ABSTRACT
Background:
Hepatitis B virus (HBV) infection is a major global health problem in the worldwide that associated with significant morbidity and mortality in cardiac surgery. The available data on HBV distribution and genotyping of HBV are very heterogeneous. Therefore in this study, we tried to indicate the prevalence of HBV infections in cardiac catheterization patients referred to health centers in the north of Iran and identified the HBV genotypes using polymerase chain reaction (PCR).
Methods:
In this cross-sectional study, we studied 2650 patients who underwent selective coronary artery angiography and coronary artery bypass grafting in Mazandaran heart center, Sari, Iran from 2011 to 2013. All serum samples were examined to detect HBsAg by ELISA test. HBV-DNA was extracted from HBsAg positive samples using Mini Elute Kit from Qiagen and determined the genotypes of HBV by PCR using the Master Mix kit with Taq-DNA polymerase enzyme and with type of specific primers. All samples were examined in the virology laboratory of Sari Medical School.
Results:
The mean age of patients was 59.7±10.9 (range, 20 to 81) year that 1590 (60%) patients were male and 1060 (40%) were female. Seventeen cases (0.08 %) were found with hepatitis B virus infection, and the highest rates of infection were reported among those aged 40–60 years old in this study. We found genotype D the predominant type in this study.
Conclusion:
This study indicates that the prevalence of HBV endemicity in the north of Iran is low and genotype D is the only genotype in patients infected with HBV.
Keywords: Hepatitis B virus, Acute Hepatitis, Genotype of HBV, PCR
1. INTRODUCTION
The acute and chronic consequence of Hepatitis B virus (HBV) infection is the major public health problem worldwide. It can cause liver diseases including chronic hepatitis, cirrhosis and hepatocellular carcinoma (1, 2). The HBV, is highly contagious and relatively easy to transmit by blood-to-blood contact, during birth, unprotected sex, and by sharing needles in the tropics. It is estimated that one third of the world population have been infected by HBV and more than 350 million of people live with chronic infection of these; and nearly one million people die annually from complications of chronic hepatitis B (3-4). Throughout the world, carrier variability rate for hepatitis B infection is estimated to be 0.1% to 20% (5), with regions classified as having low (<2%), intermediate (2-7%) and high (>8%) endemicity. In Iran, it is estimated that almost 35% of population have been exposed to HBV and the endemicity is at intermediate level, with a carrier rated 3% (6). It was reported that HBV infection indicates an intermediate rate in this country and the distribution of carrier rate of HBV infection in varies different provinces of Iran (1.3% to 6.3%) (7). After HBV vaccination program, Iran can be considered as one of the countries with low HBV infection endemicity (8). HBV screening of all patients undergoing invasive cardiac procedures can minimize the risk of transmission from one patient to another and also to the catheterization laboratory personnel. In patients with a positive result, proper precautions can be taken and the entire material used for cardiac catheterization can be discarded (9).
The HBV belongs to hepadnaviridae family and is an enveloped, double-stranded DNA genome of approximately 3200 bass pairs. So far, ten HBV genotypes (A-J) and multiple subtypes have been identified (10-16) and the genotypes of HBV have distinct geographical distribution of the world (17-19). The study was showed that Turkish patients with chronic hepatitis B infection indicated very little genotypic heterogeneity. Genotype D of HBV represented that almost the whole Turkish patient population were infected with HBV (20, 21). In Pakistan, genotype D was the predominant type found in 128 (64%) patients followed by A in 47 (23%) and mixed A/D in 26 (13%) cases (22). A study was reported that 65.34% were classified into genotype D, 26.73% were of genotype B while 4.95% had genotype A. So in 2.98% samples, multiple genotypes were detected (genotype A+B; 1.98% and genotypes B+D; 1%) (23). In India, HBV genotype D was the most predominant (56.0%) genotype followed by HBV genotype C (23.4%) and HBV genotype A (20.6%) (24). The study demonstrated that genotype D (35.67%) is the predominant genotype circulating in Afghan population and followed by genotype C (32.16%), genotype A (19.30%), and genotype B (7.02%) (25). In Iran, Genotype D of HBV made almost the whole patient population infected with HBV in different clinical forms (26-30). In the north of lran, genotype D was found in 93% of HBV positive patients followed by genotype B in 7% of HBV positive patients (31). In Kermansha province, genotype D was found in 98.8% of HBV positive patients followed by genotype B in 1.2% of HBV positive patients (32). The distribution of HBV genotypes may guide us in determining the disease burden. HBV genotypes have been shown to differ with regard to prognosis, clinical outcomes and antiviral responses (33, 34). So, it is important to recognize the epidemiology of HBV genotyping as well and respond to this question: Is necessary, routine screening performed for hepatitis B in patients undergoing cardiac catheterization? The available data of the prevalence of HBV infection and the genotype of HBV in Iran are very heterogeneous. Therefore, this study was designed to determine and analyze the distribution of HBV infections in cardiac catheterization patients referred to the health centers in the north of lran and identified the HBV genotypes applying polymerase chain reaction (PCR) during 2011-2013.
