Abstract
Purpose
The purpose of this study was to evaluate the feasibility of using a short echo time, 3D H-1 magnetic resonance spectroscopic imaging (MRSI) sequence at 7T to assess the metabolic signature of lesions for patients with glioma.
Materials and Methods
29 patients with glioma were studied. MRSI data were obtained using CHESS water suppression, spectrally-selective adiabatic inversion-recovery pulses and automatically prescribed outer-volume-suppression for lipid suppression, and spin echo slice selection (TE=30ms). An interleaved flyback echo-planar trajectory was applied to shorten the total acquisition time (~10min). Relative metabolite ratios were estimated in tumor and in normal-appearing white and gray matter (NAWM, GM).
Results
Levels of glutamine, myo-inositol, glycine and glutathione relative to total creatine (tCr) were significantly increased in the T2 lesions for all tumor grades compared to those in the NAWM (p < 0.05), while N-acetyl aspartate to tCr were significantly decreased (p < 0.05). In grade 2 gliomas, level of total choline-containing-compounds to tCr was significantly increased (p = 0.0137), while glutamate to tCr was significantly reduced (p = 0.0012).
Conclusion
The improved sensitivity of MRSI and the increased number of metabolites that can be evaluated using 7T MR scanners is of interest for evaluating patients with glioma. This study has successfully demonstrated the application of a short-echo spin-echo MRSI sequence to detect characteristic differences in regions of tumor versus normal appearing brain.
Keywords: magnetic resonance spectroscopic imaging, glioma, 7T
Introduction
It is estimated that 68,530 new cases of primary brain and central nervous system tumors were diagnosed in the United States in 2012 with 24,170 being malignant (1). Gliomas account for the majority of primary malignant brain tumors in adults. Patients have a variable prognosis, ranging from a median survival of 15 months for glioblastoma (GBM) (2), to over 10 years in the case of oligodendroglioma. Although low-grade lesions have a better prognosis, they often undergo transformation to a more malignant, higher grade at the time of recurrence. Both primary and recurrent gliomas infiltrate into adjacent brain tissue, making it difficult to define tumor margins. Proper diagnosis and grading, correct localization, and assessment of response to therapy are of great importance for all phases of treatment planning and selection.
Conventional T1- and T2-weighted MR imaging (MRI) are applied in conjunction with an injection of a Gadolinium-based contrast agent to delineate structural abnormalities in the brain and assess regions where the brain-blood barrier has been compromised. Regions of hyperintensity on T2-weighted images are used to evaluate tumors, but fail to distinguish between edema, gliosis, inflammation, cyst, and active tumor. New anti-angiogenic therapies in patients with GBM, such as agents that target vascular endothelial growth factor, have been shown to decrease the extent of contrast enhancement within the lesion (3). This makes conventional MRI even more challenging in distinguishing tumor recurrence from non-specific treatment effects (4).
Proton magnetic resonance spectroscopic imaging (MRSI) is a powerful noninvasive tool for assessing changes in the metabolic properties of the tumor and surrounding normal brain for patients with glioma. Spectra acquired with long echo time (TE) offer several major metabolites that have been widely studied in patients, including total choline containing compounds (tCho), total creatine (tCr), N-acetyl aspartate (NAA), lactate, and lipid. Elevations in levels of tCho and reductions in levels of NAA have been used as biomarkers for distinguishing regions of tumor from normal brain (5). High levels of lactate and lipid were reported to be the best predictors for poor overall survival in patients with GBM (6).
The availability of improved hardware and acquisition techniques offers the possibilities of obtaining in vivo MRSI at high field strengths, such as 7T (7–10). Previous studies have showed a linear gain in SNR with the increase of field strengths and improved spectral quantification of metabolites at 7T compared to current clinical scanners (8,11). The increase in SNR can be used to shorten the total acquisition time and/or increase the spatial resolution, and heightened spectral resolution can improve the detection of metabolites with lower concentration and/or complex J-coupling patterns such as myo-inositol (mI), glutamate (Glu), glutamine (Gln), and glutathione (GSH), which are difficult to isolate into their individual components at 3T. Ex vivo high resolution magic-angle-spinning nuclear magnetic resonance (HRMAS-NMR) spectroscopy from the image-guided tissue samples have shown that mI/Cho is of interest for differentiating between tumor and gliosis (12), and that malignant transformation in recurrent low-grade glioma is associated with changes in levels of mI, 2-hydroxygluatarate (2-HG), and/or GSH (13–15). These findings provide strong motivation for performing high field in vivo studies of patients with glioma.
The purpose of this study was to implement and test a 3D short-echo-time spin-echo MRSI sequence using a 7T whole body scanner within a clinically feasible acquisition time in patients who were undergoing treatment for glioma. After validation in phantoms, the sequence was applied to patients with glioma in order to characterize the metabolic signatures of lesions and to compare metabolite levels in tumor vs. normal appearing white matter (NAWM).
Materials and Methods
Study population
The sequence was developed using a standard GE MRS phantom (3mM Cho, 10 mM Cr, 12.5 mM NAA, 12.5 mM Glu, 7.5 mM mI and 5 mM Lac) and a mixed metabolite phantom (3mM Cho, 10 mM Cr, 12.5 mM NAA, 12.5 mM Glu, 7.5 mM Gln, 5 mM GABA, 5 mM GSH, 7.5 mM mI, 3 mM Gly and 5 mM Lac). A total of twenty-nine patients with glioma (15 males and 14 females, median age = 45 years) were referred by physicians from the neuro-oncology service at our institution and recruited to this study. The characteristics of the patient population are summarized in Table 1. Tumors had been graded by histological examination of tissue samples obtained during biopsy or surgical resection: 12 had grade II, 10 grade III, and 7 grade IV glioma. All the patients were being followed using standard clinical criteria (4,16). Twelve of the patients were newly diagnosed with stable disease (3 grade II, 7 grade III and 2 grade IV), 16 had recurrence but without any malignant transformation at the time of the scan (9 grade II, 3 grade III and 4 grade IV), and 1 was newly diagnosed grade IV examined prior to receiving any treatment.
Table 1.
