Abstract
Obesity is a crucial public health issue in developed countries, with implications for cardiovascular and brain health as we age. A number of commonly-carried genetic variants are associated with obesity. Here we aim to see whether variants in obesity-associated genes - NEGR1, FTO, MTCH2, MC4R, LRRN6C, MAP2K5, FAIM2, SEC16B, ETV5, BDNF-AS, ATXN2L, ATP2A1, KCTD15, and TNN13K - are associated with white matter microstructural properties, assessed by high angular resolution diffusion imaging (HARDI) in young healthy adults between 20–30 years of age from the Queensland Twin Imaging study (QTIM). We began with a multi-locus approach testing how a number of common genetic risk factors for obesity at the single nucleotide polymorphism (SNP) level may jointly influence white matter integrity throughout the brain and found a wide spread genetic effect. Risk allele rs2815752 in NEGR1 was most associated with lower white matter integrity across a substantial portion of the brain. Across the area of significance in the bilateral posterior corona radiata, each additional copy of the risk allele was associated with a 2.2% lower average FA. This is the first study to find an association between an obesity risk gene and differences in white matter integrity. As our subjects were young and healthy, our results suggest that NEGR1 has effects on brain structure independent of its effect on obesity.
1. Introduction
Obesity is a major public health issue facing developed countries. In the United States over a third of adults are classified as obese, and another third are considered to be overweight (Ogden et al., 2012). Obesity has well-established links to serious health issues such as diabetes, heart disease, and premature death (Must et al., 1999). High body mass index (BMI)1 in midlife is linked to poorer cognitive functioning in old age (Fitzpatrick et al., 2009; Walther et al., 2009). Greater BMI is associated with lower brain volume (Walther et al., 2009; Ward et al., 2005; Taki et al., 2008), brain atrophy (Gustafson et al., 2004), and lower gray matter density (Pannacciulli et al., 2006), and neuronal and myelin abnormalities (Gazdzinski et al., 2010). Obesity is associated with abnormalities in white matter volume (Haltia et al., 2007; Raji et al., 2009), diffusivity (Alkan et al., 2008) and integrity across many brain regions (Stanek et al., 2009; Verstynen et al., 2012; Xu et al., 2013). These brain differences in obese people may be attributable to a less healthy diet and lifestyle, which negatively affect brain health (Molteni et al., 2002; Northstone et al., 2012; Ars, 2012). They may be partly due to genetic variants with joint effects on the brain and obesity risk. A gene may directly affect the brain, and its effects on appetite and physical activity could affect obesity. Alternatively, a gene could affect vascular health, reducing cerebral blood flow, and therefore delivery of oxygen and nutrients to the brain, with concomitant effects on brain function.
Diet and lifestyle are the most readily identifiable causes of obesity, yet it is highly heritable (Wardle et al., 2008), and genetic vulnerabilities interact with lifestyle factors. A number of genes have been repeatedly associated with obesity in cohorts worldwide (Frayling et al., 2007; Loos et al., 2008; Ng et al., 2012; Okada et al., 2012; Wen et al., 2012). We previously found that elderly carriers of the FTO risk allele had lower frontal and occipital lobe volumes (Ho et al., 2010), and a recent paper found that a locus near the obesity risk gene MC4R was associated with increased amygdalar, hippocampal, and medial orbitofrontal volume, as well as differences in eating behaviors (Horstmann et al., 2013). The obesity risk gene Taq1A has been associated with decreased striatal activation in response to receiving chocolate (Stice et al., 2008). Recent genome-wide association studies (GWAS) identified a number of loci associated with BMI (Speliotes et al., 2010; Thorleifsson et al., 2008; Willer et al., 2008).
Axonal integrity is vital for efficient brain function; well-myelinated tracts propagate signals quickly, but poor or impaired myelination can decrease the speed or reliability of neuronal transmission (Purves et al., 2001). FA is a widely accepted measure of white matter integrity, and evaluates the degree to which water diffuses along the primary direction of the axon rather than across it. Lower FA has been found in many diseases, such as Alzheimer’s disease, multiple sclerosis, epilepsy, and many neuropsychiatric diseases (Ciccarelli et al., 2008). Genetic variants have also been discovered that may affect white matter integrity as measured by FA. Associations have been reported between FA and a number of genetic variants, including polymorphisms in CLU, HFE, NTRK1, and many other genes (Braskie et al., 2011; Jahanshad et al., 2012; Braskie et al., 2012). These are genes that are already closely tied to cognitive function or neuropsychiatric disorders.
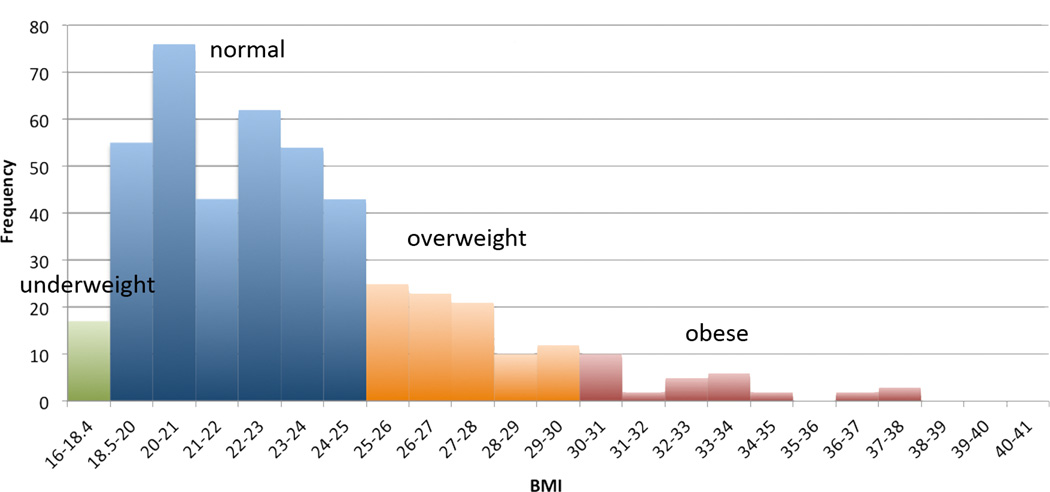
Here we investigated whether 16 common variants in obesity-related genes (NEGR1, FTO, MTCH2, MC4R, LRRN6C, MAP2K5, FAIM2, SEC16B, ETV5, BDNF-AS, ATXN2L, ATP2A1, KCTD15, and TNN13K) relate to the brain’s white matter integrity. We selected our SNPs based on three recent GWAS studies of obesity all with large sample sizes (Speliotes et al., 2010; Thorleifsson et al., 2008; Willer et al., 2008). Using a multi-locus approach to assess their combined effect, we tested whether obesity-related variants might predict differences in white matter integrity assessed using high angular resolution diffusion imaging (HARDI) (Kohannim et al., 2012). As a post hoc test, we evaluated the most promising SNP (single nucleotide polymorphism) driving the effects in the multi-locus model. Analyses were completed in 499 healthy young adults (aged 20–30), to test if there was any evidence of a link between obesity-related genetic variants and white matter integrity. While the global incidence of obesity in developed countries is typically close to 30% (Ogden et al., 2012), our population was healthy with a lower obesity incidence, with around 6% obese and 20% overweight. Therefore, we did not map the effects of this biased population’s BMI on the brain. Rather, we were interested in determining whether common genetic variants, which play a subtle role in obesity, and are also common in the general healthy population, continued to show effects on white matter integrity. We expected variants associated with increased risk of obesity would be associated with lower white matter integrity.
2. Materials and Methods
2.1 Participants
Participants were recruited as part of a 5-year project research project examining healthy Australian twins with structural MRI and diffusion-weighted imaging (de Zubicaray et al., 2008). Our analysis included 499 right-handed subjects (326 females/173 males, mean age=23.8, SD=2.5 years, range=20–30 years). This sample included 163 monozygotic (MZ) twins, 274 dizygotic (DZ) twins, and 62 non-twin siblings, from 309 families. This information is summarized in Table 1, along with BMI information for each group. A histogram of BMI is shown in Figure 1. All QTIM subjects were Caucasian, and ancestry outliers, defined as individuals more than 6 SD from the PC1/PC2 centroid after principal components analyses of the GWAS data (Medland et al., 2009), were excluded. Gene allele frequencies can differ between ethnicities, as can the risks associated with various alleles, so ethnically homogenous groups are generally preferred in genetic studies. Additionally, the three published studies (Speliotes et al., 2010; Thorleifsson et al., 2008; Willer et al., 2008) – which we used to select our SNPs of interest – were analyses of sampled populations that were 99.7% Caucasian (one of the studies [Thorleifsson et al., 2008] included a very small number of African American subjects as well).
Table 1. Subject demographics.
Number of subjects, female/male, mean BMI (body mass index) and standard deviation, across all genetic groups.
| Genetic group | QTIM Subjects | ||
| N | F/M | BMI | |
| AA | 188 | 125/63 | 23.1 (3.80) |
| AG | 233 | 154/79 | 23.4 (3.64) |
| GG | 78 | 47/31 | 23.6 (3.97) |
Figure 1. Histogram of BMIs.
Green=underweight, blue=normal weight, orange=overweight, red=obese.
