Abstract
Hepatitis C virus (HCV) infection represents a significant health problem and represents a heavy load on some countries like Egypt in which about 20% of the total population are infected. Initial infection is usually asymptomatic and result in chronic hepatitis that give rise to complications including cirrhosis and hepatocellular carcinoma. The management of HCV infection should not only be focus on therapy, but also to screen carrier individuals in order to prevent transmission. In the present, molecular detection and quantification of HCV genome by real time polymerase chain reaction (PCR) represent the gold standard in HCV diagnosis and plays a crucial role in the management of therapeutic regimens. However, real time PCR is a complicated approach and of limited distribution. On the other hand, isothermal DNA amplification techniques have been developed and offer molecular diagnosis of infectious dieses at point-of-care. In this review we discuss recombinase polymerase amplification technique and illustrate its diagnostic value over both PCR and other isothermal amplification techniques.
Keywords: Hepatitis C virus, Nucleic acid testing, Polymerase chain reaction, Point-of-care, Recombinase polymerase amplification
Core tip: Recombinase polymerase amplification (RPA) shows many advantages over both real time polymerase chain reaction and other isothermal Amplification methods. In this review we show the importance of molecular detection methods and how isothermal amplification techniques offer molecular point-of-care diagnosis. RPA shows unique characteristics among isothermal approaches that makes it a promising tool in the molecular diagnosis. Because hepatitis C virus is an endemic viral infection, we suggest that RPA may play an important role and save much time in screening infected individuals and managing the therapeutic course.
HEPATITIS C VIRUS
Hepatitis C virus (HCV) is a positive-sense single-stranded RNA virus that was first cloned in 1989 and classified as a member of the family Flaviviridae[1]. This viral infection is characterized by high replication rate. It is estimated that about 1012 virions per day are produced in a given individual[2]. In addition, its genome exhibits a high degree of sequence variation caused by its error prone RNA polymerase. However, there are 6 characterized genotypes of HCV, 52 subtypes within these genotypes[3]. Humans are the only reservoir for HCV infection; which often leads to an asymptomatic chronic state in 80% of cases with subsequent development to acute liver disease.
An estimated 2%-3% of the world’s population is living with HCV infection and each year more than 350000 die of HCV-related complications, including cirrhosis, liver failure or hepatocellular carcinoma[4].
Although hepatitis C is considered to be endemic disease worldwide, there is a high degree of geographical variation in its distribution[5-9]. The prevalence of HCV infection is low, in most European countries where it represents 0.5%-2% of the general population[10,11], Americas, Australia, and South Africa (0.2% to 0.5%)[10]. Intermediate prevalence is reported in Middle East, India and Brazil[7,10]. Egypt recorded the highest prevalence of HCV in the world with about 20% of the population[7,9].
HCV is a blood prone infection, modes of transmission that have been reported include; transfusion of contaminated blood products, organ transplantation from infected donors, intravenous drug use, sexual transmission, public shaving, acupuncture, and invasive hospital procedures with contaminated equipment[12-16]. In Egypt where the highest prevalence in the world has been recorded, the major route of HCV infection was via an antischistosomal treatment program, with more than 35 million injections given over a 20-year in the period (1960-1980)[17].
The current standard treatment for chronic hepatitis C is a combination of pegylated interferon alfa and ribavirin. Sustained Virological Response (SVR) represents the endpoint of the treatment regimen, which indicates undetectable HCV RNA 24 wk post treatment[18].
Due to the lack of a vaccine or some form of post-exposure prophylaxis, the number of infected individuals will continue to increase, and in turn HCV-related morbidity and mortality, in the absence of effective care and treatment programs. The management of hepatitis C infection should not only focus on the treatment, but also prevention of infection to reduce the reservoir of infected individuals who can transmit the virus[19,20].
HCV DIAGNOSIS TECHNIQUES
The current laboratory techniques used for HCV diagnosis include: (1) Serological assays (e.g., the enzyme-linked immunosorbent assay, recombinant immunoblot assay, etc.); and (2) Molecular assays: Depends on nucleic acid testing (NAT): qualitative [e.g., reverse transcriptase polymerase chain reaction (RT-PCR), TMA, etc.] and quantitative (e.g., real time PCR, etc.).
