (β-sheet structure) formed after 80% methanol annealing. These results indicated that the secondary structures of the silk nanofiber hydrogels could be regulated through the different annealing processes.
Fig 1.
Micromorphologies of silk hydrogels with different annealing treatments: (a) and (b) SEM and AFM images of diluted silk hydrogels. The hydrogels were diluted to below 0.1% to avoid masking the original morphology with the multilayers of silk. (c) and (d) SEM images of the freeze-dried silk hydrogels. The images of freeze-dried hydrogels at high magnification indicated the existence of nanofiber structures. The different samples were as follows: (A) silk nanofiber hydrogel without treatment, SN-H; (B) Water-annealed silk nanofiber hydrogel, WA-SN-H; (C) 50% methanol annealed silk nanofiber hydrogel, MA50-SN-H; and (D) 80% methanol annealed silk nanofiber hydrogel, MA80-SN-H.
Fig 2.
FTIR spectra (a), XRD patterns (b), CD spectra (c) and elastic modulus G' (d) of different silk nanofiber hydrogels. The samples were as follows: SN-H, silk nanofiber hydrogel; WA-SN-H, water-annealed silk nanofiber hydrogel; MA50-SN-H, 50% methanol-annealed silk nanofiber hydrogel; MA80-SN-H, 80% methanol-annealed silk nanofiber hydrogel.
Table 1.
The structural conformations in silk derived from deconvoluted amide I FTIR spectra.
| Samples | Conformation content of silk fibroin
|
|||
|---|---|---|---|---|
| Random | β-Sheet | Silk I (Type II β-turn) | Bends and Turns | |
| SN-H | 18.84±1.30 | 49.08±0.81 | 7.86±0.65 | 22.91±2.16 |
| WA-SN-H | 7.80±0.62 | 50.69±1.08 | 15.88±0.53 | 22.57±1.89 |
| MA50-SN-H | 6.51±0.98 | 50.35±1.29 | 15.57±1.08 | 26.35±1.48 |
| MA80-SN-H | 6.01±0.77 | 58.41±1.32 | 11.47±0.62 | 22.27±1.34 |
3.2 Mechanical properties of silk nanofiber hydrogels
The mechanical properties were determined by dynamic oscillatory shear rheology. The storage moduli (elastic moduli) of silk nanofiber hydrogels were about 6 kPa (Fig. 2d), significantly higher than native central nerve tissues (0.1–1 kPa).43 The moduli of the hydrogels decreased to about 5 kPa after water annealing, and was further reduced to 1.27 kPa and 570 Pa (Fig. 2d), respectively, when methanol treatment was 50% and 80%, values approaching the stiffness of central nerve tissues. These results indicated that tunable mechanical properties of silk nanofiber hydrogels could be achieved through different annealing processes without the sacrifice of the topography of the nanofibrous structures.
3.3 Migration and proliferation of NSCs on silk nanofiber hydrogels with tunable stiffness
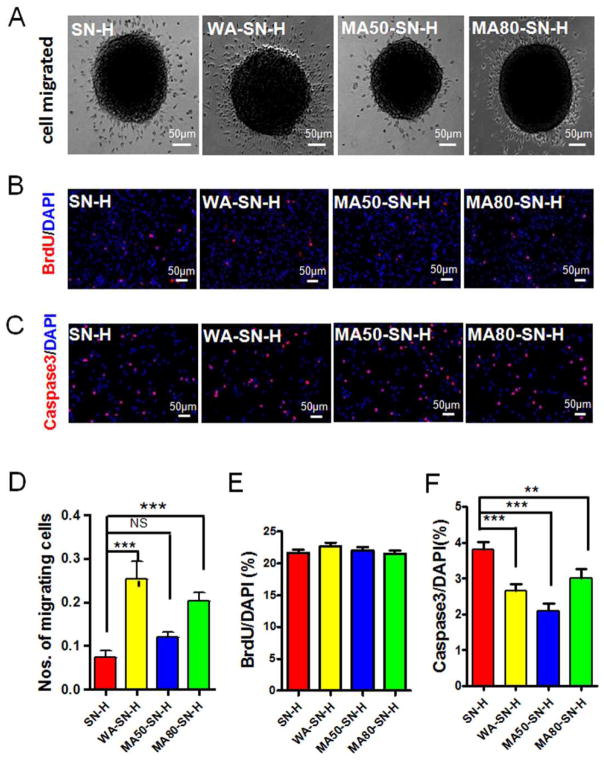
To investigate the effects of silk nanofiber hydrogels (SN-H) with different mechanical properties on the migration of NSCs, neurospheres derived from embryonic 14 day (E14) C57/BL6J mice were seeded onto the plates which were coated with SN-H, water annealed SN-H, 50% methanol annealed SN-H and 80% methanol annealed SN-H, respectively (Fig. 3A, Fig. S1). Upon omitting of basic fibroblast growth factor (bFGF) and epidermal growth factor (EGF) for 24 h, some cells migrated from the neurospheres and displayed immunoreactivity of Sex determining region Y-Box 2 (SOX2, a maker of NSC) by fluorescence staining (Fig. S2), indicating that the cells that migrated from the neurospheres were still NSCs. The numbers of cells that migrated from neurospheres were analyzed.56 Compared to untreated nanofiber hydrogels (SN-H), the neurospheres on the water annealed and 80% methanol annealed nanofibers (WA-SN-H and MA80-SN-H) had significantly more migrating cells (Fig. 3A and D). In contrast, no significant differences in the numbers of migrating cells were observed from the neurospheres on untreated and 50% methanol annealed nanofibers (SN-H and MA50-SN-H) (Fig. 3A and D). These results indicated that the mechanical properties of silk nanofibers, at least in part, modulated the migration of NSCs.