2.2 Scan Acquisition
Whole-brain anatomical and high angular resolution diffusion images (HARDI) were collected with a 4T Bruker Medspec MRI scanner. T1-weighted anatomical images were acquired with an inversion recovery rapid gradient echo sequence. Acquisition parameters were: TI/TR/TE = 700/1500/3.35ms; flip angle = 8 degrees; slice thickness = 0.9mm, with a 256×256 acquisition matrix. HARDI was also acquired using single-shot echo planar imaging with a twice-refocused spin echo sequence to reduce eddy-current induced distortions. Imaging parameters were: 23cm FOV, TR/TE 6090/91.7ms, with a 128×128 acquisition matrix. Each 3D volume consisted of 55 2-mm thick axial slices with no gap, and 1.79 × 1.79 mm2 in-plane resolution. 105 images were acquired per subject: 11 with no diffusion sensitization (i.e., T2-weighted b0 images) and 94 diffusion-weighted (DW) images (b = 1159 s/mm2) with gradient directions evenly distributed on a hemisphere in the q-space. Scan time for the 105-gradient HARDI scan was 14.2 min.
2.3 Establishing Zygosity and Genotyping
Zygosity was objectively established by typing nine independent DNA microsatellite polymorphisms (polymorphism information content > 0.7), using standard PCR methods and genotyping. Results were crosschecked with blood group (ABO, MNS, and Rh), and phenotypic data (hair, skin, and eye color), giving an overall probability of correct zygosity assignment > 99.99%, and these were subsequently confirmed by GWAS. Genomic DNA samples were analyzed on the Human610-Quad BeadChip (Illumina) according to the manufacturers protocols (Infinium HD Assay; Super Protocol Guide; Rev. A, May 2008). We imputed to Hapmap3. Information on the imputation protocols and quality control steps may be found at http://enigma.ini.usc.edu/wp-content/uploads/2010/09/ImputationProtocolsv1.2.pdf.
2.4 Diffusion Tensor Image (DTI) Processing
Non-brain regions were automatically removed from each T1-weighted MRI scan using ROBEX (Iglesias et al., 2011) a robust brain extraction program trained on manually “skull-stripped” MRI data and FreeSurfer (Fischl et al., 2004), and from a T2-weighted image from the DWI set, using the FSL tool “BET” (Smith, 2002; FMRIB Software Library, http://fsl.fmrib.ox.ac.uk/fsl/). Intracranial volume estimates were obtained from the full brain mask, and included cerebral, cerebellar, and brain stem regions. All T1-weighted images were linearly aligned using FSL flirt (with 9 DOF) (Jenkinson et al., 2002) to a common space (Holmes et al., 1998) with 1mm isotropic voxels and a 220×220×220 voxel matrix. Raw diffusion-weighted images were corrected for eddy current distortions using the FSL tool, “eddy_correct”. For each subject, the eddy-corrected images with no diffusion sensitization were averaged (11 images), linearly aligned and resampled to a downsampled version of their corresponding T1-weighted image (110×110×110 matrix, 2×2×2mm3 voxel size). Averaged b0 maps were elastically registered to the structural scan using a mutual information cost function (Leow et al., 2005) to compensate for EPI-induced susceptibility artifacts. The resulting 3D deformation fields were then applied to the remaining 94 DWI volumes. To examine subject motion, we compared the acquired and theoretical DWI at each voxel based on the reconstructed tensor with the actual gradients after eddy correction. A high degree of motion will show significant deviations between the theoretical and actual scans, particularly around the boundaries of the brain.
We compared fractional anisotropy (FA) values at each voxel across NEGR1 genotypes. Diffusion tensors were computed at each voxel using FSL software (http://fsl.fmrib.ox.ac.uk/fsl/). From the tensor eigenvalues (λ1, λ2, λ3), FA was calculated according to the following formula:
| Eq. 1 |
We also analyzed radial diffusivity (Drad = the average of λ2 and λ3), mean diffusivity (Dmean = λ̄) and axial diffusivity (Dax = λ1) to clarify the extent to which each might be contributing to the changes in FA.
2.5 MDT
The MDT (minimal deformation template) is the template that deviates least from the anatomy of the subjects, and, in some circumstances, it can improve statistical power (Leporé et al., 2007). Using a customized template from subjects in the study (rather than a standard atlas or a single optimally chosen subject) can reduce bias in the registrations. Included in the MDT were FA images from 32 randomly selected unrelated subjects (16 female/16 male) (calculated after susceptibility correction) (Jahanshad et al., 2010a). The N 3D vector fields that fluidly registered a specific individual to all other N subjects were averaged and applied to that subject, preserving the image intensities and anatomical features of the template subject. Susceptibility-corrected FA maps were registered to the final population-averaged FA-based MDT using a 3D elastic warping technique with a mutual information cost function (Leow et al., 2005) and smoothed with a Gaussian kernel (7 mm full width at half-maximum). To better align white matter regions of interest, the MDT and all whole- brain registered FA maps were thresholded at 0.25 (excluding contributions from non-white matter). In this way, the outlines of the major white matter structures are stable and have been normalized to a very fine degree of matching across subjects, greatly reducing the neuroanatomical variations in these structures across subjects.
2.6 MultiSNP Analysis
Linear mixed-effects models were used to study the joint associations of SNPs with imaging measures, while taking into account any relatedness among the subjects. For N subjects and p independent predictors (SNPs or other covariates), regression coefficients (β) were obtained, using the efficient mixed-model association (EMMA; http://mouse.cs.ucla.edu/emma/) software with restricted maximum likelihood estimation (Kang et al., 2008), according to the formula:
| Eq. 2 |
Here, y represents an n-component vector of voxel-wise FA, Dmean, Drad, Dax measures, X is a matrix of SNP genotypes (coded additively as 0, 1, or 2 for the number of minor alleles) and/ or covariates (sex and age), Z is the identity matrix, b is a vector of random effects with a variance of σ2gK, where K is the N × N kinship matrix for the twins and siblings, and ε is a matrix of residual effects with a variance of σ2eI, where I is identity matrix. A kinship matrix coefficient of 1 denoted the relationship of each subject to him/herself; the coefficient for MZ twins within the same family was 1; the coefficient for DZ twins and siblings within the same family was 0.5; and the coefficient for subjects not in the same family was 0, corresponding to the expected proportion of their shared genetic polymorphisms, respectively. Ancestry outliers were removed, so no additional modeling was used in the kinship matrix to adjust for population genetic structure between families. ε is a matrix of residual effects with a variance of σ2eI, and I is an identity matrix. P-values for the significance of individual and joint SNP associations with diffusivity were assessed using a partial F-test, according to the formula:
| Eq. 3 |
Where RSS represents the residual sum-of-squares, p is the p-value of the model, and n is the number of subjects, a reduced model includes only covariates, and a full model contains both SNPs and covariates. Further details can be seen in (Kohannim et al., 2012). For all statistical analyses, the LONI pipeline (http://pipeline.loni.usc.edu/) was used for voxel-wise parallelization on a multi-CPU grid computer. The searchlight false discovery rate method (Langers et al., 2007) was used for multiple comparisons correction across all voxels. As described in further detail in Kohannim et al., 2012, the correction for the number of SNPs input and for each statistical test performed is built into the model. As is the case with all voxel-wise neuroimaging studies, the number of tests is far greater than the number of subjects, so multiple comparisons correction across all voxels is necessary and often involves controlling the false discovery rate at a stringent threshold (Hibar et al., 2011; Jahanshad et al., 2013; Medland et al., Nature Neuroscience 2014). We also ran multi-SNP iteratively, removing the weakest SNP, to determine what panel of SNPs was maximally predictive of WM integrity.
2.7 Candidate gene follow-up
We followed up with individual voxel-wise FA analyses on all of the SNPs in the panel that comprised the “maximally predictive” SNP panel from the iterative multiSNP analysis, correcting for the number of SNPs tested. Of these 7 SNPs, only rs2815752 had associations with FA that passed correction voxel-wise and across all 7 SNPs tested (q<0.0071). The NEGR1 (rs2815752) risk allele (A) is associated with higher BMI, with a per allele change of 0.10–0.13 kg/m2 (Speliotes et al., 2010; Willer et al., 2008). The statistical model used is that listed in Eq. 2, again co-varying for age and sex, and correcting for multiple comparisons using searchlight FDR (Langers et al., 2007). BMI was not significantly associated with FA in our cohort.
2.8 Additional NEGR1 Analyses
To examine the effects that the NEGR1 gene has on white matter integrity in more depth, we next ran a gene-based test, PCReg (principal components regression) (Hibar et al., 2011). In PCReg, the entire list of genotyped SNPs within a gene can be assessed for joint association with a brain measure (here, voxel-wise FA). This is similar to the multiSNP method (Kohannim et al., 2012), but instead of focusing on uncorrelated SNPs that are hypothesized to be related, it includes all the SNPs in a gene, in an attempt to see the larger picture of genetic association with brain measures. Importantly, it can be run on SNPs that are in LD, critical for its use as a gene-based test. PCReg works by first running a principal components analysis on the SNPs, to reduce the dimensions of the analysis, and avoid the complications of collinearity. Components with the highest eigenvalues (higher proportions of explained variance) were included until 80% of the SNP variance was explained, and the rest were discarded. This was followed by a multiple partial-F test, similar to Eq. 3. As this is a gene-based test encompassing the effects of possibly hundreds of SNPs, it does not suggest a directionality for the association; it tests whether a model containing SNPs that explain at least 80% of the variance in NEGR1 are a better predictor of voxel-wise FA than a reduced model containing only age and sex. We generated a list of SNPs within 100kb of NEGR1 and filtered out those with an MAF<0.22 leaving us with 275 NEGR1 SNPs input into PCReg. In this method, the number of degrees of freedom of the F statistic accounts for the number of predictors, and corrects for the number of SNPs input into the model. Further details of this method may be found in Hibar et al., 2011.