2. MATERIALS AND METHODS
In this cross-sectional study, during 2010-2013, serum samples were collected from 2560 patients who refereed to Mazandaran heart center, Sari, Iran. Centrifuged and separated plasmas were immediately stored at -80°C. All samples had elevated serum aminotransferases a positive test for anti-HBsAg using enzyme linked immunosorbent assay (ELISA), and determined genotypes of HBV based using DNA extraction kit and appropriate protocol. A question was used to collect some information including the patients age, gender, employment status, and place of residence. These data were analyzed using descriptive statistics and SPSS V.19 and Chi-square test.
2.1. DNA extraction
DNA extraction from plasma samples was done using Mini EluteKit (Qiagen) and appropriate protocol. HBV was isolated from serum on following procedures; 200plasma of patients with 200 AL Buffer (Lyses Buffer) mixed for 15 seconds by vortex and incubated at 65°C for 15 minutes and then centrifuged quickly. Afterwards 250 Ethanol (96%-100%) was added and mixed for 15 seconds by vortex and incubated at room temperature, which was then added to QIAMP Mini Elute. Samples were centrifuged for one minute at 8000 revolutions per minute after finishing centrifuge, overlaid fluid separated and added in to the Sam volume. Next, tube content remained solution was washed with 500of AW1 Buffer and centrifuged for one minute at 8000 revolutions per minute and discharged overlaid fluid, and washed with 500of AW2 Buffer and then washed with 500of Ethanol 96%-100%. Sample tubes were centrifuged again for one minute at 8000 revolutions per minute and incubate for 3 minutes at 65°C for drying. Finally, the pellet was re-suspended in 50 sterile distilled water and DNA quantification was determined using a spectrophotometer and resulted residue was solved for next stages. All samples were examined in the virology laboratory of Sari Medical School.
2.2. PCR Test
Plasma samples from HBsAg positive were confirmed for the presence of HBV nucleic acid and determined the genotypes of HBV genome. Total DNA was isolated from serum samples and PCR-Test was done using special Kit and according to special protocol with individual primers. All steps of preparing relational mix over ice resells have been performed and primers, after diluting, were stored in -2o”c. Examination method summarized as follow: To make Master Mix 3%:1075,.U distilled water, dNTP 30)ll, PCR Buffer lOX 150)ll and MgCl2 90,.U. 11.5,.U of above reaction was mixed with 0.2,.U of Taq DNA polymerase, 40 pmol of each forward and reverse primers, 4ng of DNA sample and added dH20 that final reaction volume was 20,.U. The samples were placed into the Eppendorf Master Cycler PCR machine and amplified. PCR program for amplification consisted of95°C for 5 minutes, followed by 35 cycles of94°C for 1 minute, 58.5°C for 1 minute and 72°C for 1 minute and finally, followed by 72°C for 10 minute.
2.3. Gel Electrophoresis
Agarose gel electrophoresis is an easy way to separate and visualize DNA fragments by their sizes. It is a common diagnostic procedure used in molecular biological labs. We use 1.5% Agarose gel loaded the DNA fragment from samples then they were separated by size. This is a graphic representation of an Agarose gel made by “running” DNA molecular weight markers. These gels were visualized on a UV analyzer by staining the DNA with a fluorescent dye (Ethidium bromide which is very carcinogenic). The DNA molecular weight marker is a set of DNA fragments of known molecular sizes that are used as a standard to determine the sizes of fragments.