Characteristics of Patients
| No. | Age | Sex | Disease State | Grade | RT | Chemotherapy Received |
|---|---|---|---|---|---|---|
| 1 | 32 | F | Recurrent | 3 | Yes | TMZ, Bevacizumab |
| 2 | 39 | M | Recurrent | 2 | Yes | TMZ |
| 3 | 65 | F | Newly-Diagnosed | 2 | No | TMZ |
| 4 | 37 | M | Newly-Diagnosed | 2 | No | No |
| 5 | 43 | M | Newly-Diagnosed | 3 | Yes | TMZ |
| 6 | 28 | F | Newly-Diagnosed | 3 | Yes | TMZ |
| 7 | 55 | F | Recurrent | 2 | Yes | TMZ |
| 8 | 64 | M | Recurrent | 4 | Yes | TMZ, Toca511, 5-FC |
| 9 | 45 | F | Recurrent | 2 | Yes | TMZ |
| 10 | 45 | M | Newly-Diagnosed | 2 | No | No |
| 11 | 52 | M | Recurrent | 2 | No | TMZ |
| 12 | 40 | M | Recurrent | 4 | Yes | TMZ, Bevacizumab |
| 13 | 51 | M | Newly-Diagnosed | 3 | Yes | BCNU |
| 14 | 48 | M | Newly-Diagnosed | 3 | No | TMZ |
| 15 | 45 | M | Recurrent | 2 | Yes | No |
| 16 | 27 | F | Recurrent | 2 | No | TMZ |
| 17* | 43 | F | Newly-Diagnosed | 4 | No | No |
| 18 | 40 | F | Recurrent | 2 | No | TMZ |
| 19 | 34 | F | Newly-Diagnosed | 3 | Yes | TMZ |
| 20 | 42 | F | Recurrent | 3 | Yes | TMZ |
| 21 | 27 | M | Recurrent | 2 | No | TMZ |
| 22 | 52 | M | Recurrent | 2 | Yes | TMZ |
| 23 | 61 | F | Newly-Diagnosed | 3 | Yes | TMZ |
| 24 | 33 | M | Recurrent | 4 | Yes | TMZ, Bevacizumab, CCNU, carboplatin |
| 25 | 58 | F | Recurrent | 3 | Yes | TMZ, Poly ICLC |
| 26 | 30 | M | Newly-Diagnosed | 3 | Yes | TMZ |
| 27 | 64 | M | Newly-Diagnosed | 4 | Yes | TMZ |
| 28 | 56 | F | Newly-Diagnosed | 4 | Yes | PCV |
| 29 | 51 | F | Recurrent | 4 | Yes | HSP, TMZ |
Patient was examined prior receiving treatment.
TMZ = temozolomide; HSP = heat-shock protein.
Data Acquisition
All MR studies were performed using a 32-channel receive-only array with a volume transmit head coil (NOVA Medical, Wilmington, MA) on a GE 7T MR950 scanner (GE Healthcare, Waukesha, WI). Anatomic imaging consisted of a T1-weighted sagittal scout (repetition time [TR]/TE = 6/2 ms), 2D T2-weighted fast spin echo (FSE) (TR/TE/inversion time [TI] = 6000/86/600 ms, matrix size = 512×512, field of view (FOV) = 241×241 mm2, 19–21 slices, slice thickness/gap = 3/1 mm), 2D T2*-weighted gradient recalled echo (GRE) (TR/TE = 250/12 ms, matrix size = 512×512, FOV = 220×220 mm2, 11 slices, slice thickness/gap = 2/3 mm), and 3D T1-weighted inversion recovery-prepared spoiled gradient echo (IRSPGR) images (TR/TE/TI = 6/2/600 ms, matrix size = 256×256×192, FOV = 256×256×192 mm3, voxel size = 1×1×1 mm3).
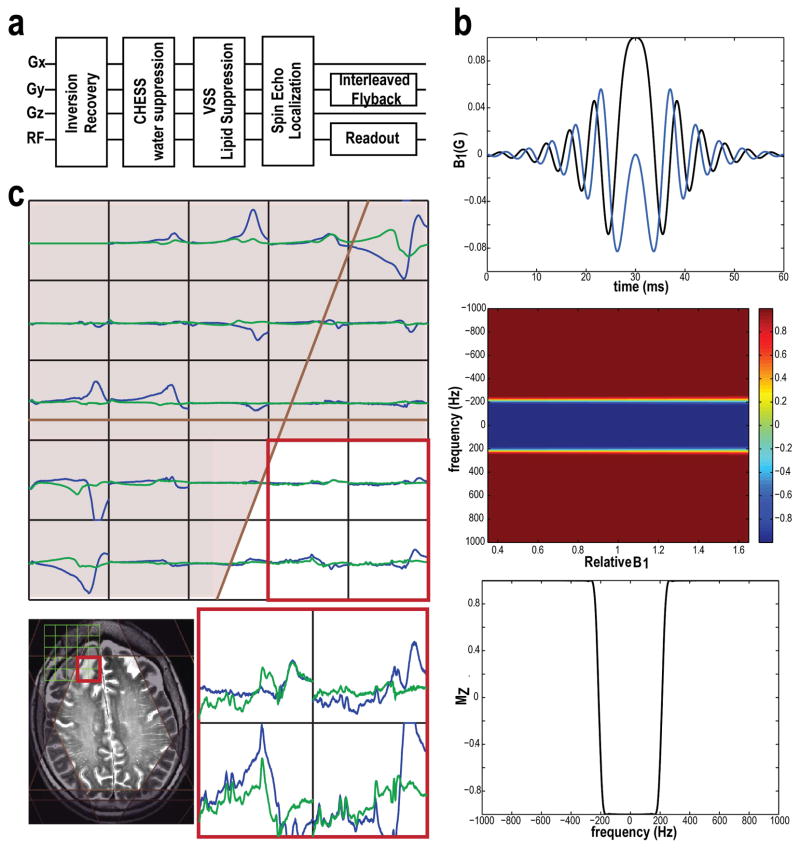
Before spectral acquisition, the manufacturer’s higher-order shimming procedure was performed. 3D H-1 MRSI data were obtained using CHESS water suppression, very selective suppression (VSS) (17) outer volume suppression, and spin echo slice selection (Figure 1a) with TE/TR = 30/2000ms, spectral array = 16–20×22×8, and spatial resolution = 1 cm3. Eight VSS suppression bands were positioned automatically to suppress the subcutaneous fat, calculated by using masks of lipid and brain that were generated from T1-weighted IRSPGR images (18,19). An example of automatically prescribed VSS is illustrated in Figure 2. To further eliminate lipid signal, a spectrally-selective adiabatic inversion recovery pulse was applied prior to the CHESS pulses with an inversion bandwidth of 400Hz (20) (Figure 1b–c, TI = 300–542 ms), and placed at 0.9 ppm to avoid modulating the GABA signals. The excited slab was 1.9 times thicker than the volume of interest, resulting in a 1.9 over-excitation factor (OE), to reduce the effects of chemical shift misregistration in the S/I direction. The edges of the volume were then suppressed using VSS pulses (see Figure 2). To shorten the total acquisition time, an interleaved flyback echo-planar trajectory (21) with a spatial resolution of 1 cm was designed and applied in the anterior/posterior (A/P) direction with a FOV of 22 cm. Each acquisition comprised 552 readout lobes with a spectral-bandwidth of 958 Hz after reconstruction using the flat portion of the flyback lobes. The second acquisition was temporally shifted by 1/(2 × spectral bandwidth). The combined data were able to provide a bandwidth of 1916 Hz and a spectral resolution of ~1.74 Hz per point. The total 3D MRSI acquisition time was 8.77 or 10.96 minutes for 16×22×8 or 20×22×8 spectral array, respectively. The variation in RF excitation due to B1 inhomogeneities was minimized by adjusting both the transmitter gain and relative RF power of the CHESS, 90° and 180° pulses.