3. Results
For our initial multiSNP analyses we selected our SNPs of interest based on the following 3 reports: Speliotes et al. conducted a genome-wide association study (GWAS) across nearly 250,000 individuals to find loci associated with BMI. Willer et al. ran a meta-analysis of 15 genome-wide association studies searching for loci reliably associated with BMI, giving them a total N>32,000, with a follow-up analysis in another dataset of around 59,000 individuals. Thorleifsson et al. also conducted a GWAS of nearly 35,000 individuals to find loci associated with weight and BMI. Some of the SNPs in the 3 GWAS papers (Speliotes et al., 2010; Thorleifsson et al., 2008; Willer et al., 2008) were not in Hapmap3. We further narrowed the list down to those with a minor allele frequency (MAF) >0.22 (to make sure that at least 5% of our subject pool of 499 were homozygous carriers of the minor allele). We additionally excluded 3 SNPs that were in high linkage disequilibrium (LD) with any other SNP we were evaluating (LD>0.4) to reduce data redundancy and avoid the multicollinearity problem for the multiSNP analysis. This resulted in a reduced list of 16 SNPs, listed in Table 2. All genetic analyses – multiSNP and individual SNP – used an additive genetic model that assessed the effect of each additional risk allele. No SNPs deviated significantly from Hardy-Weinberg equilibrium.
Table 2. SNPs included in the multiSNP model.
The gene (nearest when SNP is intergenic), context (intergenic or in intron), minor allele frequency (as listed in dbSNP), risk allele associated with obesity for each SNP, and the GWAS study each SNP was taken from.
| SNP | Nearest Gene |
Context | MAF | Risk Allele |
GWAS Study |
|---|---|---|---|---|---|
| rs10913469 | SEC16B | intron | 0.234 | C | Thorleifsson et al., 2009 |
| rs7647305 | ETV5 | intergenic | 0.2248 | C | Thorleifsson et al., 2009 |
| rs925946 | BDNF-AS | intron | 0.2285 | T | Thorleifsson et al., 2009 |
| rs10501087 | BDNF-AS | intron | 0.2436 | T | Thorleifsson et al., 2009 |
| rs8049439 | ATXN2L | intron | 0.359 | C | Thorleifsson et al., 2009 |
| rs6499640 | FTO | intron | 0.4835 | A | Thorleifsson et al., 2009 |
| rs3751812 | FTO | intron | 0.2413 | T | Thorleifsson et al., 2009 |
| rs9931989 | ATP2A1 | intron | 0.2514 | G | Willer et al., 2008 |
| rs2815752 | NEGR1 | intergenic | 0.3008 | A | Willer et al., 2008 |
| rs10838738 | MTCH2 | intron | 0.2834 | G | Willer et al., 2008 |
| rs571312 | MC4R | intergenic | 0.2372 | A | Speliotes et al., 2010 |
| rs29941 | KCTD15 | intergenic | 0.3965 | C | Speliotes et al., 2010, Thorleifsson et al., 2009 |
| rs7138803 | FAIM2 | intergenic | 0.292 | A | Speliotes et al., 2010, Thorleifsson et al., 2009 |
| rs2241423 | MAP2K5 | intron | 0.4006 | G | Speliotes et al., 2010 |
| rs1514175 | TNN13K | intron | 0.3864 | A | Speliotes et al., 2010 |
| rs10968576 | LRRN6C | intron | 0.2422 | G | Speliotes et al., 2010 |
3.1 MultiSNP Analysis
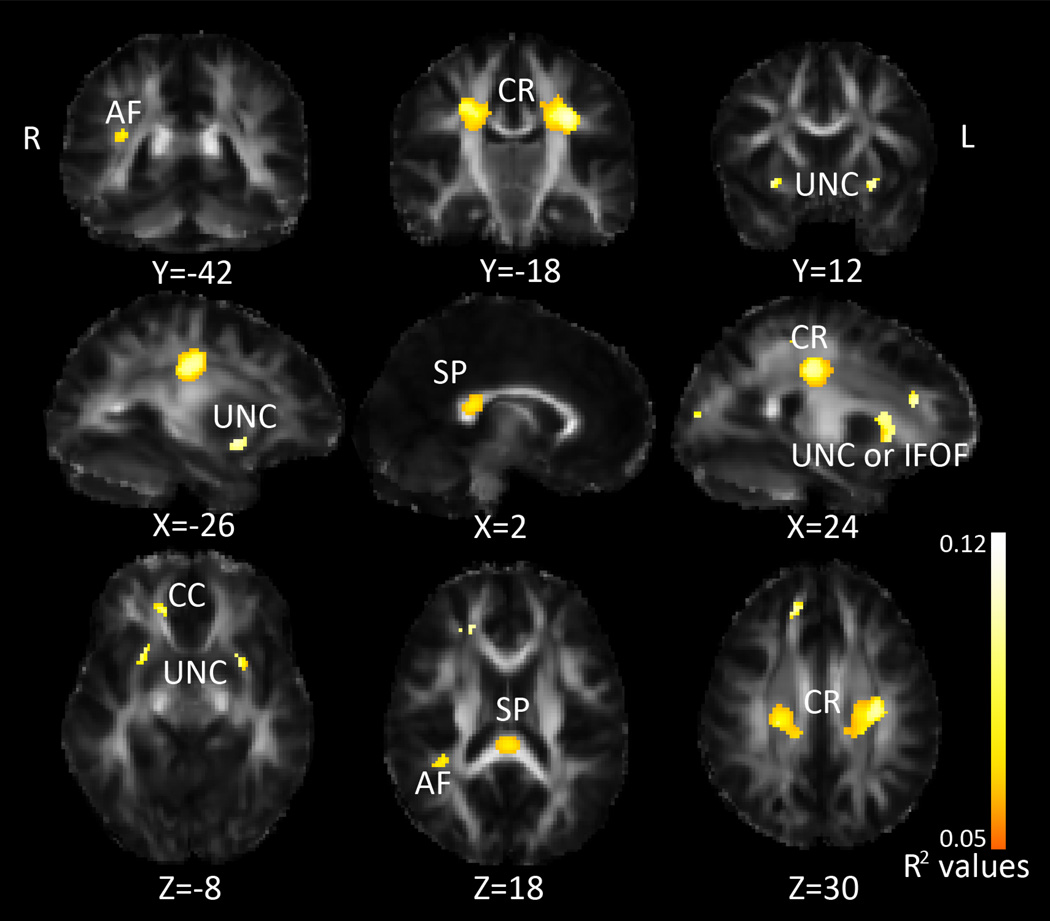
Using DTI data from 499 healthy young adults (mean age = 23.8 years, SD = 2.5, Table 1), we jointly assessed the effect of 16 BMI-related SNPs (Table 2) on FA, Dmean, Drad, and Dax. We started with the multiSNP analysis, as none of these SNPs had yet been associated with white matter connectivity so there was no reason to prioritize any one specifically. This analysis yielded associations between our SNPs and FA in the bilateral corona radiata, corpus callosum, fornix, arcuate, and an area corresponding to both the uncinate and inferior fronto-occipital fasciculus (IFOF), as shown in Figure 2. The multiSNP analysis yields an R2 coefficient, which is the predictability of our model; in Figure 2, R2 is shown only in areas where the association was declared significant after multiple comparisons correction across all voxels in the image considering all the SNPs tested (see Methods). The maximum R2 value (predictability) in these regions was 0.115. The maps for Dmean, Drad, and Dax are shown in Supplementary Figure 1. For Dmean, Drad, and Dax, there were associations with our SNP panel in an area corresponding to both the right uncinate and IFOF, and an area overlapping with the left IFOF and fornix. For Dmean and Drad, there were associations with our SNP panel in the genu, bilateral corona radiata, bilateral internal capsule, right arcuate fasciculus, cingulum, and splenium. There was additionally an area of association between the SNP panel and Dmean in the right forceps minor. The voxel-wise multiSNP method allowed us to determine where in the brain joint information on all 16 SNPs was significantly better able to predict FA than just age and sex alone by establishing significance maps from the partial F-test. We additionally explored submodels to determine if any single one of the 16 SNPs was better at predicting FA when added to the model than sex, age and the remaining 15 SNPs all together; this implies that the SNP is able to predict FA even when covarying for sex, age and all other SNPs. We found several SNPs showed borderline significant associations on their own even when covarying for the other 15 SNPs. While it is not necessary to correct across the number of SNPs tested in the multiSNP model, it is necessary to correct for them when examining the effect of the individual SNPs, if a post hoc inference is made about whether any one of them is explaining variance in the model. While their joint effect did survive voxel-wise multiple comparisons corrections across the whole brain, when covarying for all additional 15 SNPs included, none of the individual SNPs passed a multiple comparisons corrections threshold controlling the false positive rate at q<0.003125 (0.05/16). This underscores the utility of the multiSNP method. As we are covarying for the effect of all other SNPs included in the model, these results are not purely the association between the individual SNP and voxel-wise FA, but also the association controlling for the effect of all other SNPs.
Figure 2. MultiSNP results: Associations between FA and SNPs linked with BMI.
R2 values are combined predictive value of our SNPs, white areas are areas with higher R2 values, as shown by the color bar. CR=corona radiata, IFOF=inferior fronto-occipital fasciculus, CC=corpus callosum, AF=arcuate fasciculus, UNC=uncinate, SP=splenium. Left in the image is right in the brain, coordinates are in MNI space.
