Abstract
Enterohemorrhagic E. coli (EHEC) manipulate their human host through at least 39 effector proteins which hijack host processes through direct protein-protein interactions (PPIs). To identify their protein targets in the host cells, we performed yeast two-hybrid screens, allowing us to find 48 high-confidence protein-protein interactions between 15 EHEC effectors and 47 human host proteins. In comparison to other bacteria and viruses we found that EHEC effectors bind more frequently to hub proteins as well as to proteins that participate in a higher number of protein complexes. The data set includes six new interactions that involve the translocated intimin receptor (TIR), namely HPCAL1, HPCAL4, NCALD, ARRB1, PDE6D, and STK16. We compared these TIR interactions in EHEC and enteropathogenic E. coli (EPEC) and found that five interactions were conserved. Notably, the conserved interactions included those of serine/threonine kinase 16 (STK16), hippocalcin-like 1 (HPCAL1) as well as neurocalcin-delta (NCALD). These proteins co-localize with the infection sites of EPEC. Furthermore, our results suggest putative functions of poorly characterized effectors (EspJ, EspY1). In particular, we observed that EspJ is connected to the microtubule system while EspY1 appears to be involved in apoptosis/cell cycle regulation.
Enterohemorrhagic E. coli (EHEC) are pathogenic E. coli that produce shiga toxins similar to Shigella dysenteriae1,2. Aside from bloody and non-bloody self-limiting diarrhea, EHEC can cause so-called hemolytic uremic syndrome (HUS), a disease that is characterized by hemolytic anemia, acute and occasionally chronic renal failure as well as thrombocytopenia. The public health impact of EHEC infections is considerable3,4 as a consequence of their low infectious dose (<100 colony forming units) and the corresponding frequency of systemic complications in patients. EHEC colonization utilizes a type three-secretion system (T3SS) and up to 62 putative effector proteins. The T3SS system directly translocates at least 39 effectors into the cytoplasm of their human host cells5. The combined action of these effectors results in brush border remodeling and the formation of attaching and effacing (A/E) lesions in the small intestine that are typical for EHEC and enteropathogenic E. coli (EPEC) infections. Furthermore, effectors manipulate several host cell signaling pathways to allow successful colonization of the host. Especially the assembly of pedestals through actin-cytoskeleton reorganization is considered important for host cell colonization, a step that is substantially conditioned by the translocated intimin receptor (TIR)6. TIR is involved in several distinct cellular processes besides actin pedestal assembly, such as down-regulation of Map-dependent filopodia formation and suppression of inflammatory cytokine production6,7,8,9. Furthermore, TIR interacts with numerous host cellular targets such as 14-3-3tau, alpha-actinin, cortacin, CK18, IQGAP1, IRTKS, IRSp53, Nck, PI3K, talin, vinculin and AnxA210,11,12,13,14,15,16,17,18,19,20,21 as reviewed in ref 7. Only recently two novel interactions with SHP-1 and SHP-2 were published8,9. Although several other novel EHEC - host protein interactions have been described that involve the effectors EspG, EspZ, NleB and NleF8,9,22,23,24,25,26 our knowledge of interactions between effector and host cell proteins is far from complete. To fill this gap we mapped the first comprehensive, experimentally derived network of binary EHEC-host interactions which allowed us to investigate the interactions between the translocated intimin receptor TIR and its human targets in more detail.
Results and Discussion
The EHEC effector-host protein interaction map
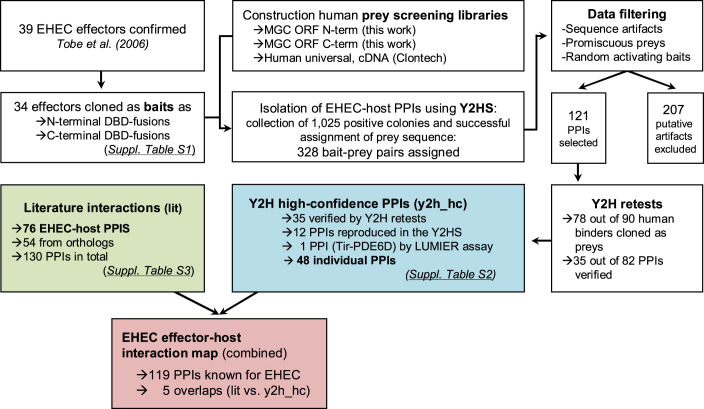
In 2006 Tobe et al. identified 62 putative EHEC effector proteins through computational analysis and experimentally confirmed that 39 effectors were indeed translocated into host cells5. While several already have been demonstrated to modify certain host functions (reviewed in ref 7), many effector proteins remain functionally poorly characterized. To investigate the global landscape of effector-host interactions we systematically screened 34 of the 39 effectors verified by Tobe et al. using a well established, semi-automated comprehensive Yeast 2-Hybrid (Y2H) screening protocol27,28 (Figure 1, Supplementary Table S1). Each effector was fused to the GAL4-DBD (DNA binding domain of the GAL4 transcription factor) on both its N- and C-terminus. Similarly, we built two different prey screening libraries from human open reading frame (ORFeome) collections29,30,31. Each consisted of a single pool of over 10,000 individual human protein-coding ORFs that were either fused C- or N-terminally to the AD (activation domain) of GAL4. The N- and C-terminally-tagged effector bait constructs were then screened individually against both ORFeome libraries to increase the detection sensitivity32,33 as well as against a cDNA prey library (Human Universal, Clontech). These screens resulted in 1,025 positive yeast colonies from which we successfully retrieved 328 unique EHEC bait-human prey pairs. We removed 207 pairs with prey sequences that were not located in the protein coding region of genes (3′-UTR artifacts), interactions involving randomly activating bait constructs (i.e. baits that returned exclusively multiple non-reproducible hits), or interactions including prey proteins known to interact non-selectively with many different bait proteins. In the latter case we excluded prey constructs known to bind to more than five bait proteins28 (Schwarz et al., unpublished). Out of the remaining 121 interactions 86 were detected only once, i.e., a prey could be assigned to the known bait protein only from one unique positive yeast colony. Using pairwise Y2H assays we retested all 121 pairs that had human full-length ORF constructs. We cloned 78 out of 90 human interactors into prey plasmids and were able to reproduce 35 out of 82 tested PPIs (42.7%). All 23 pairs that had two or more colonies in the screens were retested positive; pairs with only a single colony had a success rate of 20.7%. In the final dataset, we included all PPIs that were isolated multiple times, as well as those that were independently reproduced. In total, our approach yielded 48 individual EHEC-host PPIs of high confidence (Table 1 and for more details Supplementary Table S2) where the majority of 36 PPIs was identified by using a C-terminal ORFeome prey library. In comparison, the N-terminal ORFeome prey library and the cDNA prey libraries isolated clearly fewer interactions (seven PPIs each). Only two interactions were found in all the different ORFeome prey libraries (NleF with Caspase 9 and dihydrofolate reductase).
Figure 1. Interaction screening workflow.
Workflow of our procedure to find interactions between EHEC effector and human host proteins. See main text for a detailed explanation.