Figure 1.
The flowchart of 3D 1-H MRSI sequence (a) with the components of spectrally-selective adiabatic inversion recovery (b), CHESS water suppression, automatically prescribed outer volume suppression bands, spin echo slice selection and an interleaved flyback echo-planar trajectory sampling. The sensitivities of inversion recovery pulses to B1 variation were simulated using Matlab (b). MRSI data (with baseline, 0.3–4.3 ppm) from a patient acquired without (blue) and with inversion recovery pulses (green, TI=442 ms) are shown in Panel C. The combined application of inversion recovery pulses and VSS spatial suppression bands achieved better lipid suppression.
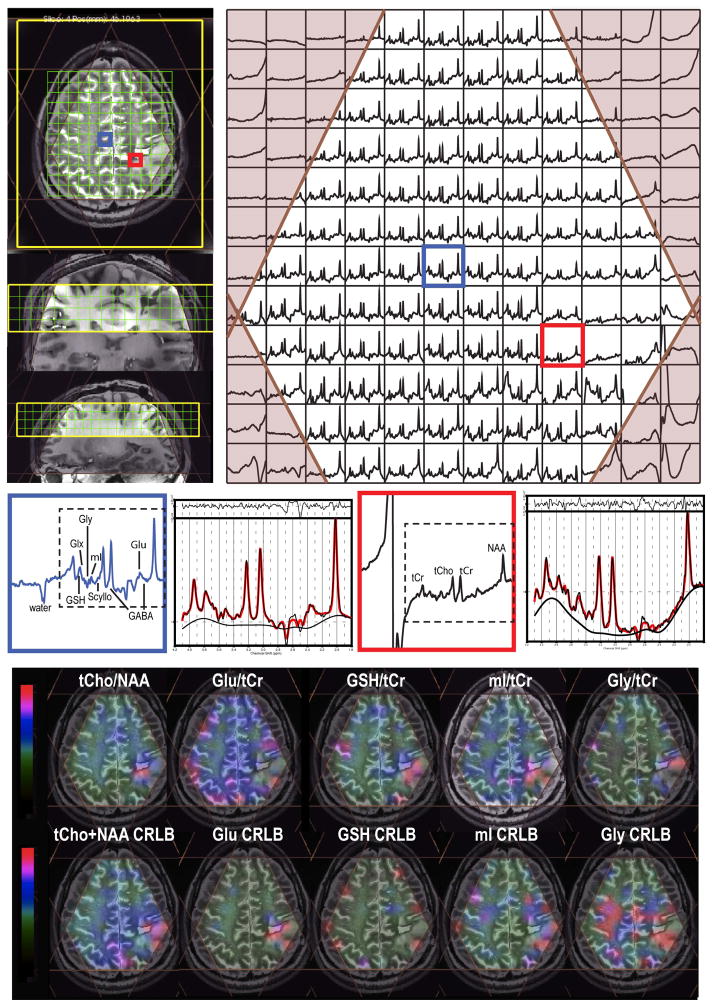
Figure 2.
One slice from a 3D MRSI dataset from a patient with a grade II astrocytoma (TE/TR = 30/2000 ms; spatial resolution = 1 cm3; total acquisition time ~ 10 min). Note that the baseline has not been removed from the spectra that are shown. 8 VSS were automatically prescribed that was based on the position of the selected. Small water signal was seen from the selected voxel in the GM, and the fitted spectra from LCModel were related to the phased spectra in the two selected voxels. The metabolite maps, tCho/NAA plotted within the range between 0 and 0.3 [0–0.3], Glu/tCr [0–2.2], GSH/tCr [0–0.5], mI/tCr [0–1.2] and Gly/tCr [0–0.4], and CRLB maps, tCho+NAA [0–10], Glu [0–20], GSH [0–30], mI [0–20] and Gly [0–30], which were quantified from LCModel, overlaid on FSE images.
Post-Processing and Analysis
The 3D MRSI datasets acquired using the interleaved flyback readout trajectory were combined and reordered to correspond to a rectilinear grid with resampling and phase correction (21), followed by frequency and additional phase corrections (22). The 32-channel data were combined by weighting with estimates of the individual channel sensitivities as determined from a separate calibration scan (23).
Metabolite levels were quantified for voxels within the brain using LCModel (24). Sixteen metabolite signals were generated for the basis set using NMR-SCOPE (25) with prior knowledge of chemical shift and J-coupling (26). Spectra were fitted between 4.1 and 1.8 ppm. Levels were expressed relative to tCr. Metabolite ratios included in the analysis were those with Cramer-Rao lower bounds (CRLB) lower than 10% for tCho, tCr and NAA, and 20% for the others. Linewidths of tCho, tCr, and NAA were computed for each voxel as described previously (22).
The 3D MRSI data were referenced to the FSE images by assuming that there was no movement between the two acquisitions. The 3D IRSPGR anatomic images were then resampled to the orientation of the FSE images. Regions of interests (ROIs) included voxels within the region of T2 hyperinstensity (T2L) on the FSE images, and NAWM and gray matter (GM) defined from the 3D SPGR brain images. Median metabolite ratios were analyzed within each of these ROIs.
Statistical analysis was performed using R version 2.9.2 (www.r-project.org). Nonparametric Wilcoxon rank sum tests were applied to assess differences in metabolite ratios between tumor grades, T2L and NAWM, or NAWM and GM. P-values of 0.05 or smaller were considered to be significant. The coefficients of variance (CVs) for metabolite ratios in the phantoms were calculated using the standard deviation divided by the mean. Due to the exploratory nature of this study, adjustment for multiple comparisons was not applied.
