Abstract
Discoveries on the toxic effects of cysteine accumulation and, particularly, recent findings on the many physiological roles of one of the products of cysteine catabolism, hydrogen sulfide (H2S), are highlighting the importance of this amino acid and sulfur metabolism in a range of cellular activities. It is also highlighting how little we know about this critical part of cellular metabolism. In the work described here, a genome-wide screen using a deletion collection of Saccharomyces cerevisiae revealed a surprising set of genes associated with this process. In addition, the yeast vacuole, not previously associated with cysteine catabolism, emerged as an important compartment for cysteine degradation. Most prominent among the vacuole-related mutants were those involved in vacuole acidification; we identified each of the eight subunits of a vacuole acidification sub-complex (V1 of the yeast V-ATPase) as essential for cysteine degradation. Other functions identified included translation, RNA processing, folate-derived one-carbon metabolism, and mitochondrial iron-sulfur homeostasis. This work identified for the first time cellular factors affecting the fundamental process of cysteine catabolism. Results obtained significantly contribute to the understanding of this process and may provide insight into the underlying cause of cysteine accumulation and H2S generation in eukaryotes.
Introduction
The concentration of free intracellular cysteine is tightly regulated in eukaryotic and prokaryotic cells, serving two opposing homeostatic requirements. Cysteine concentration must be sufficient for synthesis of proteins and other essential sulfur containing molecules such as glutathione (GSH) and coenzyme A, while on the other hand cysteine concentration must be kept below the threshold of cytotoxicity [1]. The toxicity of excess cysteine was demonstrated in yeasts [2], [3] and animal models [4], leading to growth inhibition [3], endoplasmic reticulum (ER) stress [2] and is associated with various diseases including Parkinson's and Alzheimer's [5]. A key product of cysteine catabolism is hydrogen sulfide (H2S) [1], [6], [7], increasingly recognised to be a powerful regulator of many aspects of cell physiology (Reviewed in: [8], [9]–[12]). Still, despite the increasing interest in H2S generated from cysteine, fundamental questions regarding regulation of cysteine homeostasis remain to be answered.
H2S generation is evolutionarily conserved in all three kingdoms of life [7]. In mammals, endogenous H2S is presumed to be generated through the catabolism of cysteine by two cytosolic enzymes cystathionine β-lyase (CBS) and cystathionine γ-lyase (CSE) [1], [6], [9], [13]–[15]. Regulation of expression and activity of these enzymes remain to be elucidated. Consistent with the evolutionary conservation of cysteine catabolism, mammalian CBS- and CSE-encoding genes are similar to those of yeast and bacteria [16]–[19]. Predicted yeast and human CBS proteins share 72% similarity and these have significant similarity to the predicted rat CBS protein and bacterial cysteine synthase [16]. Expression of the gene encoding a human CBS in yeast was able to recover cysteine-auxotrophy caused by deletion of the yeast native CBS [16]. The predicted yeast CSE product was also found to be closely related to rat and E. coli CSE [19]. These similarities reinforce the utility of S. cerevisiae as a model eukaryotic system to explore cellular homeostasis of cysteine and the catabolism of cysteine to release H2S. In this paper we describe a genome-wide survey using a haploid EUROSCARF S. cerevisiae gene deletion library to shed light on the cellular processes influencing cysteine catabolism.
Materials and Methods
Reagents, yeast strains and media
Chemical reagents were obtained from Sigma-Aldrich. The S. cerevisiae strains used in this study include BY4742 (MATα/his3Δ1/leu2Δ0/lys2Δ0/ura3Δ0) [20] and the gene deletion collection in the same genetic background, purchased from EUROSCARF (Frankfurt, Germany). Precultures were grown in YPD medium to be subsequently inoculated into microtiter plates or shake flasks filled with chemically defined medium, based on Winter et al. [21] with the following modifications: 100 g/L sugars (50 g/L glucose, 50 g/L fructose), addition of auxotrophic requirements (20 mg/L uracil, 20 mg/L histidine, 60 mg/L leucine and 30 mg/L lysine), and exclusion of cysteine from the media. Where specified cysteine was supplemented at 500 mg/L and adenine supplementation was 20 mg/L.
High-throughput H2S screening
Screening was performed using a novel high-throughput method that estimated H2S excreted in the growth medium during fermentation and allows generation of H2S production profiles [22]. For follow-up experiments, H2S released during fermentation was detected in the shake-flask headspace using silver nitrate selective gas detector tubes (Komyo Kitagawa, Japan), as described in [23].
Genome-wide screening for genes involved in cysteine catabolism to H2S
Cultures of strains in the S. cerevisiae BY4742 EUROSCARF deletion collection were pre-grown to stationary phase, each in 200 µl YPD with agitation (150 rpm) in a microtiter plate. The preculture was inoculated at 20 µl into 180 µl of medium supplemented with cysteine and 10% (v/v) of either H2S detection mix [22] including 100 mM citric acid and 5 mg/ml of methylene blue, hereafter referred to as MB samples or water, referred to as control samples. Each plate included a well with the wild-type strain (BY4742) as a control in addition to un-inoculated wells to account for medium contamination and non-enzymatic degradation of cysteine to H2S. Cultures were inoculated into a microtiter plate in four equal blocks (wells A1 to D6, A7 to D12, E1 to H6 and E7 to H12), representing four replicates of each strain. Following inoculation, microtiter plates were covered with Breath-Easy membranes (Astral Scientific, Australia) and were incubated at 28°C. Optical density measurements at 600 and 663 nm wavelength were carried out at intervals of three hours, for 36 hours. Inoculation and assay monitoring were handled robotically (Freedom EVO 150, Tecan, Männedorf, Switzerland; Cytomat Automated Incubator, ThermoFisher Scientific, Massachusetts, USA). Where specified, repeat experiments were conducted manually.
Data analysis was carried out using a custom script written in R [24], that constructs growth and H2S production curves for each strain as well as performing statistical analysis of replicates. Strains were assessed for ability to produce H2S via cysteine catabolism, based on the amount of H2S detected using the MBR method for MB samples and their ability to form biomass relative to the wild-type strain as measured in the control samples. A calculation of the minima value of the MB decolourisation curve minus the maxima value of the growth curve was used to classify strains as ‘low’ or ‘high’ H2S producers. Strains with scores greater than 1 and lower than 2.5 were classified as ‘low’, while strains with a score smaller than one were defined as ‘high’. As the initial OD of the MB supplemented culture was 2.5–2.7, scores above 2.5 were due to growth defects. Classification was then verified against the control wild type strain in each plate. Strains classified as either low or high producers were verified from follow-up micro-fermentation analysis. Cellular processes identified as important for cysteine catabolism to H2S were evaluated for statistical significance (P-value <0.01) with the GO Term Finder program of the Saccharomyces Genome Database (http://www.yeastgenome.org/cgi-bin/GO/goTermFinder.pl).