Fig 3.
Migration, proliferation and apoptosis of NSCs on silk nanofiber hydrogels (SN-H) with different mechanical properties. NSCs on silk nanofiber hydrogels with different mechanical properties were cultured in medium without bFGF and EGF for 24 h (A). NSCs on silk nanofiber hydrogels with different mechanical properties were incorporated BrdU for 4 h and stained for BrdU and DAPI (B). NSCs on silk nanofiber hydrogels with different mechanical properties stained for active Caspase3 and DAPI (C). The numbers of migrating cells from neurosphere were quantified and expressed as the percentage of total cells (D). The numbers of BrdU+ (E) and Caspase3+ cells (F) were quantified and expressed as the percentage of the number of DAPI+ cells. SN-H, silk nanofiber hydrogel; WA-SN-H, water-annealed silk nanofiber hydrogel; MA50-SN-H, 50% methanol-annealed silk nanofiber hydrogel; MA80-SN-H, 80% methanol-annealed silk nanofiber hydrogel. Scale bars: 50 μm. Results are presented as mean+SEM. **: p<0.01; ***: p<0.001; One-way Anova.
To investigate the effects of mechanical properties on the proliferation of NSCs, the cells were cultured in NSC culture medium containing bromodeoxyuridine (BrdU), an analog of thymidine, which incorporates into DNA during S-phase of cell cycle, on the different silk nanofibers for 4 h. The NSCs were stained for BrdU and 4',6-diamidino-2-phenylindole (DAPI), which is a fluorescent stain that binds to DNA and therefore can stain the nuclei (Fig. 3B, Fig. S3A). The proportion of cells with incorporated BrdU showed no differences among the NSCs cultured on the untreated and treated nanofibers (Fig. 3B and E, Fig. S3A). These results indicated that the mechanical properties of the silk nanofiber hydrogels did not impact the proliferation of NSCs. One concern with the transplantation of biomaterials is their effect on cell survival. The NSCs were cultured on different silk nanofibers and stained for active Caspase3, a marker of apoptosis, and DAPI. Compared to the untreated nanofibers, NSCs cultured on water annealed, 50% methanol annealed and 80% methanol annealed nanofibers displayed reduced numbers of Caspase3+ cells (Fig. 3C and F, Fig. S3B), indicating that these treatments prevented or delayed NSCs from apoptosis.