Results
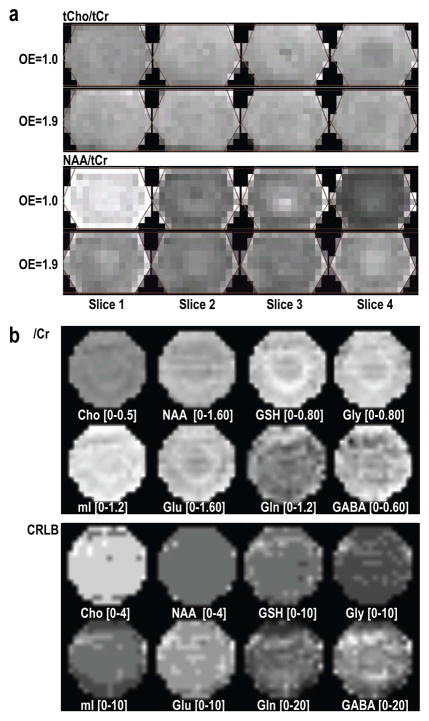
Figure 3 illustrates the results of 3D MRSI data acquired from phantoms. The metabolic maps of tCho/tCr and NAA/tCr that were quantified using LCModel are relatively uniform with an over-excitation factor of 1.9 in the S/I direction and in-plane phase encoding. For data acquired from the phantom, maps of relative metabolite ratios to Cr were smooth not only for Cho and NAA singlets, but also for those with more complex coupling patterns. The metabolite CRLBs were within quantifiable range for all of the metabolites considered. The CVs for data within the phantom were 4.3% for Cho/Cr, 6.8% for NAA/Cr, 8.5% for GSH/Cr, 9.3% for Gly/Cr, 9.2% for mI/Cr, 8.8% for Glu/Cr, 16.7% for Gln/Cr, and 18.9% for GABA/Cr, and became 3.9%, 5.8%, 7.4%, 6.7%, 7.6%, 8.7%, 14.1%, and 16.1%, respectively after removing 1 voxel on the edge of the spectral regions.
Figure 3.
Relative metabolite maps quantified using LCModel acquired from a GE MRS phantom (a) and a mixed metabolite phantom containing Cho, Cr, NAA, Glu, Gln, GABA, GSH, mI, Gly and Lac (b) to examine chemical shift misregistration (a) and accuracy of quantification (b). The spectra from the first slice have highest NAA/tCr, whereas those from the last slice have lowest levels (a, OE=1.0). Exciting larger than the encoded volume in the S/I direction (OE=1.9) significantly reduced this difference. Not only these singlets, but also the metabolites with complex coupling information, such as Glu, Gln, GABA, GSH, mI and Gly, were quantified and remarkably uniform in the spectra array with relatively small CRLB values.
An example of in vivo 3D MRSI data from a patient with grade II astrocytoma is demonstrated in Figure 2. Note that the baseline was not removed from the spectra shown and that the water signal is smaller than NAA from the GM voxel. The application of 8 automatically prescribed VSS outer volume suppression bands preserved the spectra within the region of interest. By using both 8 VSS and spectrally-selectively inversion recovery pulses (Figure 1c), good suppression of subcutaneous and cavity lipid signal was achieved. The CRLB estimates of Glu, mI, Gly and GSH from LCModel shown in Figure 2 were relatively small within the spectral array. Decreased Glu/tCr and increased mI/tCr, Gly/tCr, and GSH/tCr were observed within the T2L compared to the NAWM. The linewidths of tCho, tCr, and NAA within the NAWM for patients were 13.6±3.6, 14.1±3.1, and 16.2±3.6 Hz, respectively; and they were 13.8±3.3, 13.7±2.6, and 15.5±3.5 Hz, respectively, in the GM.
The median metabolite ratios in the NAWM, GM and T2L from the patients are summarized in Table 2. The differences in metabolite ratios between NAWM and GM were statistically significant for tCho/tCr, NAA/tCr, Glu/tCr, Glx/tCr, and Gly/tCr (p < 0.0001, = 0.0027, < 0.0001, < 0.0001, and 0.0344). Significant differences between tumor grades were found for the levels of Cho/NAA and NAA/tCr between grade III and grade IV glioma (p = 0.0431 and 0.0250). When compared to values in NAWM, the levels of tCho/tCr (p = 0.0137, >0.05 and 0.0051, in grade II, III and IV, respectively), tCho/NAA (p < 0.0001, >0.05, and < 0.0001), Gln/tCr (p = 0.0352, 0.0090 and 0.0028), mI/tCr (p = 0.0001, 0.0004 and 0.0108), Gly/tCr (p = 0.0001, 0.0060 and 0.0003), mIG/tCr (p = 0.0006, 0.0010 and 0.0319), and GSH/tCr (p < 0.0001, = 0.0224 and = 0.0062) were significantly increased in the T2L, while NAA/tCr (p = 0.0001, 0.0479 and 0.0002) and Glu/tCr (p = 0.0012, > 0.05 and > 0.05) were significantly decreased. The combination of Glu and Gln over tCr (Glx/tCr) was not significantly different from that in NAWM.
Table 2.
Metabolite ratios quantified using LCModel (mean ± standard deviation) and significance (p-value) was tested on metabolite ratios between the T2L or GM and the NAWM.
| G2 T2L (N=12) | G3 T2L (N=10) | G4 T2L (N=7) | NAWM (N=29) | GM (N=29) | |
|---|---|---|---|---|---|
| tCho/tCr | 0.31±0.08 | 0.26±0.07 | 0.29±0.04 | 0.24±0.03 | 0.18±0.04 |
| P=0.0137 | P=0.0051 | P < 0.0001 | |||
| NAA/tCr | 1.15±0.29 | 1.33±0.31 | 1.07±0.20 | 1.51±0.16 | 1.37±0.19 |
| P=0.0001 | P=0.0479 | P=0.0002 | P=0.0027 | ||
| tCho/NAA | 0.30±0.17 | 0.20±0.08 | 0.28±0.07 | 0.16±0.03 | 0.13±0.03 |
| P<0.0001 | P<0.0001 | P=0.0002 | |||
| Glu/tCr | 0.99±0.17 | 1.07±0.34 | 1.00±0.26 | 1.19±0.16 | 1.53±0.25 |
| P=0.0012 | P<0.0001 | ||||
| Gln/tCr | 0.74±0.36 (N=10) | 0.91±0.34 (N=6) | 0.80±0.21 | 0.50±0.21 (N=19) | 0.61±0.40 (N=17) |
| P=0.0352 | P=0.0090 | P=0.0028 | |||
| Glx/tCr | 1.27±0.24 | 1.49±0.55 (N=9) | 1.37±0.34 | 1.24±0.15 | 1.61±0.27 |
| P<0.0001 | |||||
| GSH/tCr | 0.33±0.05 | 0.32±0.12 | 0.30±0.06 | 0.23±0.05 | 0.21±0.05 |
| P<0.0001 | P=0.0224 | P=0.0062 | |||
| mI/tCr | 1.07±0.36 | 1.00±0.26 | 0.86±0.24 | 0.66±0.14 | 0.61±0.10 |
| P=0.0001 | P=0.0004 | P=0.0108 | |||
| Gly/tCr | 0.31±0.10 | 0.36±0.19 (N=9) | 0.33±0.08 | 0.20±0.05 | 0.17±0.03 |
| P=0.0001 | P=0.0060 | P=0.0003 | P=0.0344 | ||
| mIG/tCr | 1.16±0.29 | 1.13±0.31 | 1.02±0.22 | 0.81±0.14 | 0.75±0.09 |
| P=0.0006 | P=0.0010 | P=0.0319 | |||
| mI/tCho | 3.61±1.62 | 4.30±1.62 | 3.09±0.95 | 2.85±0.49 | 3.67±0.81 |
| P=0.0046 | P<0.0001 | ||||
| GABA/tCr | 0.48±0.08 (N=8) | 0.54±0.07 (N=6) | 0.50±0.10 (N=6) | 0.52±0.06 | 0.54±0.11 |
N = number of patients; G2 = grade II; G3 = grade III; G4 = grade IV.
