SUMMARY
Heart failure is a leading cause of mortality and morbidity in the developed world, partly because mammals lack the ability to regenerate heart tissue. Whether this is due to evolutionary loss of regenerative mechanisms present in other organisms or to an inability to activate such mechanisms is currently unclear. Here, we decipher mechanisms underlying heart regeneration in adult zebrafish and show that the molecular regulators of this response are conserved in mammals. We identified miR-99/100 and Let-7a/c, and their protein targets smarca5 and fntb, as critical regulators of cardiomyocyte dedifferentiation and heart regeneration in zebrafish. Although human and murine adult cardiomyocytes fail to elicit an endogenous regenerative response following myocardial infarction, we show that in vivo manipulation of this molecular machinery in mice results in cardiomyocyte dedifferentiation and improved heart functionality after injury. These data provide a proof-of-concept for identifying and activating conserved molecular programs to regenerate the damaged heart.
INTRODUCTION
Cardiovascular diseases remain the major contributors to the high mortality and morbidity rates in developed countries. Heart failure due to poor ventricle myocardial contractile performance has become a worldwide public health problem. Noticeably, adult mammalian hearts have been shown to elicit a primitive regenerative response upon injury (Addis and Epstein, 2013; Aguirre et al., 2013; Laflamme and Murry, 2011). Supporting this notion, mature differentiated mammalian cardiomyocytes have been shown to re-enter cell cycle upon application of chemical compounds targeting specific signaling pathways (Bersell et al., 2009). However, in both cases, the regenerative response is insufficient for the healing and complete recovery of myocardial function. Interestingly, lower vertebrates such as the zebrafish, are able to naturally activate endogenous regenerative responses that lead to complete heart regeneration throughout their entire lifetime by a process of cardiomyocyte dedifferentiation (Jopling et al., 2010; Kikuchi et al., 2010; Poss et al., 2002; Raya et al., 2003). Dedifferentiated cardiomyocytes were shown to re-enter the cell cycle and accounted for all newly formed cells repairing the heart (Jopling et al., 2010; Kikuchi et al., 2010). Different hypotheses have been considered to explain these different responses across species (Brockes and Kumar, 2008; Poss, 2010). One possibility is that regenerative mechanisms have been positively or negatively selected during evolution and are therefore absent in the mammalian genome. Alternatively, regenerative mechanisms, following injury, even though present, might remain silenced due to differences in epigenetic regulatory mechanisms when compared with species able to regenerate (Brockes and Kumar, 2008; Kragl et al., 2009; Lehoczky et al., 2011; Seifert et al., 2012; Stewart et al., 2009). Certain regenerative processes present in neonate mammals and redolent of development have been described (Porrello et al., 2011a; Xin et al., 2013a). Moreover, a recent finding has reported regeneration in several tissues upon metabolic reprogramming and manipulation of Lin28. However, despite successful in several tissues, adult murine heart regeneration could not be accomplished by this approach (Shyh-Chang et al., 2013a). Therefore, the mechanisms underlying heart regeneration and whether they can be activated in adult individuals remain obscure (Aguirre et al., 2013; Xin et al., 2013b).
Here we investigate the regenerative mechanisms controlling adult zebrafish heart regeneration and cardiomyocyte dedifferentiation and demonstrate that similar mechanisms are indeed present, yet dormant, in adult mammals. As a response to injury, we identify miR-99/100 and Let-7a/c down-regulation, and the subsequent upregulation of two of their target genes, Fntβ and Smarca5, as critical steps required for heart regeneration in the adult zebrafish. This process fails to be activated in adult mammals upon injury. Experimental modulation in mice of any of the identified molecules in zebrafish: use of anti-miRs/overexpression of its downstream targets FNTβ and SMARCA5, results in adult mammalian cardiomyocyte dedifferentiation, the activation of a proliferative response and, ultimately, facilitation of heart regeneration in vivo.
