Abstract
Most vertebrate organs are composed of epithelium surrounded by support and stromal tissues formed from mesenchyme cells, which are not generally thought to form organized progenitor pools. Here we use clonal cell labeling with multicolor reporters to characterize individual mesenchymal progenitors in the developing mouse lung. We observe a diversity of mesenchymal progenitor populations with different locations, movements, and lineage boundaries. Airway smooth muscle (ASM) progenitors map exclusively to mesenchyme ahead of budding airways. Progenitors recruited from these tip pools differentiate into ASM around airway stalks; flanking stalk mesenchyme can be induced to form an ASM niche by a lateral bud or by an airway tip plus focal Wnt signal. Thus, mesenchymal progenitors can be organized into localized and carefully controlled domains that rival epithelial progenitor niches in regulatory sophistication.
INTRODUCTION
Most vertebrate organs are composed of epithelial tubes or sacs surrounded by support and stromal tissues that form during development from a loose collection of undifferentiated progenitor cells called mesenchyme (1). Mesenchyme gives rise to a variety of cell types including cartilage, smooth muscle, pericytes, mesothelium, and fibroblasts, whose development and ultimate structural organization is intimately associated with the epithelia they surround (2, 3). Mesenchymal cells have long been of interest to developmental biologists for their role as a source of signals that induce and pattern epithelial cell types (4, 5). More recently, there has been a surge of interest in these cells because of their ability to serve as signaling centers that support stem cells and influence tumor progression (6, 7), and because mesenchyme-derived cell types contribute to many serious diseases such as fibrosis of the lung and other organs (8), and airway wall remodeling in asthma (9).
Although the rules governing the behavior and regulation of a variety of epithelial progenitor cells and their niches have begun to be elucidated (10, 11), much less is known about the identity and behavior of stromal cells and other progenitors in the mesenchyme (12). The classical view is that mesenchyme cells are highly proliferative, migratory, and multipotent cells that expand during organ development and condense around endodermal epithelium to generate diverse support and stromal cell types (13, 1). Epithelial signals including Wnt, FGF, and Hedgehog family members have been implicated in these processes (14-16), but how the pathways affect mesenchymal progenitor cells is not well understood due to the variety of signals and challenges in following the behavior of individual mesenchymal cells in vivo.
Here we adapt clonal cell labeling strategies in mice, like those recently used to elucidate epithelial progenitor and stem cell behavior in vivo (17-19), to probe the behavior and potential of individual mesenchyme cells during lung development. The developing lung is composed of a branching airway tree and corresponding networks of developing blood vessels, all surrounded by undifferentiated pulmonary mesenchyme cells. Each of these tubular networks is associated with distinct smooth muscle support layers and other intervening cell types formed from the mesenchyme (20). Our clonal analysis shows that mesenchymal progenitors are highly proliferative, as classical studies suggest, and they are also highly motile. However, their motility is not random, and we show how clonal analysis can be used to map mesenchymal progenitor niches and progenitor cell behavior at cellular resolution. This reveals a surprising diversity of mesenchymal progenitor populations with different locations, patterns of movement, and migration and lineage boundaries. We focus on the airway smooth muscle (ASM) progenitor population, which displays a remarkable degree of regulation that rivals that of epithelial stem cells, and show that a focal Wnt signal can regulate movement of ASM progenitors out of their “niche.”
RESULTS
Marking cell clones in the developing lung mesenchyme
To identify progenitor cells in mouse lung mesenchyme and characterize their behavior at single cell resolution, we carried out a clonal analysis in which individual lung mesenchyme cells were genetically marked and the structures of the resulting clones analyzed to reveal the proliferation, migration, and developmental potential of individual progenitor cells (Figure 1A, B). Two Cre recombinase-dependent clonal labeling strategies were used to mark individual lung mesenchyme cells. In one, a ubiquitously-expressed HPRT-Cre allele was used to catalyze rare interchromosomal recombination events between Mosaic Analysis with Double Markers (MADM) chromosomes, which recreate the complete coding sequence for GFP on one resultant chromosome and the complete coding sequence for DsRed on the other (21). This generates either red and green twinspot clones or yellow clones in all major lineages of the developing mouse lung (Figure 1C, E, F), and we focused on clones in the mesenchyme and its derivatives. Although HPRT-Cre is expressed from the earliest stages of embryogenesis (22), cell labeling in the lung was first observed at E11.5 and continues thereafter; we restricted our analysis to between E11.5 and E13.5 when individual clones could be resolved. All MADM samples were immunostained and analyzed in whole mount to preserve the three-dimensional arrangement of cells in the clone and their relationship to the rest of the developing lung.
Figure 1. Marking cell clones in developing lung mesenchyme.
A, Diagram of embryonic day E11.5 mouse lung (ventral view) showing airway epithelium (AE; white), mesenchyme (Me; light gray), mesothelium (Mt; dark gray), airway (ASM) and vascular (VSM) smooth muscle (red), and pulmonary arteries (PA) and vascular plexus (Pl) in blue. B, Labeled mesenchymal progenitor cell (green) in early lung (left), which at E11.5 (right) has generated a mixed lineage clone (green, dashed circle) with labeled ASM, mesothelial cells, and undifferentiated fibroblasts. C-F, Close-up of clones (pseudo-colored green) in indicated tissues of lungs of indicated ages. Clones were marked with MADM (C, E, F) or Rainbow (D) strategies and immunostained for E-cadherin (airway epithelium; blue), PECAM (vascular endothelium; blue in F) and smooth muscle α-actin (ASM and VSM; red). Cells in each clone are numbered. G, Structure of lung mesenchyme specific Cre transgene using lung mesenchyme-specific enhancer (LME) from Tbx4 locus and Phsp68 minimal promoter to drive Cre recombinase expression. H-J, Cre mRNA in situ hybridization of lungs from Tbx4LME-Cre transgenic mice of indicated ages. mRNA is robustly detected by E10.5 exclusively in lung mesenchyme (H), remains strong at E13.5 (I), and begins to downregulate by E14.5 (J). Similar results were obtained for Tbx4LME-CreER (Figure S1). K, Tiled image of transverse section through E13.5 Tbx4LME-Cre; R26R-EYFP lung immunostained for YFP lineage trace marker (white). Lineage-traced cells are confined to lung. NT, neural tube; Li, liver; St, stomach. L, Section through E13.5 Tbx4LME-Cre; R26R-EYFP lung immunostained for YFP (green) and E-cadherin (magenta). Nearly all mesenchyme cells are lineage labeled (green). Lineage label is not detected in epithelium (magenta). M-O, Tbx4LME-CreER; Rainbow embryos given limiting doses of tamoxifen (0.6 mg/dam) at E9.5 (M) or E10.5 (N, O) and analyzed at E13.5 by immunostaining for smooth muscle α-actin (SMA) and clone markers indicated. Clones are labeled by a single recombination reporter, either mCherry (M), mOrange (N) or Cerulean (O). Cells in each clone are numbered. Bars, 50 μm (C-F; H-J), 100 μm (L-O).