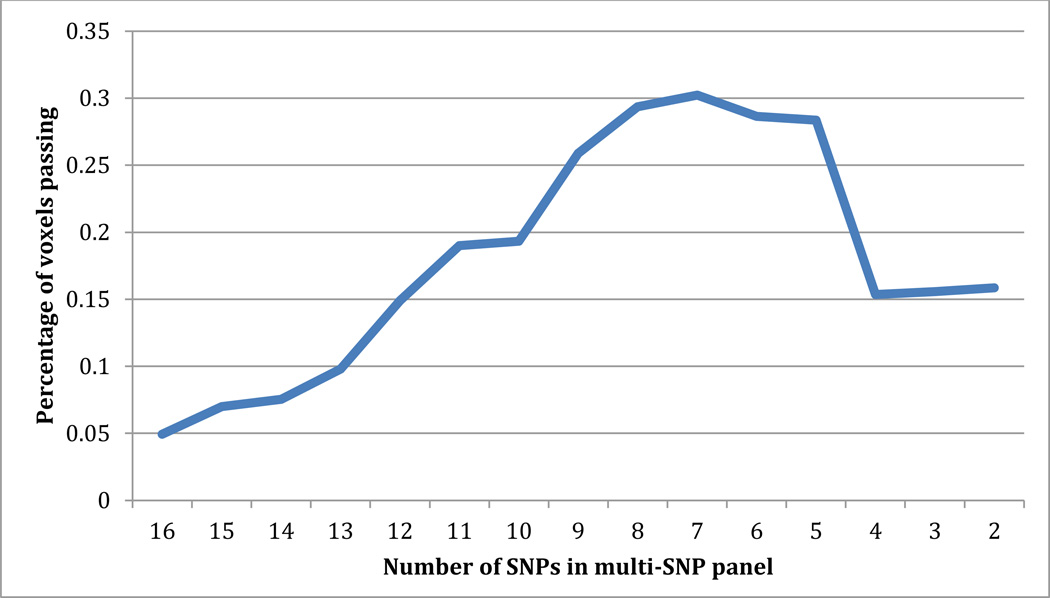
3.2 Iterative MultiSNP Analysis
To determine whether a smaller group of SNPs in the multiSNP panel explained a greater portion of variance, we ran multiSNP iteratively, removing the weakest SNP after each iteration. A graph of the number of SNPs included and the percentage of voxels passing searchlight FDR can be seen in Figure 3. As seen in this figure, the panel including 7 SNPs was most significant. These 7 SNPs were: rs2815752, rs2241423, rs571312, rs925946, rs1514175, rs10913469, and rs10968576. Rs2815752 remained the strongest signal through each iteration. We followed up on all 7 SNPs individually in the voxel-wise FA maps.
Figure 3. Results of iterative multi-SNP analysis.
Multi-SNP was run iteratively, removing the weakest SNP after each iteration to find the strongest panel of SNPs. The plot shows the number of SNPs included in the panel and the percentage of voxels passing searchlight FDR for each iteration. 7 SNPs was the strongest panel.
3.3 Candidate Gene Analyses
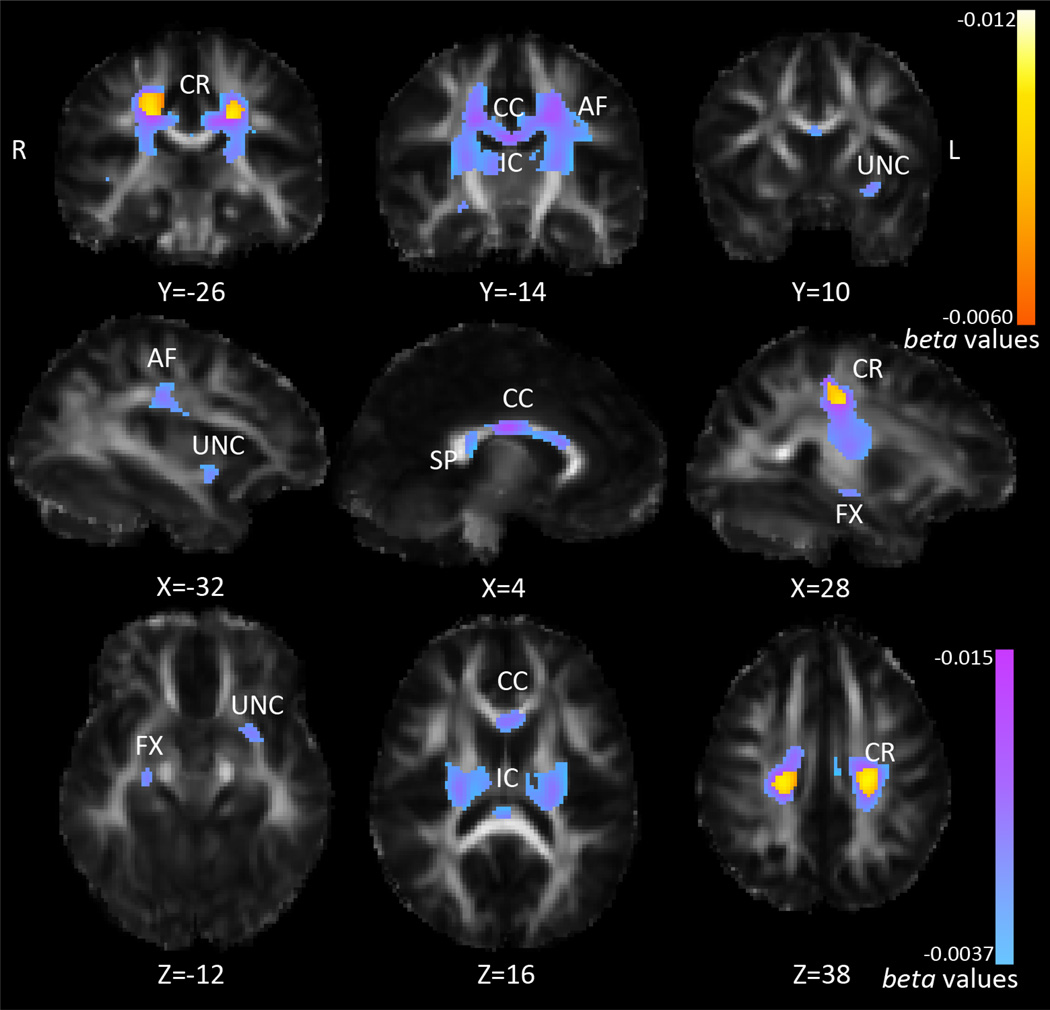
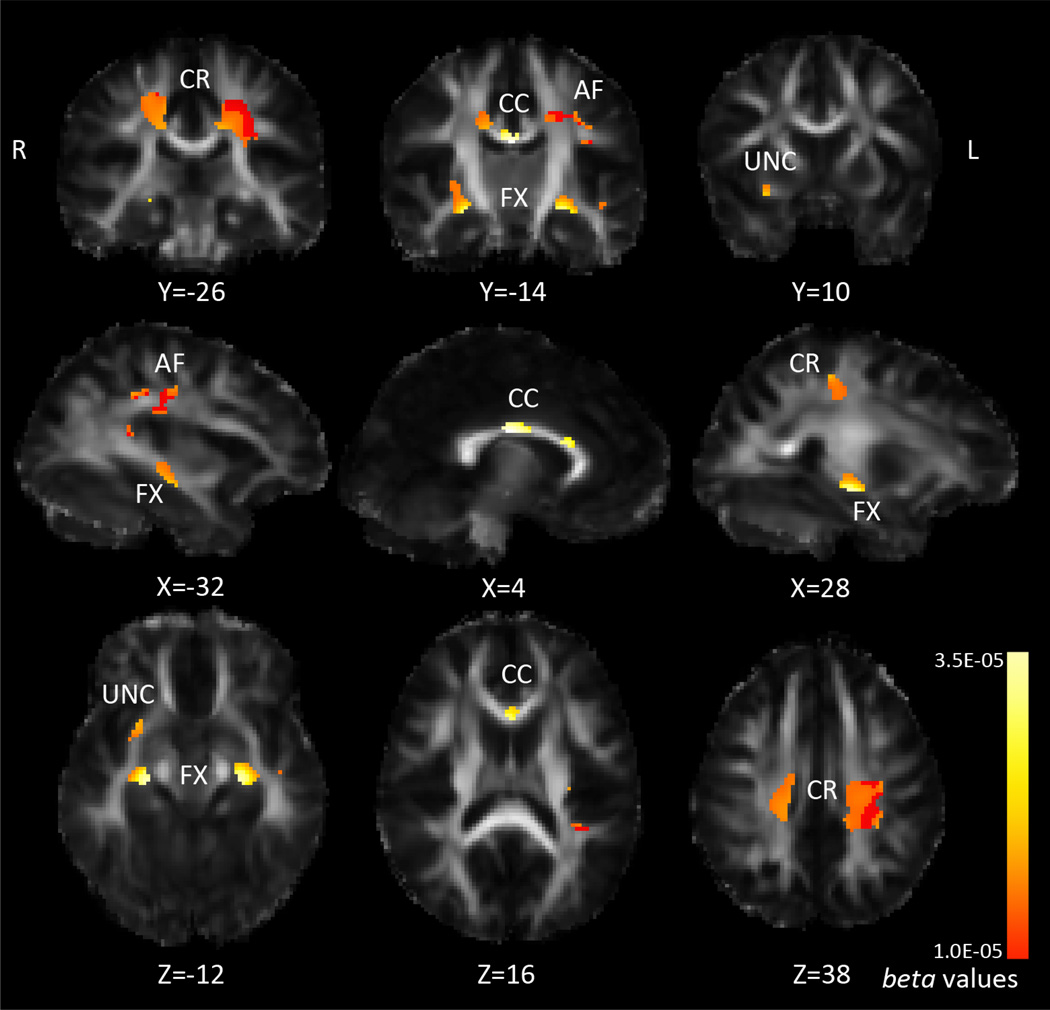
We followed up on all 7 SNPs that comprised the most significant SNP panel, from the iterative multiSNP analysis, correcting for the number of SNPs tested. Of these 7 SNPs, only rs2815752 (NEGR1) had significant associations in the FA maps when we corrected for multiple comparisons across SNPs. We then followed up on rs2815752 with analyses of Dmean, Drad, and Dax (q<0.0071). For rs2815752, 188 subjects were homozygous risk (AA), 233 were heterozygous (AG), and 78 were homozygous non-risk (GG). The minor allele (G) frequency for rs2815752 is 0.301. NEGR1 risk allele dosage was not significantly associated with BMI in our sample (p=0.30), neither was voxel-wise FA. NEGR1 risk allele dosage (A) was negatively associated with FA, as shown in Figure 4. In this figure, we show both the associations that survived corrections across the whole brain, and those that additionally survived correction across all 7 SNPs tested. The posterior body of the corpus callosum and nearby corona radiata showed strongest associations with NEGR1 risk allele dosage (in terms of lowest p-value), but the area of association covered the entire corpus callosum, large areas of the corona radiata, arcuate fasciculus, fornix, internal capsule, and areas that could be the inferior fronto-occipital fasciculus, inferior longitudinal fasciculus, and/or uncinate fasciculus. These last tracts overlap in these areas so we cannot say with confidence that one specific fasciculus is selectively affected. Across the areas of significance (only the voxels that survived whole-brain correction across all 7 SNPs), each risk allele was associated with a 2.2% decrease in average FA. Drad was also positively associated with NEGR1 risk allele dosage, across overlapping areas, as shown in Figure 5. Across the area of significance, each risk allele was associated with a 1.8% increase in average Drad. Again, we covaried for age and sex. There were no significant differences in head motion during scan acquisition across genetic groups (p=0.51), or associations between BMI and motion (p=0.70), which could have explained results as a recent study showed that inadequately accounting for head motion can artificially influence results (Yendiki et al., 2014). A table of the average FA and Drad across the area of significance for each genetic group can be seen in Table 3.