Table 1. All high confidence interactions found in this study. Previously published interactions are in bold with their source citations given under “Ref”.
| EHEC effector | Interacting human protein | ||||
|---|---|---|---|---|---|
| Name | Sakai gene id | Name | GeneID | Protein names | Ref |
| EspB | ECs4554 | RBCK1 | 10616 | RanBP-type and C3HC4-type zinc finger-containing protein 1 | |
| EspB | ECs4554 | STK16 | 8576 | Serine/threonine-protein kinase 16 | |
| EspF1 | ECs4550 | MAD2L2 | 10459 | Mitotic spindle assembly checkpoint protein MAD2B | |
| EspF1 | ECs4550 | SNX33 | 257364 | Sorting nexin-33 | |
| EspF1 | ECs4550 | SNX9 | 51429 | Sorting nexin-9 | 35 |
| EspG | ECs4590 | “Hs.658052” | UGID:2727370 | N/A | |
| EspG | ECs4590 | NMI | 9111 | N-myc-interactor | |
| EspJ | ECs2714 | CENPH | 64946 | Centromere protein H | |
| EspJ | ECs2714 | IFT20 | 90410 | Intraflagellar transport protein 20 homolog | |
| EspJ | ECs2714 | MRFAP1L1 | 114932 | MORF4 family-associated protein 1-like 1 | |
| EspJ | ECs2714 | RIC8A | 60626 | Synembryn-A | |
| EspO1-1 | ECs1567 | FEM1B | 10116 | Protein fem-1 homolog B | |
| EspY1 | ECs0061 | CAPN3 | 825 | Calpain-3 | |
| EspY1 | ECs0061 | CDKN2AIPNL | 91368 | CDKN2AIP N-terminal-like protein | |
| EspY1 | ECs0061 | CLK1 | 1195 | Dual specificity protein kinase CLK1 | |
| EspY1 | ECs0061 | DNAJC14 | 85406 | DnaJ homolog subfamily C member 14 | |
| EspY1 | ECs0061 | PCID2 | 55795 | PCI domain-containing protein 2 | |
| EspY1 | ECs0061 | PIH1D1 | 55011 | PIH1 domain-containing protein 1 | |
| EspY1 | ECs0061 | PSMC1 | 5700 | 26S protease regulatory subunit 4 | |
| EspY1 | ECs0061 | ZNHIT1 | 10467 | Zinc finger HIT domain-containing protein 1 | |
| Map | ECs4562 | RHPN1 | 114822 | Rhophilin-1 | |
| Map | ECs4562 | SLC9A3R2 | 9351 | Na(+)/H(+) exchange regulatory cofactor NHE-RF2 (NHERF-2) | 34 |
| NleA | ECs1812 | DSCR4 | 10281 | Down syndrome critical region protein 4 | |
| NleA | ECs1812 | FRMD3 | 257019 | FERM domain-containing protein 3 | |
| NleA | ECs1812 | PENK | 5179 | Proenkephalin-A | |
| NleA | ECs1812 | PTP4A1 | 7803 | Protein tyrosine phosphatase type IVA 1 | |
| NleB1 | ECs3857 | DRG2 | 1819 | Developmentally-regulated GTP-binding protein 2 | |
| NleB1 | ECs3857 | LRRC18 | 474354 | Leucine-rich repeat-containing protein 18 | |
| NleB1 | ECs3857 | POLR2E | 5434 | DNA-directed RNA polymerases I, II, and III subunit RPABC1 | |
| NleC | ECs0847 | C8orf71 | 26138 | Putative uncharacterized protein encoded by LINC00588 | |
| NleC | ECs0847 | CTDSPL2 | 51496 | CTD small phosphatase-like protein 2 | |
| NleD | ECs0850 | METTL2A | 339175 | Methyltransferase-like protein 2A | |
| NleF | ECs1815 | CASP9 | 842 | Caspase-9 | 23 |
| NleF | ECs1815 | DHFR | 1719 | Dihydrofolate reductase | |
| NleF | ECs1815 | ERI3 | 79033 | ERI1 exoribonuclease 3 | |
| NleF | ECs1815 | HMGN2 | 3151 | Non-histone chromosomal protein HMG-17 | |
| NleF | ECs1815 | LMO4 | 8543 | LIM domain transcription factor LMO4 | |
| NleF | ECs1815 | TRNT1 | 51095 | CCA tRNA nucleotidyltransferase 1, mitochondrial | |
| NleH1-2 | ECs1814 | UFC1 | 51506 | Ubiquitin-fold modifier-conjugating enzyme 1 | |
| TccP | ECs2715 | ZNF626 | 199777 | Zinc finger protein 626 | |
| Tir | ECs4561 | ARRB1 | 408 | Beta-arrestin-1 | |
| Tir | ECs4561 | BAIAP2 | 10458 | Brain-specific angiogenesis inhibitor 1-associated protein 2 | 15 |
| Tir | ECs4561 | BAIAP2L1 | 55971 | Brain-specific angiogenesis inhibitor 1-associated protein 2-like protein 1 | 14 |
| Tir | ECs4561 | HPCAL1 | 3241 | Hippocalcin-like protein 1 | |
| Tir | ECs4561 | HPCAL4 | 51440 | Hippocalcin-like protein 4 | |
| Tir | ECs4561 | NCALD | 83988 | Neurocalcin-delta | |
| Tir | ECs4561 | PDE6D | 5147 | Retinal rod rhodopsin-sensitive cGMP 3′,5′-cyclic phosphodiesterase subunit delta | |
| Tir | ECs4561 | STK16 | 8576 | Serine/threonine-protein kinase 16 | |
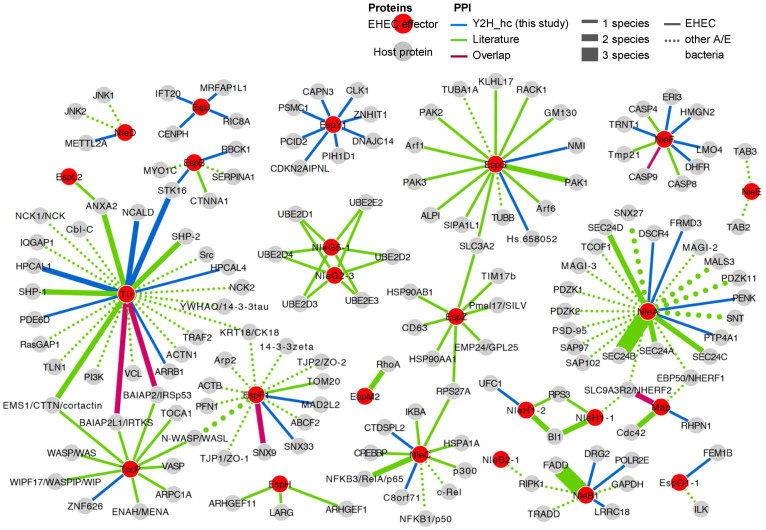
To generate a combined protein interaction network including the 48 newly found interacions, we extensively reviewed the existing literature and curated 130 effector-host interactions, including 76 EHEC-host protein interactions. Furthermore, we found a few interactions that were inferred from protein complexes although no experimental evidence for a direct interaction appears to be available.
The combined network is shown in Figure 2 (for a complete list of EHEC-host interactions and references see Supplementary Table S3). We found that five out of the discovered 48 pathogen-host pairs were previously published (Tir-BAIAP2L1 ( = IRTKS)14, Tir-BAIAP2 ( = IRSp53)15, Map-NHERF2 ( = SLC9A3R2)34, EspF-SNX935 and NleF-CASP923).
Figure 2. EHEC effector - host protein interaction map.
Combined network showing the 48 interactions detected in this study as well as 119 literature-curated interactions between EHEC effector and human host proteins. Furthermore, we added interactions between human host and effector proteins of various A/E pathogens that have orthologs with EHEC effector proteins. For a list of all interactions see Table 1 and Supplementary Table S3.
From the literature, EHEC-host interactions are known for 18 out of 39 effector proteins described by Tobe et al. Our screens allowed us to find PPIs that involved 15 out of 39 effectors including EspJ and EspY1 that previously were not reported to interact with the host. While cellular targets of the remaining 13 effectors, EspB, EspF, EspG, EspO, Map, NleA, NleB; NleC, NleD, NleF, NleH, TccP (EspFu) and TIR, were previously known, we determined 31 novel interactions.