3. RESULTS
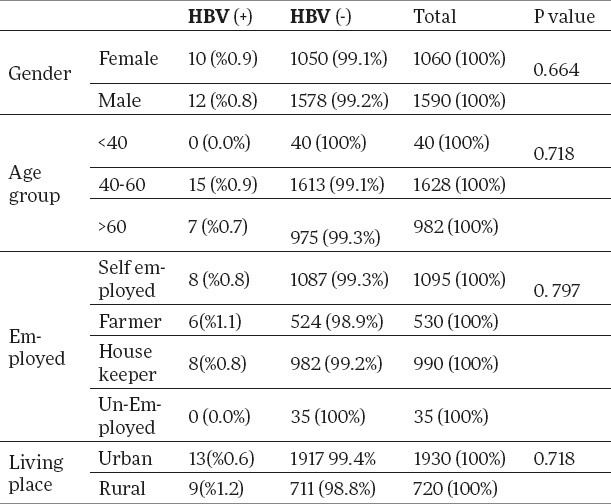
During study period, we studied 2650 cardiac catheterization patients attending Mazandaran Heart Center in the North of Iran (Mazandaran province) from 2011 to 2013 were studied. The mean age of patients was 59.7±10.9 (range, 20 to 81) year that 1590 (60%) patients were male and 1060 (40%) was female. We found no patients with chronic hepatitis or had acute hepatitis. Among the patients 89% were married and 11% were single. All samples had elevated serum aminotransferases a positive test for anti-HBsAg using enzyme linked immunosorbent assay (ELISA) using HBsAb and appropriate protocol, and determined genotypes of HBV based using DNA extraction kit and appropriate protocol. Genotyping was done using restriction fragment length polymorphism of HBV DNA positive serum samples. All of these HBV-DNA positive patients were infected with genotype of HBV-D. In this study, 17 cases (0.08 %) were found with hepatitis B virus infection, and the highest rates of infection were reported among the 40-60 years old population. There was no significant relation between HBV genotypes and gender, disease and age group (Table 1).
Table 1.
Distribution of HBV according to gender, age group, employed and living place.
HBV-DNA was extracted from HBsAg positive samples using Mini Elute Kit from Qiagen and determined the genotypes of HBV by PCR using the Master Mix kit with Taq-DNA polymerase enzyme and with type of specific primers. All samples were examined in the virology laboratory of Sari Medical School. We found that the genotype D of HBV was the only genotype in the HBsAg positive patients in this study (Figure 1).
Figure 1.

Distribution of HBV-D genotype in patients who referred to Mazandaran Heart Center during 2011-2013.
The electrophoresis pattern of HBV genotyping products were done by PCR using genotype specific primers. M is 100 bp marker, 1-4 are HBV positive samples, 5 and 6 are control positive samples.
4. DISCUSSION
The carrier variability rate for hepatitis B infection is estimated to be 0.1% to 20% throughout the world (5). In the Middle East, it has been repot1ed that HBV infection is an intermediate rate, varying between 2% and 7% in different countries (35). In blood donors, this rate was 0.8 %in Iran (36), 4% in Pakistan (37), 1.1-3.5 %in Kuwait (38), 4.19 %in Turkey (39) and 1.9% in Saudi Arabia (40). It was reported that the HBV infection indicates an intermediate rate in this country and the distribution of carrier rate of HBV infection in the different provinces of Iran was shown to be different (1.3% to 6.3%) (7). After HBV vaccination program, Iran can be considered as one of the countries with low HBV infection endemicity (8). Our results showed that the prevalence of HBV was 8 of cases in 1000 patients (%0.8), which highlights the value of HBV vaccination program. It can be concluded that, HBsAg rate among blood donors in Iran is still less in comparison with other neighboring countries of lran.