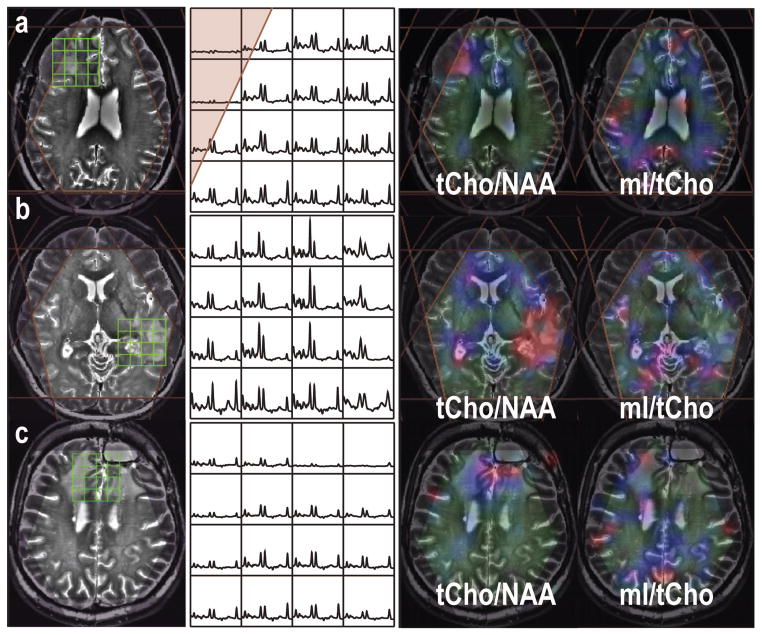
There is no difference in metabolite ratios within the T2L between newly diagnosed patients with stable disease and recurrent patients (Table 3). Examples of MR images, MRSI data and metabolite maps from three representative patients with a grade IV, II and IV glioma, who were prior to any treatment, recurrent within 10 months and over 3 years, respectively, are illustrated in Figure 4. Patients who had recently active tumors had higher tCho/NAA and relatively lower mI/tCho within the T2 lesion.
Table 3.
Metabolite ratios (95% confidence interval) within the T2L between newly diagnosed and recurrent patients.
| Newly-Diagnosed (N=12) | Recurrent (N=16) | P-value | |
|---|---|---|---|
| tCho/tCr | 0.24–0.31 | 0.26–0.34 | 0.5070 |
| NAA/tCr | 1.20–1.45 | 0.98–1.26 | 0.1330 |
| tCho/NAA | 0.18–0.24 | 0.22–0.37 | 0.1457 |
| Glu/tCr | 0.91–1.26 | 0.91–1.09 | 0.5070 |
| Gln/tCr | 0.56–1.00 (N = 7) | 0.66–1.00 (N = 15) | 0.8601 |
| Glx/tCr | 1.07–1.66 (N = 11) | 1.25–1.54 | 0.4155 |
| GSH/tCr | 0.26–0.37 | 0.29–0.36 | 0.2601 |
| mI/tCr | 0.77–1.11 | 0.89–1.19 | 0.6313 |
| Gly/tCr | 0.26–0.45 (N = 11) | 0.27–0.37 | 0.6569 |
| mIG/tCr | 0.91–1.27 | 1.00–1.25 | 0.9091 |
| mI/tCho | 2.85–4.48 | 2.95–4.57 | 0.9091 |
| GABA/tCr | 0.45–0.57 (N = 8) | 0.48–0.56 (N = 11) | 0.9039 |
N = number of patients.
Figure 4.
MRSI data (baseline removed from LCModel) from three patients with grade IV, II and IV gliomas, who were prior to any treatment (a), recurrent within 10 months (b) and over 3 years (c), respectively. The metabolite maps were quantified using LCModel and plotted with the range between 0 and 0.36 for tCho/NAA and between 0 and 6 for mI/tCho. Higher tCho/NAA in the recent active tumor lesions, while mI/tCho was elevated in the region that had been diagnosed stable for a long period.
Discussion
Improving the sensitivity of MRSI and increasing the number of metabolites that can be assessed would be an important advance for evaluating patients with glioma. This study has successfully demonstrated the application of short echo 3D MRSI to patients with glioma at 7T and has demonstrated differences in metabolite levels for regions of tumor versus normal brain, as well as between lesions.
Major concerns in obtaining data using high field strength MR scanners are increased variations in susceptibility and B1. A number of different methods for shimming have been proposed, for example, FASTERMAP shimming (27) and adding 3rd and 4th degree shims (28). In this study, the commercially available GE high-order shimming method provided good spectral quality in terms of metabolite linewidths, with a mean value of 13.8 Hz for tCr in the GM. Although this was higher than the reported tCr linewidth (8.5 Hz) in healthy controls that had been acquired with a single voxel MRS acquisition (8), the majority of our patients had large surgical cavities and hence this is a reasonable performance. Changes in the B1 field cause nonuniform excitation and reception. Although adiabatic excitation and refocusing pulses were not used in this project, the transmit gain and RF power were adjusted during the acquisition to provide a balance between brain metabolite excitation and lipid suppression. With a 32-channel receive-only array and a volume transmit head coil, the data were of good quality for both phantoms and in vivo acquisitions.
Spatial variations in metabolite ratios on the edges of the selected volume that are caused by chemical shift misregistration are exacerbated with increasing field strength. An alternative is to use sequences with RF pulses such as localized adiabatic spin-echo refocusing (LASER) (7,10), and full intensity acquired spectroscopy (SPECIAL) (8), that have large bandwidth, provide more uniform excitations, higher SNR, and shorter TE. In our study, the phantom data in Figure 3 illustrated uniform, quantifiable metabolite ratios in all directions, which allowed changes in metabolite ratios to be evaluated in different tissues. A TE of 30 ms was used in this case to avoid lipid contamination.
Automatic prescription of the volume of interest and the outer volume suppression bands (18,19), facilitates the acquisition process and offers consistent data quality, which is extremely important for obtaining reproducible results from serial studies. An interleaved flyback echo-planar trajectory was utilized in this study to shorten the total acquisition time to ~10 min and the VSS bands were prescribed automatically. As seen in Figure 1c, the combination of spectrally-selectively inversion recovery pulses and spatially selective volume suppression reduced the contamination from subcutaneous lipid. In this study, the spatial resolution was 1cm3 and the thickness of the excited slab was 4 cm, which possibly was not able to pick up all the differences due to tumor heterogeneities and treatment-induced effects in the tumor. Other fast spectroscopic imaging techniques, such as sensitivity encoding (SENSE) and GRAPPA, could be implemented to increase to the spatial resolution and coverage while maintaining a reasonable clinical acquisition time for future studies.