Figure 4. Association between FA and NEGR1 risk allele dosage.
Pink corresponds to stronger beta-values (more negative); larger blue-pink areas are those that pass FDR across brain, q<0.05, smaller yellow-orange areas are those that additionally pass correction for the 7 SNPs tested, q<0.0071. CR=corona radiata, CC=corpus callosum, IC=internal capsule, AF=arcuate fasciculus, SP=splenium, FX=fornix, UNC=uncinate. Left in the image is right in the brain, coordinates are in MNI space.
Figure 5. Voxelwise associations between NEGR1 risk allele dosage and Drad.
Yellow corresponds to stronger beta-values (more positive); only areas surviving FDR across the brain are shown. Left in the image is right in the brain, coordinates are in MNI space.
Table 3. Average measures for FA and Drad across genetic groups.
| Measure | |||
|---|---|---|---|
| Genetic group | FA | Drad | |
| rs2815752 | AA (homozygous risk) | 0.432351 | 0.00060734 |
| AG | 0.444210 | 0.00059130 | |
| GG | 0.451888 | 0.00058535 | |
3.4 Additional NEGR1 Analyses
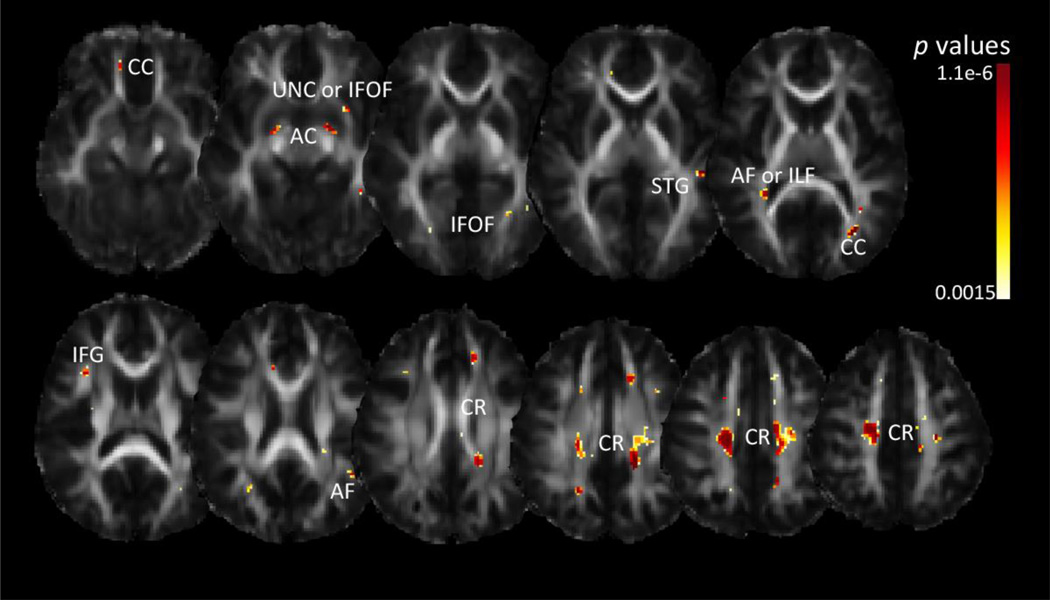
Our gene-based test, PCReg, yielded significant associations between NEGR1 and voxel-wise FA in the corpus callosum, anterior commissure, corona radiata, inferior frontal gyrus, arcuate fasciculus, superior temporal gyrus, and regions corresponding to the inferior fronto-occipital fasciculus or uncinate (Figure 6). Like the multiSNP analysis, PCReg does not yield information on the direction of the association, just the p-value. Additionally, like the multiSNP analysis, there is an implicit correction for the effective number of genetic predictors included in the model, but we avoid the need to correct for the number of SNPs included, as PCA performs data reduction and compaction (see Methods section and Hibar et al., 2011).
Figure 6. Voxelwise associations with FA from NEGR1 whole gene principal components regression.
Dark red corresponds to more significant p-values; only areas surviving FDR across the brain are shown. CR=corona radiata, AF=arcuate fasciculus, CC=corpus callosum, IFG=inferior frontal gyrus, ILF=inferior longitudinal fasciculus, STG=superior temporal gyrus, IFOF=inferior fronto-occipital fasciculus, UNC=uncinate, AC=anterior commissure. Left in the image is right in the brain, coordinates are in MNI space.
4. Discussion
Many genes have been linked to obesity, yet thus far only two studies have examined the effect these obesity genes may have on brain structure (Ho et al., 2010; Horstmann et al., 2013). Here, we revealed a joint effect of a set of obesity-associated SNPs on the brain in young adults, using a multiSNP approach we recently developed for screening brain images (Kohannim et al., 2012). The predictive power of these SNPs overlapped in the bilateral posterior corona radiata, arcuate, corpus callosum, fornix, and uncinate or IFOF (Figure 2). A further analysis of the SNPs to reveal any particular variant contributing most to this effect yielded widespread negative associations between FA and NEGR1 risk allele dosage of rs2815752 in our sample. To our knowledge this is the first paper to report an association between an obesity-related gene and white matter (WM) integrity. A recent paper by our group used this approach to find associations between WM and serum cholesterol and cholesterol-related SNPs (Warstadt et al., in press).
We began with the multiSNP analysis because it is a way to search for joint effects of a set of genetic variants on brain measures (Kohannim et al., 2012). FTO and MC4R are the only obesity-related gene previously associated with brain structural differences, so we did not have strong prior evidence to supported prioritizing a particular gene (besides FTO and MC4R). Of our 16 SNPs associated with obesity, a number of them converged in effect in the posterior corona radiata. Once our results showed that our panel of BMI-associated SNPs indeed was related to WM integrity, we delved further into determining which individual SNPs were most predictive of WM integrity. An iterative multiSNP analysis showed that the most significant panel of SNPs included 7 SNPs, indicating that our initial list of SNPs included some that were not significantly helpful in explaining variance in FA. These 7 SNPs were: rs2815752, rs2241423, rs571312, rs925946, rs1514175, rs10913469, and rs10968576. Of these 7 SNPs, only NEGR1 had significant associations with voxel-wise FA, correcting across all 7 SNPs tested. The rs2815752 SNP is just upstream of the NEGR1 gene, and the A risk allele tags a 45kb deletion (Jarick et al., 2011). NEGR1 codes for the protein NEGR1 or neurotractin - a member of the neural IgLON subgroup of the immunoglobin superfamily. Neurotractin is a cell adhesion molecule that plays a key role in neural development (Marg et al., 1999). In mice, NEGR1 is widely expressed in the brain. Mutations causing NEGR1 loss of function led to decreased body mass in mice in vivo, and decreases in cell adhesion and neurite growth in vitro (Lee et al., 2012). The NEGR1 risk allele (A) has been associated with higher BMI (per allele change 0.10–0.13 kg/m2; Speliotes et al., 2010; Willer et al., 2008).