At present, HBV has been classified into ten different genotypes (A-J) and multiple subtypes by genome sequencing of HBV strains (10-16). Genotypes of HBV have different geographic distribution in the world (41, 42). In this study, we found the genotype D of HBV in all HBsAg positive patients. Same to our study, it has been shown the HBV genotype D is distributed worldwide, and has been reported frequently (23-26, 43). Some studies from different part of lran have reported that the genotype D is the only detectable genotype in the different clinical forms of HBV infection, including carriers HBV, chronic liver disease and cirrhosis (26-30) that is the same as our results. But, in the north of Iran, genotype D was found in 93% of HBV positive patients followed by genotype Bin 7% (31), and in another study, genotype D was found in 98.8% of HBV positive patients followed by genotype Bin 1.2% (32), that was different from our result because we found the genotype D of HBV in all samples of this study. It has been reported that the distribution of HBV genotypes was not just one genotype but some samples were mixed. It has been reported that 62.2% were found genotype D, 13% were found genotype A and 12%. :Mixed genotype D+A was found in 12% of acute patients, 5.6% of chronic patients and 5.6% of carriers (43), 85.1% were genotyped as type D/E, 4.4% were genotyped as type A, 1.4% were genotyped as type C, and 0.7% were genotyped as type F (44) and other study was indicated the HBV genotype frequencies were: B, 57.9%; C, 16.0%; and BC, 26.1% (45). A study conducted among injecting drug users showed that the presence of genotype Din 62%, genotype A in 9% while 29% individuals were found to be infected with a mixture of genotype A and D (46). Genotypes A and D were most prevalent in co-infected patients with HlV and HBV and so, HBV subtype A was present among three-fourth of patients infected through sexual contact, whereas the same percentage of subtype D was isolated among injection drug users (47).
HBV genotyping may guide us in selection of the duration and type of antiviral therapy and to predict the likelihood of sustained HBV clearance after therapy. It seems that there are different types of HBV genotypes in different parts of countries, due to wide range of geographical distribution and influence of the neighbors in the abundance of different types of HBV genotypes. Furthermore, factors such as repeated blood transfusion and treatment of the patients also can be some of the most important criteria which can cause this wide range of different genotypes. In conclusion, this research describes HBV genotyping in studied patients infected with HBV. This preliminary report describes that the genotype D is not the only genotype in the patients with HBsAg, which is different from other studies. Thus, further studies are needed to achieve the confirmation and so to determine the distribution of genotype of HBV subtype in patients with HBV positive.
Acknowledgments
This study was granted (grant number: 91-53) by Vice-Chancellor for Research of Mazandaran University of Medical Sciences. The authors highly thank all coworkers in Mazandaran heart center, Sari, Iran.
Footnotes
CONFLICT OF INTEREST: NONE DECLARED.
REFERENCES
- 1.Lavanchy D. Hepatitis B virus epidemiology, disease burden, treatment, and current and emerging prevention and control measures. J Viral Hepat. 2004;11(2):97–107. doi: 10.1046/j.1365-2893.2003.00487.x. [DOI] [PubMed] [Google Scholar]
- 2.Maynard JE. Hepatitis B: global importance and need for control. Vaccine. 1990;8:18–20. doi: 10.1016/0264-410x(90)90209-5. [DOI] [PubMed] [Google Scholar]