A global analysis of EHEC-host interactions compared to other pathogens
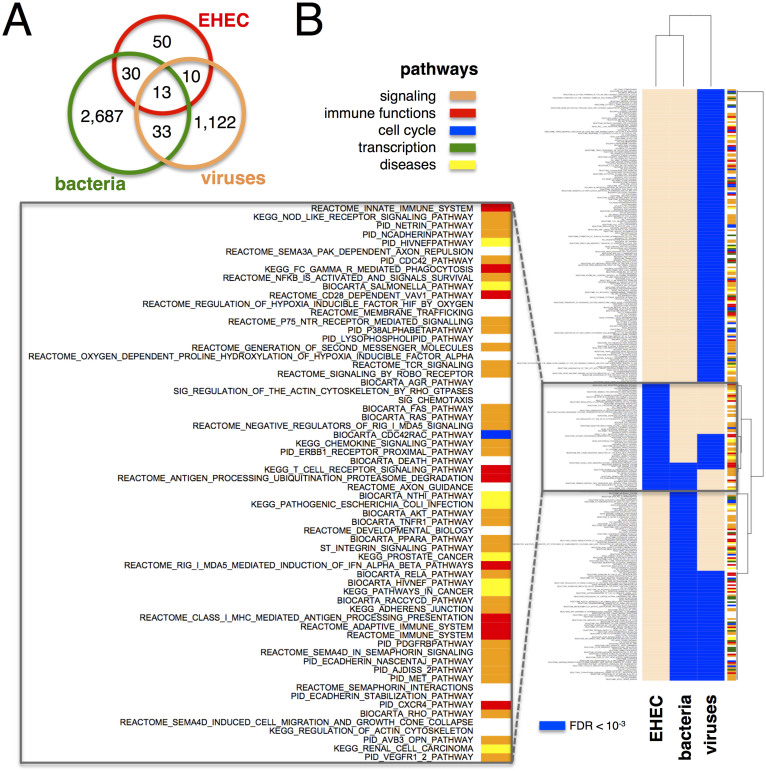
To compare proteins targeted by EHEC with those of other pathogens, we compared our data to 8,297 protein interactions between proteins of B. anthracis, F. tularensis and Y. pestis and 3,363 human host proteins36. Furthermore, we compiled a set of 3,156 interactions between viral proteins of the Epstein-Barr, HIV, Hepatitis, Herpes, Influenza, Papilloma and Vaccinia virus and 1,778 human host proteins37. We find that many human host proteins were targeted by both bacterial and viral proteins (Figure 3A).
Figure 3. Pathway comparison of pathogen-host interaction networks.
(a) Overlap of various host-pathogen interactions. Numbers indicate host proteins targeted by the pathogen group. (b) Pathways that were significantly enriched among human EHEC targets (Fisher's exact test, FDR < 10−3). In particular, we observed a small set of mostly signalling pathways that were significantly targeted by EHEC effector proteins.
To investigate the impact on pathways, we determined the enrichment of proteins targeted by EHEC effectors in pathways that we collected from the molecular signature database38. We observed a significant enrichment of a small set of mostly signaling as well as infection-related pathways (Figure 3B).
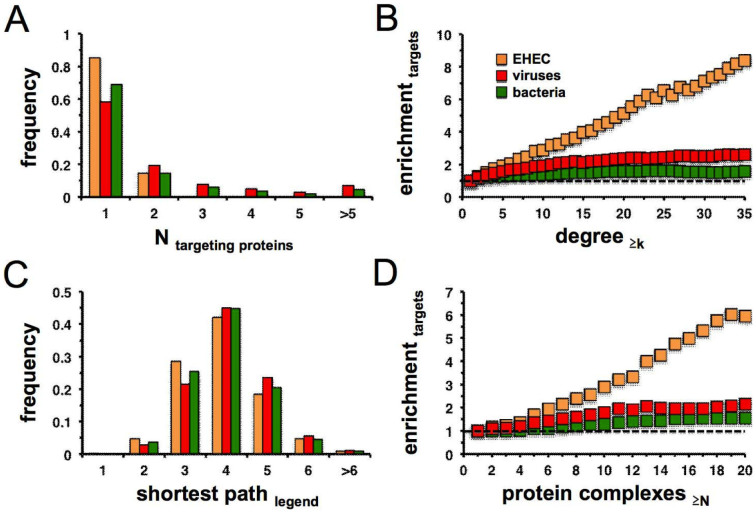
In general, the majority of human target proteins interact with one effector protein, a characteristic that is shared with bacterial and viral targets (Figure 4A). Next, we wondered whether pathogen proteins interact with highly connected human proteins (hubs) or rather focus on less frequently interacting proteins in the human interactome. Figure 4B indicates that bacterial as well as viral proteins are predominately enriched in groups of highly connected host proteins. In comparison, targets of EHEC effector proteins surprisingly show a significantly stronger enrichment signal. Calculating shortest path lengths (Figure 4C) we observed that proteins targeted by EHEC effector proteins had shorter paths to other human proteins than the corresponding targets of other bacteria and of viruses (Student's t-test, p < 10−20). A similar trend for binding to highly connected proteins is apparent in the enrichment of targeted proteins in protein complexes, as shown in Figure 4D. In comparison to bacterial and viral targets we observed that targets of EHEC effector proteins appear significantly more frequently in protein complexes.
Figure 4. Topological characteristics of EHEC effector – host protein interactions.
(a) Counting the number of EHEC effector – host protein interactions, we found that the vast majority of host proteins were targeted by one EHEC effector protein. Such an observation resembles targets of viruses and bacteria. (b) We calculated the enrichment of targeted proteins as a function of their numbers of interaction partners in a human interaction network. Focusing on targets of viruses and bacteria, respectively, targeted host proteins were predominately enriched in groups of highly connected proteins. In comparison, targets of EHEC effector proteins showed a significantly reinforced enrichment signal. (c) Proteins that were targeted by EHEC effector proteins had shorter paths to other human proteins than the corresponding targets of bacteria and viruses (Student's t-test, p < 10−30). (d) We determined the enrichment of targeted proteins as a function of their appearance in different protein complexes. In comparison to bacterial and viral targets EHEC effector proteins interact with host proteins that occurred in an increasing number of complexes.
EspY1 interacts with proteins involved in apoptosis and cell cycle regulation
While neither a function nor a human interacting protein was previously known for EspY1, we detected eight novel interactions that involved this effector protein. EspY1 appears to be primarily involved in apoptosis and cell cycle regulation as these functions are shared among its host targets calpain-3, 26S protease regulatory subunit 4, and PCI domain-containing protein 2. Such interactions may complement other EHEC effector proteins that target host proteins known to block apoptosis while assuring host cell survival. For instance, NleF interacts with caspases 4,8, and 923, while NleH1/NleH2 inhibit the anti-apoptotic protein Bax inhibitor-1 (BI-139). Furthermore, NleD cleaves JNK1 and JNK2, potentially affecting AP-1 activity in EHEC and EPEC-infected cells40.
EspJ and its interactions with the cytoskeleton and phagocytosis
EspJ is another poorly characterized effector that we found to interact with four proteins in our screens. Manually reviewing the function of its binding partners we found that EspJ was functionally linked to the cytoskeleton in three cases: (i) EspJ binds to the guanine nucleotide exchange factor Synembryn-A (RIC8A) that may be involved in signaling and the cytoskeleton41 (ii) Furthermore, EspJ interacts with intraflagellar transport protein 20 homolog (IFT20) that may play a role in the trafficking of ciliary membrane proteins from the Golgi complex to the cilium42. (iii) EspJ also interacts with centromere protein H (CENHP), a component of the nucleosome-associated complex that plays a role in assembly of kinetochore proteins, mitotic progression and chromosome segregation43,44,45.