Advantages and limitation of serologic assays
The ease of automation and cost-effectiveness made serologic assays the most practical tool in HCV diagnosis[21]. However, antibody detection exhibits many disadvantages including that; detection is limited during the early stages of infection, poor sensitivity (false negative) in hemodialysis patients, immunocompromised patients[22-25], an abundance of false-positives[26] (because recovered patients may stay anti-HCV positive for years) and variability in accuracy between deferent commercial kits.
NAT
NAT detect and quantify HCV RNA and are now considered the gold standard in the diagnosis of HCV infection. In this approach, HCV RNA is extracted from the sample and reverse transcribed into the complementary DNA, which is then amplified into a large number of detectable copies by the polymerase chain reaction (PCR). Unlike antibody detection that could be positive for years after resolving infection, the presence of HCV RNA indicates active infection and it can be detected in 1-2 wk post-infection[27,28]. NAT offers accurate and sensitive diagnosis of HCV without any additional confirmatory test and can be used to diagnose individuals with acute HCV infection. In addition, NAT play a crucial role in the management of antiviral therapies by monitoring HCV RNA level. It determines the basal viral load and monitors the treatment response[29]. Till now, fully automated real-time PCR is the most promising approach in NAT as it is faster, more sensitive and is not prone to contamination.
ADVANTAGES AND LIMITATION OF NAT
The importance of NAT arises from its ability to detect and quantify HCV RNA and in turn detecting the active infection (in contrast to anti-HCV). In addition, it can determine the level of the virus replication. Furthermore, it plays an important role in the antiviral treatment regimens and determines whether a virological response has been occurred or not[30].
However, molecular techniques for HCV diagnosis have many limitations including that; it is of complex procedures, time consuming and technically demanding as it cannot be carried out except in a highly equipped molecular biology laboratory (high cost analytical instruments).
ADVANCES IN HCV DIAGNOSIS
Every day the world takes a step towards NAT which becomes more practical than it was before. The competition between the commercial products enforces the companies to produce more simple, easy to use and cheap assays. In addition to the growing dependence on NAT, the significant advances in HCV diagnosis include using point-of-care (POC) alternatives instead of the routine venous puncture. POC can use specimen matrices such as oral fluid or finger-stick blood. Most existing POC are immunoassays and are now widely used for different applications. POC represent an ideal approach for the management of hepatitis C infection as it can reaches remote areas where the high equipped molecular biology laboratories are limited and in turn shorten the time of results which extend HCV screening. For instance, the development of a molecular point-of-care assay would represent a significant improvement in the field of HCV diagnosis.
ISOTHERMAL DNA AMPLIFICATION
Molecular analytical techniques gain a growing interest. According to the mentioned limitation combining conventional molecular methods, especially real time PCR, there was a demand to develop a simple, sensitive and cost effective technology.
Isothermal DNA amplification is an alternative to PCR-based technique and developed for point-of-care diagnosis[31,32]. In isothermal techniques, amplification reactions are performed at a constant temperature and hence there is no need for expensive thermal cycling instrument. Major practiced isothermal amplification techniques include; nucleic acid sequence-based amplification, loop-mediated isothermal amplification (LAMP)[33], strand-displacement amplification (SDA)[34], rolling circle amplification (RCA)[35], helicase-dependent amplification (HDA)[36] and recombinase polymerase amplification (RPA)[37]. Isothermal DNA amplification techniques are simple, rapid and cost effective with equivalent specificity and sensitivity to PCR, enabling point-of-care diagnostics without the need to high costing equipment[31,32]. However, isothermal amplification approaches differed from each other in terms of operating temperature, reaction duration, mechanism, strengths and weaknesses. Table 1 summarizes the characters of the major practiced isothermal amplification methods.
Table 1.