- 3.Kane Global program for control of hepatitis B infection. Vaccine. 1995;13:S47–S9. doi: 10.1016/0264-410x(95)80050-n. [DOI] [PubMed] [Google Scholar]
- 4.Shepard CW, Simard EP, Finelli L, Fiore AE, Bell BP. Hepatitis B virus infection: epidemiology and vaccination. Epidemiology Review. 2006;28(1):112–125. doi: 10.1093/epirev/mxj009. [DOI] [PubMed] [Google Scholar]
- 5.Lee WM. Hepatitis B virus infection. N Engl J Med. 1997;337(24):1733–1745. doi: 10.1056/NEJM199712113372406. [DOI] [PubMed] [Google Scholar]
- 6.Alavian SM. Hepatitis B virus infection in Iran;Changing the epidemiology. Iran J Clin Infect Dis. 2010;5(1):51–61. [Google Scholar]
- 7.Alavian SM, Hajarizadeh B, Ahmadzad-Asl M, Kabir A, Bagheri-Lankarani K. Hepatitis B virus infection in Iran: A Systematic Review. Hepatitis monthly. 2008;8(4):281–294. [Google Scholar]
- 8.Alavian SM. Ministry of Health in Iran Is Serious about Controlling Hepatitis B. Hepatitis Monthly. 2007;7(1):3–5. [Google Scholar]
- 9.Satish OS, Raghu C, Lakshmi V, Rao DS. Routine screening for HIV and hepatitis B in patients undergoing cardiac catheterisation: the need to make it mandatory. Indian Heart J. 1999;51(3):285–288. [PubMed] [Google Scholar]
- 10.Ganguly A, Rock MJ, Prockop DJ. Conforrnation-sensitive gel electrophoresis for rapid detection of single-base differences in double-stranded PCR products and DNA fragments: evidence for solvent-induced bends in DNA heteroduplexes. Proc Natl Acad Sci USA. 1993;90(21):10325–10329. doi: 10.1073/pnas.90.21.10325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Norder H, Courouce AM, Magnius LO. Complete genomes, phylogenetic relatedness, and structural proteins of six strains of the hepatitis B virus, four of which represent two new genotypes. Virology. 1994;198(2):489–503. doi: 10.1006/viro.1994.1060. [DOI] [PubMed] [Google Scholar]
- 12.Sakai T, Shiraki K, Inoue H, Okano H, Deguchi M, Sugimoto K, et al. HBV subtype as a marker of the clinical course of chronic HBV infection in Japanese patients. J Med Virol. 2002;68(2):175–181. doi: 10.1002/jmv.10180. [DOI] [PubMed] [Google Scholar]
- 13.okamoto H, Tsuda F, Sakugawa H, Sastrosoewignjo RI, Imai M, Miyakawa Y, et al. Typing hepatitis B virus by homology in nucleotide sequence: comparison of surface antigen subtypes. J Gen Virol. 1988;69(pt 10):2575–2583. doi: 10.1099/0022-1317-69-10-2575. [DOI] [PubMed] [Google Scholar]
- 14.Tong W, He J, Sun L, He S, Qi Q. Hepatitis B virus with a proposed genotype I was found in Sichuan Province, China. J Med Virol. 2012;84(6):866–870. doi: 10.1002/jmv.23279. [DOI] [PubMed] [Google Scholar]
- 15.Huy TTT, Ngoc TT, Abe K. New .Complex Recombinant Genotype of Hepatitis B Virus Identified in Vietnam. Journal of Virology. 2008;82(11):5657–5663. doi: 10.1128/JVI.02556-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tatematsu K, Tanaka Y, Kurbanov F, Sugauchi F, Mano S, Maeshiro T, et al. A Genetic Variant of Hepatitis B Virus Divergent from Known Human and Ape Genotypes Isolated from a Japanese Patient and Provisionally Assigned to New Genotype J. J Viral. 2009;8(20):10538–10547. doi: 10.1128/JVI.00462-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Alavian SM, Keyvani H, Rezai M, Ashayeri N, Sadeghi HM. Preliminary report of hepatitis B virus genotype prevalence in Iran. World J Gastroenterol. 2006;12(32):5211–5213. doi: 10.3748/wjg.v12.i32.5211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Norder H, Courouce AM, Coursaget P, Echevarria JM, Lee SD, Mushahwar IK, et al. Genetic diversity ofhepatitis B virus strains derived worldwide: genotypes, subgenotypes, and HBsAg subtypes. Intervirology. 2000;47(6):289–309. doi: 10.1159/000080872. [DOI] [PubMed] [Google Scholar]