Generation of respiratory deficient petite cells
Cultures of AWRI1522 (Δhis3, Δleu2, Δtrp1, Δlys2 mat α) were grown in YPD with shaking (100 rpm) at 25°C. Mitochondrial mutants (AWRI1520, mat α, Δhis3, Δleu2, Δtrp1, Δlys2, rho –ve) were isolated by treating cells for 8 hours in synthetic complete medium (0.17% (wt/vol) yeast nitrogen base without amino acids, 0.5% (wt/vol) ammonium sulfate, 2% (wt/vol) glucose) containing 10 µg/ml ethidium bromide. Cells were then diluted in water and, due to their inability to perform aerobic metabolism and utilise glycerol as a carbohydrate source, mitochondrial mutants were revealed by petite colony growth on YPDG (1% w/v yeast extract, 2% w/v peptone, 3% w/v glycerol and 0.1% w/v glucose) [25]. Loss of respiratory function in these petites was then confirmed by their inability to grow on YPG media (1% w/v yeast extract, 2% w/v peptone, 3% w/v glycerol).
In-vivo V-ATPase downregulation
Cells were grown at 28°C with agitation (150 rpm) in chemically defined medium supplemented with cysteine until mid-log phase (OD 0.5). While producing H2S, cells were centrifuged and washed with distilled water prior to transfer into chemically defined medium containing 0.112 M glucose or 0.112 M galactose as carbon source, with or without cysteine supplementation. Cells were then incubated with shaking (150 rpm) at 28°C. Following 15 hours of incubation glucose was added to a final concentration of 0.112 M in medium previously containing galactose as a sole carbon source. Cysteine-generated H2S was measured throughout the assay using H2S detection tubes. Biomass formation was monitored by measuring the absorbance at 600 nm wavelength.
Results
Screen background and methodology
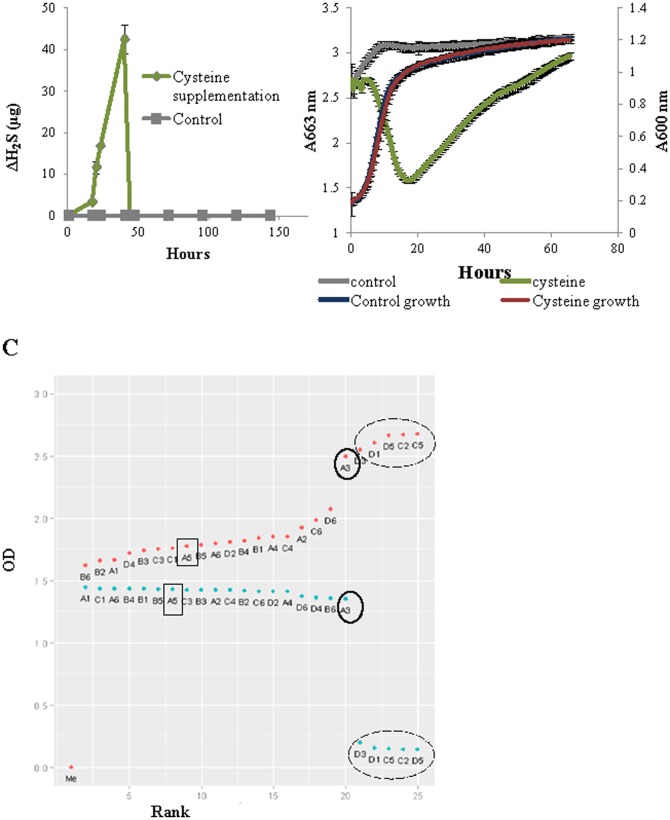
While developing a novel high-throughput assay for H2S detection [22], we noted that S. cerevisiae laboratory strain BY4742 did not produce detectable concentrations of H2S when grown in a chemically defined medium. In the current work, after supplementation of medium with cysteine, we detected high concentrations of H2S, suggesting this H2S was generated by BY4742 solely from cysteine catabolism (Fig. 1). Fig. 1b shows the discolouration of H2S detection-dye when cysteine was added to the medium, indicative of H2S formation. Notably, H2S formation occurred during yeast logarithmic phase of growth and cysteine addition did not affect biomass formation (Fig. 1b).
Figure 1. Cysteine catabolism to release H2S.
Screening process A H2S production profile generated during a shake flask (200 ml) growth of strain BY4742 with or without cysteine supplementation to the medium. H2S was measured using lead acetate detection tubes. Error bars represent standard deviation of triplicate experiments. B H2S production profile and growth curves generated during micro-scale (200 µl) growth of strain BY4742. H2S formation was measured through the degradation of methylene blue measured at 663 nm and biomass formation measured at 600 nm with and without cysteine. Experiments were carried out in a microtiter plate. Error bars represent the standard deviation of quadruplicate experiments. C Illustrative graph of cysteine-generated H2S screen analysis. Graph summarises two replicative microtiter plates assays carried out using cysteine-supplemented media. One measured for growth at 600 nm (blue circles) and the second measured for H2S detection dye degradation at 663 nm, as an indication for H2S formation (red circles). Each plate was inoculated in quadruplicates and data here represents the average of each replicate, labelled with the first replicate well position. Yeasts were ranked based on their methylene blue decolouration measurements (low value indicates dye discolouration and H2S formation). An example of a mutant strain that did not produce H2S while being able to grow under the experimental conditions is circled. Data highlighted in a dashed circle represents either empty wells or strains that were not able to grow. Each plate contained quadruplicates of the background strain (highlighted in rectangle).
Because detectable H2S in the cysteine supplemented culture of BY4742 is derived from cysteine catabolism, it provides a useful marker to identify genetic factors influencing this cellular process. It was therefore used in a genome-wide screen of S. cerevisiae deletion strains [26] to identify cellular processes involved in cysteine catabolism and H2S formation. H2S production was normalised against biomass formation by plotting the minima for each strain in the assay plate, supplemented with H2S detection dye (indicative of maximum H2S production) against the maxima for each strain in the growth plate (indicative of maximum biomass formation) (Fig. 1c). Deletants able to grow under the experimental conditions were classified as low or high producers of H2S from cysteine based upon their H2S minima value in the assay plate, in comparison to the wild-type strain. Fig. 1c shows an example of a strain classified as low producer of H2S from cysteine (see well A3, circled).
Genome-wide classification of mutants that lead to low or high H2S production from cysteine
A total of 226 deletion strains displayed differences in their H2S accumulation profiles compared to wild type. These were classified as low (188 strains) and high (38 strains) producers of H2S from cysteine. Strains were grouped according to gene ontology of the encoded gene product, as defined in the Saccharomyces Genome Database (www.yeastgenome.org), using Gorilla [27]. Most deletions that impacted on H2S production were for genes from one or more of nine functional groups (Table 1, a comprehensive list is available at S1 Supporting Information). Interestingly, in most cases genes within a group displayed the same trend with respect to H2S production (where deletions led to either low or high H2S production), indicative of the specificity of the cellular processes involved in catabolism of cysteine to H2S. Functional groups of deletions causing high H2S production included mitochondrial iron-sulfur homeostasis, mitochondrial translation, and cellular response to ionic iron. Deletions causing low H2S production were associated with purine base metabolism process, cellular aromatic compound metabolic process, vacuole acidification, vesicle trafficking to the vacuole, and translation and RNA processing functional groups. Selected deletants from the main functional groups implicated in this screen were further verified using H2S detection tubes for an independent H2S measurement (S2 Supporting Information). All deletants identified as low H2S producers produced significantly lower amounts of H2S in comparison to the wild type. For the high producers – H2S production was significantly increased during growth of three deletants, one deletants did not show significant difference in H2S production in comparison to the wild type and an additional deletants produced significantly lower amounts of H2S compared to the wild type. The method's limitation in identification of high H2S producers is discussed below. Still an 80% successful validation rate (100% for the low producers) as well as the multiple positive confirmations for major processes implicated in screen reinforces the findings of this work.