EspJ was shown to inhibit FCYR and CR3-mediated phagocytosis (immune globulin and complement system-mediated, respectively)46, thereby avoiding the internalization of the bacterial cell and degradation by macrophages. However, the mechanism as well as any human binding partners that could explain these observations remained unknown. Another effector, EspH, is known to counteract FCYR phagocytosis by binding to Rho-GEFs (ARHGEF11, ARHGEF1, LARG) while blocking Rho activation through actin cytoskeletal remodeling46. Finally, EspF, inhibits phagocytosis probably through its actin cytoskeleton-related binding partners (actin, profilin, sorting nexin 9; reviewed in ref 7). The microtubule system is essential for optimal FCYR-mediated phagocytosis47. We speculate that EspJ may specifically affect the microtubule system to block phagocytosis while EspH and EspF interfere with such processes through targeting the actin cytoskeleton.
EspO1 and EspF interact with cell cycle regulators and apoptosis proteins
EspO1-1: The Shigella flexneri effector OspE (an ortholog of EspO1-1) was shown to bind to integrin-linked kinase (ILK) at focal adhesions while reinforcing host cell adherence to the basement membrane48,49. As for EHEC EspO1-1 we discovered another interaction with fem-1 homolog b (Fem1b), a component of an E3 ubiquitin-protein ligase complex acting as substrate recognition subunit50. Fem1b interacts with the checkpoint kinase CHEK1 to sense replication stress51 and can act as a proapoptotic protein in cancer cells52,53,54. EHEC EspO1-1 may be another candidate that could act anti-apoptotically through its Fem1b interaction. In particular, targeting of the ubiquitin system by type III secreted factors is a common theme in pathogenesis of many bacteria (reviewed in ref 55).
EspF1: EspF1 is a highly multifunctional effector that localizes to mitochondria, the cytosol, nucleus, apical and lateral membrane and tight junctions56. EPEC EspF1 interacts with 14-3-3-zeta, ABCF2, actin, ARP2, CK-18, N-WASP, profilin, ZO-1/ZO-2 and SNX9 (sorting nexin 9). These interactions potentially allow EPEC to disrupt mitochondria, tight junctions, nucleolus, intermediate filaments, PI3K-dependent phagocytosis, and membrane remodeling. Such processes are involved in apoptosis and counteract phagocytosis7,35,57,58,59,60,61,62. Specifically, we found that EHEC EspF1 interacts with sorting nexin 33 (SNX33) and mitotic arrest deficient like 2 (MAD2L2) as well as with SNX9, an interaction that was previously found for EPEC and EHEC EspF. Sorting nexins are a large evolutionary conserved protein family involved in cargo sorting through the endosomal network63. SNX9 and SNX33 - both members of the SNX9 subfamily - are (together with SNX18) required for progression and completion of mitosis64. This role in cell cycle regulation is in line with EspF's binding to MAD2L2, a protein participating in the spindle assembly checkpoint65, translesional DNA synthesis in S phase cells66,67 and in Cdc20 homolog 1 (Cdh1) regulation68.
Notably, MAD2L2 is also targeted by the Shigella effector IpaB, leading to a cell cycle arrest in the G2/M phase69, a cellular event that is also induced during EPEC infection70.
TIR interactions in vitro and localization in vivo
To validate the quality of our data we focused on interactions isolated for the translocated intimin receptor (TIR). Our Y2H screen found eight human proteins interacting with TIR, namely arrestin beta 1 (ARRB1), BAI1-associated protein 2 (BAIAP2), BAI1-associated protein 2-like 1 (BAIAP2L1), hippocalcin-like 1 (HPCAL1), neurocalcin delta (NCALD), serine/threonine kinase 16 (STK16), hippocalcin like 4 (HPCAL4) and phosphodiesterase 6D (PDE6D).
Interaction validation: To validate, we tested these interactions using LUMIER assays71,72 (Figure 5A): The effector proteins were purified using a Protein A tag and co-purified binding partners fused to luciferase were detected by luminescence. This confirmed all Y2H interactions, including the interactions of TIR with BAIAP2 and BAIAP2L1 that have been previously found with EHEC and EPEC TIR14,15.
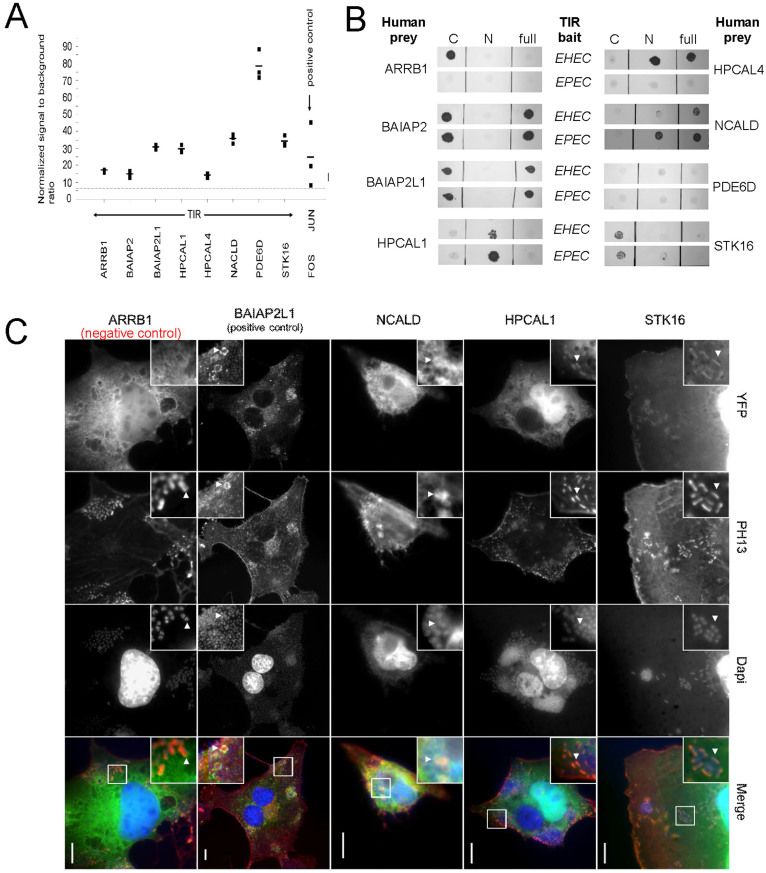
Figure 5. Validation and conservation of TIR-host interactions.
(a) TIREHEC interactions that were detected in our Y2H screens were confirmed by LUMIER assays, using full-length TIR as protein A fusion and the human test partners co-purified as luciferase-tagged fusions. Black squares represent individual measurements of the co-purified luciferase by luminescence, while their averages are shown as black horizontal bars. We considered values above the dashed threshold line (signal to background ratio >6) as a positive binding signal. As a positive control we used JUN/FOS. (b) Comparison of homologous interactions of TIR in EHEC and EPEC by pairwise Y2H tests. In particular, we probed full-length constructs (full) as well as the N- and C-terminal cytosolic domains (not shown) as baits against human preys. (c) We tested the co-localization of TIR interactors with EPEC infection sites on COS-7 cells and used BAIAP2L1 that interacts with TIR as positive control. Co-localization sites are indicated by white arrows. Specifically, we fused (YFP) human binders C-terminally to YFP and used (PH13) F-actin staining to visualize pedestals and (Dapi) DNA staining.
Five interactions are conserved in EHEC and EPEC: Since the TIRs of EHEC and EPEC share 58% of their amino acid sequence73 and carry out similar though not identical functions in these bacteria, we wondered if the isolated TIREHEC interactors would also bind TIREPEC. We used the N- and C-terminal cytosolic domains and a full-length construct of TIR to compare TIR of EHEC and EPEC (Figure 5B). We used BAIAP2 and BAIAP2L1 as positive controls as they are known to interact with both TIREPEC and TIREHEC14,15. HPCAL1, NCALD and STK16 interacted with TIRs from both EPEC and EHEC, representing novel conserved targets of TIR. However, we could not detect any interactions between TIR and ARRB1 and HPCAL4 in EPEC.