3.4 Differentiation of NSCs on silk nanofiber hydrogels with tunable stiffness
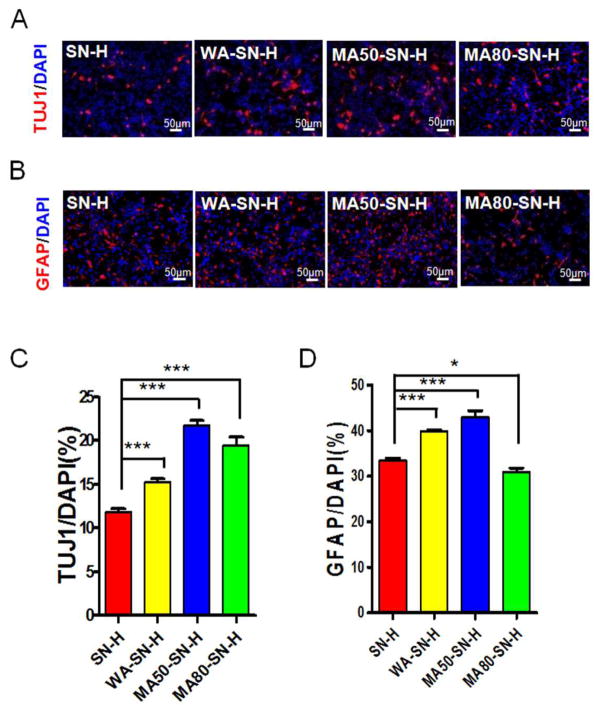
To investigate the effects of different mechanical properties of the silk nanofiber hydrogels on the differentiation of NSCs, the cells cultured on the different silk nanofiber hydrogels were induced to differentiate by withdrawal of bFGF and EGF. The cells were stained for either βIII-tubulin (TUJ1, a marker of neuron; Fig. 4A, Fig. S4A) or glial fibrillary acidic protein (GFAP, a marker of astrocyte; Fig. 4B, Fig. S4B) and DAPI. NSCs cultured on water annealed, 50% methanol annealed and 80% methanol annealed silk nanofiber hydrogels differentiated into more neurons compared to on the untreated silk nanofiber hydrogels, as indicated that the proportion of TUJ1+ cells increased in these three groups (Fig. 4A and C, Fig. S4A). More GFAP+ cells were differentiated from the NSCs on the water annealed and 50% methanol annealed hydrogels compared to on the untreated hydrogels (Fig. 4B and D, Fig. S4B). In contrast, the NSCs cultured on 80% methanol annealed hydrogels showed decreased numbers of GFAP+ cells compared to on the untreated hydrogels (Fig. 4B and D, Fig. S4B). These results indicated that water annealed or 50% methanol annealed hydrogels promoted NSCs to differentiate to astrocytes while 80% methanol annealed hydrogel inhibited NSCs to differentiation to astrocytes.
Fig 4.
Differentiation of NSCs on silk nanofiber hydrogels (SN-H) with different mechanical properties. NSCs seeded on silk nanofiber hydrogels with different mechanical properties were cultured for 3~5 days in vitro. The cells were stained for TUJ1 (A) or GFAP (B) and DAPI. The numbers of TUJ1+ (C) or GFAP+ (D) cells were quantified and expressed as the percentage of the number of DAPI+ cells. SN-H, silk nanofiber hydrogel; WA-SN-H, water-annealed silk nanofiber hydrogel; MA50-SN-H, 50% methanol-annealed silk nanofiber hydrogel; MA80-SN-H, 80% methanol-annealed silk nanofiber hydrogel. Scale bars: 50 μm. Results are presented as mean+SEM. *: p<0.05; **: p<0.01; ***: p<0.001; One-way Anova.
4. Discussion
Effectively controlling the differentiation of NSCs to specific nerve cells is a critical step for reconstruction of damaged neural circuitry. Recently, silk was utilized as a material platform to control nerve regeneration because of its neuro-biocompatibility and the curative effect of silk nerve conduits on peripheral-nerve repairs analogous to that seen with autografts.33–36 Different strategies encompassing physical approaches, architectural designs, bioactive molecule release, genetic engineering and synergism among these features have been evaluated to achieve control over cell fate.4,41–45,57,58
A first attempt to couple nanofiber fabrication and stiffness control of silk hydrogels was studied in the present work. A three-dimensional network of nanofibers formed by regulating the self-assembly of silk was pursued. Different to previous reported silk nanofibers with diamter of about 5 nm due to different self-assembly processes,59,60 the nanofibers in our study had high aspect ratios and high surface areas, were 10 to 20 nm in diameter and had lengths of few micrometers, similar to previous peptide amphiphile molecule nanofibers that induced rapid differentiation of cells into neurons.61,62 Higher beta-sheet content endowed silk nanofibers enough hydrophobicity, to generate hydrogels. Considering that the stiffness of silk-based materials with high beta-sheet structure is usually higher than nerve tissues, the mechanical properties of the nanofiber hydrogels were further regulated by different annealing processes to achieve suitable mechanical stimuli needed for neuronal regeneration. The annealing methods were previously used to induce the silk I or silk II formation of amorphous silk scaffolds and films.52,63 Methanol/water penetrated the silk aggregates to increase molecular motility and promoted the conformational transitions to beta sheet. In the present study, beta-turns slightly increased at the expense of random coil in water annealed and 50% methanol annealed scaffolds, and then decreased in 80% methanol annealed scaffolds because of beta-sheet formation, which caused a significant decrease of stiffness, approaching that of nerve tissues. The result seems contradictory, since silk-based materials with significant crystalline content usually stiffer. Based on our previous study, the degradation of random structures generally resulted in an increase of silk II content but a mechanical loss of the materials,54 since the random structures between the silk II crystals are digested. In contrast, the stiffness of the nanofiber hydrogels was significantly lower than previous silk hydrogels with similar silk II content, because the further aggregation of the nanofibers through hydrophilic interaction of random structures were restrained by negative charge repulsion during processing. These results implied that random structures of silk are critical to achieve higher stiffness and the decrease in stiffness of the treated hydrogels may be due to the loss of random structure. On the other hand, since the relationships between mechanical properties and structures in silk materials is not fully understood,64,65 further research is necessary to clarify why small changes in secondary conformations lead to the variations in stiffness of the silk hydrogels. Although the results seem surprising, the feasibility of tuning mechanical properties to mimic the elastic modulus of the extracellular environment of nerve tissues based on these self-assembled silk nanofiber hydrogel systems is a useful option for the studies performed here.