Table 1. Cellular processes influencing cysteine catabolism identified by H2S production phenotype.
| Functional group/cellular process | No. of genes | H2S production phenotype* |
| Vacuole related | 37 | Low |
| Purine base metabolic process | 19 | Low |
| Cellular aromatic compound metabolic process | 11 | Low |
| Translation | 32 | Low |
| RNA Processing | 22 | Low |
| Mitochondria related | 28 | High/Low |
| Mitochondrion translation | 9 | High |
| Cellular response to iron ion | 2 | High |
| Unknown function | 22 | High/Low |
| Miscellaneous | 64 | High/Low |
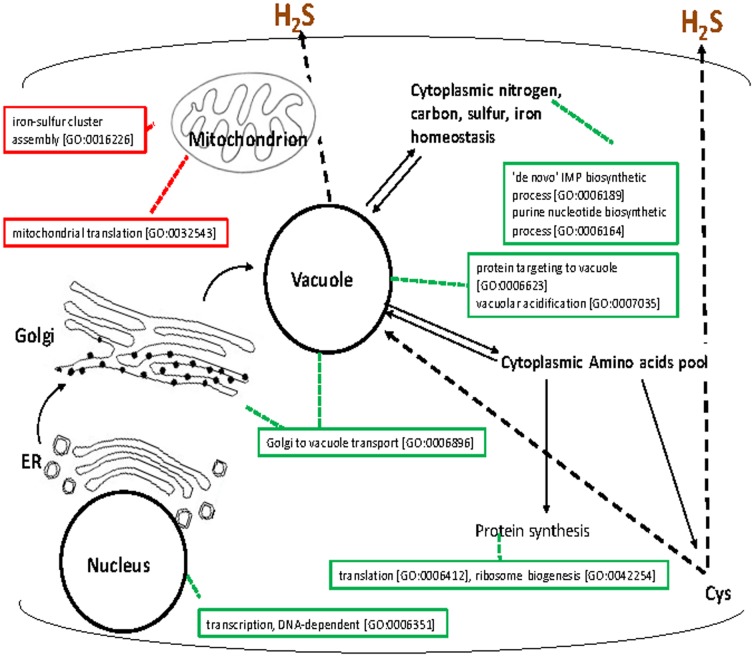
Results of this screen provide insight into metabolic and regulatory networks that influence formation of H2S from cysteine. A schematic representation of the data is provided in Fig. 2.
Figure 2. Overview of the cellular processes influencing cysteine catabolism to release H2S in S. cerevisiae.
The figure depicts cysteine catabolism cellular pathway with known and putative transport routes that may mediate the cysteine flux. Boxed annotations indicate some of the cellular processes identified as important for cysteine catabolism to H2S and evaluated for statistical significance (P-value <0.01) with the GO Term Finder program of the Saccharomyces Genome Database (http://www.yeastgenome.org/cgi-bin/GO/goTermFinder.pl).
Role of the mitochondrion in catabolism of cysteine to release H2S
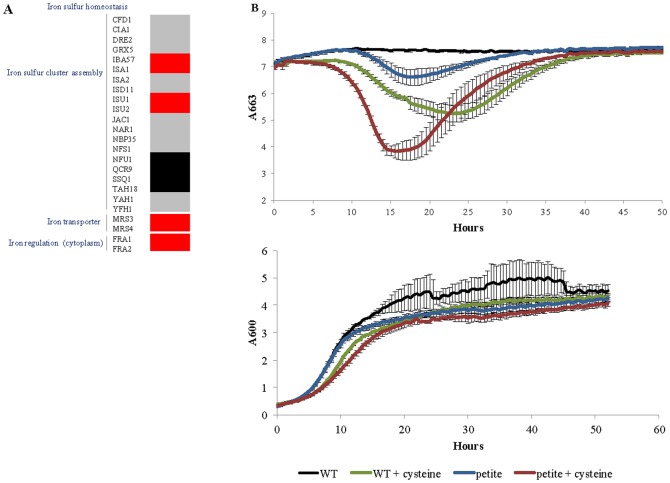
A large number of deletants affected in mitochondrial function generated high concentrations of H2S from cysteine relative to the wild-type strain; none generated H2S at detectable concentrations in the absence of cysteine. Amongst these H2S producers were strains with deletions in genes involved in mitochondrial maintenance of iron-sulfur (Fe-S) homeostasis (Fig. 3a). ‘Maintenance of Fe-S homeostasis’ includes genes involved in Fe-S cluster synthesis, iron transport into mitochondria, and a cytosolic protein complex involved in regulating transcription of the iron regulon in response to mitochondrial Fe-S cluster synthesis. Whilst for most genes involved in these processes null mutation leads to inviability [28], there were some viable deletants from each sub-group, and these were ‘high’ producers of H2S from cysteine (Fig. 3a), highlighting the role of Fe-S homeostasis in cysteine catabolism to release H2S.
Figure 3. Influence of mitochondrial gene deletions on cysteine catabolism to release H2S.
A Representation of genes involved in cellular iron-sulfur homeostasis with indication of inviable mutants (grey), mutants that lead to ever-secretion of H2S (red) and mutants that do not affect H2S release from cysteine (black). B Extracellular release of cysteine-generated H2S (upper graph) and growth curves (lower graph) for wt and petite cells lacking respiratory function, with and without cysteine supplementation. H2S formation was measured at 663 nm using the MBR method growth was measured at 600 nm. Experiments were carried out in a microtiter plate. Error bars represent the standard deviation for quadruplicate experiments.
An additional class of deletants overproducing H2S from cysteine were those impaired in mitochondrial translation (Δimg2, Δrml2, Δmrps35, Δmrpl20, Δmrpl4, Δmrpl24, Δmrpl10), suggesting that de novo synthesis of mitochondrial proteins is important for cysteine catabolism. The connection between mitochondrial function and H2S generation from cysteine was further examined using respiratory deficient petite mutants. These strains were shown to produce considerably higher concentrations of cysteine-derived H2S relative to the wild-type strain, and to produce detectable amounts of H2S even in the absence of cysteine (Fig. 3b). These results reinforce the importance of mitochondrial function in H2S production resulting from cysteine catabolism. The small, but nonetheless detectable, levels of H2S in the absence of supplemented cysteine may be due to catabolism of endogenous sources of this amino acid. Further experimental work is required to test this.