To gain temporal and spatial control of clone generation, we combined a lung-specific mesenchyme enhancer element from the mouse Tbx4 locus (23) with an Hsp68 minimal promoter to drive Cre expression (Tbx4LME-Cre; Figure 1G) exclusively in the lung mesenchyme of transgenic mice (Figure 1K). Cre mRNA expression is robustly detected in early lung buds by E10.5, continues through E14.5, but is undetectable by birth (Figure 1H-J). When crossed to a Cre reporter, recombination is observed in all lineages of the lung mesenchyme but excluded from both epithelium (Figure 1L) and endothelium (24). Virtually all lung mesenchyme and its derivatives are labeled by Tbx4LME-Cre by E13.5, including undifferentiated mesenchyme, ASM, vascular smooth muscle (VSM) and mesothelium. This Cre mouse line thus allows targeted labeling and other genetic manipulations specifically in lung mesenchyme.
We also constructed a Tbx4LME-CreER transgenic mouse (Figure S1) to allow temporal control of clone marking and the extent of cell labeling. We used the CreER transgene with the multicolor Rainbow reporter, in which an individual recombination event is marked by expression of one of three possible fluorescent proteins (25). We titrated the dose of tamoxifen such that just one or at most two recombination events occurred in the lung, indicated by the presence of marked cells of a single or two different colors (Figure 1M-O). Lungs in which more than two colors were detected were excluded from analysis, and all cells of the same color within a lung were considered a clone. Because of the uncertainty about when MADM clones were induced, they are not included in the major clone table (Table 1); however, below we use examples of clones generated with both the Rainbow and MADM clone-marking systems (and separately tabulate the MADM examples) because our findings were similar for both.
Table 1.
Tbx4LME-CreERT2;Rainbow mesenchyme clones1,2

|
Clonal analysis reveals cell dynamics of developing mesenchyme
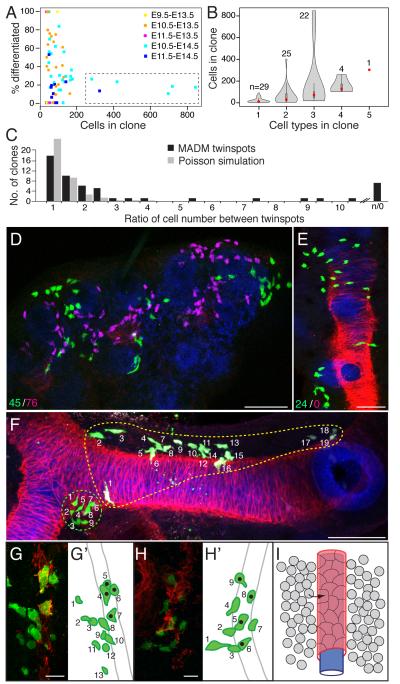
Principles governing cell behavior in the developing mesenchyme can be deduced by examining random clones (26). Comparison of cell number in each clone showed that proliferation rates vary substantially among mesenchyme cells. Rainbow clones induced at E10.5 and analyzed at E13.5 showed a range of 4 to 134 cells, giving calculated doubling times of 9 to 33 hours, assuming a 12 hour lag between tamoxifen administration and recombination (Table 1). Comparison of the E13.5 clones with ones analyzed at E14.5 identified a striking example of proliferative regulation (Figure 2A). At E13.5, all clones, regardless of chase time or degree of differentiation, were composed of fewer than 200 cells, whereas at E14.5 a second class of clones was also observed (boxed in Figure 2A) that contained many more cells (257-840 cells) with various differentiated mesenchymal cells but dominated by undifferentiated mesenchyme ("fibroblasts") (Table 1; Figure S2A). Simulations indicate that these large, fibroblast-dominated clones are unlikely to be misassigned polyclones (Figure S2B), but rather identify a subpopulation of highly proliferative fibroblasts that arise just after E13.5.
Figure 2. Cell dynamics of mesenchymal clones and structure of vascular smooth muscle clones.
A, Scatter plots of each Tbx4LME-CreER; Rainbow mesenchyme clone (Table 1) showing percent differentiated cells (cells expressing airway smooth muscle, vascular smooth muscle, mesothelial, or cartilage markers) in each clone as function of clone size (number of cells in clone). Clones harvested at E13.5 (dots) have < 200 cells and range from undifferentiated to fully differentiated, whereas clones harvested at E14.5 (squares) show the E13.5 pattern or are large (> 200 cells) and mostly undifferentiated clones (dashed box), indicating emergence of highly proliferative fibroblast population soon after E13.5. Clone size is greater (P = 9 × 10−7) and degree of differentiation less (P = 9 × 10−5) for clones harvested at E14.5 compared to E13.5. B, Violin plots of clones as above showing clone size (red dot, mean) as function of cell type complexity (number of mesenchymal cell types). n, number of clones analyzed. C, Ratio of cell number in E13.5 lungs of "twinspot" mesenchyme clones (n = 53 clones; Table S1) generated by HPRT-Cre; MADM, which differentially labels the two daughter cells of the original recombined cell with GFP or DsRed. Ratios were binned in increments of 0.5 (black bars) and shown with data from 10,000 simulated twinspots (gray bars; clone frequency values divided by 250) assuming a Poisson distribution in cell number. Note overrepresentation of clones with strong asymmetries (ratios > 5) relative to clones in Poisson simulation (P = 3 × 10−101 for all ratios >5, P = 1 × 10−18 with solo twinspots excluded). D, E, Projections of confocal z-stacks of twinspot clones 22 (D) and 47 (E) as above immunostained for GFP (clone mark, green), DsRed (sibling cell clone mark, magenta) and for E-cadherin (epithelium, blue), SMA (smooth muscle, red) and in D for PECAM (endothelium, also in blue). Values at bottom left show cell numbers in each twinspot; note asymmetry (D) and outright loss of one twinspot (E). No pattern was observed between loss of a twinspot and the clone’s location in the lung or position along a branch. F, Mesenchyme clones generated as above showing how arrangement of labeled cells in clones varies widely, from tight clusters (green-circled clone) to amorphous clouds of cells (yellow-circled clone, and clone in panel D). A slight bias was noted for clones arranged along the length of a branch, as in the yellow-circled clone, but no correlation with specific lobes or branches formed by a particular mode was apparent. G, H, Clones including VSM cells. In schematics (G', H'), cells in each clone are numbered. Note similar arrangement of cells, with undifferentiated mesenchyme cells (green in schematics) adjacent to SMA-expressing VSM cells (green with black dot in schematics). I, Model of vascular smooth muscle recruitment. Progenitors (gray) in mesenchyme directly surrounding endothelial cells (blue) are recruited to form VSM of artery wall (red). Bars, 50 μm (C, D, F), 10 μm (G-H).