Electrospinning is a widely studied means of fabricating ECM-mimetic nanofibrous structures from different biomaterials including silk for neural tissue engineering.4 Further fabrication methods have been developed to achieve the control of dimensions, morphology and orientation of silk nanofibers, and the addition of growth factors for synergistic control of stem cell fate and functional repair of nerve tissues has also been reported.41–45 Many positive results based on in vitro and in vivo studies have demonstrated the suitability of electrospun silk nanofibers as support matrix for neuronal development. However, electrospun silk scaffolds have been mostly studied for the peripheral nerves, and the repair of central nerves remains a challenge partly because of the inability to fine-tune biomaterial stiffness to mimic the elasticity of these nerves. Although compelling evidence has confirmed the critical influence of scaffold stiffness on the differentiation of stem cells, the combination of control of rigidity and nanofiber topography for many synthetic and natural biomaterials remains challenging.4 In contrast, self-assembly is an effective way to form nanoscale features.61,62 Unfortunately, nanoscale structures as well as stiffness of self-assembled materials usually depend on the inherent nature of the materials, which make it difficult to regulate the modulus. Unlike other biomaterials, silk has tunable mechanical properties that are regulated by secondary conformations, hierarchical nanostructures and hydrophobic-hydrophilic interactions.44,52,63,66,67 Recently, silk hydrogels with stiffness similar to nerve tissues but without nanostructure fabrication were prepared through the ultrasonication of silk solution with different concentrations.44 We have also reported silk nanofibers with stiffness higher than nerve tissues that were also self-assembled from aqueous solution.51 Although silk nanofibers with tunable mechanical properties were not achieved, the properties of silk generated under these conditions suggested the potential to form nanofibers and then tune the stiffness, as realized in the present work.
The mechanical properties of silk nanfiber hydrogels modulated the migration and differentiation of NSCs, while it did not affect the proliferation of NSCs (Table 2). NSCs showed similar growth ability but significant different migration and differentiation behavior on the silk nanofiber hydrogels with tunable stiffness. The migration behavior was improved on the water-annealed nanofibers with a stiffness of about 5 kPa, significantly higher than that of nerve matrices. This finding is not unexpected, as in previous studies improved neurite extension and migration was found on electrospun silk nanofibers with high stiffness.34,39 Interestingly, significant promotion of cell migration also occurred on methanol-annealed nanofibers with stiffness levels similar to nerve matrices, implying a biphasic response of nerve cells to mechanical properties. A similar biphasic migration response for nerve cells was reported in many recent studies and also found for fibroblasts, endothelial cells, hepatocytes and myocytes.17,68,69 Although a biphasic migration response is universal, mechanical properties similar to specific tissues are still preferred, since mechanical matches to specific tissues is critical for cell behavior and for minimizing mechanical mismatches at host-implant interfaces.17 Therefore, silk nanofibers with stiffness similar to nerve matrices are a logical choice for a biomaterial system for nerve regeneration.
Table 2.
Summary of the migration, proliferation, differentiation of NSCs on silk nanofiber hydrogels (SN-H) with different mechanical properties.