RESULTS
Mechanisms controlling zebrafish heart regeneration are conserved in the mammalian genome
We and others have previously described that adult zebrafish heart regeneration occurs at the cellular level by dedifferentiation of mature cardiomyocytes followed by proliferation and further re-differentiation (Jopling et al., 2010; Kikuchi et al., 2010). More recently, it has been shown that epigenetic remodeling is a key step controlling regenerative processes (Kragl et al., 2009; Zhang et al., 2013). To further investigate the pathways underlying heart regeneration we decided to focus on microRNAs (given their wide potential to regulate gene expression changes) differentially regulated during zebrafish heart regeneration (Eulalio et al., 2012; Ivey and Srivastava, 2010). Three days post amputation (dpa) of the ventricular apex, we observed significant changes in the expression of ~60 microRNAs (Figure S1A). We focused our attention on those microRNAs presenting significant expression changes that were additionally conserved to a high degree across vertebrates, both in microRNA sequence and 3′ UTR binding sites. This approach identified two microRNA families (miR-99/100, let-7a/c), which were clustered in two well-defined genomic locations (miR-99/Let-7c and miR-100/Let-7a, Figure S1B) and strongly downregulated during regeneration (Figures 1A and S1A). Bioinformatic analysis for GO processes controlled by these two microRNA clusters indicated significant enrichment in proliferation pathways, as well as processes related to chromatin remodeling and morphogenesis (Han et al., 2011; Paige et al., 2012) (Figure S1C and Table S1). Interestingly, Let-7a/c upregulation has been previously reported as an important regulator of stem cell differentiation, and Lin28-mediated Let-7a/c downregulation has been linked to regenerative processes (Shyh-Chang et al., 2013a, b). A recent report indicates a common role for the miR-99a/let-7c cluster in regulating cardiomyogenesis (Coppola et al., 2014). Together, suggesting that Let-7a/c might be an interesting candidate for controlling heart regeneration in combination with miR-99/100. Next, we sought to identify downstream targets for miR-99/100. MIRANDA based microRNA-UTR binding predictions suggested downstream targets, and a strong interaction was found for microRNA-99/100 with zebrafish fntb (beta subunit of farnesyl-transferase) and smarca5 (SWI/SNF-related matrix associated actin-dependent regulator of chromatin subfamily a, member 5), (Figures 1A, 1B and S1D). ClustalX alignment of the 3′UTRs on the three species showed that the interactions sites were highly evolutionarily conserved in all of them (data not shown). Bioinformatic predictions allow for the pre-screening of potential miRNA target sequences, however they do not unequivocally identify target genes and potential targets need to be validated by further biochemical approaches. As reported in other species, luciferase reporter assays confirmed microRNA binding predictions to both the human and zebrafish fntb and smarca5 3′ UTRs (Mueller et al., 2012; Brenner et al., 2012; Chen et al., 2013; Liang et al., 2014; Sun et al., 2011) (Figures S1D and S1E). Together, these results demonstrate that both fntb and smarca5 mRNAs are direct targets of miR-99/100.
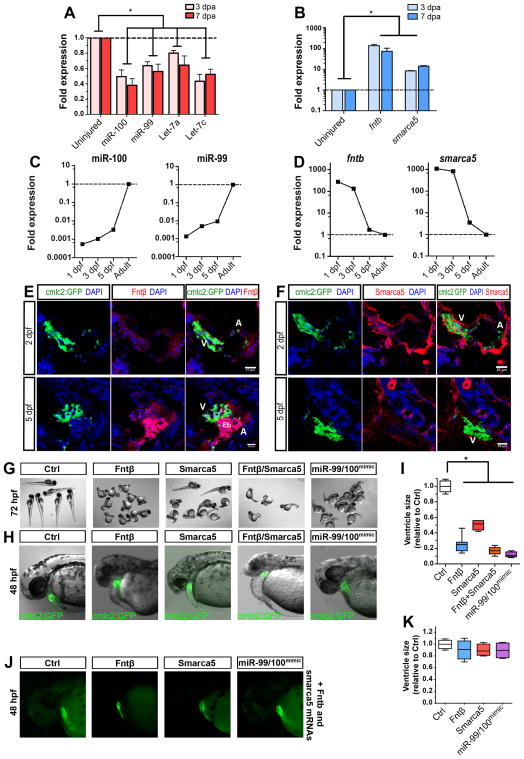
Figure 1. miR-99/100 and its direct targets contribute to zebrafishheartregeneration and development.