The MADM labeling system allows comparison of the behavior of sister cells in twinspot clones, red or green marked clones generated from a single parent cell (Figure 2C-F). Such twinspot clones showed frequent and in some cases dramatic asymmetries in number of cells generated from each sister cell (Figure 2D,E): the ratio of cell number in twinspots ranged from 1 to 11 (n = 53 twinspots, Table S1), with 68% of twinspots differing by more than 1.5 fold (Figure 2C) and in extreme cases (n = 7 of 53) the complete absence of a red or green twinspot (Figure 2E). Simulations indicated that such differences in sibling cell proliferation rates are unlikely to result solely from stochastic variation (Figure 2C, P = 3 × 10−101 vs. Poisson simulation), implying important biological regulation of sister cell proliferation and/or survival.
Clonal analysis also revealed a large amount of migration and intermixing among cells of the mesenchyme. Only 26% of labeled cells (n = 575 cells scored from five randomly selected clones) remained in contact with a sibling (Figure 1D; Figure 2D,E). These dispersed clones spanned as much as 1700 μm along the proximal-distal axis over a chase period of four days (Table 1). Although 81% of clones (n = 70) were confined to a single lung lobe, clones induced as late as E11.5 were able to contribute to multiple lobes (Table 1), implying that divisions between lobes are not complete until after that stage. However, clones spanning left and right lungs were never observed, even among clones induced at E9.5 when lung buds first emerge, indicating that the division between left and right is established early and cell movement between them occurs rarely or not at all (Table 1). MADM samples analyzed in whole mount showed that most mesenchyme clones were found in cell clouds of no regular shape (Figure 2D), but when clones showed a directional bias, cells were arrayed lengthwise along airway branches rather than around a branch’s circumference (Figure 2F), presumably reflecting cell movements associated with branch elongation.
An important exception to the above rules were clusters of squamous cells on the surface of the lung, in which 91% of labeled cells (n = 181 cells scored from five randomly selected MADM clones) remained tightly grouped, touching at least one other sibling cell (Figure 1C). In an extreme MADM clone of this type (clone 11, Table S2), clonally-related and tightly-grouped clusters were found distributed across multiple lobes. We infer that these cells are mesothelium, the mesodermally-derived squamous epithelium forming the outermost cell layer of the embryonic lung. This implies that mesothelium is specified early and disperses broadly across the lung surface, and then the distributed cells proliferate locally. No clones analyzed at E13.5 or earlier that contained mesothelial cells also contained other cell types (n = 3 Rainbow clones, Table 1; n = 11 MADM clones, Table S2), suggesting that early in development the mesothelium is a lineage largely separate from the rest of the mesenchyme. However, 85% of mesothelial Rainbow clones examined at E14.5 (n = 13 clones) were mixed clones that also contained fibroblasts or other cell types (Table 1) indicating mesothelium does not remain a separate lineage at later time points, consistent with recent mesothelial lineage tracing experiments (27, 28).
Using clonal analysis to map mesenchyme progenitors and track progenitor behavior
Rules governing cell differentiation in the developing lung can also be inferred from clonal analysis. Given the common characterization of mesenchyme as a homogeneous tissue of broad potential (3) and the extensive movements revealed by the clonal analysis described above, our expectation was that mesenchyme clones would each be a random mix of multiple mesenchymal derivatives (29), as illustrated in Figure 1B. Such mixed clones were indeed recovered, especially in larger clones (Figure 2B), indicating that fixed single-potential lineages like early mesothelium do not dominate development of other mesenchymal cell types. However, most (82%) Rainbow clones containing differentiated cell types at E13.5 (n = 27) showed a much more limited potential, containing just a single differentiated cell type (Table 1; Figure S3).
By focusing on clones that contained fibroblasts (mesenchymal cells with spindle-shaped morphology that expressed none of the tested cell type specific differentiation markers) plus cells of a particular cell type, and analyzing large numbers of clones, specific patterns emerged from which the location of progenitor cells for each cell type and the mechanism of progenitor recruitment and migration could be inferred. A simple example can be seen in clones containing fibroblasts and VSM, which wraps the pulmonary arteries. In all such clones analyzed at E13.5 (n = 8, Table 1), undifferentiated fibroblasts flanking the arteries were also labeled (Figure 2G,H), supporting a direct lineage relationship between flanking fibroblasts and artery walls. This implies that arteries recruit cells to form VSM from mesenchyme immediately surrounding the vessel (Figure 2I; 30). Below, we use this approach to map progenitors of airway smooth muscle.
Airway smooth muscle progenitors reside at branch tips
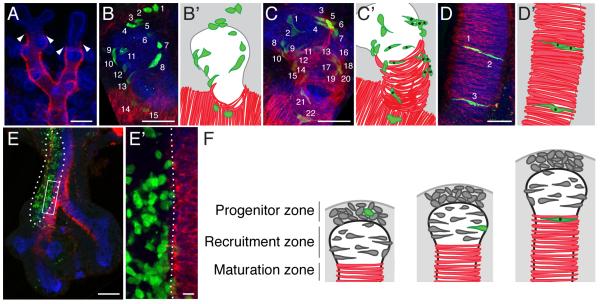
Double labeling with cell type specific antibodies to highlight both the airway epithelium and developing smooth muscle (Figure 3A) shows that ASM forms first along bronchial branch stalks and only later at their tips, long implicating mesenchyme surrounding the stalk as the likely source of ASM progenitors (31, 14), as in the model above for VSM. However, an analysis of the expression pattern of an FGF10-regulated lacZ transgene suggested movement of tip mesenchyme along branch stalks (32), and the clone data and grafting experiments detailed below demonstrate that stalk mesenchyme does not contribute to ASM and ASM progenitors are located exclusively at tips.
Figure 3. Clonal analysis reveals airway smooth muscle progenitor niche, dynamics, and lineage boundaries.
A, Airway branches of whole mount E13.5 mouse lung immunostained for E-cadherin (epithelium, blue) and smooth muscle α-actin (smooth muscle, red). SMA-positive airway smooth muscle (ASM) fibers cover airway stalks but are excluded from tips. Arrowheads, distal boundary of smooth muscle fibers. B-D, ASM-containing clones in E13.5 lungs immunostained for tissue markers as in A and for GFP clone marker (green). Cells in clone are numbered. In schematics (B’-D’), clone cells are green and those that are SMA-positive are marked with black dots. ASM clones fall into three classes: those with undifferentiated mesenchyme at distal tip plus elongating but SMA-negative cells around tip epithelium (B, B’); those with elongated but SMA-negative cells plus SMA-positive ASM cells (C, C’); and those with all cells SMA-positive (D, D’). Examples shown were chosen to illustrate the morphologies and distribution of clone cells; differences in cell number between clones are arbitrary. E, A large clone (outlined) with extensive labeling of mesenchyme along trachea and right primary airway stalk at E11.5. E’, Close up of boxed region. Note lack of recruitment into ASM layer from labeled mesenchyme at branch side, with lineage boundary marked by dotted line. F, Three step model of ASM formation. Fate of one progenitor (green) is highlighted. Proliferating progenitors are located in mesenchyme at branch tip (progenitor zone), initiate differentiation through interaction with epithelium (recruitment zone), and mature along airway stalk (maturation zone) as branch extends. Bars, 100 μm (A, E), 50 μm (B-D), 10 μm (E’).