Characters of some isothermal amplification techniques[51]
| NASBA | LAMP | SDA | RCA | HDA | RPA | |
| Template | DNA, RNA | DNA1 | DNA1 | DNA1 | DNA1 | DNA1 |
| No. of primers | 2 | 4-6 | 4 | 1 | 2 | 2 |
| No. of enzymes | 3 | 1 | 2 | 2 | 2 | 2 |
| Temperature (°C) | 41 | 60-65 | 37 | 37 | 65 | 30-42 |
| Reaction duration (min) | 90-120 | 60-90 | 120 | 60 | 75-90 | 20 |
| Denaturation step | Y | N | Y | N | N | N |
| Inhibition tolerance | N | Y | N | N | Y | Y |
| Product detection | GE, RT | GE, RT, TE | GE, RT | GE | GE, RT | RT |
| Multiplex | Y | N | Y | N | Y | Y |
| Point-of-care | Y | Y | Y | N | Y | Y |
RNA can be amplified after the introduction of a reverse transcription step. NASBA: Nucleic acid sequence-based amplification; LAMP: Loop-mediated isothermal amplification; SDA: Strand-displacement amplification; RCA: Rolling circle amplification; HDA: Helicase-dependent amplification; RPA: Recombinase polymerase amplification; GE: Gel Electrophoreses; RT: Real Time; TE: Turbidity; Y: Yes; N: No.
As a competition between isothermal amplification techniques to perform molecular diagnosis at point-of-care, RCA will be kicked out of the race because it is incompatible with point-of-care diagnosis. Complex primer designing and the inability to perform multiplex amplification eliminates LAMP. The need to a denaturation step and the inability to tolerate inhibitory biological components exit both NABSA and SDA. Finally, RPA beats HAD in being faster and cheaper. Table 2 shows advantages and disadvantages of the major practiced isothermal amplification techniques.
Table 2.
Advantages/disadvantages of some isothermal amplification methods
| Technique | Advantages | Disadvantages |
| NASBA | Specifically designed to detect RNA and in turn RNA viruses | Denaturation step |
| Power saving (41 °C) | Less efficient in Amplifying RNA targets out of the range 120-250 bp | |
| LAMP | Highly specific (utilizes 4-6 primers spanning 6-8 distinct sequences) | Primer design is complex |
| Tolerance to biological substances | Unable to perform multiplex amplification | |
| Could be detected by a cheap turbidity-meter | ||
| SDA | Power saving (37 °C) | Sample prep. needed |
| Nuclease selection is complex | ||
| Inefficient in long target sequences | ||
| RCA | Power saving (37 °C) | Primer is complex |
| Specific enough to allow SNP analysis | RNA amplification is complex | |
| Works only with a circular nucleic acid template | ||
| HDA | Simple primer design | Expensive enzymes |
| Robust to biological substances | ||
| No initial heating step | ||
| RPA | Power saving (37 °C) | |
| Simple primer design | ||
| Extremely quick (20 min) | ||
| No initial heating step | ||
| Robust to biological substances |
NASBA: Nucleic acid sequence-based amplification; LAMP: Loop-mediated isothermal amplification; SDA: Strand-displacement amplification; RCA: Rolling circle amplification; HDA: Helicase-dependent amplification; RPA: Recombinase polymerase amplification.
RPA
RPA is an isothermal DNA amplification and detection method[37]. The amplification depends on a specific combination of enzymes and proteins (recombinase, single strand binding protein, and strand displacing DNA polymerase) used at a constant temperature and yielding a result in maximum 10 min. At first, RecA coat a single-stranded DNA (primers) to form nucleoprotein filaments. These filaments can then scan targeted double-stranded DNA for sequences complementary to those of coated primers. Then, the nucleoprotein filaments initiate a 5’-strand invasion at the site of homology (Figure 1) forming what is known as D-loop. The strand invasion is stabilized by single strand binding protein. After that, strand extension takes place at the free 3’-end of the nucleoprotein filaments by a strand displacing DNA polymerase to synthesize a new complementary strand. During strand extension, the new synthesized strand displaces the originally paired strand.
Figure 1.