The tCho peak that is observed in vivo at 3.22 ppm includes contributions from choline, phosphocholine (PC), glycerophosphocholine (GPC) and acetylcholine. The level of tCho in tumors is elevated compared to normal tissue due to increased cell density and membrane turnover. On the other hand, NAA is decreased in pathology due to the loss of neurons or function. Changes in the relative levels of NAA and tCho have been widely studied for patients with brain tumors but the results on using the ratio of Cho-to-NAA to differentiate tumor recurrence from treatment effects have been quite variable (29–32). This suggests that information provided by other metabolites may be required to more fully evaluate disease progression. The increased spectral resolution and improved accuracy of quantification at 7T compared to clinical 1.5T and 3T scanners (8,11,33) provides the opportunity to investigate a larger range of metabolites in patients with brain tumors. Results from ex vivo spectroscopy indicated that Cho, mI, Gly, Glu, Gln, GSH and 2-hydroxyglutarate (2-HG) vary with tumor grade and during therapy (12–14,34,35). This information could be used for interpreting disease processes and directing treatment in vivo since MRSI is a non-invasive method. Patients recruited in the study were at different stage of disease and had received varying treatment, which may explain why only the Cho/NAA and NAA/tCr were found to be significantly different between grade III and IV.
Previous ex vivo studies in tissue samples obtained from patients with tumor have shown that mI/tCho and tCho/NAA could be used to differentiate treatment effects from active tumor (12), and that mI/tCho clearly distinguished recurrent grade II gliomas from other grades (13). mI is predominately located within astrocytes and the increases of mI/tCr and mI/tCho in the T2L compared to those in the NAWM could be a reflection of the presence of gliosis. There were no differences in mI/tCho in T2L detected in this study between newly diagnosed with stable disease and recurrent patients, which may have been due to the limited population size. On clinical scanners, the mI peaks often overlap with the peak of Gly, which is an inhibitory neurotransmitter. Elevations in mIG (sum of mI and Gly) relative to Cr from in vivo MRS data at 1.5T have been observed in low-grade astrocytomas but were shown to decrease in high grade lesions (36). As shown in Figure 2, Gly has been successfully separated from mI at 7T. Both the mI/tCr and mIG/tCr in our study showed a similar pattern with no significance was found between grades. The elevation in Gly/tCr observed within the T2L in this study was consistent with the previous reports (34,37), but the role of Gly in brain tumors is still not clear.
Glu is the main excitatory neurotransmitter in the brain and is converted to Gln by the re-absorption by neurons or reuptake by astrocytes in order to avoid Glu excitotoxicity. Glioma cells have been found to secrete Glu resulting in an increase in extra-cellular Glu (38,39). Gln is not only a precursor for Glu and GSH, but is also a suppressor of apoptosis (40); thus it may protect cells from external stimuli such as radiation therapy (41). GSH is an antioxidant that prevents damage from reactive oxygen species (42) and its metabolism was found to play an important role in tumor proliferation, invasion, and resistance to therapy in many tumors (43–48). The complex roles of Glu, Gln and GSH in tumors make it difficult to interpret the changes in such a small population. Interestingly, the Glx (sum of Glu and Gln peaks) was found to not be significantly different from that in the NAWM, suggesting the importance to evaluate these two metabolites separately in future studies.
Another important metabolite to consider in evaluating glioma is the onco-metabolite 2-HG, which is associated with the presence of mutated isocitrate dehydrogenase (IDH) enzyme (49). It has been reported that more than 70% of patients with low-grade glioma and secondary GBM have IDH mutations (50), and that the presence of such mutations is associated with longer overall survival (51–53). While we did include 2-HG in the basis set for LCModel fitting, we did not attempt to interpret the differences in intensity that were observed, because we did not have tissue available to obtain information about the IDH mutation status for this study population. Based on the previous ex vivo studies in tissue samples (13,15), it is anticipated that an increase in 2HG, along with elevated GSH and decreased mI/tCho would indicate malignant progression. Future studies will investigate data acquisition parameters for 7T MRSI that have been optimized to observe 2-HG and will apply these methods to patients with glioma who have known IDH mutation status.
In conclusion, the results of our study demonstrated differences in tCho/NAA, Glu/tCr, Gln/tCr, mI/tCr, Gly/tCr, and GSH/tCr in the T2L compared to those in NAWM. These data reflect the complexity of processes associated with disease progression and treatment effects, and further studies in a more specific study population, such as patients with newly diagnosed GBM, would be helpful in interpreting changes in metabolite ratios. In terms of data quality, our results were also encouraging and offer the opportunity to study other brain diseases using 7T H-1 MRSI.
Acknowledgments
Grant support: A St. Louis Fall Festival Committee American Brain Tumor Association Fellowship, an NCI training grant in translational brain tumor research (T32 CA151022), NIH research grants (RO1 CA127612, 1S10RR026845-01) and a technology development research grant from GE Healthcare.
Footnotes
Presented at the Annual Meeting of International Society for Magnetic Resonance in Medicine, Melbourne, Australia, May 2012 and Annual Scientific Meeting of the society for Neuro-Oncology, Washington, DC, Nov 2012.