HPCAL1, NCALD and STK16 co-localize with EPEC infection sites in COS-7 cells
To test the in vivo-relevance of the conserved TIREPEC interactors we expressed the human binders as YFP-tagged fusion proteins in COS-7 cells and infected the cells with EPEC afterwards. Subsequently, we assessed co-localization of the YFP constructs with the infection sites. As expected, BAIAP2L1 (Figure 5C) and BAIAP2 (data not shown) clearly co-localized with EPEC infection sites14,15. As for HPCAL1, STK16 and NCALD, that interact with TIREPEC, we found that only HPCAL1 and STK16 clearly co-localized with EPEC infection sites despite stronger signals obtained for BAIAP2 and BAIAP2L1. In case of NCALD, the signal seems to be enhanced in the vicinity of EPEC infection sites, but not directly where bacteria and TIR are located. The localization is somewhat reminiscent of NHERF2 and Annexin A2, which also localize to membrane areas surrounding the bacteria21. As expected, ARRB1 and HPCAL4 (not shown) do not co-localize with EPEC infection sites, as they interact specifically with TIREHEC.
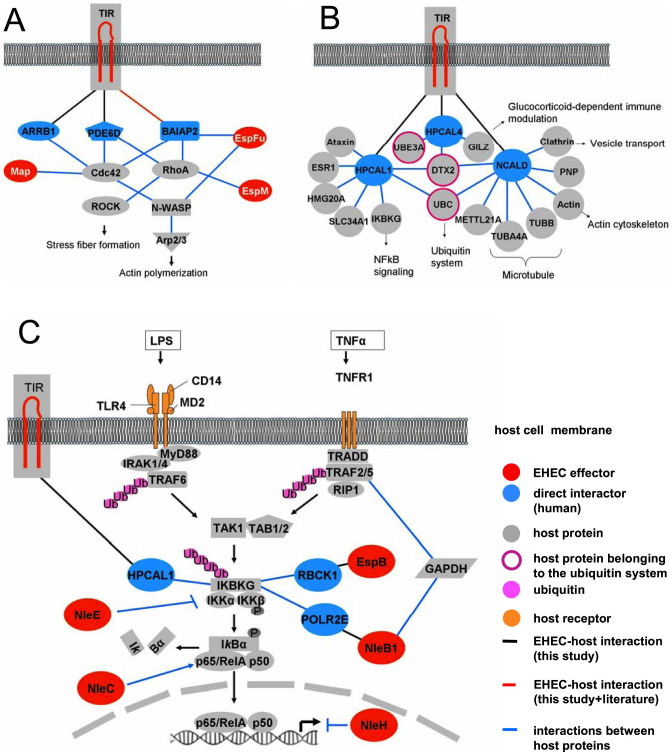
In summary, we discovered another group of TIREHEC interactors, including three calcium binders of the VILIP family, namely HPCAL1, HPCAL4 and NCALD with highly similar amino acid sequences (62% identity). Although mainly known as proteins involved in neuronal signaling in the central nervous system74,75, each gene is expressed in tissues of the digestive tract76,77. The interactions of TIREHEC with the three calcium binding proteins are noteworthy, since the roles of Ca2+ signaling during EPEC infection are contentious78. Some studies found evidence supporting a role of Ca2+ in pedestal formation16,79,80, others did not81. The interactions of TIR are summarized in Figure 6, including previously published interactions.
Figure 6. Tir subnetworks.
(a) TIR and other effectors are highly interconnected with human proteins. Detected, published and detected/published interactions are depicted in black, blue and red, respectively. BAIAP2L1 (not depicted) also interacts with EspFu and TIR and thus plays a homologous role as BAIAP2. (b) EHEC TIR directly targets three calcium binding proteins of the VILIP family. Possible targeted pathways are indicated. (c) IKBKG ( = NEMO) is indirectly targeted by three effector proteins, TIR, EspB and NleB1. In addition, the NF-kB pathway is targeted by several effector proteins such as the metalloprotease NleC. Furthermore, NleH inhibits NF-kB signaling, and NleB1 targets glyceraldehyde 3-phosphate dehydrogenase (GAPDH) while interfering with the signaling cascade through TRAF2. For previously published interactions see Suppl. Table S3.
Conclusions
In this study we determined 43 novel and 5 previously published pathogen-host interactions of EHEC effector proteins. Including data from the literature, our network comparisons showed that EHEC effectors attack the host interactome more specifically than bacteria and viruses.
The quality of the interactome was exemplarily confirmed for six novel TIR interactions by LUMIER assay. Furthermore, we provide evidence for the biological relevance of three interactions, involving NCALD, HPCAL1 and STK16, based on their recruitment to EPEC infection sites.
Notably, these three TIR interactors are small calcium binding proteins that are composed of four consecutive tandem repeated EF-hand motifs. They all belong to the superfamily of neuronal calcium sensors82,83 and were found in TIR-dependent signaling complexes, in addition to AnxA221, IQGAP and calmodulin16. Early upon pathogen contact, microvilli effacement is induced, which was also shown to involve calcium signaling and calcium binding proteins such as calpain84. Calcium has been implicated in microvilli- effacement and disruption84,85 but requires detailed further analysis for a better understanding of EHEC/EPEC pathogenicity.
Methods
Cloning
Effector ORFs of EHEC5 were cloned from E. coli O157:H7 str. Sakai DNA using the Gateway® Technology (Invitrogen) (see Supplementary Table S1). Each effector was amplified in a two-step PCR using gene specific primers and the universal second primers attB1 and attB2 for the attachment of the Gateway attB sites. Primer design, PCR and cloning followed manufacturer's instructions. Resulting PCR products were shuttled into the pDONR221 entry vector via Gateway® BP reactions. Correctness of entry clones was confirmed by sequencing. Then the ORFs were shuttled into Gateway-compatible pGBT9 and pGBKCg plasmids27,32 using an LR reaction (Invitrogen). For the LUMIER tests, TIREHEC was shuttled into pTREX-dest30-ntPrA72. Human ORFs of the TIR binders were shuttled from entry clones (collections29,30,31) into pcDNA3-Rluc-GW72 and for the infection/localization assay into pEYFP-N186.
For the amplification of the TIREHEC amino-and carboxy-terminal fragments the gene specific primers 5′-A GGC TCC ACC ATG AAA ATA ACA AAC TAT ATA CTG CC-3′ (forw) + 5′-C TGG GTG GAT YCA ACT TTC CCT CCC GTA AAG-3′ (rev) and 5′-A GGC TCC ACC ATG GGC GGG CAG CAG TTT ATT TGG-3′ (forw) + 5′-C TGG GTG GAT YCA TTT CGA TGC ATT TAC CAT TGA G-3′ (rev) were used, respectively.
TIREPEC was cloned from E. coli O127:H6 str. E2348/69. The amino- and carboxy terminal regions as well as full length constructs were amplified using the gene specific primer pairs 5′-A GGC TCC ACC ATG CCT ATT GGT AAC CTT GG-3′ (forw) + 5′-C TGG GTG GAT YCA ATC GGT GGT TGT AGG ATC-3′ (rev), 5′-A GGC TCC ACC ATG GAA AGC AAT GCA CAG GCG C-3′ (forw) + 5′-C TGG GTG GAT YCA AAC GAA ACG TAC TGG TCC C-3 (rev) and 5′-A GGC TCC ACC ATG CCT ATT GGT AAC CTT GG-3′ (forw) + 5′-C TGG GTG GAT YCA AAC GAA ACG TAC TGG TCC C-3′ (rev), respectively. Depending on whether a stop is required or not Y can be either T or C (stop or no stop). The second PCR and cloning were performed as described for the EHEC effectors.
Y2H prey libraries
The Mammalian Gene Collection (MGC)30 and the human ORF collection from the German Center for Genome Research (RZPD)29,30,31 were shuttled as pools into pGAD424-GW (Clontech, Gateway-compatible variant) and pGADCg (Clontech32), using an LR reaction. After counter selection in E. coli DH10B prey plasmids were then transfected into yeast strain Y187 (Clontech). The cDNA prey library was purchased from Clontech (Human Universal, Clontech 638874).