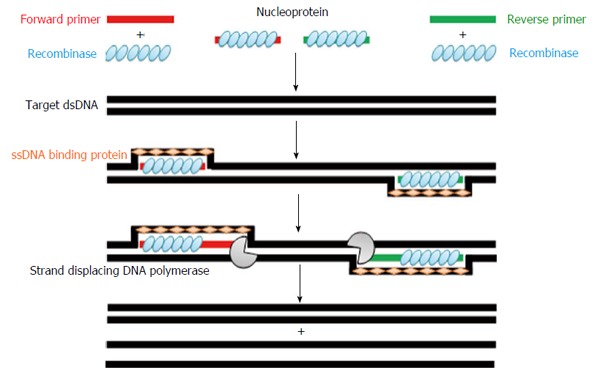
Recombinase polymerase amplification technology amplification cycle (for details, see the text above). dsDNA: Double-stranded DNA.
Real-time detection of RPA amplicons is possible via specific probes (Figure 2). Development of fluorescence depends on the separation of fluorophore and quencher via Exonuclease III cleaving at an internal abasic site mimic [tetrahydrofuran (THF)] of the hybridized exo-probe (Figure 2)[38,39]. Fluorescence signal can be measured in real-time via a simple point-of-care scanner.
Figure 2.
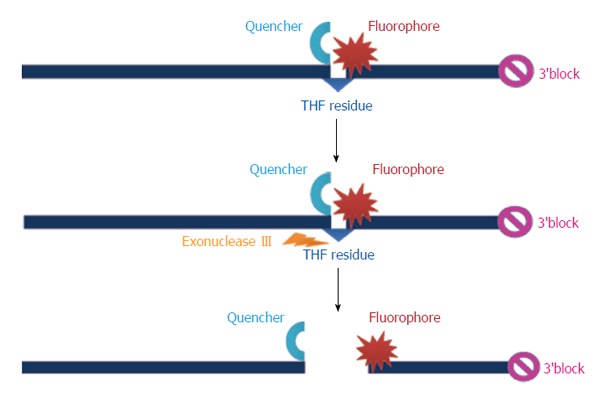
Example for the specific probe of the recombinase polymerase amplification assay. THF: Tetrahydrofuran.
RPA technique is not restricted for amplification of the double stranded DNA targets, but also it could be used for amplification of RNA targets, as in the case with RT-PCR. Ahmed Abd El Wahed et al[40], 2013 had developed reverse transcriptase RPA (RT-RPA) assay for the detection of corona virus. The assay showed rapid kinetics with equal sensitivity and specificity of the real-time RT-PCR. The author suggested the diagnostic importance of the RT-RPA assay during the Hajj for the point-of-care detection of MERS-CoV infected cases to prevent the spread of the virus. Euler et al[39], 2012 have developed a qualitative real-time RPA assay for detection of Francisella tularensis and the assay showed results comparable to real-time PCR. In another wider study by Euler et al[41], 2013 RPA based assays were developed for the detection of Gram-negative (Francisella tularensis and Yersinia pestis) and Gram-positive bacteria (Bacillus anthracis), DNA viruses (variola virus), whereas RT-RPA assays were developed for RNA viruses including Rift Valley fever virus, Ebola virus, Sudan virus and Marburg virus. The authors found analytical sensitivity and specificity equal to PCR with no cross-detection among respective targets. Also, Ahmed et al[42], 2014 have developed RPA based assay for the detection of Leptospira and the method showed fast and less sensitivity to amplification inhibitors. Another competitive character compared to PCR based protocols had been reported by Kersting et al[43], 2014 study in which RPA have been used for multiplex detection (detection multiple targets in the same reaction) of Neisseria gonorrhoeae, Salmonella enterica and Staphylococcus aureus. The author concluded that the kinetic performance of RPA was faster than PCR with no loss in sensitivity and specificity[43]. In the light of the above mentioned results it is clear that, RPA show competitive results as compared with PCR as regard to sensitivity and specificity, whereas RPA exceeds PCR as regard to the reaction kinetics.
In another study, RPA showed an impressive results in which the amplification reaction was conducted under a broad range of conditions from 30 °C-45 °C with high inhibitory concentration of known PCR inhibitors in just 15 min[44].
ADVANTAGES AND DISADVANTAGES OF RPA
RPA overcomes the technical difficulties posed by current molecular techniques.