- 19.Kramvis A, Weitzmann L, Owiredu WK, Kew MC. Analysis of the complete genome of subgroup A’ hepatitis B virus isolates from South Africa. J Gen Virol. 2002;83(pt 4):835–839. doi: 10.1099/0022-1317-83-4-835. [DOI] [PubMed] [Google Scholar]
- 20.Bozdayi G, Tiirkyilmaz AR, Idilman R, Karatayli E, Rota S, Yurdaydin C, et al. Complete genome sequence and phylogenetic analysis of hepatitis B virus isolated from Turkish patients with chronic HBV infection. J Med Virol. 2005;76(4):476–481. doi: 10.1002/jmv.20386. [DOI] [PubMed] [Google Scholar]
- 21.Bozdayi AM, Asian N, Bozdayi G, Tiirkyilmaz AR, Sengezer T, Wend U, et al. Molecular epidemiology of hepatitis B, C and D viruses in Turkish patients. Arch Virol. 2004;149(11):2115–2129. doi: 10.1007/s00705-004-0363-2. [DOI] [PubMed] [Google Scholar]
- 22.Baig S, Siddiqui AA, Chakravarty R, Moatter T, Unnissa T, Hasnain N. Phylogenetic analysis of hepatitis B virus in Pakistan. J Coli Physicians Surg Pak. 2008;18(11):688–694. [PubMed] [Google Scholar]
- 23.Alam MM, Zaidi SZ, Malik SA, Shaukat S, Naeem A, Sharif S, et al. Molecular epidemiology of Hepatitis B virus genotypes in Pakistan. BMC Infect Dis. 2007;8(7):115. doi: 10.1186/1471-2334-7-115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Biswas A, Chandra PK, Datta S, Panigrahi R, Banerjee A, Chakrabarti S, et al. Frequency and distribution of hepatitis B virus genotypes among eastern Indian voluntary blood donors: Association with precore and basal core promoter mutations. Hepatol Res. 2009;39(1):53–59. doi: 10.1111/j.1872-034X.2008.00403.x. [DOI] [PubMed] [Google Scholar]
- 25.Attaullah S, Rehman SU, Khan S, Ali I, Ali S, Khan SN. Prevalence of Hepatitis B virus genotypes in HBsAg positive individuals of Afghanistan. Virol J. 2001;7(8):281. doi: 10.1186/1743-422X-8-281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mohebbi SR, Amini-Bavil-Olyaee S, Zali N, Noorinayer B, Derakhshan F, Chiani M, et al. Molecular epidemiology of hepatitis B virus in Iran. Clin Microbial Infect. 2008;14(9):858–866. doi: 10.1111/j.1469-0691.2008.02053.x. [DOI] [PubMed] [Google Scholar]
- 27.Bahramali G, Sadeghizadeh M, Amini-Bavil-Olyaee S, Alavian SM, Behzad-Behbahani A, Adeli A, et al. Clinical, virologic and phylogenetic features of hepatitis B infection in Iranian patients. World J Gastroenterol. 2008;14(35):5448–5453. doi: 10.3748/wjg.14.5448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Amini-Bavil-Olyaee S, Sarrami-Forooshani R, Adeli A, Sabahi F, Abachi M, Azizi M, et al. Complete genomic sequence and phylogenetic relatedness of hepatitis B virus isolates from Iran. J Med Virol. 2005;76(3):318–326. doi: 10.1002/jmv.20362. [DOI] [PubMed] [Google Scholar]
- 29.Mojiri A, Behzad-Behbahani A, Saberifirozi M, Ardabili M, Beheshti M, Rahsaz M, et al. Hepatitis B virus genotypes in southwest Iran: molecular, serological and clinical outcomes. World J Gastroenterol. 2008;14(10):1510–1513. doi: 10.3748/wjg.14.1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vaezjalali M, Alavian SM, Jazayeri SM, Nategh R, Mahmoodi M, Hajibeigi B, et al. Genotype of Hepatitis B Virus Isolates from Iranian Chronic Carriers of the Virus. Hepattis Monthly. 2008;8(2):97–100. [Google Scholar]
- 31.Haghshenas MR, Mousavi T, Rafiei AR, Hosseini V, Hosseinikha Z. prevalence of Hepatitis B virus genotypes with HBsAg positive patients in the Northern oflran (Mazandaran) during 2010-2011. HealthMED. 2012;6(5):1568–1573. [Google Scholar]