Y2H screening
Y2H pool screening was done as described in refs 27, 28 (Figure 1).
Pairwise Y2H tests
In binary Y2H assays, a single bait and a single prey plasmid were transformed in the bait and prey yeast strains, CG-1945 and Y187, respectively (Clontech). Bait and prey were mated on YPDA plates at 30°C over night or for 3 h at 100 rpm in liquid YPDA containing 40% PEG. Diploids were harvested and grown in ΔLW medium for 2 days and then plated on selective agar plates (ΔLWH). To adjust stringency, various concentrations ranging from 0 to 150 mM of 3-amino-1, 2, 4-triazole were added to the selective agar plates. To check for auto-activation all baits were tested against Y187 harboring empty prey vector (negative control). Details can be found in ref 87.
LUMIER assay
Luciferase based pull down assays were performed as described in ref 23.
Infection assay
Transfection of the YFP constructs, infection assays, and image analysis was performed as described in ref 15.
Computational analysis
Human protein interactions
We utilized a total of 28,627 high quality protein interactions between 8,495 human proteins from the HINT database88, accounting for both binary and co-complex interactions.
Human canonical pathway information
As a source of canonical pathway information, we used 1,320 pathways from the Molecular Signatures Database38 that were largely obtained from the NCI Pathway Interaction Database, Reactome and Biocarta.
Bacteria-host and Virus-host protein interactions
As for Bacteria we utilized 2,989 interactions of B. anthracis, 1,347 interactions of F. tularensis and 3,961 interactions of Y. pestis with human host proteins36. Utilizing the IntAct database37, we collected 330 interactions of the Epstein-Barr virus, 557 interactions of the HIV virus, 766 interactions of the Hepatitis virus, 332 interactions of the Herpes virus, 756 interactions of the Influenza virus, 238 interactions from the Papilloma virus and 177 interactions of the Vaccinia virus with human host proteins.
Human protein complexes
We utilized 1,843 protein complexes in H. sapiens from the CORUM database89, that collects information about experimentally determined protein complexes from the literature.
Enrichment analysis as a function of degree
We grouped human proteins according to their number of interactions in an underlying human protein interaction network. We represented each group by N≥k proteins that had at least k interactions and calculated the number of targeted proteins i, Ni,≥k in each group. Randomly picking targeted genes we defined 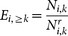 as their enrichment where
as their enrichment where 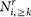 was the corresponding random number of targeted proteins among all Ni,≥k proteins. After averaging Ei over 10,000 randomizations Ei > 1 pointed to an enrichment and vice versa, while Ei ~ 1 indicated a random process90.
was the corresponding random number of targeted proteins among all Ni,≥k proteins. After averaging Ei over 10,000 randomizations Ei > 1 pointed to an enrichment and vice versa, while Ei ~ 1 indicated a random process90.
Author Contributions
The experiments were performed by S.B., S.A., G.S and M.A.S. Experiments were designed and conceived by S.B., M.K., P.U., T.S. and P.A. Data was analyzed by S.B., R.H., A.C., S.W. and F.S. The paper was written by S.B., R.H., P.U., S.W. and M.K.
Supplementary Material
Supplementary Tables S1-S3
Acknowledgments
We are grateful to Kerstin Mohr, Sara Burmester and Nadja Wermke for their help with yeast two-hybrid screens. We thank Ute Ernst for sequencing the entry clones, Barbara Leuchs for assisting with safety issues and Michael S. Donnenberg for kindly providing E. coli O127:H6 str. E2348/69 DNA.
Note The protein interactions from this publication have been submitted to the IMEx (http://www.imexconsortium.org) consortium through the IntAct database37 and assigned the identifier IM-23549.
Funding The work was supported by the Seventh Research Framework Programme of the European Union (“AntiPathoGN” - EU grant HEALTH-F3-2009-223101), the German Research Council DFG (grant STR666/6-1) to T.S., and NIH grant 8U54HD080784.
References
- O'Brien A. O., Lively T. A., Chen M. E., Rothman S. W. & Formal S. B. Escherichia coli O157:H7 strains associated with haemorrhagic colitis in the United States produce a Shigella dysenteriae 1 (SHIGA) like cytotoxin. Lancet 1, 702 (1983). [DOI] [PubMed] [Google Scholar]
- Levine M. M. et al. A DNA probe to identify enterohemorrhagic Escherichia coli of O157:H7 and other serotypes that cause hemorrhagic colitis and hemolytic uremic syndrome. J Infect Dis 156, 175–182 (1987). [DOI] [PubMed] [Google Scholar]
- Tilden J. Jr et al. A new route of transmission for Escherichia coli: infection from dry fermented salami. Am J Public Health 86, 1142–1145 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karch H., Tarr P. I. & Bielaszewska M. Enterohaemorrhagic Escherichia coli in human medicine. Int J Med Microbiol 295, 405–418 (2005). [DOI] [PubMed] [Google Scholar]
- Tobe T. et al. An extensive repertoire of type III secretion effectors in Escherichia coli O157 and the role of lambdoid phages in their dissemination. Proc Natl Acad Sci U S A 103, 14941–14946 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campellone K. G. Cytoskeleton-modulating effectors of enteropathogenic and enterohaemorrhagic Escherichia coli: Tir, EspFU and actin pedestal assembly. FEBS J 277, 2390–2402 (2010). [DOI] [PubMed] [Google Scholar]
- Wong A. R. et al. Enteropathogenic and enterohaemorrhagic Escherichia coli: even more subversive elements. Mol Microbiol 80, 1420–1438 (2011). [DOI] [PubMed] [Google Scholar]
- Yan D. et al. Enteropathogenic Escherichia coli Tir recruits cellular SHP-2 through ITIM motifs to suppress host immune response. Cell Signal (2013). [DOI] [PubMed] [Google Scholar]
- Yan D., Wang X., Luo L., Cao X. & Ge B. Inhibition of TLR signaling by a bacterial protein containing immunoreceptor tyrosine-based inhibitory motifs. Nat Immunol 13, 1063–1071 (2012). [DOI] [PubMed] [Google Scholar]
- Goosney D. L., DeVinney R., Pfuetzner R. A., Frey E. A., Strynadka N. C. & Finlay B. B. Enteropathogenic E. coli translocated intimin receptor, Tir, interacts directly with alpha-actinin. Curr Biol 10, 735–738 (2000). [DOI] [PubMed] [Google Scholar]
- Patel A. et al. Host protein interactions with enteropathogenic Escherichia coli (EPEC): 14-3-3tau binds Tir and has a role in EPEC-induced actin polymerization. Cell Microbiol 8, 55–71 (2006). [DOI] [PubMed] [Google Scholar]
- Cantarelli V. V. et al. Tyrosine phosphorylation controls cortactin binding to two enterohaemorrhagic Escherichia coli effectors: Tir and EspFu/TccP. Cell Microbiol 9, 1782–1795 (2007). [DOI] [PubMed] [Google Scholar]
- Batchelor M. et al. Involvement of the intermediate filament protein cytokeratin-18 in actin pedestal formation during EPEC infection. EMBO Rep 5, 104–110 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vingadassalom D. et al. Insulin receptor tyrosine kinase substrate links the E. coli O157:H7 actin assembly effectors Tir and EspF(U) during pedestal formation. Proc Natl Acad Sci U S A 106, 6754–6759 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss S. M. et al. IRSp53 links the enterohemorrhagic E. coli effectors Tir and EspFU for actin pedestal formation. Cell Host Microbe 5, 244–258 (2009). [DOI] [PubMed] [Google Scholar]
- Brown M. D., Bry L., Li Z. & Sacks D. B. Actin pedestal formation by enteropathogenic Escherichia coli is regulated by IQGAP1, calcium, and calmodulin. J Biol Chem 283, 35212–35222 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruenheid S. et al. Enteropathogenic E. coli Tir binds Nck to initiate actin pedestal formation in host cells. Nat Cell Biol 3, 856–859 (2001). [DOI] [PubMed] [Google Scholar]