At first; it demonstrates a rapid kinetics, the process begins operating the immediately when the sample is contacted to the reagents and there is no need for melting the double-stranded DNA target.
Second, it operates at a constant temperature (30-42 °C, optimum nearly 37 °C), which give the advantage of being an energy saving technique and cost saving (there is no need for thermal cycler).
In addition, the target can be either DNA, or RNA, making RPA suitable for the detection and diagnosis of RNA viruses, like HCV.
Furthermore, the combination of probe-based detection, RPA represents a significant advance in the development of portable and accessible nucleic acid-based tests.
Unlike PCR in which the amplification reaction is controlled by temperature, digital RPA suffers from undesired reactions because the amplification could proceeds at room temperature if the nucleic acid sample is premixed with initiation reagents prior to compartmentalization and thus increasing the target count. However, any low-temperature non-specific pre-amplification reaction can be eliminated by compartmentalization of the nucleic acid template prior to adding initiation reagents[45,46].
Another drawback of low temperature amplification results from the interaction between primers even when well-designed. These interactions can create noise that defeats the analysis. However, this drawback could be avoided by using Self Avoiding Molecular Recognition System (SAMRS)[47]. SAMRS are nucleotide analogues that can bind to natural DNA but not to other SAMRS species. Therefore, primers built from SAMRS not interfere with each other. The concept of SAMRS was introduced over a decade ago[48,49] and was exploited to fix interactions between primers in PCR and multiplex PCR[50].
CONCLUSION
Early diagnosis and treatment of HCV infection can reduce the risk of long-term complications and prevent further transmission as well. NAT represent the gold standard for the diagnosis of HCV infection. Detection of HCV RNA level is an important factor in antiviral regimens especially for determination of SVR. RPA combines the advantages of serologic and Molecular techniques and overcome limitations of both. It represents a simple, accurate and cost effective diagnostic tool and can be carried out at remote areas. In turn, it could improve the management of HCV infection by screening carrier individuals and stop transmission. The demand for the development of nucleic acid based point-of-care assay is increasing alongside with increasing the number of HCV infected patients. RPA based HCV diagnosis would represent a significant advance in the management of HCV.
Footnotes
P- Reviewer: He JY, Koch TR S- Editor: Tian YL L- Editor: A E- Editor: Liu SQ
References
- 1.Purcell R. The hepatitis C virus: overview. Hepatology. 1997;26:11S–14S. doi: 10.1002/hep.510260702. [DOI] [PubMed] [Google Scholar]
- 2.Neumann AU, Lam NP, Dahari H, Gretch DR, Wiley TE, Layden TJ, Perelson AS. Hepatitis C viral dynamics in vivo and the antiviral efficacy of interferon-alpha therapy. Science. 1998;282:103–107. doi: 10.1126/science.282.5386.103. [DOI] [PubMed] [Google Scholar]
- 3.Joyce MA, Tyrrell DL. The cell biology of hepatitis C virus. Microbes Infect. 2010;12:263–271. doi: 10.1016/j.micinf.2009.12.012. [DOI] [PubMed] [Google Scholar]
- 4.Averhoff FM, Glass N, Holtzman D. Global burden of hepatitis C: considerations for healthcare providers in the United States. Clin Infect Dis. 2012;55 Suppl 1:S10–S15. doi: 10.1093/cid/cis361. [DOI] [PubMed] [Google Scholar]