Labeled ASM cells in mesenchyme clones did not have siblings in the undifferentiated mesenchyme along the branch stalk, but rather sibling cells were located around and just ahead of the tip of the budding branch. These clones could be grouped into three categories based on the location and differentiation status of cells in the clones (Figure 3B-D). In the first category (18% of Rainbow clones analyzed at E13.5; clones 28, 32, 40 and 59, Table 1) some cells in the clone, those furthest from the differentiated ASM cells, are undifferentiated mesenchyme at the branch tip several cell diameters away from the epithelium, whereas intervening cells in the clone are directly adjacent to the airway epithelium, elongating into the crescent shape characteristic of ASM cells but not expressing smooth muscle differentiation markers (Figure 3B). In the second category (55% of E13.5 ASM-containing Rainbow clones; clones 29-31, 37, 41, 44, 46, 55, 58, 60, 64 and 65), some cells of the clone are elongated but marker negative as above whereas other members of the clone are integrated into the ASM layer and express smooth muscle α-actin (SMA; Figure 3C). In the third category (27% of E13.5 ASM-containing Rainbow clones; clones 14-19), all cells in the clone are definitive ASM cells expressing SMA (Figure 3D). We propose that these categories represent three stages in a developmental series in which undifferentiated smooth muscle progenitors reside just ahead of the budding airway branch tip and are recruited to the ASM lineage. During recruitment by the airway epithelium, ASM progenitors adopt an elongated, crescent morphology and orient circumferentially around the bronchial branch and later turn on smooth muscle markers as the airway tip extends, leaving in its wake the differentiated contractile rings of smooth muscle cells along the airway stalk (Figure 3F).
The tip location of ASM progenitors was tested ex vivo by micrografting a tiny patch (~200 cells) of mesenchyme from a branch tip from ZsGreen-expressing E11.5 and E12.5 donor lungs into tip positions in unlabeled littermate host lungs (Figure 4A). Host lungs were cultured at 37°C for four days and grafted cells monitored by ZsGreen fluorescence. During the culture period, grafted tip cells moved proximally along the airway epithelium, with cells in 86% of cultures migrating 200 μm or more (Figure 4B, Figure S5A-D). Many of the migrating cells near the airway epithelium differentiated into SMA-positive ASM cells (Figure 4C). This confirms that ASM progenitors are located just ahead of branch tips, and shows that they can migrate along the tip to reach the stalk and begin differentiating into ASM.
Figure 4. Tissue grafts and lineage tracing show ASM progenitors are located in tip mesenchyme and form anew with new domain branches.
A, Graft strategy. Mesenchyme excised from stalk or tip of ZsGreen-expressing donor E11.5 lung were grafted into tip of an unlabeled littermate host lung, and grafted cells (green) were traced in culture. B, Quantification of progenitor cell migration from the graft in tip-to-tip (T>T) or stalk-to-tip (S>T) mesenchyme grafts, measuring the distance from the graft margin to the furthest invading graft cell, along the shortest inferred migration path. n, number of grafts. Note robust invasion of tip-derived cells (all grafts showed > 100 μm migration) relative to stalk-derived cells (11% showed > 100 μm migration). C, C’, Stereomicroscope image showing recipient lung four days after tip-to-tip micrograft (C), and single confocal optical slice of grafted region (C’). Note in C’ that tip cells have migrated away from lung margin (dotted line) along the airways (dashed lines) and some graft cells (arrowheads) are expressing ASM differentiation marker smooth muscle actin (red). D, Stalk-to-tip mesenchyme graft. Grafted cells fail to leave graft site, and do not migrate along airways or express smooth muscle actin (D’). E, Strategy of lineage trace of smooth muscle of parent branch of L.M1, a late-forming domain branch. Budding L.M1 (highlighted in insets) is first medial domain branch formed along left primary airway; it buds through existing ASM layer of parent branch and forms its own ASM layer. Saturating tamoxifen (3 mg) was administered at E11.5 to SM-MHC (smooth muscle myosin heavy chain)-CreER; mTmG to label differentiated ASM of parent branch and early forming lateral branches (L.L1 – L.L3) with heritable GFP expression (green bar on timeline, with tamoxifen assumed active for 24 hours) before emergence of L.M1 bud at E12.5. Lungs were analyzed at E14.5 after L.M1 has budded and formed its ASM (“new” ASM, black bar in inset). F, Confocal z-stack projections (ventral view) showing close-up of boxed region (F’) of SMMHC-CreER;mTmG E14.5 embryo treated as above and immunostained for lineage trace of smooth muscle of parent branch (GFP, green) and all airway smooth muscle (SMA, red). Note ASM of L.M1 daughter branch is not lineage labeled (border highlighted with dashed line in F’ merge) indicating a new progenitor pool formed ahead of new branch that is lineally independent of existing ASM. There is no cell mixing across smooth muscle lineage boundary (dashed line), with exception of two lineage-labeled cells (arrowheads in F') in ASM of L.M1. Dots, lineage labeled non-ASM cells. Bars, 50 μm.
A lineage boundary between stalk mesenchyme and airway smooth muscle
The failure of mesenchyme from airway flank to contribute to ASM is illustrated by large clones in which stalk mesenchyme was extensively labeled but mesenchyme at branch tips was not (Figure 3E). Labeled cells in these clones surrounded and contacted the developing ASM, but they did not contribute to it; there was a lineage boundary at the edge of the ASM layer that flanking cells did not penetrate. Together with the above results, these findings support a model in which: (i) progenitors reside in a mobile progenitor pool near the distal tip of lung branches that moves with the growing branch tip; (ii) progenitor cells are progressively recruited from this pool onto the growing branch to form the ASM; and (iii) stalk mesenchyme cells are blocked from joining the ASM from the branch side.