Low levels of production of H2S from cysteine
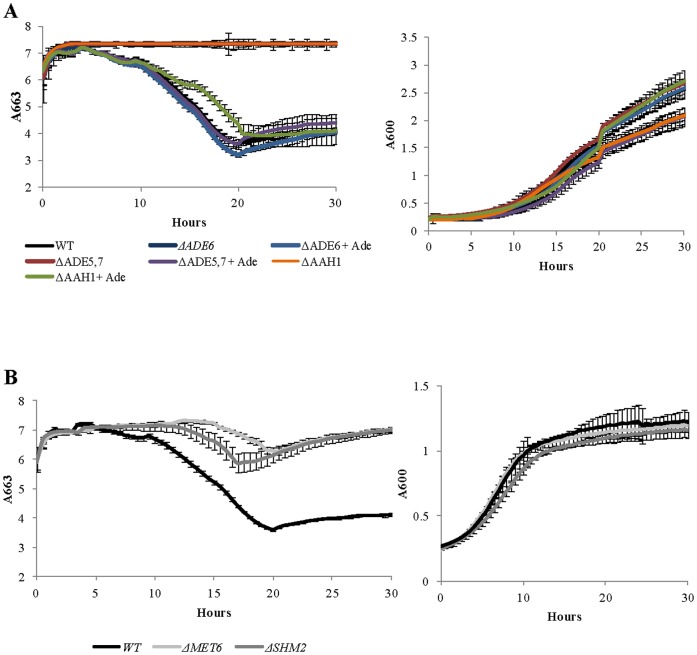
A total of 188 deletants were shown to release less H2S into cysteine-supplemented medium than the wild-type strain. Included in this number were deletions of genes involved in purine base metabolism and, more specifically, in the de novo synthesis of inosine monophosphate (IMP); Δade1, Δade5,7, Δade6, Δade8, Δado1, Δaah1, Δapt2. Strains with these deletions also grew significantly slower than the wild type, with or without cysteine addition. It is worth noting that mutants for purine metabolism have an auxotrophic requirement for adenine [29]. The medium used for the screen described here did not contain adenine, however mutants were still able to reach late logarithmic phase of growth (Fig. 4). Upon supplementation of adenine, mutants were able to grow and catabolise cysteine to produce H2S at a rate similar to the wild type (Fig. 4). Purine synthesis is linked to folate one-carbon-group metabolism and the metabolism of methionine [30]. Interestingly, deletion of genes involved in these pathways (Δshm2, Δmet6) resulted in low H2S release from cysteine, although these strains were able to grow at a rate similar to the wild type (Fig. 4).
Figure 4. Influences of mutants in purine metabolism on cysteine catabolism to H2S.
A Extracellular release of cysteine-generated H2S (left panel) and growth curves (right panel) from wt and mutants in purine biosynthesis pathway, with and without adenine supplementation. B Extracellular release of cysteine-generated H2S (left panel) and growth curves (right panel) from wt and mutants in folate derived one-carbon metabolism. H2S formation was measured at 663 nm using detection-dye; discolouration of the dye represents release of H2S. Experiments were carried out in a microtiter plate. Error bars represent the standard deviation for quadruplicate experiments.
Deletants affected in vacuolar function
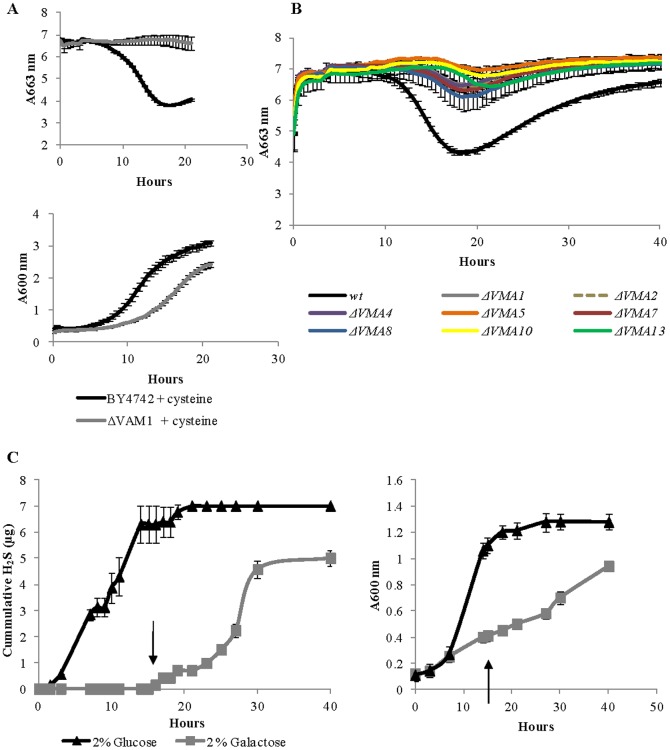
A large group of 39 deletants impaired in the release of H2S from cysteine-supplemented medium included those affected in vacuolar function and vesicle transport to the vacuole. Amongst these were deletions in genes involved in vacuole acidification, transport to the vacuole and vesicle fusion (Table 2). Cells deficient in vacuole biogenesis (Δvam1) did not produce H2S when supplemented with cysteine. Moreover, cysteine supplementation resulted in growth inhibition for this deletant (Fig. 5a). Notably, while the mutant grew more slowly than wild-type, comparison of H2S production at the same growth stage shows H2S accumulation by the wild type strain and not by the deletant.
Table 2. Vacuole function impaired deletants classified as low H2S producers.
| Functional Group | Deletants |
| Vacuole acidification | Δvps3, Δvma3, Δvps45, Δvps16, Δvps34, Δvma22, Δvph2, Δmeh1, Δvma13, Δvma1, Δvma2, Δvma4, Δvma7, Δvma10, Δvma13, Δvma21 |
| Transport to the vacuole | Δccz1, Δypt10, Δvps52, Δvma3, Δvma8, Δvps45, Δmon1, Δcog1, Δgtr2, Δavt7, Δmeh1, Δvts1, Δvma13, Δshp1, Δsym2, Δatg18 |
| Vesicle fusion | Δvam7, Δpep12, Δvps45, Δvts1, Δyck3, Δcog1, Δmon1, Δccz1, Δpep7 |
Figure 5. Central role of the vacuole in cysteine catabolism to H2S.
A Extracellular release of cysteine-generated H2S (upper panel) and biomass formation (lower panel) profiles of wt and mutant in vacuole biogenesis (Δvam1). Experiments were carried out in quadruplicates. B Extracellular release of cysteine-generated H2S for mutants deleted in each of the sub-units comprising V1 of the V-ATPase complex. Experiments were carried out in quadruplicates. C in-vivo downregulation of the yeast V-ATPase complex through nutrient limitation. H2S generation profile measured with H2S detection tubes (left panel) and biomass formation profile (right panel) for BY4742 cells grown with glucose or galactose as carbon source. An addition of glucose to a final concentration of 0.112 M was made to the galactose medium at the time indicated by arrows.
An important feature of the vacuole is its acidic pH. Vacuole acidification is regulated by a vacuolar-type ATPase (V-ATPase) protein complex. This protein complex is comprised of two multi subunits domains: a membrane integral V0 domain and a peripheral V1 domain [31]. Interaction between the two domains is essential for ATP-driven transport to maintain acidic vacuolar pH. Deletions of any of the eight subunits of the V1 complex were defective in generating H2S from cysteine (Fig. 5b) although these strains were able to grow under the experimental conditions (data not shown). Furthermore, deletion of genes encoding each of the three proteins responsible for V-ATPase assembly in the endoplasmic reticulum led to a similar phenotype of minimal H2S generation from cysteine (S1 Supporting Information). Interestingly, deletants for V0 subunits were able to generate H2S from cysteine supplemented medium at a rate similar to the wild-type strain.
The activity of yeast V-ATPase can be manipulated in vivo through growth medium composition [32]. When cells are transferred to a poor carbon source, up to 75% of existing V-ATPase complexes are disassembled into cytoplasmic (V1) and membrane-bound (Vo) sectors, and this disassembly is completely reversible. We used this in vivo down-regulation mechanism to test whether V-ATPase assembly and activity is essential for cysteine degradation leading to H2S production or whether disassembled individual sectors have an effect on the process. H2S-producing cells grown on glucose were transferred into a growth medium containing glucose or galactose as sole carbon source. Fig. 5c shows that upon transfer to galactose-containing medium cells did not generate H2S, whereas those transferred to glucose-containing medium continued to produce H2S. Addition of 0.112 M glucose to the galactose medium restored H2S formation (Fig. 5c). In all, these findings are consistent with poor carbon source driven disassembly of the V-ATPase complex preventing cysteine catabolism to release H2S.