No prior studies have linked NEGR1 risk allele dosage to brain differences in humans. However, its role in mouse brain neural development makes it a plausible candidate. Healthy adults carrying the risk allele had lower FA across a wide swath of central white matter (Figure 4). Combined with the results of increased Drad in risk allele carriers, our results point to lower white matter integrity with NEGR1 risk allele dosage. Across the area of significance, the difference in mean FA per risk allele was a 2.2% decrease. Alzheimer’s disease has been associated with decreases up to 33% in FA (Nir et al., 2013), so this is a modest but perhaps eventually significant difference among young, healthy individuals. Future studies will hopefully be able to test this association in independent samples. For example, we recently created a worldwide consortium dedicated to replicating genetic effects on the brain (Stein et al., 2012; Hibar et al., 2013; Thompson et al., 2013), and a multi-site GWAS of diffusion images is underway (Jahanshad et al., 2013; Kochunov et al., 2014). Obese individuals have significantly decreased volume in the corona radiata, where we detected significant associations (Alkan et al., 2008). Although there are exceptions, lower FA and higher MD are usually signs of decreased myelination or fiber coherence (Thomason & Thompson et al., 2011; Dennis & Thompson et al., 2013). Middle-aged obese patients show widespread increases in ADC (apparent diffusion coefficient, equivalent to mean diffusivity – Dmean) in middle-aged obese patients (Alkan et al., 2008). As NEGR1 plays a role in neural development, we could be detecting effects of lower myelination in NEGR1 risk allele carriers. We did not find any significant associations between BMI and FA in our cohort, and our subjects were aged 20–30, so it is highly unlikely that these results are chronic effects of obesity and lifestyle factors. BMI-related SNPs could also affect the brain in ways not mediated by obesity. In other words, they could have a direct effect on the brain (e.g. influencing motivation/personality, etc.). We did have some overweight and obese subjects in our sample, as noted in Figure 1, but did not find any significant differences in overweight or obese groups. While obesity rates in developed countries are typically close to 30%, our sample was quite a bit healthier, with only 6% obese and 20% overweight. We believe this is a strength of our paper, as it demonstrates that our results are more gene-related, rather than a consequence of obesity. With the makeup of our sample, our results indicate that NEGR1 can have a negative effect on white matter integrity independent of its effects on obesity risk. We can investigate whether this association holds in a sample including more obese subjects. ENIMGA-DTI is a consortium including over 2000 subjects that will allow us to address this question (Kochunov, Jahanshad et al., 2014). With this data we can test whether there are interactions between SNPs and obesity.
We also conducted a second NEGR1 analysis, running a gene-based test (called ‘PCReg’) on 275 SNPs in NEGR1 (Hibar et al., 2011). We found a large cluster of significant association in the bilateral posterior corona radiata, where we found associations in our multiSNP analysis and in our analysis of rs2815752. PCReg does not output a beta value summed across SNPs used in the model, it shows areas where the effects on a brain measure within a gene aggregate. In other smaller clusters, voxel-wise FA was significantly associated with NEGR1. The fact that we found a large association in the same area as the rs2815752 analysis suggests that there are other variations within NEGR1 that are associated with FA in the posterior corona radiata. The aim of PCReg is to see the bigger picture of genetic association of a single gene with brain measures, as we know that SNPs are not isolated variants causing brain changes. PCReg shows the associations of the SNPs in aggregate; many may have effects too small to detect individually, and rs2815752 may not be the main effect SNP within NEGR1. These results strengthen the idea that the proteins encoded by NEGR1 may play a role in WM integrity. PCReg allows us to see small effects summed, and gives us greater confidence in our rs2815752 results.
Obesity (BMI > 30 kg/m2) in midlife is associated with an increased risk of dementia later in life (Fitzpatrick et al., 2009). Our subjects did not show any associations between BMI and FA, and NEGR1 risk allele dosage was not associated with BMI. Our young adult subjects may not have had a chance for the obesity genes to have an effect, and we only had 499 subjects, which is very large for a brain imaging study, but small for a genetics study. The original studies finding an effect of these genes on obesity did so in sample sizes >30,000 with an average age around 50. We are examining a younger cohort, so brain changes may pre-date any clinical effects on BMI. The three GWAS studies (Speliotes et al., 2010; Thorleifsson et al., 2008; Willer et al., 2008) all included cohorts with average ages largely between 30–80, and were heavily weighted towards middle-aged subjects (∼50 years old). Obese subjects may have lower white matter integrity in the corpus callosum (Mueller et al., 2011; Xu et al., 2013; Marks et al., 2011) and fornix (Marks et al., 2011). One reason for this may be inflammation, as one group has found a positive association between a marker of inflammation and apparent diffusion coefficient (same as mean diffusivity) (Cazettes et al., 2011). Verstynen et al., (2013) similarly found that inflammation was a significant mediating factor in the association between adiposity and FA. Obesity is now recognized as an inflammatory disease, causing chronic, subacute inflammation (Shoelson et al., 2007). We did not find any areas of significant association between FA and BMI, but the areas found by others are generally those where we found our gene associations. The regional overlap with previous studies of BMI associations with white matter integrity suggests that NEGR1 may be one of many factors contributing to the association between BMI and white matter integrity of the corpus callosum and fornix. Our results indicate that the NEGR1 A-allele was associated with negative effects on white matter integrity in healthy, young adults, independent of effects on BMI. While in our mostly healthy-BMI sample, we found no BMI-associations with white matter integrity, genes previously found to be associated with BMI and lead to an increased risk of obesity maintained an association with brain structure. When controlling for all other tested variants, NEGR1 showed the strongest individual effect. This association may suggest a genetic relation to brain structure that is independent of obesity. Further evaluation is needed to determine if the neuroanatomical pathways compromised by this obesity-risk gene themselves indicate a mechanistic pathway for obesity.
5. Conclusions
In this study we used an innovative multi-locus approach to examine the joint effect of obesity-associated SNPs on white matter integrity in young, healthy adults. We found a panel of SNPs that jointly influenced central white matter integrity. We found the most extensive effects with NEGR1, which was associated with a lowered FA, 2.2% per allele across the area of significance. Our results indicate that the obesity risk gene NEGR1 is associated with lowered white matter integrity in young healthy individuals, mostly without obesity-related complications. Our results may help uncover mechanisms through which NEGR1 has its effects on the brain. To what degree the link between genetics and brain effects is mediated by diet and lifestyle choices is still an open and complex question.
Supplementary Material
Acknowledgments
This study was supported by the National Institute of Child Health and Human Development (R01 HD050735), and the National Health and Medical Research Council (NHMRC 486682, 1009064), Australia. Genotyping was supported by NHMRC (389875). Additional support for algorithm development was provided by NIH R01 grants EB008432, EB008281, EB007813 and P41 RR013642. ED was funded, in part, by an NIH Training Grant in Neurobehavioral Genetics (T32 MH073526-06), and by the Betty B. and James B. Lambert Scholarship from the Kappa Alpha Theta Foundation.
Footnotes
Body mass index is a ratio of weight to height, intended as an approximate but readily computed assessment of fat mass. The equation to calculate BMI (in SI units) is BMI=mass (kg)/(height (m))2
Author Disclosure Statement: The authors have no competing financial interests.
References
- Alkan A, Sahin I, Keskin L, Cikim AS, Karakas HM, Sigirci A, Erdem G. Diffusion-weighted imaging features of brain in obesity. Magnetic Resonance Imaging. 2008;26(4):446–450. doi: 10.1016/j.mri.2007.10.004. [DOI] [PubMed] [Google Scholar]
- Ars C. The Effects of Prenatal and Early Postnatal Fat and Sugar Intake on Hippocampal Volume and Memory Performance in Healthy Dutch Children aged 6–8 (Doctoral dissertation) 2012 Retrieved from http://arno.unimaas.nl/show.cgi?fid=26241.