| SN-H | WA-SN-H | MA50-SN-H | MA80-SN-H | |
|---|---|---|---|---|
| Cell migration | — | ↑↑↑ | — | ↑↑↑ |
| BrdU / DAPI (%) | — | — | — | — |
| TUJ1 / DAPI (%) | — | ↑↑↑ | ↑↑↑ | ↑↑↑ |
| GFAP / DAPI (%) | — | ↑↑↑ | ↑↑↑ | ↓ |
| Caspase3 / DAPI (%) | — | ↓↓↓ | ↓↓↓ | ↓↓ |
↑: *; —: NS
The regulation of differentiation of neural stem/precursor cells is a key concern for treating neural diseases or injuries. A useful strategy is to specifically promote differentiation of these progenitors into neurons and to inhibit glial differentiation since increased astrocytes exacerbate neuroinflammation and neurodegeneration in the brains of Alzheimer’s diease patients or in the injuried spinal cord.70,71 Many investigators are concentrating on designing systems to control these outcomes using a combination of biomaterials, different growth factors and ECM proteins in which growth factors generally are considered indispensable.17,20–23,62 However, the sensitivity of growth factors and the complexity of these systems make it difficult to generate reliable microenvironments for nerve tissue regeneration. In the present study, by mimicking both the nanostructure and modulus of the extracellular matrix of the nerve tissues, differentiation into neurons and not to glial cells was achieved with these silk nanofiber hydrogel systems (MA80-SN-H) without the addition of growth factors (Table 2). Following stiffness modulation after the different annealing processes, charge distribution on the nanofiber hydrogels was also changed, where zeta potential decreased from −48.3 mV to −44.8 mV after water annealing and was further decreased to −35.5 mV and −33.9 mV after 50% methanol annealing and 80% methanol annealing, respectively. This change of charge distribution may also influence cell behavior, which should be further investigated. Regardless, the mechanical properties of the silk nanofiber hydrogels, in part, regulated the differentiation of NSCs. Although the differentiation rate in our system is still below what can be achieved using growth factors, considering the ease of preparation of silk nanofiber hydrogels with tunable stiffness, this approach enables a reliable biomaterial system to establish a microenvironment for nerve tissue regeneration. Further modification to the system including hierarchical microstructure design and specific growth factor delivery to expand utility for nerve tissues as well as other tissue systems is a logical extension of the studies reported here.
5. Conclusions
Self-assembling silk nanofibers with tunable rigidity were prepared with different annealing processes to simulate nanostructure and mechanical properties of the extracellular matrix of central nerve tissues. Nerve stem cells grown on these nanofibers expressed preferred neuron differentiation and inhibition of glial differentiation without the addition of growth factors. The present results demonstrate the possibility of generating growth factor-free central nerve regeneration systems with the combination of topographical and mechanical simulation in silk-based biommaterials.
Supplementary Material
Acknowledgments
We thank grants from National Basic Research Program of China (973 Program 2013CB934400, 2012CB22302), and NSFC (21174097, 81272106) for support of this work. We also thank grants from the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD), the Excellent Youth Foundation of Jiangsu Province (BK2012009), the Youth Foundation of Jiangsu Province (BK20140397), the NIH (P41 EB002520), and the Key Natural Science Foundation of the Jiangsu Higher Education Institutions of China (11KGA430002) for support of this work. We thank grants to Quan-hong Ma from National Programmm on Key Basic Research Project (2013CB945602), National Natural Science Foundation of China (31171313, 81271424), Research Fund for the Doctoral Program of Higher Education of China (20113201120018) and Soochow University Start up foundation (Q421500110).