Discussion
Cysteine is maintained at a low intracellular concentration by regulation of its production and, when in excess, its efficient removal. Amongst cysteine catabolic pathways is cysteine desulfhydration in which sulfur is cleaved from cysteine to produce H2S. In mammals, desulfhydration of cysteine leads to the production of H2S, which, in turn, may impact on cellular functions [1]. The current study used a EUROSCARF S. cerevisiae genome-wide deletion library for a functional genomics approach to identifying genes and cellular processes associated with the release of H2S from cysteine.
S. cerevisiae deletion libraries have been applied in a broad range of studies on biological processes including cellular response to Na+ [33], metal toxicity [34], oxidative stress [35], GSH homeostasis [36] and H2S formation through the sulfate assimilation pathway [37]. The latter utilised BiGGy agar [38] as an indicator for sulfite reductase activity, with follow up fermentation experiments on selected strains using gas collection tubes [39]. In this study, screening of a haploid yeast deletion library revealed a large number of genes (226) whose deletion altered H2S formation via cysteine catabolism. The caveat to this genetic approach is the relatively high level of H2S production from cysteine observed for the wild-type strain, which is close to the method's upper detection limit (Fig. 1). For this reason, the screen was aimed more at the identification of mutants impaired in H2S-generating catabolism of cysteine and discoveries made here regarding high H2S producers require further validation. An additional consideration is the genetic background of strain BY4742 that includes a number of auxotrophies. The possibility that discoveries observed here may be influenced by the genetic background of this deletion collection must be acknowledged.
Two PLP-dependent enzymes, highly conserved across prokaryotes and eukaryotes, were previously implicated in H2S-generating cysteine catabolism: CBS and CSE [1], [13], [14], [40]. The S. cerevisiae genes encoding these enzymes are CYS4 and CYS3, respectively. In this genomic screen, deletion of CYS4 did not affect H2S production from cysteine (S1 Supporting Information). Growth of a Δcys3 strain was delayed under cysteine supplementation, however cells still reached stationary phase of growth and were able to catabolise cysteine to release H2S (S1 Supporting Information). These results are rather surprising considering these enzymes are central in cysteine catabolism to release H2S in higher eukaryotes. However yeast differ from other eukaryotes by having an additional pathway for H2S generation, through sulfate assimilation. It has been previously reported that deletion or inhibition of cys3/cys4 results in increased H2S formation through the sulfate assimilation pathway [41], [42], which may compensate for the decrease in cysteine catabolism to release H2S. The regulatory mechanism and relative contribution of the two enzymes to H2S generation and cysteine metabolism is not yet understood [40], [43] and further study is needed to understand the catalytic and regulatory role of these enzymes. Additionally, the possibility that other PLP-dependent enzymes play some role in this process must be considered.
Cysteine in Fe-S cluster biogenesis
An additional PLP-dependent enzyme, Nfs1p, which is similar to its bacterial orthologs NifS and IscS, produces sulfur from cysteine for incorporation into Fe-S proteins [44], [45]. In eukaryotes, including S. cerevisiae, the biosynthesis of Fe-S clusters is mostly carried out in the mitochondrion [46]–[48]. Amongst other physiological roles, the synthesis of Fe-S clusters serves to maintain mitochondrial iron homeostasis [48]–[50]. Disruption of this process leads to mitochondrial iron overload, which is detrimental to the cell [48]. Accordingly, null mutation of many of the genes involved in Fe-S cluster biogenesis, including NFS1, results in cell death [28].
In this study mutants involved in all aspects of iron-sulfur homeostasis maintenance produced large amounts of H2S in comparison to the wild type strain (Fig. 3A). Increased production of H2S was also observed in a respiratory deficient petite mutant (Fig. 3B), although these mutants produced more H2S than the wild-type cells even in the absence of exogenous cysteine. The link between mitochondrial function and H2S formation was observed in Linderholm et al. (2008) [37], where deletion of gene YIA6, involved in NAD+ transport to mitochondria, resulted in high H2S production when cells were grown on BiGGy agar. The fact that deletants involved in Fe-S cluster assembly were not identified as H2S over-producers in Linderholm et al. (2008) [37] suggests that this phenotype appears only when cysteine is in excess, and reinforces the connection between iron-sulfur homeostasis and cysteine catabolism. Taken together these results lend weight to the hypothesis that disruption of Fe-S clusters leads to increased sulfur release associated with cysteine catabolism. However, the downstream metabolism of H2S must be considered as well and further studies are needed to understand the mitochondria role in H2S biogenesis and clearance.
Cysteine link to purine biosynthesis and folate derived one-carbon metabolism
Acting as a sulfur source for Fe-S biogenesis is one of the many cellular functions of cysteine. Cysteine is one of the twenty amino acids required for protein synthesis and is a precursor for synthesis of several other essential molecules including GSH, coenzyme A, taurine, and inorganic sulfur [1]. Additionally, cysteine concentration regulates transcription of genes associated with yeast sulfur assimilation and metabolism [41], [51]. A link between purine metabolism and H2S production was previously made by Linderholm et al. (2008) [37]. In that study S. cerevisiae mutants with deletions in genes involved in purine metabolism produced darker colour colonies when grown on BiGGY agar, indicative of high concentrations of H2S. Here, the same deletants produced less H2S than wild-type cells upon cysteine supplementation during fermentation. The difference between the two results may simply reflect the different pathways examined; Linderholm et al. (2008) [37] studied H2S formation through the sulfate assimilation pathway while here the release of H2S from cysteine was studied. Alternatively, differences in the medium composition between the two experiments could have led to contrasting outcomes. In this study, addition of adenine resulted in increased H2S production, at a similar level to the wild-type strain (Fig. 4), suggesting the observed low H2S production was due to auxotrophic requirements of the deletants. Though this aspect was not tested in Linderholm, et al., (2008) [37] further studies are needed to identify the extent of a link between sulfur and purine metabolism.
A possible connection between the two is through folate derived one-carbon metabolism, necessary for synthesis of purines and methionine [30]. We report here, somewhat surprisingly, that disruption of methionine biosynthesis through the methyl cycle results in decreased catabolism of cysteine to release H2S, without affecting growth rate (Fig. 4B). Under these conditions one pathway for cysteine catabolism (conversion to methionine) is inhibited, thus the fact that cysteine was not catabolised to produce H2S under these conditions requires further investigation.