References
- 1.Dolecek TA, Propp JM, Stroup NE, Kruchko C. CBTRUS Statistical Report: Primary Brain and Central Nervous System Tumors Diagnosed in the United States in 2005–2009. Neuro Oncol. 2012;14 (Suppl 5):v1–v49. doi: 10.1093/neuonc/nos218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stupp R, Mason WP, van den Bent MJ, et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med. 2005;352(10):987–996. doi: 10.1056/NEJMoa043330. [DOI] [PubMed] [Google Scholar]
- 3.van den Bent MJ, Vogelbaum MA, Wen PY, Macdonald DR, Chang SM. End point assessment in gliomas: novel treatments limit usefulness of classical Macdonald’s Criteria. J Clin Oncol. 2009;27(18):2905–2908. doi: 10.1200/JCO.2009.22.4998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wen PY, Macdonald DR, Reardon DA, et al. Updated response assessment criteria for high-grade gliomas: response assessment in neuro-oncology working group. J Clin Oncol. 2010;28(11):1963–1972. doi: 10.1200/JCO.2009.26.3541. [DOI] [PubMed] [Google Scholar]
- 5.McKnight TR, Noworolski SM, Vigneron DB, Nelson SJ. An automated technique for the quantitative assessment of 3D-MRSI data from patients with glioma. J Magn Reson Imaging. 2001;13(2):167–177. doi: 10.1002/1522-2586(200102)13:2<167::aid-jmri1026>3.0.co;2-k. [DOI] [PubMed] [Google Scholar]
- 6.Li Y, Lupo JM, Parvataneni R, et al. Survival analysis in patients with newly diagnosed glioblastoma using pre- and postradiotherapy MR spectroscopic imaging. Neuro Oncol. 2013;15(5):607–617. doi: 10.1093/neuonc/nos334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Andronesi OC, Gagoski BA, Sorensen AG. Neurologic 3D MR spectroscopic imaging with low-power adiabatic pulses and fast spiral acquisition. Radiology. 2012;262(2):647–661. doi: 10.1148/radiol.11110277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mekle R, Mlynarik V, Gambarota G, Hergt M, Krueger G, Gruetter R. MR spectroscopy of the human brain with enhanced signal intensity at ultrashort echo times on a clinical platform at 3T and 7T. Magn Reson Med. 2009;61(6):1279–1285. doi: 10.1002/mrm.21961. [DOI] [PubMed] [Google Scholar]
- 9.Henning A, Fuchs A, Murdoch JB, Boesiger P. Slice-selective FID acquisition, localized by outer volume suppression (FIDLOVS) for (1)H-MRSI of the human brain at 7 T with minimal signal loss. NMR Biomed. 2009;22(7):683–696. doi: 10.1002/nbm.1366. [DOI] [PubMed] [Google Scholar]
- 10.Scheenen TW, Heerschap A, Klomp DW. Towards 1H-MRSI of the human brain at 7T with slice-selective adiabatic refocusing pulses. Magma. 2008;21(1–2):95–101. doi: 10.1007/s10334-007-0094-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Otazo R, Mueller B, Ugurbil K, Wald L, Posse S. Signal-to-noise ratio and spectral linewidth improvements between 1. 5 and 7 Tesla in proton echo-planar spectroscopic imaging. Magn Reson Med. 2006;56(6):1200–1210. doi: 10.1002/mrm.21067. [DOI] [PubMed] [Google Scholar]
- 12.Srinivasan R, Phillips JJ, Vandenberg SR, et al. Ex vivo MR spectroscopic measure differentiates tumor from treatment effects in GBM. Neuro Oncol. 2010;12(11):1152–1161. doi: 10.1093/neuonc/noq075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Elkhaled A, Jalbert LE, Yoshihara HA, et al. Comparison of Glioma Sub-Populations Using In-Vivo ADC Values and Ex-Vivo 1H HR-MAS Spectroscopy. Proceedings of the 18th Annual Meetings of ISMRM; Stockholm, Sweden. 2010; p. Abstract 2203. [Google Scholar]
- 14.Elkhaled A, Jalbert LE, Phillips JJ, et al. Magnetic resonance of 2-hydroxyglutarate in IDH1-mutated low-grade gliomas. Sci Transl Med. 2012;4(116):116ra115. doi: 10.1126/scitranslmed.3002796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jalbert LE, Elkhaled A, Phillips JJ, Chang SM, Nelson SJ. Image-Guided Metabolomic Analysis of 2-Hydroxyglutarate in IDH-Mutant Gliomas. Proceedings of the 21th Annual Meetings of ISMRM; Salt Lake City, Utah, USA. 2013; p. Abstract 0510. [Google Scholar]
- 16.Macdonald DR, Cascino TL, Schold SC, Jr, Cairncross JG. Response criteria for phase II studies of supratentorial malignant glioma. J Clin Oncol. 1990;8(7):1277–1280. doi: 10.1200/JCO.1990.8.7.1277. [DOI] [PubMed] [Google Scholar]
- 17.Tran TK, Vigneron DB, Sailasuta N, et al. Very selective suppression pulses for clinical MRSI studies of brain and prostate cancer. Magn Reson Med. 2000;43(1):23–33. doi: 10.1002/(sici)1522-2594(200001)43:1<23::aid-mrm4>3.0.co;2-e. [DOI] [PubMed] [Google Scholar]
- 18.Ozhinsky E, Vigneron DB, Chang SM, Nelson SJ. Automated prescription of oblique brain 3D magnetic resonance spectroscopic imaging. Magn Reson Med. 2013;69(4):920–930. doi: 10.1002/mrm.24339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ozhinsky E, Vigneron DB, Nelson SJ. Improved spatial coverage for brain 3D PRESS MRSI by automatic placement of outer-volume suppression saturation bands. J Magn Reson Imaging. 2011;33(4):792–802. doi: 10.1002/jmri.22507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Balchandani P, Spielman D. Fat suppression for 1H MRSI at 7T using spectrally selective adiabatic inversion recovery. Magn Reson Med. 2008;59(5):980–988. doi: 10.1002/mrm.21537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cunningham CH, Vigneron DB, Chen AP, et al. Design of flyback echo-planar readout gradients for magnetic resonance spectroscopic imaging. Magn Reson Med. 2005;54(5):1286–1289. doi: 10.1002/mrm.20663. [DOI] [PubMed] [Google Scholar]
- 22.Nelson SJ. Analysis of volume MRI and MR spectroscopic imaging data for the evaluation of patients with brain tumors. Magn Reson Med. 2001;46(2):228–239. doi: 10.1002/mrm.1183. [DOI] [PubMed] [Google Scholar]
- 23.Pruessmann KP, Weiger M, Scheidegger MB, Boesiger P. SENSE: sensitivity encoding for fast MRI. Magn Reson Med. 1999;42(5):952–962. [PubMed] [Google Scholar]
- 24.Provencher SW. Estimation of metabolite concentrations from localized in vivo proton NMR spectra. Magn Reson Med. 1993;30(6):672–679. doi: 10.1002/mrm.1910300604. [DOI] [PubMed] [Google Scholar]
- 25.Graverondemilly D, Diop A, Briguet A, Fenet B. Product-Operator Algebra for Strongly Coupled Spin System. J Magn Reson, Series A. 1993;101(3):233–239. [Google Scholar]
- 26.Govindaraju V, Young K, Maudsley AA. Proton NMR chemical shifts and coupling constants for brain metabolites. NMR Biomed. 2000;13(3):129–153. doi: 10.1002/1099-1492(200005)13:3<129::aid-nbm619>3.0.co;2-v. [DOI] [PubMed] [Google Scholar]