References
- 1.Lampe KJ, Antaris AL, Heilshorn SC. Acta Biomater. 2013;9:5590–5599. doi: 10.1016/j.actbio.2012.10.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Leipzig ND, Wylie RG, Kim H, Shoichet MS. Biomaterials. 2011;32:57–64. doi: 10.1016/j.biomaterials.2010.09.031. [DOI] [PubMed] [Google Scholar]
- 3.Li YC, Tsai LK, Wang JH, Young TH. Biomaterials. 2014;35:1192–1204. doi: 10.1016/j.biomaterials.2013.10.066. [DOI] [PubMed] [Google Scholar]
- 4.Purcell EK, Naim Y, Yang A, Leach MK, Velkey JM, Duncan RK, Corey JM. Biomacromolecules. 2012;13:3427–3438. doi: 10.1021/bm301220k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xie H, Li J, Li L, Dong Y, Chen GQ, Chen KC. Acta Biomater. 2013;9:7845–7854. doi: 10.1016/j.actbio.2013.04.038. [DOI] [PubMed] [Google Scholar]
- 6.Guan J, Zhu ZH, Zhao RC, Xiao ZF, Wu CX, Han Q, Chen L, Tong W, Zhang J, Han Q, Gao J, Feng M, Bao X, Dai JW, Wang R. Biomaterials. 2013;34:5937–5946. doi: 10.1016/j.biomaterials.2013.04.047. [DOI] [PubMed] [Google Scholar]
- 7.Meamar R, Nasr-Esfahani MH, Mousavi SA, Basiri K. J Clin Neurosci. 2013;20:1659–1663. doi: 10.1016/j.jocn.2013.04.024. [DOI] [PubMed] [Google Scholar]
- 8.Gage FH, Temple S. Neuron. 2013;80:588–601. doi: 10.1016/j.neuron.2013.10.037. [DOI] [PubMed] [Google Scholar]
- 9.Sandoe J, Eggan K. Nat Neurosci. 2013;16:780–789. doi: 10.1038/nn.3425. [DOI] [PubMed] [Google Scholar]
- 10.Neirinckx V, Marquet A, Coste C, Rogister B, Wislet-Gendebien S. Plos One. 2013;8:e64723. doi: 10.1371/journal.pone.0064723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yoo J, Kim HS, Hwang DY. J Cell Biochem. 2013;114:743–753. doi: 10.1002/jcb.24427. [DOI] [PubMed] [Google Scholar]
- 12.Joddar B, Ito Y. J Biotechnol. 2013;168:218–228. doi: 10.1016/j.jbiotec.2013.04.021. [DOI] [PubMed] [Google Scholar]
- 13.Prewitz MC, Seib FP, Bonin MV, Friedrichs J, Stibel A, Niehage C, Muller K, Anastassiadis K, Waskow C, Hoflack B, Bornhauser M, Werner C. Nat Methods. 2013;10:788–794. doi: 10.1038/nmeth.2523. [DOI] [PubMed] [Google Scholar]
- 14.Eng G, Lee BW, Parsa H, Chin CD, Schneider J, Linkov G, Sia SK, Vanjak-Novakovic G. Proc Natl Acad Sci USA. 2013;110:4551–4556. doi: 10.1073/pnas.1300569110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Giobbe GG, Zagallo M, Riello M, Serena E, Mari G, Barzon L, Camillo BD, Elvassore N. Biotechnol Bioeng. 2012;109:3119–3132. doi: 10.1002/bit.24571. [DOI] [PubMed] [Google Scholar]
- 16.Lee J, Li M, Milwid J, Dunham J, Vinegoni C, Gorbatov R, Iwamoto Y, Wang F, Shen K, Hatfield K, Enger M, Shafiee S, McCormack E, Ebert BL, Weissleder R, Yarmush ML, Parekkadan B. Proc Natl Acad Sci USA. 2012;109:19638–19643. doi: 10.1073/pnas.1208384109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stabenfeldt SE, Laplaca MC. Acta Biomater. 2011;7:4102–4108. doi: 10.1016/j.actbio.2011.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Georgiou M, Bunting SCJ, Davies HA, Loughlin AJ, Golding JP, Phillips JB. Biomaterials. 2013;34:7335–7343. doi: 10.1016/j.biomaterials.2013.06.025. [DOI] [PubMed] [Google Scholar]
- 19.Mawad D, Stewart E, Officer DL, Romeo T, Wagner P, Wagner K, Wallace GG. Adv Funct Mater. 2012;22:2692–2699. [Google Scholar]
- 20.Volpato FZ, Fuhrmann T, Migliaresi C, Hutmacher DW, Dalton PD. Biomaterials. 2013;34:4945–4955. doi: 10.1016/j.biomaterials.2013.03.057. [DOI] [PubMed] [Google Scholar]
- 21.Li YC, Lin YC, Young TH. Acta Biomater. 2012;8:3035–3048. doi: 10.1016/j.actbio.2012.04.036. [DOI] [PubMed] [Google Scholar]
- 22.Wei GJ, Yao M, Wang YS, Zhou CW, Wan DY, Lei PZ, Wen J, Lei HW, Dong DM. Int J Nanomed. 2013;8:3217–3225. doi: 10.2147/IJN.S43681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Callahan LAS, Xie S, Barker IA, Zheng J, Reneker DH, Dove AP, Becker ML. Biomaterials. 2013;34:9089–9095. doi: 10.1016/j.biomaterials.2013.08.028. [DOI] [PubMed] [Google Scholar]