Central role of the vacuole in cysteine catabolism
Cysteine accumulation was previously shown to have a cytotoxic effect [2], [3]. With that consideration, the central role discovered here for the vacuole in catabolism of cysteine to release H2S is reasonable. The yeast vacuole is an acidic compartment that shares a great deal of functional and morphological similarity with the mammalian lysosome and supports detoxification, protein degradation and ion and metabolite storage [52], [53]. Our genome-wide screen identified a strong link between the vacuole and degradation of cysteine to release H2S (Fig. 5). Of particular importance, deletion of each of the subunits comprising the V1 sub complex of V-ATPase, responsible for vacuolar acidic pH, resulted in low (to undetected) H2S production from cysteine. V-ATPases are evolutionary conserved multisubunit enzymes responsible for acidification of the vacuole in yeast and the lysosome in mammals [53]. Deletion of genes encoding ubiquitous V-ATPase subunits is lethal in all eukaryotes except fungi [53], due to their ability to take proton from the extracellular environment and thus maintain acidic intracellular pH [53]. Therefore, yeast impaired in V-ATPase activity are able to grow at a similar rate to the wild type under acidic conditions (pH<5), making studies of this complex in S. cerevisiae particularly important. Serendipitously, the medium used for our experiments was at pH 4.5, based on the optimal pH needed for H2S detection using the MBR method [22]. It is possible that the role of V-ATPase in H2S formation from cysteine is to encourage conversion of HS to H2S, downstream of enzymatic release from cysteine. This aspect requires further investigation.
Observations made in this study infer a major role of V-ATPase in the catabolism of excess cysteine to H2S and highlight the peripheral V1 sub-complex as the active unit. Most interestingly, deletants impaired in V-ATPase activity were previously classified as high H2S producers when evaluated for sulfate reductase activity using BiGGy agar [37]. While their classification was not validated, the observations differ from those described in this study possibly due to the pleiotropy of the V-ATPase. Impaired V-ATPase activity has been reported from studies utilising genomic screens to assess sensitivity to various drugs, metal ions, multiple forms of oxidative and other stresses (these studies are summarised in [54]. This suggests multiple roles for the V-ATPase complex, depending on the conditions under which yeast is grown. It is therefore plausible that activation of the sulfate reduction pathway in these deletants promotes high H2S production through a mechanism unrelated to cysteine catabolism. As the use of different environmental conditions leads to different H2S production patterns [22], the role of yeast V-ATPase should be evaluated under these environmental conditions using a yeast strain that is able to produce large amounts of H2S through both sulfate reduction and cysteine catabolism, possibly utilising labelled sulfur sources.
Regardless, the nature of the role of V-ATPase in cysteine catabolism requires further elucidation. V-ATPase generates a proton gradient across the vacuolar membrane that drives transport of ions and small molecules into the vacuole [55], [56]. It is possible that cysteine transport into the vacuole is facilitated by this proton gradient, which would explain our identification of the proton-generating ATP hydrolysis sub-complex (V1) as the active contributor to cysteine degradation. Supporting this, GSH transport to the vacuole is also partially mediated through a V-ATPase coupled system [57]. Alternatively, the observation that deletants impaired in vesicle formation and fusion with the vacuole (Table 1) are impaired in cysteine catabolism to release H2S, suggests that V-ATPase activity may facilitate cysteine transport to the vacuole in a vesicle-mediated manner. This is in line with recent studies demonstrating vesicle fusion with the vacuole requires the activity of the V-ATPase complex [53], [58]. Interestingly, cysteine metabolism was indirectly linked with V-ATPase, through the discovery that Δcys4 mutants display an in vivo loss of vacuole acidification, due to inactivation of the V-ATPase complex [59]. It has been suggested that V-ATPase activity is regulated by cytosolic redox state; a concept supported by other studies from which it has been proposed that the thiol/disulphide ratio may serve as a “third messenger” [60]. These findings indicate that, in addition to being regulated by redox state, V-ATPase may operate to maintain cytosolic redox balance by removing excess cysteine from the cytosolic pool.
Cysteine catabolism as a detoxification mechanism
Incorporation into GSH must be considered as a potential mechanism of cysteine exclusion from the cytosolic pool. GSH synthesis facilitates cysteine homeostasis by acting as the cellular reservoir for this amino acid. However, GSH homeostasis is tightly regulated [36] and the capacity for it to act as cysteine reservoir is limited. Thus it is unlikely to be the main destination in cysteine exclusion from the cytosolic pool. Supporting this view we observed catabolism of cysteine to release H2S in mutants impaired in GSH synthesis (Δgsh1, Δgsh2), at a similar level to the wild-type.
Two genome-wide studies have explored GSH homeostasis, analysing both its intra- and extracellular concentration [36], [61]. Our findings suggest that deletants with perturbed cysteine catabolism generally differ from those affecting GSH homeostasis. Most notably, there was considerable similarity between our findings and results from genome-wide screens for metal tolerance [34], [62]; deletants impaired in V-ATPase activity are sensitive to metal toxicity and have compromised cysteine catabolism leading to release of H2S. In addition, mutants reported to confer metal resistance were similar to those conferring high H2S production. These points of similarity reinforce that degradation of excess cysteine to H2S is a detoxification mechanism.
Conclusions
Cysteine catabolism generates reduced sulfur and is critical for cysteine homeostasis in eukaryotes [1]. There is a substantial body of evidence supporting the physiological roles of cysteine degradation to release H2S, highlighting the need for a better understanding of the genetic factors influencing this part of metabolism. In this study we identified genes associated with cysteine catabolism that have not been previously linked with this process. The vacuolar role that has been established here and particularly the role of yeast V-ATPase point to new directions for future studies in cysteine homeostasis using S. cerevisiae as a model organism.
Acknowledgments
We thank Laffort Australia and in particular Dr. Tertius Van der Westhuizen for continued support. Drs Paul Chambers and Paul Henschke are thanked for critical review of the manuscript. Jane McCarthy is thanked for organisation of the yeast deletion collection. Jenny Bellon is acknowledged for preparation of respiratory deficient petite yeast cells and Chiara Bozzinni for assistance with shake-flask H2S profile generation. We thank Angus Forgan and Dr Simon Schmidt for their help with programming of robotics for high-throughput screening experiments. The research was supported by an Industry Partnership grant. Research at The Australian Wine Research Institute is supported by Australia's grapegrowers and winemakers through their investment agency the Grape and Wine Research and Development Corporation, with matching funds from the Australian Government. The Australian Wine Research Institute is a member of the Wine Innovation Cluster.
Supporting Information
Genes classified as low or high H2S producers in a genome-wide screen for cysteine catabolism.
(DOCX)
H2S accumulation during growth of selected deletants classified as high/low H2S producers. H2S accumulation in wild type (white column) and selected deletants classified as high (red columns: Δfra1, Δisu1, Δmrs3, Δmtm1, Δisa1) or low (green columns: Δvma5, Δvam7, Δvam1, Δshm2) H2S producers following cysteine catabolism to release H2S as measured after 16 hours of cultivation when it plateaued. Culture medium was based on [22] without the addition of sulfate and with supplementation of 500 mg/L cysteine. Strains cultivation and H2S measurements are described in Materials and Methods section. Error bars represent standard deviation of triplicate experiments and different letters denote significance at p<0.05 (Tuckey test).
(DOCX)
Data Availability
The authors confirm that all data underlying the findings are fully available without restriction. All relevant data are within the paper and its Supporting Information files.
Funding Statement
This study was funded by Laffort Australia. Gal Winter was a PhD student at the University of Western Sydney and The Australian Wine Research Institute. Antonio G. Cordente and Chris Curtin are employed by The Australian Wine Research Institute. The Australian Wine Research Institute is supported by Australia's grape growers and winemakers through their investment agency the Grape and Wine Research and Development Corporation, with matching funds from the Australian Government. The funder had no role articulated in study design, data collection and analysis, decision to publish, or preparation of the manuscript. The Australian Wine Research Institute provided support in the form of salaries for authors AGC and CC, but did not have any additional role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript. The specific roles of these authors are in the "author contributions" section.