- Sason H. et al. Enteropathogenic Escherichia coli subverts phosphatidylinositol 4, 5-bisphosphate and phosphatidylinositol 3, 4, 5-trisphosphate upon epithelial cell infection. Mol Biol Cell 20, 544–555 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantarelli V. V. et al. Talin, a host cell protein, interacts directly with the translocated intimin receptor, Tir, of enteropathogenic Escherichia coli, and is essential for pedestal formation. Cell Microbiol 3, 745–751 (2001). [DOI] [PubMed] [Google Scholar]
- Freeman N. L. et al. Interaction of the enteropathogenic Escherichia coli protein, translocated intimin receptor (Tir), with focal adhesion proteins. Cell Motil Cytoskeleton 47, 307–318 (2000). [DOI] [PubMed] [Google Scholar]
- Munera D., Martinez E., Varyukhina S., Mahajan A., Ayala-Sanmartin J. & Frankel G. Recruitment and membrane interactions of host cell proteins during attachment of enteropathogenic and enterohaemorrhagic Escherichia coli. Biochem J 445, 383–392 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clements A., Smollett K., Lee S. F., Hartland E. L., Lowe M. & Frankel G. EspG of enteropathogenic and enterohemorrhagic E. coli binds the Golgi matrix protein GM130 and disrupts the Golgi structure and function. Cell Microbiol 13, 1429–1439 (2011). [DOI] [PubMed] [Google Scholar]
- Blasche S. et al. The E. coli Effector Protein NleF Is a Caspase Inhibitor. PLoS One 8, e58937 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shames S. R., Croxen M. A., Deng W. & Finlay B. B. The type III system-secreted effector EspZ localizes to host mitochondria and interacts with the translocase of inner mitochondrial membrane 17b. Infect Immun 79, 4784–4790 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao X. et al. NleB, a bacterial effector with glycosyltransferase activity, targets GAPDH function to inhibit NF-kappaB activation. Cell Host Microbe 13, 87–99 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen R. L., Echtenkamp F., Cheranova D., Deng W., Finlay B. B. & Hardwidge P. R. The enterohemorrhagic Escherichia coli effector protein NleF binds mammalian Tmp21. Vet Microbiol 164, 164–170 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albers M. et al. Automated yeast two-hybrid screening for nuclear receptor-interacting proteins. Mol Cell Proteomics 4, 205–213 (2005). [DOI] [PubMed] [Google Scholar]
- Mohr K. & Koegl M. High-throughput yeast two-hybrid screening of complex cDNA libraries. Methods Mol Biol 812, 89–102 (2012). [DOI] [PubMed] [Google Scholar]
- Strausberg R. L. et al. Generation and initial analysis of more than 15,000 full-length human and mouse cDNA sequences. Proc Natl Acad Sci U S A 99, 16899–16903 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Temple G. et al. The completion of the Mammalian Gene Collection (MGC). Genome Res 19, 2324–2333 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brasch M. A., Hartley J. L. & Vidal M. ORFeome cloning and systems biology: standardized mass production of the parts from the parts-list. Genome Res 14, 2001–2009 (2004). [DOI] [PubMed] [Google Scholar]
- Stellberger T., Hauser R., Baiker A., Pothineni V. R., Haas J. & Uetz P. Improving the yeast two-hybrid system with permutated fusions proteins: the Varicella Zoster Virus interactome. Proteome Sci 8, 8 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y. C., Rajagopala S. V., Stellberger T. & Uetz P. Exhaustive benchmarking of the yeast two-hybrid system. Nat Methods 7, 667–668; author reply 668 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez E. et al. Binding to Na(+)/H(+) exchanger regulatory factor 2 (NHERF2) affects trafficking and function of the enteropathogenic Escherichia coli type III secretion system effectors Map, EspI and NleH. Cell Microbiol 12, 1718–1731 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marches O. et al. EspF of enteropathogenic Escherichia coli binds sorting nexin 9. J Bacteriol 188, 3110–3115 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyer M. D. et al. The human-bacterial pathogen protein interaction networks of Bacillus anthracis, Francisella tularensis, and Yersinia pestis. PLoS One 5, e12089 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orchard S. et al. The MIntAct project–IntAct as a common curation platform for 11 molecular interaction databases. Nucleic Acids Res 42, D358–363 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A. et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 102, 15545–15550 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemrajani C. Berger C. N., Robinson K. S., Marches O., Mousnier A. & Frankel G. NleH effectors interact with Bax inhibitor-1 to block apoptosis during enteropathogenic Escherichia coli infection. Proc Natl Acad Sci U S A 107, 3129–3134 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baruch K. et al. Metalloprotease type III effectors that specifically cleave JNK and NF-kappaB. EMBO J 30, 221–231 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruisu K. et al. Ablation of RIC8A function in mouse neurons leads to a severe neuromuscular phenotype and postnatal death. PLoS One 8, e74031 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Follit J. A., Tuft R. A., Fogarty K. E. & Pazour G. J. The intraflagellar transport protein IFT20 is associated with the Golgi complex and is required for cilia assembly. Mol Biol Cell 17, 3781–3792 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saffery R. et al. Transcription within a functional human centromere. Molecular cell 12, 509–516 (2003). [DOI] [PubMed] [Google Scholar]
- Orthaus S., Ohndorf S. & Diekmann S. RNAi knockdown of human kinetochore protein CENP-H. Biochemical and biophysical research communications 348, 36–46 (2006). [DOI] [PubMed] [Google Scholar]
- Okada M. et al. The CENP-H-I complex is required for the efficient incorporation of newly synthesized CENP-A into centromeres. Nat Cell Biol 8, 446–457 (2006). [DOI] [PubMed] [Google Scholar]
- Dong N., Liu L. & Shao F. A bacterial effector targets host DH-PH domain RhoGEFs and antagonizes macrophage phagocytosis. EMBO J 29, 1363–1376 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison R. E. & Grinstein S. Phagocytosis and the microtubule cytoskeleton. Biochem Cell Biol 80, 509–515 (2002). [DOI] [PubMed] [Google Scholar]
- Kim M. et al. Bacteria hijack integrin-linked kinase to stabilize focal adhesions and block cell detachment. Nature 459, 578–582 (2009). [DOI] [PubMed] [Google Scholar]
- Qin J. & Wu C. ILK: a pseudokinase in the center stage of cell-matrix adhesion and signaling. Curr Opin Cell Biol 24, 607–613 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamura T. et al. VHL-box and SOCS-box domains determine binding specificity for Cul2-Rbx1 and Cul5-Rbx2 modules of ubiquitin ligases. Genes Dev 18, 3055–3065 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun T. P. & Shieh S. Y. Human FEM1B is required for Rad9 recruitment and CHK1 activation in response to replication stress. Oncogene 28, 1971–1981 (2009). [DOI] [PubMed] [Google Scholar]
- Subauste M. C., Sansom O. J., Porecha N., Raich N., Du L. & Maher J. F. Fem1b, a proapoptotic protein, mediates proteasome inhibitor-induced apoptosis of human colon cancer cells. Mol Carcinog 49, 105–113 (2010). [DOI] [PubMed] [Google Scholar]
- Chan S. L. et al. F1Aalpha, a death receptor-binding protein homologous to the Caenorhabditis elegans sex-determining protein, FEM-1, is a caspase substrate that mediates apoptosis. J Biol Chem 274, 32461–32468 (1999). [DOI] [PubMed] [Google Scholar]
- Chan S. L., Yee K. S., Tan K. M. & Yu V. C. The Caenorhabditis elegans sex determination protein FEM-1 is a CED-3 substrate that associates with CED-4 and mediates apoptosis in mammalian cells. J Biol Chem 275, 17925–17928 (2000). [DOI] [PubMed] [Google Scholar]