Reversal of the stalk lineage boundary to form new tip progenitor pools
The lung forms by three main modes of branching, domain branching, planar bifurcation, and orthogonal bifurcation (Figure S4; 33), and each of these different types of branches is associated with a tip progenitor pool. As branch number increases, so do the number of tip progenitor pools. In the case of branches formed by airway tip bifurcation, we infer that the tip progenitor pool simply splits with the bifurcation of the epithelial portion of the branch. However, late-forming domain branches emerge from the side of the parental branch after the smooth muscle layer along the parent branch has formed, so these branches must bud through an existing smooth muscle layer (34; Figure 4E). Rainbow clones generated with an ASM-specific CreER were found to be both multicellular and dispersed (Figure S6) showing that differentiated ASM cells of the parent branch remain proliferative and migratory. To test whether the smooth muscle layer that forms along the new branches comes from recruitment of these differentiated ASM cells of the parent branch or from undifferentiated stalk mesenchyme we used a smooth muscle specific CreER (35) and a saturating dose of tamoxifen at E11.5 to label virtually all of the existing ASM cells of the parent branch just prior to budding of new domain branches. We then allowed the new branches to grow out through the labeled smooth muscle and harvested the embryos three days later, after the smooth muscle layer had formed around the new branches. We found that the daughter branch smooth muscle was unlabeled (Figure 4F), demonstrating that parental branch ASM is not a source of new smooth muscle cells along the daughter branch nor does it significantly mix with daughter branch ASM. Rather, the daughter branch induces a new tip progenitor pool from non-lineage labeled cells, presumably the stalk mesenchyme that flanks the ASM of the parent branch, overcoming the previous lineage restriction. Thus, with each new domain branching event a new source of ASM progenitors is induced, and a lineage boundary forms between the ASM layers of parental and daughter branches. The maintenance of this ASM lineage boundary also implies that any proliferation and cell movement within the formed ASM remains local and rarely crosses branch boundaries.
A focal Wnt signal can prime stalk mesenchyme to form tip progenitor pool
To determine if all signals required to convert naïve mesenchyme into a tip progenitor pool emanate from the epithelial branch tip, we exposed stalk mesenchyme to epithelial branch tips by ex vivo grafting. Small patches of stalk mesenchyme from ZsGreen-expressing donor lungs were micrografted into tip positions of unlabeled littermate hosts (Figure 4A), thereby exposing grafted cells to any signals coming from tip epithelium or mesenchyme flanking the graft site. During the four day culture period, the grafted stalk cells in nearly all grafts (89%, n = 27 grafts) remained at the graft site with little or no movement along the airway (Figure 4B) or differentiation to ASM (Figure 4D). Thus, signals from tip epithelium and flanking mesenchyme are not sufficient to induce stalk mesenchyme to form a tip progenitor pool.
This failure of grafted stalk mesenchyme to respond to tip epithelial cues suggests that factors normally found in tip mesenchyme are required to induce or maintain the tip progenitor pool. To identify signals capable of replacing this required tip mesenchyme factor, we grafted small pieces of labeled stalk mesenchyme to branch tips as above, then implanted a ligand-soaked bead at the distal tip of the grafted tissue. Host lungs were placed into culture and the position of the engrafted cells was monitored for four days, after which samples were fixed and stained for differentiated cell types. Grafted stalk cells in nearly all grafts implanted with PBS-soaked control beads (88%, n = 8 grafts) remained at the graft site, as expected (Figure 5A, D). However, grafted cells in grafts with beads soaked in WNT1 (89%, n = 19 grafts) migrated 100 μm or more along the growing airway and contributed to the smooth muscle layer (Figure 5C, D, Figure S5E), an effect that was partially blocked by inclusion in the culture medium of extracellular Wnt antagonists Dkk1 or Sfrp1 (Figure 5D). Indeed, stalk mesenchyme grafts exposed to WNT1 beads were indistinguishable from grafts of tip mesenchyme (Figure 5C, D; Figure 4C), demonstrating that a distal Wnt signal is sufficient to overcome the barrier to migration and smooth muscle recruitment observed in untreated stalk mesenchyme grafts. This effect was not seen with FGF10-soaked beads (n = 8 grafts; Figure 5B, D), though FGF10 is a factor prominently expressed in tip mesenchyme and has been implicated in smooth muscle recruitment (32, 36).
Figure 5. Effect of Wnt bead implants and transgenic activation of Wnt pathway show that Wnt signaling primes ASM progenitors.
A-C, Stereoscope images of stalk-to-tip mesenchyme grafts performed as in Figure 4D, except also implanted with control (A) or ligand-soaked (B, C) agarose beads as indicated. Asterisk, position of bead. A'-C' show single confocal slices of graft region after immunostaining for GFP (graft marker, green), E-cadherin (epithelium, blue), and SMA (ASM, red). Graft cells exposed to PBS- or FGF10-soaked beads showed little or no migration along airways (A', B'), whereas those exposed to WNT1-soaked beads (C') migrated along airways where some (arrowheads) differentiated to ASM (separate channels shown in Figure S5E). D, Quantification of graft cell invasion in untreated grafts (reproduced from Figure 4B), and following exposure to ligand-soaked beads and Wnt inhibitors, as indicated. Note that WNT1 beads (column 5) induce stalk mesenchyme invasion at levels comparable to that seen in tip-to-tip grafts (column 1), and the effect is reduced in the presence of 100 ng/ml Wnt inhibitors DKK1 or SFRP1 (column 6). E-H, Right middle lobes of E12.5 control (E) and Tbx4LME-Cre;βcatex3/+ lungs that express stabilized β-catenin to activate canonical Wnt pathway throughout mesenchyme (F-H) immunostained for E-cadherin (epithelium, green), SMA (smooth muscle, red), and PECAM (endothelium, blue). Wnt pathway activation results in disorganized smooth muscle around airways (F) and ectopic wrapping of vascular plexus by cells continuous with ASM (arrowheads in G). (H) Single confocal slice of branch as in G showing that the ectopic smooth muscle wrapping the plexus (arrowhead) expresses vascular smooth muscle marker NG2 (magenta). Bars, 50 μm (A-E), 25 μm (F, G).
To examine the molecular effects of Wnt signaling on stalk mesenchyme, we used oligonucleotide microarrays to transcriptionally profile stalk mesenchyme cultured with or without WNT1, and identified a Wnt response signature of 306 genes whose expression changed two-fold or more (Table S3). The Wnt response signature was enriched for “WNT signaling” genes as expected, and also for “cell motility and migration”, “cell projection”, and “contractile fibers” genes (Table S4), which presumably mediate the observed migratory and differentiative effects of WNT1. Comparison of the expression profiles of untreated stalk and tip mesenchyme showed that Wnt response signature genes were expressed at higher levels in tip mesenchyme (Figure S7), supporting a role for Wnt signaling in defining tip mesenchyme potential in vivo.
Because a Wnt ligand was sufficient to transform grafted stalk mesenchyme into cells whose behavior is indistinguishable from that of the tip progenitor pool, and because higher expression of Wnt response signature genes distinguishes tip mesenchyme, we tested the effect of increased levels of canonical Wnt signaling in the mesenchyme along branch stalks using Tbx4LME-Cre to express a constitutively active form of β-catenin in the mesenchyme. In such lungs, the distal vascular plexus, which in control lungs of the same stage is a simple endothelial net, became wrapped with ectopic smooth muscle cells expressing the VSM marker NG2 (Figure 5F-H), an arrangement reminiscent of the pericytes surrounding mature capillaries. This precocious recruitment of VSM to the distal plexus is accompanied by more subtle patterning changes in the smooth muscle layer around airways: the normally strictly parallel fibers oriented orthogonal to the branch axis (Figure 5E) were disorganized in the mutant (Figure 5F), and the cells were less tightly apposed resulting in a thicker, less condensed ASM layer. In addition, smooth muscle cells were observed connecting the ASM layer with the ectopic VSM layer wrapping the vascular plexus (Figure 5G), a feature never observed in control lungs at any stage.