References
- 1. Stipanuk MH (2004) Sulfur amino acid metabolism: pathways for production and removal of homocysteine and cysteine. Ann Rev Nutr 24:539–577. [DOI] [PubMed] [Google Scholar]
- 2. Kumar A, John L, Alam MM, Gupta A, Sharma G, et al. (2006) Homocysteine- and cysteine-mediated growth defect is not associated with induction of oxidative stress response genes in yeast. Biochem J 396:61–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ono B-I, Naito K, Shirahige Y-I, Yamamoto M (1991) Regulation of cystathionine γ-lyase in Saccharomyces cerevisiae . Yeast 7:843–848. [DOI] [PubMed] [Google Scholar]
- 4. Lee JI, Londono M, Hirschberger LL, Stipanuk MH (2004) Regulation of cysteine dioxygenase and Î3-glutamylcysteine synthetase is associated with hepatic cysteine level. J Nutr Biochem 15:112–122. [DOI] [PubMed] [Google Scholar]
- 5. Heafield MT, Fearn S, Steventon GB, Waring RH, Williams AC, et al. (1990) Plasma cysteine and sulphate levels in patients with motor neurone, Parkinson's and Alzheimer's disease. Neurosci Lett 110:216–220. [DOI] [PubMed] [Google Scholar]
- 6. Chiku T, Padovani D, Zhu W, Singh S, Vitvitsky V, et al. (2009) H2S Biogenesis by human cystathionine Î3-Lyase leads to the novel sulfur metabolites lanthionine and homolanthionine and is responsive to the grade of hyperhomocysteinemia. J Biol Chem 284:11601–11612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kabil O, Banerjee R (2010) Redox biochemistry of hydrogen sulfide. J Biol Chem 285:21903–21907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kimura H (2002) Hydrogen sulfide as a neuromodulator. Mol Neurobiol 26:13–19. [DOI] [PubMed] [Google Scholar]
- 9. Wang RUI (2002) Two is company, three is a crowd: can H2S be the third endogenous gaseous transmitter? FASEB Journal 16:1792–1798. [DOI] [PubMed] [Google Scholar]
- 10. Kimura H (2010) Hydrogen sulfide: from brain to gut. Antioxid Redox Signal 12:1111–1123. [DOI] [PubMed] [Google Scholar]
- 11. Olson KR (2011) The therapeutic potential of hydrogen sulfide: separating hype from hope. Am J Physiol 301(2):R297–312. [DOI] [PubMed] [Google Scholar]
- 12. Bengtsson L, Roeckner E, Stendel M (1999) Why is the global warming proceeding much slower than expected? J Geophys Res Atmos 104:3865–3876. [Google Scholar]
- 13. Singh S, Banerjee R (2011) PLP-dependent H2S biogenesis. Biochim Biophys Acta 1814:1518–1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Chen X, Jhee K-H, Kruger WD (2004) Production of the neuromodulator H2S by Cystathionine β-Synthase via the condensation of cysteine and homocysteine. J Biol Chem 279:52082–52086. [DOI] [PubMed] [Google Scholar]
- 15. Beard RS, Bearden SE (2011) Vascular complications of cystathionine Î2-synthase deficiency: future directions for homocysteine-to-hydrogen sulfide research. Am J Physiol Heart Circ Physiol 300:H13–H26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kruger WD, Cox DR (1994) A yeast system for expression of human cystathionine beta-synthase: structural and functional conservation of the human and yeast genes. Proc Nat Acad Sci 91:6614–6618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kruger WD, Cox DR (1995) A yeast assay for functional detection of mutations in the human cystathionine β-synthase gene. Hum Mol Genet 4:1155–1161. [DOI] [PubMed] [Google Scholar]
- 18. Shan X, Kruger WD (1998) Correction of disease-causing CBS mutations in yeast. Nat Genet 19:91–93. [DOI] [PubMed] [Google Scholar]
- 19. Barton AB, Kaback DB, Clark MW, Keng T, Ouellette BFF, et al. (1993) Physical localization of yeast CYS3, a gene whose product resembles the rat γ-cystathionase and Escherichia coli cystathionine γ-synthase enzymes. Yeast 9:363–369. [DOI] [PubMed] [Google Scholar]
- 20. Brachmann C, Davies A, Cost G, Caputo E, Li J, et al. (1998) Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast 14:115–132. [DOI] [PubMed] [Google Scholar]
- 21. Winter G, Van Der Westhuizen T, Higgins VJ, Curtin C, Ugliano M (2011) Contribution of cysteine and glutathione conjugates to the formation of the volatile thiols 3-mercaptohexan-1-ol (3MH) and 3-mercaptohexyl acetate (3MHA) during fermentation by Saccharomyces cerevisiae . Aus J Grape Wine Res 17:285–290. [Google Scholar]
- 22. Winter G, Curtin C (2012) In situ high throughput method for H2S detection during fermentation. J Microbiol Methods 91:165–170. [DOI] [PubMed] [Google Scholar]
- 23. Winter G, Henschke P, Higgins V, Ugliano M, Curtin C (2011) Effects of rehydration nutrients on H2S metabolism and formation of volatile sulfur compounds by the wine yeast VL3. AMB Express 1:36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.R Development Core Team (2008) R: A language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing. [Google Scholar]
- 25.Sherman F, Fink G, Hicks J (1986) Laboratory course manual for methods in yeast genetics. Cold Spring Harbor, NY: Cold Spring Harbour Laboratory Press. [Google Scholar]
- 26. Winzeler EA, Shoemaker DD, Astromoff A, Liang H, Anderson K, et al. (1999) Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science 285:901–906. [DOI] [PubMed] [Google Scholar]
- 27. Eden E, Navon R, Steinfeld I, Lipson D, Yakhini Z (2009) GOrilla: a tool for discovery and visualization of enriched GO terms in ranked gene lists. BMC Bioinformatics 10:48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Giaever G, Chu AM, Ni L, Connelly C, Riles L, et al. (2002) Functional profiling of the Saccharomyces cerevisiae genome. Nature 418:387–391. [DOI] [PubMed] [Google Scholar]
- 29. Roberts RL, Metz M, Monks DE, Mullaney ML, Hall T, et al. (2003) Purine synthesis and increased Agrobacterium tumefaciens transformation of yeast and plants. Proc Nat Acad Sci 100:6634–6639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Cossins EA, Chen L (1997) Folates and one-carbon metabolism in plants and fungi. Phytochemistry 45:437–452. [DOI] [PubMed] [Google Scholar]
- 31. Kane PM (2006) The where, when, and how of organelle acidification by the yeast vacuolar H+-ATPase. Microbiol Mol Biol Rev 70:177–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kane PM, Smardon AM (2003) Assembly and regulation of the yeast vacuolar H+-ATPase. J Bioenerg Biomembr 35:313–321. [DOI] [PubMed] [Google Scholar]