- 5.Alter MJ. Epidemiology of hepatitis C virus infection. World J Gastroenterol. 2007;13:2436–2441. doi: 10.3748/wjg.v13.i17.2436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mohd Hanafiah K, Groeger J, Flaxman AD, Wiersma ST. Global epidemiology of hepatitis C virus infection: new estimates of age-specific antibody to HCV seroprevalence. Hepatology. 2013;57:1333–1342. doi: 10.1002/hep.26141. [DOI] [PubMed] [Google Scholar]
- 7.Wasley A, Alter MJ. Epidemiology of hepatitis C: geographic differences and temporal trends. Semin Liver Dis. 2000;20:1–16. doi: 10.1055/s-2000-9506. [DOI] [PubMed] [Google Scholar]
- 8.Sy T, Jamal MM. Epidemiology of hepatitis C virus (HCV) infection. Int J Med Sci. 2006;3:41–46. doi: 10.7150/ijms.3.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yen T, Keeffe EB, Ahmed A. The epidemiology of hepatitis C virus infection. J Clin Gastroenterol. 2003;36:47–53. doi: 10.1097/00004836-200301000-00015. [DOI] [PubMed] [Google Scholar]
- 10.Martins T, Narciso-Schiavon JL, Schiavon Lde L. [Epidemiology of hepatitis C virus infection] Rev Assoc Med Bras. 2011;57:107–112. doi: 10.1590/s0104-42302011000100024. [DOI] [PubMed] [Google Scholar]
- 11.Hepatitis C--global prevalence (update) Wkly Epidemiol Rec. 1999;74:425–427. [PubMed] [Google Scholar]
- 12.Alter MJ. Prevention of spread of hepatitis C. Hepatology. 2002;36:S93–S98. doi: 10.1053/jhep.2002.36389. [DOI] [PubMed] [Google Scholar]
- 13.Recommendations for prevention and control of hepatitis C virus (HCV) infection and HCV-related chronic disease. Centers for Disease Control and Prevention. MMWR Recomm Rep. 1998;47:1–39. [PubMed] [Google Scholar]
- 14.Puro V, Petrosillo N, Ippolito G. Risk of hepatitis C seroconversion after occupational exposures in health care workers. Italian Study Group on Occupational Risk of HIV and Other Bloodborne Infections. Am J Infect Control. 1995;23:273–277. doi: 10.1016/0196-6553(95)90056-x. [DOI] [PubMed] [Google Scholar]
- 15.Roberts EA, Yeung L. Maternal-infant transmission of hepatitis C virus infection. Hepatology. 2002;36:S106–S113. doi: 10.1053/jhep.2002.36792. [DOI] [PubMed] [Google Scholar]
- 16.Terrault NA. Sexual activity as a risk factor for hepatitis C. Hepatology. 2002;36:S99–105. doi: 10.1053/jhep.2002.36797. [DOI] [PubMed] [Google Scholar]
- 17.Frank C, Mohamed MK, Strickland GT, Lavanchy D, Arthur RR, Magder LS, El Khoby T, Abdel-Wahab Y, Aly Ohn ES, Anwar W, et al. The role of parenteral antischistosomal therapy in the spread of hepatitis C virus in Egypt. Lancet. 2000;355:887–891. doi: 10.1016/s0140-6736(99)06527-7. [DOI] [PubMed] [Google Scholar]
- 18.NIH Consensus Statement on Management of Hepatitis C: 2002. NIH Consens State Sci Statements. 2002;19:1–46. [PubMed] [Google Scholar]
- 19.Schafer DF, Sorrell MF. Conquering hepatitis C, step by step. N Engl J Med. 2000;343:1723–1724. doi: 10.1056/NEJM200012073432309. [DOI] [PubMed] [Google Scholar]
- 20.Lauer GM, Walker BD. Hepatitis C virus infection. N Engl J Med. 2001;345:41–52. doi: 10.1056/NEJM200107053450107. [DOI] [PubMed] [Google Scholar]
- 21.Pawlotsky JM, Lonjon I, Hezode C, Raynard B, Darthuy F, Remire J, Soussy CJ, Dhumeaux D. What strategy should be used for diagnosis of hepatitis C virus infection in clinical laboratories? Hepatology. 1998;27:1700–1702. doi: 10.1002/hep.510270632. [DOI] [PubMed] [Google Scholar]
- 22.Zhu WF, Lei SY, Li LJ. Hepatitis C virus infection and biological false-positive syphilis test: a single-center experience. Hepatobiliary Pancreat Dis Int. 2011;10:399–402. doi: 10.1016/s1499-3872(11)60067-2. [DOI] [PubMed] [Google Scholar]