DISCUSSION
Our lung mesenchyme-specific clonal analysis and micrografting experiments show that ASM progenitors are not broadly distributed, but rather are confined to dynamic local pools around branch tips that move and bifurcate with the tips and from which progenitors are continuously recruited onto airway stalks where they differentiate into smooth muscle. Clonal analysis also revealed a lineage boundary that prevents mesenchymal cells surrounding the stalks from becoming ASM from branch sides. However, new progenitor pools are induced in these regions around late-forming domain branches, creating a smooth muscle progenitor pool for each new branch whose progeny do not intermix with those of the parent branch. These progenitor pools require a budding airway tip and a priming signal that can be provided by a focal Wnt source, and delocalized Wnt pathway activity in vivo expands or alters the progenitor pool, causing ectopic smooth muscle formation on nearby plexus endothelial cells. Thus, ASM progenitors are organized into localized pools that are carefully regulated by surrounding signals.
These localized mesenchymal progenitor pools share several features with classical epithelial stem cell niches such as those that maintain adult hair follicles and gut epithelium. Each is a focal collection of progenitors that resides in a specialized signaling environment that controls their proliferation as well as their recruitment out of the niche and differentiation into specialized cells (37, 11), thereby coordinating the generation of a critical cell type with organ growth or maintenance. But unlike classical epithelial niches, which are typically located at fixed positions with stem cells segregated from other cell types (38), these mesenchymal progenitor pools are highly dynamic and not bounded in any obvious way. A new smooth muscle progenitor pool is established with each new budding branch, and it moves and splits as the tip extends and bifurcates, with rapidly cycling and highly motile progenitors leaving the tip pool after only short and variable stays. And, whereas epithelial stem cells self-renew (39, 40), the smooth muscle progenitor pools are gradually depleted during embryogenesis, although some might remain at the end of branching to serve as adult stem cells in airway wall maintenance and remodeling.
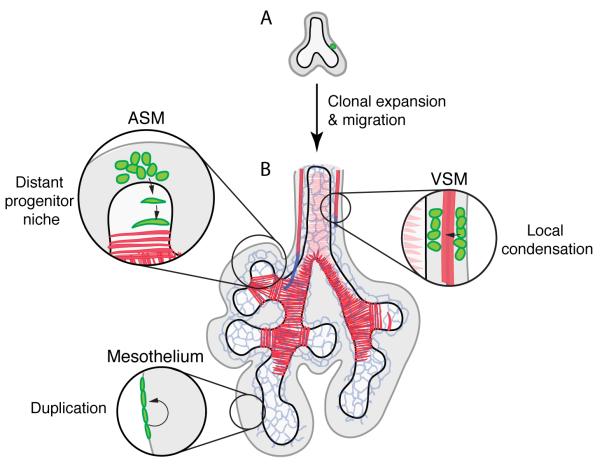
Progenitors in lung mesenchyme are not all organized into such focal progenitor niches (Figure 6). Mesothelial progenitors populate the full mesenchymal surface and clones remained coherent and restricted to the surface and the mesothelial lineage early in lung development (prior to E13.5); later this barrier is lifted as dispersed clones containing mesothelium and other mesenchymal derivatives were recovered. VSM progenitors are even more broadly distributed, matching closest the conventional view of delocalized mesenchymal progenitors. However, even in this case there is precise temporal and spatial control of progenitor recruitment, as VSM begins to form only at positions where the early vascular plexus has been replaced by the larger and more regular arterial and venous endothelial tubes. And, the mixed clones containing multiple differentiated cell types (clones 54-78, Table 1) demonstrate that daughter cells of early progenitors can disperse to seed multiple progenitor pools. Thus, lung mesenchyme is neither a large homogenous progenitor pool as it has classically been viewed, nor is it a collection of discrete, isolated, and unchanging progenitor niches. Rather, each differentiated cell type has a distinct mode of progenitor recruitment and regulated boundaries, which are modulated temporally and spatially, such that delocalization of the control signals disrupts the progenitor pools and therefore patterning of stromal and support tissues. It will be important to elucidate the full set of signals that controls individual cells in each of these pools, including the specific Wnt(s) and other signals that control ASM progenitors, and how these signals change during development and adult life. It will also be valuable to use the clonal and other approaches described here to explore the full diversity and dynamics of mesenchyme progenitor pools in other organs and disease contexts.
Figure 6. Diverse mechanisms generate mesenchymal cell derivatives.
(A) Single labeled mesenchyme cells (green) proliferate (clonal expansion) and daughter cells disperse to seed progenitor "niches" that generate stromal and support cells by different mechanisms. (B) ASM (airway smooth muscle) progenitors are recruited to branch stalks from a niche distal to the branch tip. VSM (vascular smooth muscle) progenitors are recruited by local condensation of mesenchyme immediately surrounding the vessel. Mesothelium forms by self-duplication and limited spreading of progenitors. Black, airway epithelium; grey, mesenchyme; dark gray, mesothelium; red, smooth muscle; blue, endothelium.
MATERIALS AND METHODS
Mice
Mouse lines used and details of the cloning of the Tbx4LME-Cre and -CreERT2 transgenes and construction of the transgenic lines are described in Supplemental Methods. All animal experiments were performed in accordance with national and institutional guidelines.
Clonal analysis
Clones were visualized using two independent multicolor cell labeling systems, MADM (21) and Rainbow (25). In MADM, Cre-mediated interchromosomal recombination between MADM-RG and MADM-GR chromosomes in HPRT-Cre; MADM-GR/MADM-RG embryos generates MADM-GG (full length GFP) and MADM-RR (full length myc-epitope-tagged DsRed) chromosomes, which can either segregate to the same daughter cell to give "yellow" clones, or to separate sibling daughter cells to form "twinspots" with "green" GFP daughter cells and "red" myc-epitope-tagged DsRed daughter cells. Recombination is extremely rare and not temporally controlled, however no labeled cells were detected in lung prior to E11.5. Lungs were harvested between E11.5 and E13.5, the latest stage at which individual clones could be resolved. Whole mount lungs were fixed and stained with antisera specific for clone markers GFP and myc-epitope-tagged DsRed plus a panel of tissue-specific markers, as described in Supplemental Methods. Whole mount immunostained lungs were imaged by confocal fluorescence microscopy. When recombination of MADM chromosomes was driven with Tbx4LME-Cre, labeled cells were not observed until after the developmental window considered here.