- 33. Warringer J, Ericson E, Fernandez L, Nerman O, Blomberg A (2003) High-resolution yeast phenomics resolves different physiological features in the saline response. Proc Nat Acad Sci 100:15724–15729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ruotolo R, Marchini G, Ottonello S (2008) Membrane transporters and protein traffic networks differentially affecting metal tolerance: a genomic phenotyping study in yeast. Genome Biol 9:R67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Thorpe GW, Fong CS, Alic N, Higgins VJ, Dawes IW (2004) Cells have distinct mechanisms to maintain protection against different reactive oxygen species: Oxidative-stress-response genes. PNAS 101:6564–6569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Perrone GG, Grant CM, Dawes IW (2005) Genetic and environmental factors influencing glutathione homeostasis in Saccharomyces cerevisiae . Mol Biol Cell 16:218–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Linderholm AL, Findleton CL, Kumar G, Hong Y, Bisson LF (2008) Identification of genes affecting hydrogen sulfide formation in Saccharomyces cerevisiae . Appl Environ Microbiol 74:1418–1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Nickerson WJ (1953) Reduction of inorganic substances by yeast. I. Extracellular reduction of sulfite by species of Candida . J Infectious Dis 93:43–48. [DOI] [PubMed] [Google Scholar]
- 39. Park SK (2008) Development of amethod to measure hydrogen sulfide in wine fermentation. J Microbiol Biotechnol 18:1550–1554. [PubMed] [Google Scholar]
- 40. Stipanuk MH, Beck PW (1982) Characterization of the enzymic capacity for cysteine desulphhydration in liver and kidney of the rat. Biochem J 206:267–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Hansen J, Francke Johannesen P (2000) Cysteine is essential for transcriptional regulation of the sulfur assimilation genes in Saccharomyces cerevisiae . Mol Gen Genet 263:535–542. [DOI] [PubMed] [Google Scholar]
- 42. Kim HS, Huh J, Fay JC (2009) Dissecting the pleiotropic consequences of a quantitative trait nucleotide. FEMS Yeast Res 9:713–722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Singh S, Padovani D, Leslie RA, Chiku T, Banerjee R (2009) Relative contributions of cystathionine Î2-synthase and Î3-cystathionase to H2S biogenesis via alternative trans-sulfuration reactions. J Biol Chem 284:22457–22466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Mühlenhoff U, Balk J, Richhardt N, Kaiser JT, Sipos K, et al. (2004) Functional characterization of the eukaryotic cysteine desulfurase Nfs1p from Saccharomyces cerevisiae . J Biol Chem 279:36906–36915. [DOI] [PubMed] [Google Scholar]
- 45. Mühlenhoff U, Richhardt N, Gerber J, Lill R (2002) Characterization of iron-sulfur protein assembly in isolated mitochondria. J Biol Chem 277:29810–29816. [DOI] [PubMed] [Google Scholar]
- 46. Balk J, Lill R (2004) The cell's cookbook for iron–sulfur clusters: recipes for fool's gold? Chembiochem 5:1044–1049. [DOI] [PubMed] [Google Scholar]
- 47. Lill R, Kispal G (2000) Maturation of cellular Fe-S proteins: an essential function of mitochondria. Trends Biochem Sci 25:352–356. [DOI] [PubMed] [Google Scholar]
- 48. Rouault TA, Tong W-H (2005) Iron-sulphur cluster biogenesis and mitochondrial iron homeostasis. Nat Rev Mol Cell Biol 6:345–351. [DOI] [PubMed] [Google Scholar]
- 49. Kispal G, Csere P, Prohl C, Lill R (1999) The mitochondrial proteins Atm1p and Nfs1p are essential for biogenesis of cytosolic Fe/S proteins. EMBO J 18:3981–3989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Li J, Kogan M, Knight SAB, Pain D, Dancis A (1999) Yeast mitochondrial protein, Nfs1p, coordinately regulates iron-sulfur cluster proteins, cellular iron uptake, and iron distribution. J Biol Chem 274:33025–33034. [DOI] [PubMed] [Google Scholar]
- 51. Ono B-I, Kijima K, Ishii N, Kawato T, Matsuda A, et al. (1996) Regulation of sulphate assimilation in Saccharomyces cerevisiae . Yeast 12:1153–1162. [DOI] [PubMed] [Google Scholar]
- 52. Klionsky DJ, Herman PK, Emr SD (1990) The fungal vacuole: composition, function, and biogenesis. Microbiol Mol Biol Rev 54:266–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Li SC, Kane PM (2009) The yeast lysosome-like vacuole: Endpoint and crossroads. Biochim Biophys Acta 1793:650–663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Derikx PJL, Op Den Camp HJM, van der Drift C, van Griensven LJLD, Vogels GD (1990) Odorous sulfur compounds emitted during production of compost used as a substrate in mushroom cultivation. Appl Environ Microbiol 56:176–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Arai H, Pink S, Forgac M (1989) Interaction of anions and ATP with the coated vesicle proton pump. Biochemistry 28:3075–3082. [DOI] [PubMed] [Google Scholar]
- 56. Ohsumi Y, Anraku Y (1981) Active transport of basic amino acids driven by a proton motive force in vacuolar membrane vesicles of Saccharomyces cerevisiae . J Biol Chem 256:2079–2082. [PubMed] [Google Scholar]
- 57. Penninckx MJ (2002) An overview on glutathione in Saccharomyces versus non-conventional yeasts. FEMS Yeast Res 2:295–305. [DOI] [PubMed] [Google Scholar]
- 58. Baars TL, Petri S, Peters C, Mayer A (2007) Role of the V-ATPase in regulation of the vacuolar fission - fusion equilibrium. Mol Biol Cell 18:3873–3882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Oluwatosin YE, Kane PM (1997) Mutations in the CYS4 gene provide evidence for regulation of the yeast vacuolar H+-ATPase by oxidation and reduction in vivo. J Biol Chem 272:28149–28157. [DOI] [PubMed] [Google Scholar]
- 60. Walters DW, Gilbert HF (1986) Thiol/disulfide exchange between rabbit muscle phosphofructokinase and glutathione. Kinetics and thermodynamics of enzyme oxidation. J Biol Chem 261:15372–15377. [PubMed] [Google Scholar]
- 61. Suzuki T, Yokoyama A, Tsuji T, Ikeshima E, Nakashima K, et al. (2011) Identification and characterization of genes involved in glutathione production in yeast. J Biosci Bioeng 112:107–113. [DOI] [PubMed] [Google Scholar]
- 62. Arita A, Zhou X, Ellen T, Liu X, Bai J, et al. (2009) A genome-wide deletion mutant screen identifies pathways affected by nickel sulfate in Saccharomyces cerevisiae . BMC Genomics 10:524. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Genes classified as low or high H2S producers in a genome-wide screen for cysteine catabolism.
(DOCX)
H2S accumulation during growth of selected deletants classified as high/low H2S producers. H2S accumulation in wild type (white column) and selected deletants classified as high (red columns: Δfra1, Δisu1, Δmrs3, Δmtm1, Δisa1) or low (green columns: Δvma5, Δvam7, Δvam1, Δshm2) H2S producers following cysteine catabolism to release H2S as measured after 16 hours of cultivation when it plateaued. Culture medium was based on [22] without the addition of sulfate and with supplementation of 500 mg/L cysteine. Strains cultivation and H2S measurements are described in Materials and Methods section. Error bars represent standard deviation of triplicate experiments and different letters denote significance at p<0.05 (Tuckey test).
(DOCX)
Data Availability Statement
The authors confirm that all data underlying the findings are fully available without restriction. All relevant data are within the paper and its Supporting Information files.