- Braskie MN, Jahanshad N, Stein JL, Barysheva M, Johnson K, McMahon KL, et al. Relationship of a variant in the NTRK1 gene to white matter microstructure in young adults. J Neuroscience. 2012;32(17):5964–5972. doi: 10.1523/JNEUROSCI.5561-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braskie MN, Jahanshad N, Stein JL, Barysheva M, McMahon KL, de Zubicaray GI, et al. Common Alzheimer’s disease risk variant within the CLU gene affects white matter microstructure in young adults. J Neuroscience. 2011;31(18):6764–6770. doi: 10.1523/JNEUROSCI.5794-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cazettes F, Cohen JI, Yau PL, Talbot H, Convit A. Obesity-mediated inflammation may damage the brain circuit that regulates food intake. Brain Research. 2011;1373:101–109. doi: 10.1016/j.brainres.2010.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciccarelli O, Catani M, Johansen-Berg H, Clark C, Thompson A. Diffusion-based tractography in neurological disorders: concepts, applications and future developments. The Lancet Neurology. 2008;7(8):715–727. doi: 10.1016/S1474-4422(08)70163-7. [DOI] [PubMed] [Google Scholar]
- Dennis EL, Thompson PM. Mapping connectivity in the developing brain. International Journal of Developmental Neuroscience. 2013;31(7):525–542. doi: 10.1016/j.ijdevneu.2013.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Zubicaray GI, Chiang M-C, McMahon KL, Shattuck DV, Toga AW, Martin NG, Wright MJ, Thompson PM. Meeting the challenges of neuroimaging genetics. Brain Imaging and Behavior. 2008;2:258–263. doi: 10.1007/s11682-008-9029-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duffy DL, Iles MM, Glass D, Zhu G, Barrett JH, Höiom V, et al. IRF4 Variants have age-specific effects on nevus count and predispose to melanoma. Am J Hum Genetics. 2010;87:6–16. doi: 10.1016/j.ajhg.2010.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischl B, van der Kouwe A, Destrieux C, Halgren E, Ségonne F, Salat DH, et al. Automatically parcellating the human cerebral cortex. Cerebral Cortex. 2004;14(1):11–22. doi: 10.1093/cercor/bhg087. [DOI] [PubMed] [Google Scholar]
- Fitzpatrick AL, Kuller LH, Lopez OL, Diehr P, O’Meara ES, Longstreth WT, Luchsinger JA. Midlife and late-life obesity and the risk of dementia: cardiovascular health study. Archives of Neurology. 2009;66(3):336–342. doi: 10.1001/archneurol.2008.582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frayling TM, Timpson NJ, Weedon MN, Zeggini E, Freathy RM, Lindgren CM, et al. A common variant in the FTO gene is associated with body mass index and predisposes to childhood and adult obesity. Science. 2007;316:889–894. doi: 10.1126/science.1141634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gazdzinski S, Millin R, Kaiser LG, Durazzo TC, Mueller SG, Weiner MW, Meyerhoff DJ. BMI and neuronal integrity in healthy, cognitively normal elderly: A proton magnetic resonance spectroscopy study. Obesity. 2010;18(4):743–748. doi: 10.1038/oby.2009.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustafson D, Lissner L, Bengtsson C, Björkelund C, Skoog I. A 24-year follow-up of body mass index and cerebral atrophy. Neurology. 2004;63(10):1876–1881. doi: 10.1212/01.wnl.0000141850.47773.5f. [DOI] [PubMed] [Google Scholar]
- Haltia LT, Viljanen A, Parkkola R, Kemppainen N, Rinne JO, Nuutila P, Kaasinen V. Brain white matter expansion in human obesity and the recovering effect of dieting. Journal of Clinical Endocrinology & Metabolism. 2007;92(8):3278–3284. doi: 10.1210/jc.2006-2495. [DOI] [PubMed] [Google Scholar]
- Hibar DP, Stein JL, Kohannim O, Jahanshad N, Saykin AJ, Shen L, et al. Voxelwise gene-wide association study (vGeneWAS): Multivariate gene-based association testing in 731 elderly subjects. NeuroImage. 2011;56(4):1875–1891. doi: 10.1016/j.neuroimage.2011.03.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hibar DP + 200 authors for the ENIGMA Consortium. ENIGMA2: Genome-wide scans of subcortical brain volumes in 16,125 subjects from 28 cohorts worldwide. Seattle, WA: OHBM; 2013. Jun, [Google Scholar]
- Ho AJ, Stein JL, Hua X, Lee S, Hibar DP, Leow AD, et al. A commonly carried allele of the obesity-related FTO gene is associated with reduced brain volume in the healthy elderly. Proceedings of the National Academy of Sciences. 2010;107(18):8404–8409. doi: 10.1073/pnas.0910878107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes CJ, Hoge R, Collins L, Woods R, Toga AW, Evans AC. Enhancement of MR Images Using Registration for Signal Averaging. Journal of Computer Assisted Tomography. 1998;22(2):324–333. doi: 10.1097/00004728-199803000-00032. [DOI] [PubMed] [Google Scholar]
- Horstmann A, Kovacs P, Kabisch S, Boettcher Y, Schloegl H, Tönjes A, et al. Common genetic variation near MC4R has a sex-specific impact on human brain structure and eating behavior. PLoS ONE. 2013;8(9):e74362. doi: 10.1371/journal.pone.0074362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua X, Leow AD, Lee S, Klunder AD, Toga AW, Lepore N, et al. 3D characterization of brain atrophy in Alzheimer’s disease and mild cognitive impairment using tensor-based morphometry. NeuroImage. 2008;41(1):19–34. doi: 10.1016/j.neuroimage.2008.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iglesias JE, Liu C-Y, Thompson PM, Tu Z. Robust extraction across datasets and comparison with publicly available methods. IEEE Trans Med Imaging. 2011;30:1617–1634. doi: 10.1109/TMI.2011.2138152. [DOI] [PubMed] [Google Scholar]
- Jahanshad N, Kochunov PV, Sprooten E, Mandl RC, Nichols TE, Almasy L, et al. Multi-site genetic analysis of diffusion images and voxelwise heritability analysis: A pilot project of the ENIGMA-DTI working group. Neuroimage. 2013;81:455–469. doi: 10.1016/j.neuroimage.2013.04.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahanshad N, Kohannim O, Hibar DP, Stein JL, McMahon KL, de Zubicaray GI, et al. Brain structure in healthy adults is related to serum transferrin and the H63D polymorphism in the HFE gene. PNAS. 2012;109(14):E851–E859. doi: 10.1073/pnas.1105543109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahanshad N, Lee A, Barysheva M, McMahon K, de Zubicaray GI, Martin NG, Wright MJ, Toga AW, Thompson PM. Genetic influences on brain asymmetry: A DTI study of 374 twins and siblings. NeuroImage. 2010a;52:455–469. doi: 10.1016/j.neuroimage.2010.04.236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahanshad N, Rajagopalan P, Hua X, Hibar DP, Nir TM, Toga AW, et al. Genome-wide scan of healthy human connectome discovers SPON1 variant influencing dementia severity. PNAS. 2013 doi: 10.1073/pnas.1216206110. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarick I, Vogel CIG, Scherag S, Schäfer H, Hebebrand J, Hinney A, Scherag A. Novel common copy number variation for early onset extreme obesity on chromosome 11q11 identified by a genome-wide analysis. Human Molecular Genetics. 2011;20(4):840–852. doi: 10.1093/hmg/ddq518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkinson M, Bannister P, Brady M, Smith S. Improved optimization for the robust and accurate linear registration and motion correction of brain images. NeuroImage. 2002;17:825–841. doi: 10.1016/s1053-8119(02)91132-8. [DOI] [PubMed] [Google Scholar]
- Kang HM, Zaitlen NA, Wade CM, Kirby A, Heckerman D, Daly MJ, Eskin E. Efficient control of population structure in model organism association mapping. Genetics. 2008;178(3):1709–1723. doi: 10.1534/genetics.107.080101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kochunov P, Jahanshad N, Sprooten E, Nichols TE, Mandl RC, Almasy L, et al. Multi-site study of additive genetic effects on fractional anisotropy of cerebral white matter: comparing meta and mega analytical approaches for data pooling. NeuroImage. 2014 doi: 10.1016/j.neuroimage.2014.03.033. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohannim O, Jahanshad N, Braskie MN, Stein JL, Chiang M-C, Reese AH, et al. Predicting White Matter Integrity from Multiple Common Genetic Variants. Neuropsychopharmacology. 2012;37(9):2012–2019. doi: 10.1038/npp.2012.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langers DR, Jansen JF, Backes WH. Enhanced signal detection in neuroimaging by means of regional control of the global false discovery rate. NeuroImage. 2007;38:43–56. doi: 10.1016/j.neuroimage.2007.07.031. [DOI] [PubMed] [Google Scholar]
- Lee AWS, Hengstler H, Schwald K, Berriel-Diaz M, Loreth D, Kirsch M, et al. Functional inactivation of the genome-wide association study obesity gene neuronal growth regulator 1 in mice causes a body mass phenotype. PLoS ONE. 2012;7(7):e41537. doi: 10.1371/journal.pone.0041537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leow A, Huang S-C, Geng A, Becker J, Davis S, Toga A, Thompson P. Inverse consistent mapping in 3D deformable image registration: its construction and statistical properties. Processing in Medical Imaging. 2005;3565:23–57. doi: 10.1007/11505730_41. [DOI] [PubMed] [Google Scholar]
- Leow AD, Yanovsky I, Chang M-C, Lee AD, Klunder AD, Lu A, et al. Statistical properties of Jacobian maps and the realization of unbiased large-deformation nonlinear image registration. IEEE Trans Med Imaging. 2007;26:822–832. doi: 10.1109/TMI.2007.892646. [DOI] [PubMed] [Google Scholar]
- Leporé N, Brun C, Pennec X, Chou Y-Y, Lopez OL, Aizenstein HJ, et al. Proc. 10th. Australia: MICCAI, Brisbane; 2007. Mean template for tensor-based morphometry using deformation tensors; pp. 826–833. [DOI] [PubMed] [Google Scholar]