- 23.Senevirathna D, Amuduwage S, Weerasingam S, Jayasinghe S, Fernandopulle N. Hepatitis C virus in healthy blood donors in Sri Lanka. Asian J Transfus Sci. 2011;5:23–25. doi: 10.4103/0973-6247.75976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thio CL, Nolt KR, Astemborski J, Vlahov D, Nelson KE, Thomas DL. Screening for hepatitis C virus in human immunodeficiency virus-infected individuals. J Clin Microbiol. 2000;38:575–577. doi: 10.1128/jcm.38.2.575-577.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kalantar-Zadeh K, Miller LG, Daar ES. Diagnostic discordance for hepatitis C virus infection in hemodialysis patients. Am J Kidney Dis. 2005;46:290–300. doi: 10.1053/j.ajkd.2005.05.006. [DOI] [PubMed] [Google Scholar]
- 26.Leon R, de Medina M, Schiff ER. Diagnostic tools in the evaluation of patients with viral hepatitis undergoing liver transplantation. Liver Transpl Surg. 1998;4:94–103. doi: 10.1002/lt.500040114. [DOI] [PubMed] [Google Scholar]
- 27.Young KK, Archer JJ, Yokosuka O, Omata M, Resnick RM. Detection of hepatitis C virus RNA by a combined reverse transcription PCR assay: comparison with nested amplification and antibody testing. J Clin Microbiol. 1995;33:654–657. doi: 10.1128/jcm.33.3.654-657.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Busch MP, Kleinman SH, Jackson B, Stramer SL, Hewlett I, Preston S. Committee report. Nucleic acid amplification testing of blood donors for transfusion-transmitted infectious diseases: Report of the Interorganizational Task Force on Nucleic Acid Amplification Testing of Blood Donors. Transfusion. 2000;40:143–159. doi: 10.1046/j.1537-2995.2000.40020143.x. [DOI] [PubMed] [Google Scholar]
- 29.Hendricks DA, Friesenhahn M, Tanimoto L, Goergen B, Dodge D, Comanor L. Multicenter evaluation of the VERSANT HCV RNA qualitative assay for detection of hepatitis C virus RNA. J Clin Microbiol. 2003;41:651–656. doi: 10.1128/JCM.41.2.651-656.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Komurian-Pradel F, Paranhos-Baccalà G, Sodoyer M, Chevallier P, Mandrand B, Lotteau V, André P. Quantitation of HCV RNA using real-time PCR and fluorimetry. J Virol Methods. 2001;95:111–119. doi: 10.1016/s0166-0934(01)00300-7. [DOI] [PubMed] [Google Scholar]
- 31.Kim J, Easley CJ. Isothermal DNA amplification in bioanalysis: strategies and applications. Bioanalysis. 2011;3:227–239. doi: 10.4155/bio.10.172. [DOI] [PubMed] [Google Scholar]
- 32.Craw P, Balachandran W. Isothermal nucleic acid amplification technologies for point-of-care diagnostics: a critical review. Lab Chip. 2012;12:2469–2486. doi: 10.1039/c2lc40100b. [DOI] [PubMed] [Google Scholar]
- 33.Notomi T, Okayama H, Masubuchi H, Yonekawa T, Watanabe K, Amino N, Hase T. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 2000;28:E63. doi: 10.1093/nar/28.12.e63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Walker GT, Fraiser MS, Schram JL, Little MC, Nadeau JG, Malinowski DP. Strand displacement amplification--an isothermal, in vitro DNA amplification technique. Nucleic Acids Res. 1992;20:1691–1696. doi: 10.1093/nar/20.7.1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lizardi PM, Huang X, Zhu Z, Bray-Ward P, Thomas DC, Ward DC. Mutation detection and single-molecule counting using isothermal rolling-circle amplification. Nat Genet. 1998;19:225–232. doi: 10.1038/898. [DOI] [PubMed] [Google Scholar]
- 36.Vincent M, Xu Y, Kong H. Helicase-dependent isothermal DNA amplification. EMBO Rep. 2004;5:795–800. doi: 10.1038/sj.embor.7400200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Piepenburg O, Williams CH, Stemple DL, Armes NA. DNA detection using recombination proteins. PLoS Biol. 2006;4:e204. doi: 10.1371/journal.pbio.0040204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Euler M, Wang Y, Nentwich O, Piepenburg O, Hufert FT, Weidmann M. Recombinase polymerase amplification assay for rapid detection of Rift Valley fever virus. J Clin Virol. 2012;54:308–312. doi: 10.1016/j.jcv.2012.05.006. [DOI] [PubMed] [Google Scholar]