- 32.Dokanehifard S, Bidmeshkipour A. A study of hepatitis B virus(HBV) genotypes in kermanshahprovince, west of iran. Journal ofbiological sciences. 2009;1(1):113–120. [Google Scholar]
- 33.Yuen MF, Tanaka Y, Mizokami M, Yuen JC, Wong DK, Yuan HCJ, et al. Role of hepatitis B virus genotypes Ba and C, core promoter and precore mutations on hepatocellular carcinoma: a case control study. Carcinogenesis. 2004;25(9):1593–1598. doi: 10.1093/carcin/bgh172. [DOI] [PubMed] [Google Scholar]
- 34.Thakur V, Sarin SK, Rehman S, Guptan RC, Kazim SN, Kumar S. Role ofHBV genotype in predicting response to lamivudine therapy in patients with chronic hepatitis B. Indian J Gastroenterol. 2005;24(1):12–15. [PubMed] [Google Scholar]
- 35.Qirbi N, Hall AJ. Epidemiology of hepatitis B virus infection in the Middle East.East.Mediterr. Health J. 2001;7(6):1034–1045. [PubMed] [Google Scholar]
- 36.Mahmoodian-Shooshtari M, Pourfathollah A. An overview analysis of blood donation in the Islamic Republic of lran. Arch lran Med. 2006;9(3):200–203. [PubMed] [Google Scholar]
- 37.Alam MM, Zaidi SZ, Malik SA, Naeem A, Shaukat S, Sharif S, et al. Serology based disease status of Pakistani population infected with Hepatitis B virus. BMC Jnfec Dis. 2007;27(7):64. doi: 10.1186/1471-2334-7-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ameen R, Sanad N, Al-Shemmari S, Siddique I, Chowdhury RI, Al-Hamdan S, et al. Prevalence of viral markers among first-time Arab blood donors in Kuwait. Transfusion. 2005;45(12):1973–1980. doi: 10.1111/j.1537-2995.2005.00635.x. [DOI] [PubMed] [Google Scholar]
- 39.Gurol E, Saban C, Oral O, Cigdem A, Armagan A. Trends in hepatitis Band hepatitis C virus among blood donors over 16 years in Turkey. Eur J Epidemiol. 2006;21(4):299–305. doi: 10.1007/s10654-006-0001-2. [DOI] [PubMed] [Google Scholar]
- 40.Panhotra BR, Al-Bahrani A, Ul-Hassan Z. Epiderniology of antibody to hepatitis B core antigen screening among blood donors in Eastern Saudi Arabia. Need to replace the test by HBV DNA testing. Saudi Med J. 2005;26(2):270–273. [PubMed] [Google Scholar]
- 41.Khawaja RA, Khawaja AA. Hepatitis B virus Genotypes: “Clinical & amp: Therapeutic Implications. J Pak Med Assoc. 2009;59(2):101–104. [PubMed] [Google Scholar]
- 42.Miyakawa Y, Mizokarni M. Classitying hepatitis B virus genotypes. Intervirology. 2003;46(6):329–338. doi: 10.1159/000074988. [DOI] [PubMed] [Google Scholar]
- 43.Baig S, Siddiqui AA, Ahmed W, Qureshi H, Arif A. The association of complex liver disorders with HBV genotypes prevalent in Pakistan. Virol J. 2007;27(4):128. doi: 10.1186/1743-422X-4-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kaya S, Cetin ES, Aridogan BC, Onal S, Dernirci M. Distribution of Hepatitis B Virus (HBV) Genotypes among HBV Carriers in sparta. Iran Biomed J. 2007;11(1):59–63. [PubMed] [Google Scholar]
- 45.Li DD, Ding L, Wang J, Meilang QC, Lu XJ, Song XB, et al. Prevalence of Hepatitis B Virus Genotypes and their Relationship to Clinical Laboratory Outcomes in Tibetan and Han Chinese. J Int Med Res. 2010;38(1):195–201. doi: 10.1177/147323001003800122. [DOI] [PubMed] [Google Scholar]
- 46.Alam MM, Zaidi SZ, Shaukat S, Sharif S, Mehar Angez, Naeem A, et al. Common Genotypes of Hepatitis B virus prevalent in Injecting drug abusers (addicts) of North West Frontier Province of Pakistan. Virol J. 2007;28(4):63. doi: 10.1186/1743-422X-4-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Olmeda MP, Nunez M, Samaniego JG, Rios P, Lahoz JG, Soriano V. Distribution ofHBV genotype in HIV-Infected patients with chronic HBV. Therapeutic implications. Aids Res Hum Retroviruses. 2004;19(8):657–659. doi: 10.1089/088922203322280874. [DOI] [PubMed] [Google Scholar]