In the Rainbow system (25), pregnant Tbx4LME-CreER; Rosa26R-Rainbow females were given a single intraperitoneal injection of 0.5 - 0.7 mg tamoxifen (Sigma) dissolved in corn oil (Sigma) at the indicated gestational ages and embryos were harvested several days later, as indicated. The low doses of tamoxifen induced rare recombination events; most lungs contained no labeled cells (n = 177/351 lungs assayed). Embryos were dissected and their lungs were fixed, cryosectioned and stained with SMA directly conjugated to Cy5 as described below and examined using fluorescent filters specific to each fluorophore. Only lungs containing one or at most two of the potential three-colored recombination products were used for further analysis (n = 56). The number, location and differentiation state of the cells in each clone were recorded and selected clones (n = 70) were analyzed by confocal fluorescence microscopy.
Grafting, bead implantation and organ culture
Small blocks of mesenchyme (< 0.5 mm) containing roughly 200 ± 50 cells were dissected from ZsGreen-expressing donor lungs at the indicated positions and micrografted to branch tips of wild type littermate host lungs at E11.5-E12.5. Grafted lungs were placed in organ culture in DMEM/F12 with 10% FBS for 3-4 days at 37°C and the invasion and differentiation of grafted cells was assessed without fixation by fluorescence stereomicroscopy and then by confocal fluorescence microscopy following fixation and immunohistochemistry. For bead implantation, after healing of the graft site overnight in culture, an agarose bead soaked in the indicated ligand was implanted at the graft site. For inhibitor treatments, one hour after bead implantation organ cultures were transferred to growth medium containing the indicated concentrations of inhibitor.
Transcriptional profiling and statistics
Mesenchyme blocks from tip and stalk were dissected as above, total RNA was isolated, amplified, labeled and used to probe Mouse Genome 430 2.0 Arrays (Affymetrix). To define the Wnt response signature, stalk mesenchyme pieces dissected as above were cultured overnight in either control media or media containing 400 ng/ml WNT1 before RNA isolation and analysis, and genes with expression differences of two-fold or greater between the conditions were identified using Affymetrix software. Details of the experimental and statistical analysis are given in Supplementary Methods.
Supplementary Material
ACKNOWLEDGEMENTS
This work was supported by NRSA postdoctoral fellowship F32HL083645 (MEK), NIH grants R01HL075769 and U01HL099995, and the Vera M. Wall Center for Pulmonary Vascular Disease at Stanford University (MAK), Damon Runyon and Parker B. Francis postdoctoral fellowships (FHE), NRSA postdoctoral fellowship F32HD048006 (DBM), and NIH Center of Excellence in Genomic Studies grant 5P50HG2568 (DMK). MAK and DMK are investigators of the Howard Hughes Medical Institute. MEK and MAK designed the experiments, performed data analysis and wrote the manuscript. MEK performed the experiments. MEK and PEB made the Tbx4LME transgenic mice. FHE analyzed gene expression at branch tips. DBM and DMK isolated the Tbx4 enhancer element. The data reported in this paper are tabulated in the Supporting Online Material and expression profiling data sets have been deposited to the Gene Expression Omnibus (www.ncbi.nlm.nih.gov/geo) (will provide ascession numbers once available). The authors thank D. Riordan for help with statistical analysis and L. Luo and I. Weissman for generously sharing mouse lines prior to publication.
REFERENCES
- 1.MacCord K. Mesenchyme. Embryo Project Encyclopedia. 2012 http://embryo.asu.edu/handle/10776/13941.
- 2.Prentiss CW. A Laboratory Manual and Text-book of Embryology. W.B. Saunders Company; Philadelphia London: 1915. p. 2. l., 7-400. [Google Scholar]
- 3.Arey LB. Developmental Anatomy, a Textbook and Laboratory Manual of Embryology. 5th W.B. Saunders Company; Philadelphia, London: 1946. pp. ix–616. [Google Scholar]
- 4.Grobstein C. Inductive epitheliomesenchymal interaction in cultured organ rudiments of the mouse. Science. 1953;118:52–55. doi: 10.1126/science.118.3054.52. [DOI] [PubMed] [Google Scholar]
- 5.Spooner BS, Wessells NK. Mammalian lung development: interactions in primordium formation and bronchial morphogenesis. The Journal of Experimental Zoology. 1970;175:445–454. doi: 10.1002/jez.1401750404. [DOI] [PubMed] [Google Scholar]
- 6.Bhowmick NA, Neilson EG, Moses HL. Stromal fibroblasts in cancer initiation and progression. Nature. 2004;432:332–337. doi: 10.1038/nature03096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sneddon JB, Borowiak M, Melton DA. Self-renewal of embryonic-stem-cell-derived progenitors by organ-matched mesenchyme. Nature. 2012;491:765–768. doi: 10.1038/nature11463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rock JR, et al. Multiple stromal populations contribute to pulmonary fibrosis without evidence for epithelial to mesenchymal transition. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:E1475–1483. doi: 10.1073/pnas.1117988108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Berair R, Saunders R, Brightling CE. Origins of increased airway smooth muscle mass in asthma. BMC Medicine. 2013;11:145. doi: 10.1186/1741-7015-11-145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Blanpain C, Fuchs E. Epidermal homeostasis: a balancing act of stem cells in the skin. Nature Reviews. Molecular Cell Biology. 2009;10:207–217. doi: 10.1038/nrm2636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Clevers H. The intestinal crypt, a prototype stem cell compartment. Cell. 2013;154:274–284. doi: 10.1016/j.cell.2013.07.004. [DOI] [PubMed] [Google Scholar]
- 12.Humphreys BD, et al. Fate tracing reveals the pericyte and not epithelial origin of myofibroblasts in kidney fibrosis. The American Journal of Pathology. 2010;176:85–97. doi: 10.2353/ajpath.2010.090517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Taderera JV. Control of lung differentiation in vitro. Developmental Biology. 1967;16:489–512. doi: 10.1016/0012-1606(67)90061-9. [DOI] [PubMed] [Google Scholar]
- 14.Weaver M, Batts L, Hogan BL. Tissue interactions pattern the mesenchyme of the embryonic mouse lung. Developmental Biology. 2003;258:169–184. doi: 10.1016/s0012-1606(03)00117-9. [DOI] [PubMed] [Google Scholar]
- 15.Mao J, Kim BM, Rajurkar M, Shivdasani RA, McMahon AP. Hedgehog signaling controls mesenchymal growth in the developing mammalian digestive tract. Development. 2010;137:1721–1729. doi: 10.1242/dev.044586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Goss AM, et al. Wnt2 signaling is necessary and sufficient to activate the airway smooth muscle program in the lung by regulating myocardin/Mrtf-B and Fgf10 expression. Developmental Biology. 2011;356:541–552. doi: 10.1016/j.ydbio.2011.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Snippert HJ, et al. Intestinal crypt homeostasis results from neutral competition between symmetrically dividing Lgr5 stem cells. Cell. 2010;143:134–144. doi: 10.1016/j.cell.2010.09.016. [DOI] [PubMed] [Google Scholar]