- 39.Euler M, Wang Y, Otto P, Tomaso H, Escudero R, Anda P, Hufert FT, Weidmann M. Recombinase polymerase amplification assay for rapid detection of Francisella tularensis. J Clin Microbiol. 2012;50:2234–2238. doi: 10.1128/JCM.06504-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Abd El Wahed A, Patel P, Heidenreich D, Hufert FT, Weidmann M. Reverse transcription recombinase polymerase amplification assay for the detection of middle East respiratory syndrome coronavirus. PLoS Curr. 2013;5 doi: 10.1371/currents.outbreaks.62df1c7c75ffc96cd59034531e2e8364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Euler M, Wang Y, Heidenreich D, Patel P, Strohmeier O, Hakenberg S, Niedrig M, Hufert FT, Weidmann M. Development of a panel of recombinase polymerase amplification assays for detection of biothreat agents. J Clin Microbiol. 2013;51:1110–1117. doi: 10.1128/JCM.02704-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ahmed A, van der Linden H, Hartskeerl RA. Development of a recombinase polymerase amplification assay for the detection of pathogenic Leptospira. Int J Environ Res Public Health. 2014;11:4953–4964. doi: 10.3390/ijerph110504953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kersting S, Rausch V, Bier FF, von Nickisch-Rosenegk M. Multiplex isothermal solid-phase recombinase polymerase amplification for the specific and fast DNA-based detection of three bacterial pathogens. Mikrochim Acta. 2014;181:1715–1723. doi: 10.1007/s00604-014-1198-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kersting S, Rausch V, Bier FF, von Nickisch-Rosenegk M. Rapid detection of Plasmodium falciparum with isothermal recombinase polymerase amplification and lateral flow analysis. Malar J. 2014;13:99. doi: 10.1186/1475-2875-13-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Paul N, Shum J, Le T. Hot start PCR. Methods Mol Biol. 2010;630:301–318. doi: 10.1007/978-1-60761-629-0_19. [DOI] [PubMed] [Google Scholar]
- 46.Shen F, Davydova EK, Du W, Kreutz JE, Piepenburg O, Ismagilov RF. Digital isothermal quantification of nucleic acids via simultaneous chemical initiation of recombinase polymerase amplification reactions on SlipChip. Anal Chem. 2011;83:3533–3540. doi: 10.1021/ac200247e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sharma N, Hoshika S, Hutter D, Bradley KM, Benner SA. Recombinase-based isothermal amplification of nucleic acids with self-avoiding molecular recognition systems (SAMRS) Chembiochem. 2014;15:2268–2274. doi: 10.1002/cbic.201402250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kutyavin IV, Rhinehart RL, Lukhtanov EA, Gorn VV, Meyer RB, Gamper HB. Oligonucleotides containing 2-aminoadenine and 2-thiothymine act as selectively binding complementary agents. Biochemistry. 1996;35:11170–11176. doi: 10.1021/bi960626v. [DOI] [PubMed] [Google Scholar]
- 49.Gamper HB, Arar K, Gewirtz A, Hou YM. Unrestricted hybridization of oligonucleotides to structure-free DNA. Biochemistry. 2006;45:6978–6986. doi: 10.1021/bi0600392. [DOI] [PubMed] [Google Scholar]
- 50.Hoshika S, Chen F, Leal NA, Benner SA. Artificial genetic systems: self-avoiding DNA in PCR and multiplexed PCR. Angew Chem Int Ed Engl. 2010;49:5554–5557. doi: 10.1002/anie.201001977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Karami A, Gill P, Motamedi MH, Saghafinia M. A review of the current isothermal amplification techniques: Applications, advantages and disadvantages. J Global Infect Dis. 2011;3:293–302. [Google Scholar]