- 24.Omenetto FG, Kaplan DL. Science. 2010;329:528–531. doi: 10.1126/science.1188936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shao ZZ, Vollrath F. Nature. 2002;418:741–741. doi: 10.1038/418741a. [DOI] [PubMed] [Google Scholar]
- 26.Vollrath F, Knight DP. Nature. 2001;410:541–548. doi: 10.1038/35069000. [DOI] [PubMed] [Google Scholar]
- 27.Kasoju N, Bora U. Adv Healthcare Mater. 2012;1:393–412. doi: 10.1002/adhm.201200097. [DOI] [PubMed] [Google Scholar]
- 28.Kundu B, Rajkhowa R, Kundu SC, Wang X. Adv Drug Deliv Rev. 2013;65:457–470. doi: 10.1016/j.addr.2012.09.043. [DOI] [PubMed] [Google Scholar]
- 29.Altman GH, Diaz F, Jakuba C, Calabro T, Horan RL, Chen JS, Lu H, Richmond J, Kaplan DL. Biomaterials. 2003;24:401–416. doi: 10.1016/s0142-9612(02)00353-8. [DOI] [PubMed] [Google Scholar]
- 30.Altman GH, Horan RL, Lu HH, Moreau J, Martin I, Richmond JC, Kaplan DL. Biomaterials. 2002;23:4131–4141. doi: 10.1016/s0142-9612(02)00156-4. [DOI] [PubMed] [Google Scholar]
- 31.Mauney JR, Volloch V, Kaplan DL. Tissue Eng. 2005;11:787–802. doi: 10.1089/ten.2005.11.787. [DOI] [PubMed] [Google Scholar]
- 32.Meinel L, Fajardo R, Hofmann S, Langer R, Chen J, Snyder B, Vunjak-Novakovic G, Kaplan DL. Bone. 2005;37:688–698. doi: 10.1016/j.bone.2005.06.010. [DOI] [PubMed] [Google Scholar]
- 33.Tang X, Ding F, Yang Y, Hu N, Wu H, Gu X. J Biomed Mater Res A. 2009;91:166–174. doi: 10.1002/jbm.a.32212. [DOI] [PubMed] [Google Scholar]
- 34.Yang Y, Ding F, Wu J, Hu W, Hui W, Gu X. Biomaterials. 2007;28:5526–5535. doi: 10.1016/j.biomaterials.2007.09.001. [DOI] [PubMed] [Google Scholar]
- 35.Madduri S, Papaloizos M, Gander B. Biomaterials. 2010;31:2323–2334. doi: 10.1016/j.biomaterials.2009.11.073. [DOI] [PubMed] [Google Scholar]
- 36.Zhao Y, Zhao W, Yu S, Guo Y, Gu X, Yang Y. Biomed Mater Eng. 2013;23:545–554. doi: 10.3233/BME-130775. [DOI] [PubMed] [Google Scholar]
- 37.Zhang XH, Reagan MR, Kaplan DL. Adv Drug Deliver Rev. 2009;61:988–1006. doi: 10.1016/j.addr.2009.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jin HJ, Fridrikh SV, Rutledge GC, Kaplan DL. Biomacromolecules. 2002;3:1233–1239. doi: 10.1021/bm025581u. [DOI] [PubMed] [Google Scholar]
- 39.Qu J, Wang D, Wang H, Dong Y, Zhang F, Zuo B, Zhang H. J Biomed Mater Res A. 2013;101:2667–2678. doi: 10.1002/jbm.a.34551. [DOI] [PubMed] [Google Scholar]
- 40.Huang W, Begum R, Barber T, Ibba V, Tee NCH, Hussain M, Arastoo M, Yang Q, Robson LG, Lesage S, Gheysens T, Skaer NJV, Knight DP, Priestley JV. Biomaterials. 2012;33:59–71. doi: 10.1016/j.biomaterials.2011.09.030. [DOI] [PubMed] [Google Scholar]
- 41.Wittmer CR, Claudepierre T, Reber M, Wiedemann P, Garlick JA, Kaplan D, Egles C. Adv Funct Mater. 2011;21:4232–4242. doi: 10.1002/adfm.201190103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Madduri S, Papaloizos M, Gander B. Biomaterials. 2010;31:2323–2334. doi: 10.1016/j.biomaterials.2009.11.073. [DOI] [PubMed] [Google Scholar]
- 43.Engler AJ, Sen S, Sweeney HL, Discher DE. Cell. 2006;126:677–689. doi: 10.1016/j.cell.2006.06.044. [DOI] [PubMed] [Google Scholar]
- 44.Hopkins AM, Laporte LD, Tortelli F, Spedden E, Staii C, Atherton TJ, Hubbell JA, Kaplan DL. Adv Funct Mater. 2013;23:5140–5149. [Google Scholar]
- 45.Hu X, Tang-Schomer MD, Huang W, Xia X, Weiss AS, Kaplan DL. Adv Funct Mater. 2013;23:3875–3884. doi: 10.1002/adfm.201202685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bai SM, Liu SS, Zhang CC, Xu WA, Lu Q, Han HY, Kaplan DL, Zhu HS. Acta Biomater. 2013;9:7806–7813. doi: 10.1016/j.actbio.2013.04.033. [DOI] [PubMed] [Google Scholar]
- 47.Numata K, Yamazaki S, Katashima T, Chuah JA, Naga N, Sakai T. Macromol Biosci. 2014;14:799–806. doi: 10.1002/mabi.201300482. [DOI] [PubMed] [Google Scholar]