(A, B) Real time RT-PCR for microRNA candidates miR-99/100 and Let-7a/c (A) and downstream targets fntb and smarca5 (B) in regenerating zebrafish hearts 3 and 7 days post-amputation (dpa) (n=8). (C) Real time RT-PCR revealed that expression of miR-99/100 is very low during the first stages of development and dramatically increased at 3 days post-fertilization (dpf) in zebrafish (n=10). (D) fntb and smarca5 expression inversely correlates with miR-99/100 in developing embryos (n=10). Note that in C and D, analyzes were done in whole embryos at 1, 3, and 5dpf, and whole heart at the adult stage. (E, F) Representative pictures demonstrating that both Fntβ and Smarca5 are present at high levels in the ventricles of developing hearts at 2 and 5dpf (n=10). V: ventricle; A: atrium; Eb: erythroblasts. (G–I) Knock-down of Fntβ and/or Smarca5 in zebrafishembryos resulted in abnormally small animals (G) and reduced ventricle size in cmlc2:GFP animals (H–I). The same phenotypes were observed upon injection of miR-99/100 mimics (G–I) (n>50). Rescue experiments for the conditions tested in G were performed by co-injecting in vitro transcribed mRNAs with modified 5′UTRs with corresponding morpholinos to determine the specificity of the observed phenotypes (J) (n=30). (K) Quantification of the ventricle size for the rescue experiments. Data are represented as mean +/− s.e.m. *p<0.05. See also Figure S1 and Table S1.
Regenerative pathways are involved in zebrafish and mammalian heart development but fail to be activated in the injured mammalian heart
To shed new light onto the biological role of miR-99/100 we investigated their potential functional implication in heart development as well as during heart regeneration. Due to its involvement in controlling stem cell differentiation and regeneration as well as its connections to miR-99/100 (Shyh-Chang et al., 2013a; Coppola et al., 2014),, we also estudied the role of Let-7a/c. qRT-PCR and immunofluorescence analyses demonstrated low levels of miR-99/100 expression during early zebrafish heart development concomitantly with high levels of Smarca5 and Fntβ (Figures 1C–F). Next, we sought to establish a mechanistic link between the identified microRNAs and the predicted target proteins, Fntβ and Smarca5. To do this we performed injection of antago-miRs blocking miR-99/100 and let7 in adult uninjured zebrafish. As shown in supplementary Figure 1F, injection of the antago-miRs resulted in the concomitant overexpression of fntβ and smarca5 (Figure S1F). Furthermore, functional analyses by injection of miR-99/100 mimics as well as fntb and/or smarca5 translation-blocking morpholinos (MOs) in one-cell stage cmlc2:GFP transgenic fish embryos resulted in a significantly reduced ventricle size in spite of the presence of all, apparently normal, heart structures (ventricle, atrium, valve) as compared to animals injected with scrambled control MOs (Figures 1G–I), indicating a developmental role during heart growth. To confirm the specificity of the MOs, and to rule out global gastrulation defects, we next attempted rescue experiments where the corresponding MOs were co-injected with in vitro transcribed mRNAs encoding for the human SMARCA5 and FNTB sequences, but lacking their endogenous 3′ UTR with the miR binding sites. Co-injection of MOs alongside FNTB and SMARCA5 mRNAs rescued the phenotypes observed when MOs were injected alone (Figures 1J and 1K). Together, these results confirmed the specificity of the MOs used in these studies as well as demonstrate a causal link in controlling SMARCA5 and FNTβ expression either by stabilizing their respective mRNAs or upon modulation of other factors controlling their transcription. Next, we monitored miR-99/100 as well as fntβ and smarca5 