- 27.Shen J, Rycyna RE, Rothman DL. Improvements on an in vivo automatic shimming method [FASTERMAP] Magn Reson Med. 1997;38(5):834–839. doi: 10.1002/mrm.1910380521. [DOI] [PubMed] [Google Scholar]
- 28.Pan JW, Lo KM, Hetherington HP. Role of very high order and degree B0 shimming for spectroscopic imaging of the human brain at 7 tesla. Magn Reson Med. 2012;68(4):1007–1017. doi: 10.1002/mrm.24122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Plotkin M, Eisenacher J, Bruhn H, et al. 123I-IMT SPECT and 1H MR-spectroscopy at 3. 0 T in the differential diagnosis of recurrent or residual gliomas: a comparative study. J Neurooncol. 2004;70(1):49–58. doi: 10.1023/b:neon.0000040810.77270.68. [DOI] [PubMed] [Google Scholar]
- 30.Rock JP, Scarpace L, Hearshen D, et al. Associations among magnetic resonance spectroscopy, apparent diffusion coefficients, and image-guided histopathology with special attention to radiation necrosis. Neurosurgery. 2004;54(5):1111–1117. doi: 10.1227/01.neu.0000119328.56431.a7. discussion 1117–1119. [DOI] [PubMed] [Google Scholar]
- 31.Zeng QS, Li CF, Liu H, Zhen JH, Feng DC. Distinction between recurrent glioma and radiation injury using magnetic resonance spectroscopy in combination with diffusion-weighted imaging. Int J Radiat Oncol Biol Phys. 2007;68(1):151–158. doi: 10.1016/j.ijrobp.2006.12.001. [DOI] [PubMed] [Google Scholar]
- 32.Traber F, Block W, Flacke S, et al. 1H-MR Spectroscopy of brain tumors in the course of radiation therapy: Use of fast spectroscopic imaging and single-voxel spectroscopy for diagnosing recurrence. Rofo. 2002;174(1):33–42. doi: 10.1055/s-2002-19541. [DOI] [PubMed] [Google Scholar]
- 33.Tkac I, Oz G, Adriany G, Ugurbil K, Gruetter R. In vivo 1H NMR spectroscopy of the human brain at high magnetic fields: metabolite quantification at 4T vs. 7T. Magn Reson Med. 2009;62(4):868–879. doi: 10.1002/mrm.22086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kinoshita Y, Kajiwara H, Yokota A, Koga Y. Proton magnetic resonance spectroscopy of astrocytic tumors: an in vitro study. Neurol Med Chir (Tokyo) 1993;33(6):350–359. doi: 10.2176/nmc.33.350. [DOI] [PubMed] [Google Scholar]
- 35.Righi V, Roda JM, Paz J, et al. 1H HR-MAS and genomic analysis of human tumor biopsies discriminate between high and low grade astrocytomas. NMR Biomed. 2009;22(6):629–637. doi: 10.1002/nbm.1377. [DOI] [PubMed] [Google Scholar]
- 36.Castillo M, Smith JK, Kwock L. Correlation of myo-inositol levels and grading of cerebral astrocytomas. AJNR Am J Neuroradiol. 2000;21(9):1645–1649. [PMC free article] [PubMed] [Google Scholar]
- 37.Righi V, Andronesi OC, Mintzopoulos D, Black PM, Tzika AA. High-resolution magic angle spinning magnetic resonance spectroscopy detects glycine as a biomarker in brain tumors. Int J Oncol. 2010;36(2):301–306. doi: 10.3892/ijo_00000500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ye ZC, Sontheimer H. Glioma cells release excitotoxic concentrations of glutamate. Cancer Res. 1999;59(17):4383–4391. [PubMed] [Google Scholar]
- 39.Takano T, Lin JH, Arcuino G, Gao Q, Yang J, Nedergaard M. Glutamate release promotes growth of malignant gliomas. Nat Med. 2001;7(9):1010–1015. doi: 10.1038/nm0901-1010. [DOI] [PubMed] [Google Scholar]
- 40.Chang WK, Yang KD, Chuang H, Jan JT, Shaio MF. Glutamine protects activated human T cells from apoptosis by up-regulating glutathione and Bcl-2 levels. Clin Immunol. 2002;104(2):151–160. doi: 10.1006/clim.2002.5257. [DOI] [PubMed] [Google Scholar]
- 41.Mates JM, Perez-Gomez C, Nunez de Castro I, Asenjo M, Marquez J. Glutamine and its relationship with intracellular redox status, oxidative stress and cell proliferation/death. Int J Biochem Cell Biol. 2002;34(5):439–458. doi: 10.1016/s1357-2725(01)00143-1. [DOI] [PubMed] [Google Scholar]
- 42.Pompella A, Visvikis A, Paolicchi A, De Tata V, Casini AF. The changing faces of glutathione, a cellular protagonist. Biochem Pharmacol. 2003;66(8):1499–1503. doi: 10.1016/s0006-2952(03)00504-5. [DOI] [PubMed] [Google Scholar]
- 43.Arrick BA, Nathan CF. Glutathione metabolism as a determinant of therapeutic efficacy: a review. Cancer Res. 1984;44(10):4224–4232. [PubMed] [Google Scholar]
- 44.Balendiran GK, Dabur R, Fraser D. The role of glutathione in cancer. Cell Biochem Funct. 2004;22(6):343–352. doi: 10.1002/cbf.1149. [DOI] [PubMed] [Google Scholar]
- 45.Ogunrinu TA, Sontheimer H. Hypoxia increases the dependence of glioma cells on glutathione. J Biol Chem. 2010;285(48):37716–37724. doi: 10.1074/jbc.M110.161190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhong W, Yan T, Lim R, Oberley LW. Expression of superoxide dismutases, catalase, and glutathione peroxidase in glioma cells. Free Radic Biol Med. 1999;27(11–12):1334–1345. doi: 10.1016/s0891-5849(99)00181-1. [DOI] [PubMed] [Google Scholar]
- 47.Gatti L, Zunino F. Overview of tumor cell chemoresistance mechanisms. Methods Mol Med. 2005;111:127–148. doi: 10.1385/1-59259-889-7:127. [DOI] [PubMed] [Google Scholar]
- 48.Estrela JM, Ortega A, Obrador E. Glutathione in cancer biology and therapy. Crit Rev Clin Lab Sci. 2006;43(2):143–181. doi: 10.1080/10408360500523878. [DOI] [PubMed] [Google Scholar]
- 49.Dang L, White DW, Gross S, et al. Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature. 2009;462(7274):739–744. doi: 10.1038/nature08617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yan H, Parsons DW, Jin G, et al. IDH1 and IDH2 mutations in gliomas. N Engl J Med. 2009;360(8):765–773. doi: 10.1056/NEJMoa0808710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sanson M, Marie Y, Paris S, et al. Isocitrate dehydrogenase 1 codon 132 mutation is an important prognostic biomarker in gliomas. J Clin Oncol. 2009;27(25):4150–4154. doi: 10.1200/JCO.2009.21.9832. [DOI] [PubMed] [Google Scholar]
- 52.Ohno M, Narita Y, Miyakita Y, et al. Secondary glioblastomas with IDH1/2 mutations have longer glioma history from preceding lower-grade gliomas. Brain Tumor Pathol. 2013 doi: 10.1007/s10014-013-0140-6. [DOI] [PubMed] [Google Scholar]
- 53.Leu S, von Felten S, Frank S, et al. IDH/MGMT-driven molecular classification of low-grade glioma is a strong predictor for long-term survival. Neuro Oncol. 2013;15(4):469–479. doi: 10.1093/neuonc/nos317. [DOI] [PMC free article] [PubMed] [Google Scholar]