- 48.Han FF, Liu SS, Liu X, Pei YZ, Bai SM, Zhao HJ, Lu Q, Ma FG, Kaplan DL, Zhu HS. Acta Biomater. 2014;10:921–930. doi: 10.1016/j.actbio.2013.09.026. [DOI] [PubMed] [Google Scholar]
- 49.Ma QH, Futagawa T, Yang WL, Jiang XD, Zeng L, Takeda Y, Xu RX, Bagnard D, Schachner M, Furley AJ, Karagogeos D, Watanabe K, Dawe GS, Xiao ZC. Nat Cell Biol. 2008;10:283–294. doi: 10.1038/ncb1690. [DOI] [PubMed] [Google Scholar]
- 50.Lu GZ, Liu SS, Lin SS, Kaplan DL, Lu Q. Colloid Surface B. 2014;120:28–37. doi: 10.1016/j.colsurfb.2014.03.027. [DOI] [PubMed] [Google Scholar]
- 51.Lu Q, Zhu HS, Zhang CC, Zhang F, Zhang B, Kaplan DL. Biomacromolecules. 2012;13:826–832. doi: 10.1021/bm201731e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jin HJ, Park J, Karageorgiou V, Kim UJ, Valluzzi R, Cebe P, Kaplan DL. Adv Funct Mater. 2005;15:1241–1247. [Google Scholar]
- 53.Lu SZ, Wang XQ, Lu Q, Zhang XH, Kluge JA, Uppal N, Omenetto F, Kaplan DL. Biomacromolecules. 2010;11:143–150. doi: 10.1021/bm900993n. [DOI] [PubMed] [Google Scholar]
- 54.Lu Q, Zhang B, Li M, Zuo B, Kaplan DL, Huang Y, Zhu HS. Biomacromolecules. 2011;12:1080–1086. doi: 10.1021/bm101422j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lu Q, Wang XL, Lu SZ, Li MZ, Kaplan DL, Zhu HS. Biomaterials. 2011;32:1059–1067. doi: 10.1016/j.biomaterials.2010.09.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kong H, Fan Y, Xie J, Ding JH, Sha LL, Shi XR, Sun XL, Hu G. J Cell Sci. 2008;121:4029–4036. doi: 10.1242/jcs.035758. [DOI] [PubMed] [Google Scholar]
- 57.Hronik-Tupaj M, Raja WK, Tang-Schomer M, Omenetto FG, Kaplan DL. J Biomed Mater Res A. 2013;101:2559–2572. doi: 10.1002/jbm.a.34565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Benfenati V, Stahl K, Gomis-Perez C, Toffanin S, Sagnella A, Torp R, Kaplan DL, Ruani G, Omenetto FG, Zamboni R, Muccini M. Adv Funct Mater. 2012;22:1871–1884. doi: 10.5301/JABFM.2012.10448. [DOI] [PubMed] [Google Scholar]
- 59.Gong ZG, Yang YH, Huang L, Chen X, Shao ZZ. Soft Matter. 2010;6:1217–1223. [Google Scholar]
- 60.Gong ZG, Huang L, Yang YH, Chen X, Shao ZZ. Chem Commun. 2009;0:7506–7508. doi: 10.1039/b914218e. [DOI] [PubMed] [Google Scholar]
- 61.Silva GA, Czeisler C, Niece KL, Beniash E, Harrington DA, Kessler JA, Stupp SL. Science. 2004;303:1352–1355. doi: 10.1126/science.1093783. [DOI] [PubMed] [Google Scholar]
- 62.Tysseling-Mattiace VM, Sahni V, Niece KL, Birch D, Czeisler C, Fehlings MG, Stupp SI, Kessler JA. J Neurosci. 2008;28:3814–3823. doi: 10.1523/JNEUROSCI.0143-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hu X, Shmelev K, Sun L, Gil ES, Park SH, Cebe P, Kaplan DL. Biomacromolecules. 2011;12:1686–1696. doi: 10.1021/bm200062a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Keten S, Xu ZP, Ihle B, Buehler MJ. Nat Mater. 2010;9:359–367. doi: 10.1038/nmat2704. [DOI] [PubMed] [Google Scholar]
- 65.Giesa T, Arslan M, Pugno NM, Buehler MJ. Nano Lett. 2011;11:5038–5046. doi: 10.1021/nl203108t. [DOI] [PubMed] [Google Scholar]
- 66.Yao DY, Dong S, Lu Q, Hu X, Kaplan DL, Zhang B, Zhu HS. Biomacromolecules. 2012;13:3723–3729. doi: 10.1021/bm301197h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lu Q, Hu X, Wang XQ, Kluge JA, Lu SZ, Cebe P, Kaplan DL. Acta Biomater. 2010;6:1380–1387. doi: 10.1016/j.actbio.2009.10.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Guo WH, Frey MT, Burnham NA, Wang YL. Biophys J. 2006;90:2213–2220. doi: 10.1529/biophysj.105.070144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Engler AJ, Griffin MA, Sen S, Bonnemann CG, Sweeney HL, Discher DE. J Cell Biol. 2004;166:877–887. doi: 10.1083/jcb.200405004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hensley K. J Alzheimers Dis. 2010;21:1–14. doi: 10.3233/JAD-2010-1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Shibuya S, Yamamoto T, Itano T. Cell Adhes Migr. 2009;3:1–8. doi: 10.4161/cam.3.1.7372. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.