- Angot A., Vergunst A., Genin S. & Peeters N. Exploitation of eukaryotic ubiquitin signaling pathways by effectors translocated by bacterial type III and type IV secretion systems. PLoS Pathog 3, e3 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes A., Muhlen S., Roe A. J. & Dean P. The EspF effector, a bacterial pathogen's Swiss army knife. Infect Immun 78, 4445–4453 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viswanathan V. K., Lukic S., Koutsouris A., Miao R., Muza M. M. & Hecht G. Cytokeratin 18 interacts with the enteropathogenic Escherichia coli secreted protein F (EspF) and is redistributed after infection. Cell Microbiol 6, 987–997 (2004). [DOI] [PubMed] [Google Scholar]
- Nougayrede J. P., Foster G. H. & Donnenberg M. S. Enteropathogenic Escherichia coli effector EspF interacts with host protein Abcf2. Cell Microbiol 9, 680–693 (2007). [DOI] [PubMed] [Google Scholar]
- Peralta-Ramirez J. et al. EspF Interacts with nucleation-promoting factors to recruit junctional proteins into pedestals for pedestal maturation and disruption of paracellular permeability. Infect Immun 76, 3854–3868 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean P., Maresca M., Schuller S., Phillips A. D. & Kenny B. Potent diarrheagenic mechanism mediated by the cooperative action of three enteropathogenic Escherichia coli-injected effector proteins. Proc Natl Acad Sci U S A 103, 1876–1881 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muza-Moons M. M., Schneeberger E. E. & Hecht G. A. Enteropathogenic Escherichia coli infection leads to appearance of aberrant tight junctions strands in the lateral membrane of intestinal epithelial cells. Cell Microbiol 6, 783–793 (2004). [DOI] [PubMed] [Google Scholar]
- Alto N. M. et al. The type III effector EspF coordinates membrane trafficking by the spatiotemporal activation of two eukaryotic signaling pathways. J Cell Biol 178, 1265–1278 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullen P. J. & Korswagen H. C. Sorting nexins provide diversity for retromer-dependent trafficking events. Nat Cell Biol 14, 29–37 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma M. P. & Chircop M. SNX9, SNX18 and SNX33 are required for progression through and completion of mitosis. J Cell Sci 125, 4372–4382 (2012). [DOI] [PubMed] [Google Scholar]
- Reimann J. D., Gardner B. E., Margottin-Goguet F. & Jackson P. K. Emi1 regulates the anaphase-promoting complex by a different mechanism than Mad2 proteins. Genes Dev 15, 3278–3285 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakumo Y., Mizutani S., Yamaguchi M., Ichihara M. & Takahashi M. Analyses of ultraviolet-induced focus formation of hREV1 protein. Genes to cells: devoted to molecular & cellular mechanisms 11, 193–205 (2006). [DOI] [PubMed] [Google Scholar]
- Cheung H. W. et al. Inactivation of human MAD2B in nasopharyngeal carcinoma cells leads to chemosensitization to DNA-damaging agents. Cancer Res 66, 4357–4367 (2006). [DOI] [PubMed] [Google Scholar]
- Pfleger C. M., Salic A., Lee E. & Kirschner M. W. Inhibition of Cdh1-APC by the MAD2-related protein MAD2L2: a novel mechanism for regulating Cdh1. Genes Dev 15, 1759–1764 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwai H. et al. A bacterial effector targets Mad2L2, an APC inhibitor, to modulate host cell cycling. Cell 130, 611–623 (2007). [DOI] [PubMed] [Google Scholar]
- Nougayrede J. P. et al. Type III secretion-dependent cell cycle block caused in HeLa cells by enteropathogenic Escherichia coli O103. Infect Immun 69, 6785–6795 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrios-Rodiles M. et al. High-throughput mapping of a dynamic signaling network in mammalian cells. Science 307, 1621–1625 (2005). [DOI] [PubMed] [Google Scholar]
- Blasche S. & Koegl M. Analysis of protein-protein interactions using LUMIER assays. Methods Mol Biol 1064, 17–27 (2013). [DOI] [PubMed] [Google Scholar]
- DeVinney R., Stein M., Reinscheid D., Abe A., Ruschkowski S. & Finlay B. B. Enterohemorrhagic Escherichia coli O157:H7 produces Tir, which is translocated to the host cell membrane but is not tyrosine phosphorylated. Infect Immun 67, 2389–2398 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgoyne R. D. Neuronal calcium sensor proteins: generating diversity in neuronal Ca2+ signalling. Nat Rev Neurosci 8, 182–193 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jheng F. F., Wang L., Lee L. & Chang L. S. Functional contribution of Ca2+ and Mg2+ to the intermolecular interaction of visinin-like proteins. Protein J 25, 250–256 (2006). [DOI] [PubMed] [Google Scholar]
- Uhlen M. et al. Towards a knowledge-based Human Protein Atlas. Nat Biotechnol 28, 1248–1250 (2010). [DOI] [PubMed] [Google Scholar]
- Kapushesky M. et al. Gene Expression Atlas update–a value-added database of microarray and sequencing-based functional genomics experiments. Nucleic Acids Res 40, D1077–1081 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goosney D. L., Gruenheid S. & Finlay B. B. Gut feelings: enteropathogenic E. coli (EPEC) interactions with the host. Annu Rev Cell Dev Biol 16, 173–189 (2000). [DOI] [PubMed] [Google Scholar]
- Ide T., Michgehl S., Knappstein S., Heusipp G. & Schmidt M. A. Differential modulation by Ca2+ of type III secretion of diffusely adhering enteropathogenic Escherichia coli. Infect Immun 71, 1725–1732 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldwin T. J., Ward W., Aitken A., Knutton S. & Williams P. H. Elevation of intracellular free calcium levels in HEp-2 cells infected with enteropathogenic Escherichia coli. Infect Immun 59, 1599–1604 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bain C., Keller R., Collington G. K., Trabulsi L. R. & Knutton S. Increased levels of intracellular calcium are not required for the formation of attaching and effacing lesions by enteropathogenic and enterohemorrhagic Escherichia coli. Infect Immun 66, 3900–3908 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgoyne R. D. & Weiss J. L. The neuronal calcium sensor family of Ca2+-binding proteins. Biochem J 353, 1–12 (2001). [PMC free article] [PubMed] [Google Scholar]
- Braunewell K. H. & Klein-Szanto A. J. Visinin-like proteins (VSNLs): interaction partners and emerging functions in signal transduction of a subfamily of neuronal Ca2+ -sensor proteins. Cell Tissue Res 335, 301–316 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potter D. A. et al. Calpain regulates enterocyte brush border actin assembly and pathogenic Escherichia coli-mediated effacement. J Biol Chem 278, 30403–30412 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrary E. et al. In vivo, villin is required for Ca(2+)-dependent F-actin disruption in intestinal brush borders. J Cell Biol 146, 819–830 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson J. C., Wellenreuther R., Poustka A., Pepperkok R. & Wiemann S. Systematic subcellular localization of novel proteins identified by large-scale cDNA sequencing. EMBO Rep 1, 287–292 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cagney G. & Uetz. P. High-throughput screening for protein-protein interactions using yeast two-hybrid arrays. Curr Protoc Protein Sci Chapter 19, Unit 19 16 (2001). [DOI] [PubMed] [Google Scholar]
- Das J. & Yu H. HINT: High-quality protein interactomes and their applications in understanding human disease. BMC systems biology 6, 92 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruepp A. et al. CORUM: the comprehensive resource of mammalian protein complexes–2009. Nucleic Acids Res 38, D497–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wuchty S. Evolution and topology in the yeast protein interaction network. Genome Res 14, 1310–1314 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Tables S1-S3