- 18.Mascre G, et al. Distinct contribution of stem and progenitor cells to epidermal maintenance. Nature. 2012;489:257–262. doi: 10.1038/nature11393. [DOI] [PubMed] [Google Scholar]
- 19.Desai TJ, Brownfield DG, Krasnow MA. Alveolar progenitor and stem cells in lung development, renewal and cancer. Nature. 2014;507:190–194. doi: 10.1038/nature12930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Morrisey EE, Hogan BL. Preparing for the first breath: genetic and cellular mechanisms in lung development. Developmental Cell. 2010;18:8–23. doi: 10.1016/j.devcel.2009.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zong H, Espinosa JS, Su HH, Muzumdar MD, Luo L. Mosaic analysis with double markers in mice. Cell. 2005;121:479–492. doi: 10.1016/j.cell.2005.02.012. [DOI] [PubMed] [Google Scholar]
- 22.Tang SH, Silva FJ, Tsark WM, Mann JR. A Cre/loxP-deleter transgenic line in mouse strain 129S1/SvImJ. Genesis. 2002;32:199–202. doi: 10.1002/gene.10030. [DOI] [PubMed] [Google Scholar]
- 23.Menke DB, Guenther C, Kingsley DM. Dual hindlimb control elements in the Tbx4 gene and region-specific control of bone size in vertebrate limbs. Development. 2008;135:2543–2553. doi: 10.1242/dev.017384. [DOI] [PubMed] [Google Scholar]
- 24.Bogard P. Stanford University; 2012. [Google Scholar]
- 25.Rinkevich Y, Lindau P, Ueno H, Longaker MT, Weissman IL. Germ-layer and lineage-restricted stem/progenitors regenerate the mouse digit tip. Nature. 2011;476:409–413. doi: 10.1038/nature10346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Crick FH, Lawrence PA. Compartments and polyclones in insect development. Science. 1975;189:340–347. doi: 10.1126/science.806966. [DOI] [PubMed] [Google Scholar]
- 27.Cano E, Carmona R, Munoz-Chapuli R. Wt1-expressing progenitors contribute to multiple tissues in the developing lung. American Journal of Physiology. Lung Cellular and Molecular Physiology. 2013;305:L322–332. doi: 10.1152/ajplung.00424.2012. [DOI] [PubMed] [Google Scholar]
- 28.Dixit R, Ai X, Fine A. Derivation of lung mesenchymal lineages from the fetal mesothelium requires hedgehog signaling for mesothelial cell entry. Development. 2013;140:4398–4406. doi: 10.1242/dev.098079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pearse RV, 2nd, Scherz PJ, Campbell JK, Tabin CJ. A cellular lineage analysis of the chick limb bud. Developmental Biology. 2007;310:388–400. doi: 10.1016/j.ydbio.2007.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Greif DM, et al. Radial construction of an arterial wall. Developmental Cell. 2012;23:482–493. doi: 10.1016/j.devcel.2012.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Flint JM. The development of the lungs. American Journal of Anatomy. 1906;6:1–137. [Google Scholar]
- 32.Mailleux AA, et al. Fgf10 expression identifies parabronchial smooth muscle cell progenitors and is required for their entry into the smooth muscle cell lineage. Development. 2005;132:2157–2166. doi: 10.1242/dev.01795. [DOI] [PubMed] [Google Scholar]
- 33.Metzger RJ, Klein OD, Martin GR, Krasnow MA. The branching programme of mouse lung development. Nature. 2008;453:745–750. doi: 10.1038/nature07005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Metzger R. Stanford University; 2007. [Google Scholar]
- 35.Wirth A, et al. G12-G13-LARG-mediated signaling in vascular smooth muscle is required for salt-induced hypertension. Nature Medicine. 2008;14:64–68. doi: 10.1038/nm1666. [DOI] [PubMed] [Google Scholar]
- 36.De Langhe SP, Carraro G, Warburton D, Hajihosseini MK, Bellusci S. Levels of mesenchymal FGFR2 signaling modulate smooth muscle progenitor cell commitment in the lung. Developmental Biology. 2006;299:52–62. doi: 10.1016/j.ydbio.2006.07.001. [DOI] [PubMed] [Google Scholar]
- 37.Tumbar T, et al. Defining the epithelial stem cell niche in skin. Science. 2004;303:359–363. doi: 10.1126/science.1092436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sato T, et al. Paneth cells constitute the niche for Lgr5 stem cells in intestinal crypts. Nature. 2011;469:415–418. doi: 10.1038/nature09637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Blanpain C, Lowry WE, Geoghegan A, Polak L, Fuchs E. Self-renewal, multipotency, and the existence of two cell populations within an epithelial stem cell niche. Cell. 2004;118:635–648. doi: 10.1016/j.cell.2004.08.012. [DOI] [PubMed] [Google Scholar]
- 40.Barker N, et al. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature. 2007;449:1003–1007. doi: 10.1038/nature06196. [DOI] [PubMed] [Google Scholar]
- 41.Muzumdar MD, Tasic B, Miyamichi K, Li L, Luo L. A global double-fluorescent Cre reporter mouse. Genesis. 2007;45:593–605. doi: 10.1002/dvg.20335. [DOI] [PubMed] [Google Scholar]
- 42.Harada N, et al. Intestinal polyposis in mice with a dominant stable mutation of the beta-catenin gene. The EMBO Journal. 1999;18:5931–5942. doi: 10.1093/emboj/18.21.5931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Madisen L, et al. A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nature Neuroscience. 2010;13:133–140. doi: 10.1038/nn.2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Srinivas S, et al. Cre reporter strains produced by targeted insertion of EYFP and ECFP into the ROSA26 locus. BMC Developmental Biology. 2001;1:4. doi: 10.1186/1471-213X-1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.DiLeone RJ, Russell LB, Kingsley DM. An extensive 3' regulatory region controls expression of Bmp5 in specific anatomical structures of the mouse embryo. Genetics. 1998;148:401–408. doi: 10.1093/genetics/148.1.401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gu H, Zou YR, Rajewsky K. Independent control of immunoglobulin switch recombination at individual switch regions evidenced through Cre-loxP-mediated gene targeting. Cell. 1993;73:1155–1164. doi: 10.1016/0092-8674(93)90644-6. [DOI] [PubMed] [Google Scholar]
- 47.Feil R, et al. Ligand-activated site-specific recombination in mice. Proceedings of the National Academy of Sciences of the United States of America. 1996;93:10887–10890. doi: 10.1073/pnas.93.20.10887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Furuta Y, Piston DW, Hogan BL. Bone morphogenetic proteins (BMPs) as regulators of dorsal forebrain development. Development. 1997;124:2203–2212. doi: 10.1242/dev.124.11.2203. [DOI] [PubMed] [Google Scholar]
- 49.Seita J, et al. Gene Expression Commons: an open platform for absolute gene expression profiling. PloS One. 2012;7:e40321. doi: 10.1371/journal.pone.0040321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nature Protocols. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 51.Huang da W, et al. In: Current Protocols in Bioinformatics. Baxevanis AD, et al., editors. Wiley; New York: 2009. Chapter 13, Unit 13 11. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.