- Loos RJF, Lindgren CM, Li S, Wheeler E, Zhao JH, Prokopenko I, et al. Common variants near MC4R are associated with fat mass, weight and risk of obesity. Nature Genetics. 2008;40:768–775. doi: 10.1038/ng.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marg A, Sirim P, Spaltmann F, Plagge A, Kauselmann G, Buck F, Rathjen FG, Brümmendorf T. Neurotractin, a novel neurite outgrowth-promoting Ig-like protein that interacts with CEPU-1 and LAMP. The Journal of Cell Biology. 1999;145(4):865–876. doi: 10.1083/jcb.145.4.865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marks BL, Katz LM, Styner M, Smith JK. Aerobic fitness and obesity: relationship to cerebral white matter integrity in the brain of active and sedentary older adults. Br J Sports Med. 2011;45:1208–1215. doi: 10.1136/bjsm.2009.068114. [DOI] [PubMed] [Google Scholar]
- Medland SE, Jahanshad N, Neale BM, Thompson PM. Whole-genome analyses of whole-brain data: working within an expanded search space. Nature Neuroscience. 2014;17:791–800. doi: 10.1038/nn.3718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medland SE, Nyholt DR, Painter JN, McEvoy BP, McRae AF, Zhu G, et al. Common variants in the trichohyalin gene are associated with straight hair in europeans. Am J Hum Gen. 2009;85(5):750–755. doi: 10.1016/j.ajhg.2009.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molteni R, Barnard RJ, Ying Z, Roberts CK, Gomez-Pinilla F. A high-fat, refined sugar diet reduces hippocampal brain-derived neurotrophic factor, neuronal plasticity, and learning. Neuroscience. 2002;112(4):803–814. doi: 10.1016/s0306-4522(02)00123-9. [DOI] [PubMed] [Google Scholar]
- Mueller K, Anwander A, Möller HE, Horstmann A, Lepsien J, Busse F, et al. Sex-dependent influences of obesity on cerebral white matter investigated by diffusion-tensor imaging. PLoS ONE. 2011;6(4):e18544. doi: 10.1371/journal.pone.0018544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Must A, Spadano J, Coakley EH, Field AE, Colditz G, Dietz WH. The disease burden associated with overweight and obesity. JAMA. 1999;282(16):1523–1529. doi: 10.1001/jama.282.16.1523. [DOI] [PubMed] [Google Scholar]
- Ng MCY, Hester JM, Wing MR, Li J, Xu J, Hicks PJ, et al. Genome-wide association of BMI in African Americans. Obesity. 2012;20(3):622–627. doi: 10.1038/oby.2011.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nir TM, Jahanshad N, Villalon-Reina JE, Toga AW, Jack CR, Weiner MW, et al. Effectiveness of regional DTI measures in distinguishing Alzheimer’s disease, MCI and, normal aging. NeuroImage: Clinical. 2013 doi: 10.1016/j.nicl.2013.07.006. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Northstone K, Joinson C, Emmett P, Ness A, Paus T. Are dietary patterns in childhood associated with IQ at 8 years of age? A population-based cohort study. Journal of Epidemiology & Community Health. 2012;66:624–628. doi: 10.1136/jech.2010.111955. [DOI] [PubMed] [Google Scholar]
- Ogden CL, Carroll MD, Kit BK, Flegal KM. Prevalence of obesity in the United States, 2009–2010. NCHS Data Brief. 2012:82. [PubMed] [Google Scholar]
- Okada Y, Kubo M, Ohmiya H, Takahashi A, Kumasaka N, Hosono N, et al. Common variants at CDKAL1 and KLF9 are associated with body mass index in east Asian populations. Nature Genetics. 2012;44:302–306. doi: 10.1038/ng.1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pannacciulli N, Del Parigi A, Chen K, Le DSNT, Reiman EM, Tataranni PA. Brain abnormalities in human obesity: A voxel-based morphometric study. NeuroImage. 2006;31:1419–1425. doi: 10.1016/j.neuroimage.2006.01.047. [DOI] [PubMed] [Google Scholar]
- Potkin SG, Guffanti G, Lakatos A, Turner JA, Kruggel F, Fallon JH, et al. Hippocampal atrophy as a quantitative trait in a genome-wide association study identifying novel susceptibility genes for Alzheimer’s disease. PLoS ONE. 2009;4(8):e6501. doi: 10.1371/journal.pone.0006501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purves D, Augustine GJ, Fitzpatrick D, et al. Neuroscience. 2nd edition. Sunderland (MA: Sinauer Associates; 2001. Increased Conduction Velocity as a Result of Myelination.http://www.ncbi.nlm.nih.gov/books/NBK10921/ Available from. [Google Scholar]
- Raji CA, Ho AJ, Parikshak NN, Becker JT, Lopez OL, Kuller LH, et al. Brain structure and obesity. Human Brain Mapping. 2009;31:353–364. doi: 10.1002/hbm.20870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoelson SE, Herrero L, Naaz A. Obesity, inflammation, and insulin resistance. Gastroenterology. 2007;132(6):2169–2180. doi: 10.1053/j.gastro.2007.03.059. [DOI] [PubMed] [Google Scholar]
- Smith SM. Fast robust automated brain extraction. Human Brain Mapping. 2002;17(3):143–155. doi: 10.1002/hbm.10062. 41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speliotes EK, Willer CJ, Berndt SI, Monda KL, Thorleifsson G, Jackson AU, et al. Association analyses of 249,796 individuals reveal 18 new loci associated with body mass index. Nature Genetics. 2010;42(11):937–948. doi: 10.1038/ng.686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanek KM, Grieve SM, Brickman AM, Korgaonkar MS, Paul RH, Cohen RA, Gunstad JJ. Obesity is associated with reduced white matter integrity in otherwise healthy adults. Obesity. 2009;19:500–504. doi: 10.1038/oby.2010.312. [DOI] [PubMed] [Google Scholar]
- Stice E, Spoor S, Bohon C, Small DM. Relation between obesity and blunted striatal response to food is moderated by Taq1A A1 allele. Science. 2008;322(5900):449–452. doi: 10.1126/science.1161550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein JL, Medland SE, Vasquez AA, Hibar DP, Senstad RE, Winkler AM, et al. Identification of common variants associated with human hippocampal and intracranial volumes. Nature Genetics. 2012;44(5):552–562. doi: 10.1038/ng.2250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taki Y, Kinomura S, Sato K, Inoue K, Goto R, Okada K, Uchida S, Kawashima R, Fukuda H. Relationship between body mass index and gray matter volume in 1,428 healthy individuals. Obesity. 2008;16:119–124. doi: 10.1038/oby.2007.4. [DOI] [PubMed] [Google Scholar]
- Thomason ME, Thompson PM. Diffusion imaging, white matter, and psychopathology. Annual Review of Clinical Psychology. 2011;7:63–85. doi: 10.1146/annurev-clinpsy-032210-104507. [DOI] [PubMed] [Google Scholar]
- Thompson PM, Stein JL, Medland SE, Vasquez AA, Hibar DP, Renteria M, et al. for the ENIGMA Consortium#. The ENIGMA Consortium: Large-scale Collaborative Analyses of Neuroimaging and Genetic Data. In: John D, van Horn , editors. Brain Imaging & Behavior, Special Issue on Imaging Genetics. 2013. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorleifsson G, Walters GB, Gudbjartsson DF, Steinthorsdottir V, Sulem P, Helgadottir A, et al. Genome-wide association yields new sequence variants at seven loci that associate with measures of obesity. Nature Genetics. 2008;41(1):18–24. doi: 10.1038/ng.274. [DOI] [PubMed] [Google Scholar]
- Verstynen TD, Weinstein A, Erickson KI, Sheu LK, Marsland AL, Gianaros PJ. Competing physiological pathways link individual differences in weight and abdominal adiposity to white matter microstructure. NeuroImage. 2013;79:129–137. doi: 10.1016/j.neuroimage.2013.04.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verstynen TD, Weinstein AM, Schneider WW, Jakicic JM, Rofey DL, Erickson KI. Increased body mass index is associated with a global and distributed decrease in white matter microstructural integrity. Psychosom Med. 2012;74(7):682–690. doi: 10.1097/PSY.0b013e318261909c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walther K, Birdsill AC, Glisky EL, Ryan L. Structural brain differences and cognitive functioning related to body mass index in older females. Human Brain Mapping. 2009;31:1052–1064. doi: 10.1002/hbm.20916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward MA, Carlsson CM, Trivedi MA, Sager MA, Johnson SC. The effect of body mass index on global brain volume in middle-aged adults: a cross sectional study. BMC Neurology. 2005;5(23) doi: 10.1186/1471-2377-5-23. 10.1186/1471-2377-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wardle J, Carnell S, Haworth CMA, Plomin R. Evidence for a strong genetic influence on childhood adiposity despite the force of the obesogenic environment. Am J Clin Nutr. 2008;87(2):398–404. doi: 10.1093/ajcn/87.2.398. [DOI] [PubMed] [Google Scholar]
- Warstadt NM, Dennis EL, Jahanshad N, Kohannim O, Nir TM, McMahon KL, et al. Serum cholesterol and variant in cholesterol-related gene CETP predict white matter microstructure. Neurobiology of Aging. 2014 doi: 10.1016/j.neurobiolaging.2014.05.024. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen W, Cho Y-S, Zheng W, Dorajoo R, Kato N, Qi L, et al. Meta-analysis identified common variants associated with body mass index in east Asians. Nature Genetics. 2012;44:307–311. doi: 10.1038/ng.1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willer CJ, Speliotes EK, Loos RJF, Li S, Lindgren CM, Heid IM, et al. Six new loci associated with body mass index highlight a neuronal influence on body weight regulation. Nature Genetics. 2008;41(1):25–34. doi: 10.1038/ng.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Li Y, Lin H, Sinha R, Potenza MN. Body mass index correlates negatively with white matter integrity in the fornix and corpus callosum: A diffusion tensor imaging study. Hum Brain Mapp. 2013;34:1044–1052. doi: 10.1002/hbm.21491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yendiki A, Koldewyn K, Kakunoori S, Kanwisher N, Fischl B. Spurious group differences due to head motion in a diffusion MRI study. NeuroImage. 2014;88:79–90. doi: 10.1016/j.neuroimage.2013.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.