expression in the regenerating zebrafish heart. Confirming our initial observations, miR-99/100 expression was high and confined to cardiomyocytes in uninjured hearts, and became almost undetectable upon injury (Figures 2A, 2B, S2A and S2B). As expected, miR-99/100 downregulation was paralleled by upregulation of their targets Fntβ and Smarca5 at the RNA (Figures 1A and 1B) and protein levels (Figures 2A, 2B, S2A and S2B). Immunofluorescence analyses further correlated Fntβ and Smarca5 expression with increased Histone 3 phosphorylation (H3P) and PCNA re-expression, both indicative of cellular proliferation and cardiomyocyte dedifferentiation (Jopling et al., 2010; Kikuchi et al., 2010) (Figures 2C–F). Let-7a/c expression was also maximal in adult uninjured zebrafish hearts (Figure S2C). Following amputation we observed Let-7a/c downregulation (Figures 1A and S2C) accompanied by the upregulation of its previously described target genes, Ras and c-Myc (Figures S2D–G). miR-99/100 and Let-7a/c expression, as well as that of their respective protein targets, returned to basal levels 30 dpa, once regeneration of the ventricle was nearly completed (Figures S3A and S3B). Next, we designed a series of in vivo experiments to functionally dissect the role of miR-99/100 in the adult regenerating zebrafish heart. Notably, we first injected miR-99/100 mimics and evaluated Fntb and Smarca5 expression during regeneration. As expected, avoiding downregulation of miR99-100 during regeneration significantly impaired Fntb and Smarca5 expression (Figure S3C), confirming further that the miR-99/100 cluster does influence the activity of its targets in vivo. Intra-cardiac injection of miR-99/100 mimics efficiently blocked the regenerative response in all amputated animals (Figures 3A and 3B). Reduced BrdU incorporation accompanied confirmed that cardiomyocyte proliferation was significantly disrupted in mimic-treated animals (Figures 3A and 3C). Inversely, microRNA inhibition in uninjured adult animals led to significantly enlarged zebrafish hearts suggesting a regenerative proliferative response even in the absence of injury and therefore a direct role for miR-99/100 in controlling heart mass (Figures 3D–F). Most interestingly, chemical inhibition of Fnt with the specific antagonist tipifarnib significantly impaired heart regeneration by decreasing the number of proliferating cardiomyocytes, further suggesting an active role for miR-99/100 targets during regeneration (Figures 3G–I). Taken together, these results demonstrate a functional role for miR-99/100, fntβ and smarca5 in zebrafish heart development and adult heart regeneration.
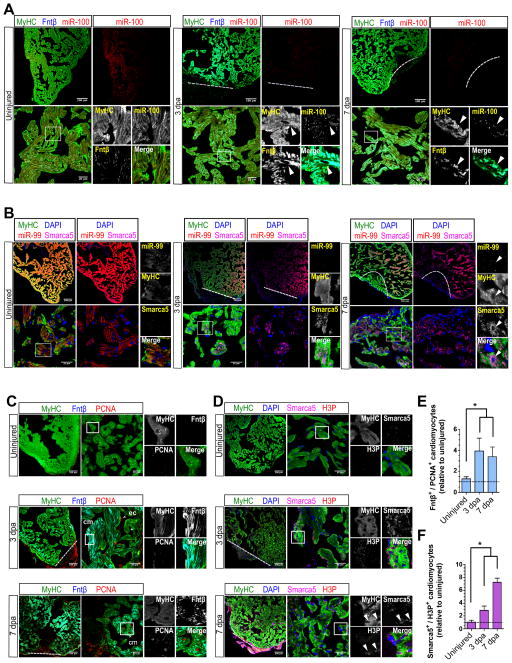
Figure 2. miR-99/100 and its downstream targets are differentially regulated in dedifferentiated cardiomyocytes during heart regeneration.
(A, B) FISH/immunofluorescence were used to determine cardiomyocyte specific expression (MyHC) of miR-99/100, Fntβ and Smarca5 in uninjured and regenerating zebrafish hearts (3 and 7dpa). Cardiomyocytes in regenerating hearts exhibited low levels of miR-99/100, and inversely correlating high levels of Fntβ and Smarca5 (n=8). (C–D) Immunofluorescence analysis demonstrating high levels of Fntβ (C) and Smarca5 (D) alongside markers indicative of proliferation, PCNA (C) and H3P (D), in dedifferentiating cardiomyocytes (n=5). (E–F) Quantitative analysis confirming the significantly higher number of MyHC+ cardiomyocytes co-expressing Fntβ/PCNA (E) and Smarca5/H3P (F) in the regenerating zebrafish heart (n=5 animals, three different sections per animal). Dashed line: amputation plane. Boxed area: magnified field. Arrows indicate cells of interest. Data are represented as mean ± s.e.m. *p<0.05. See also Figure S2 and S3.
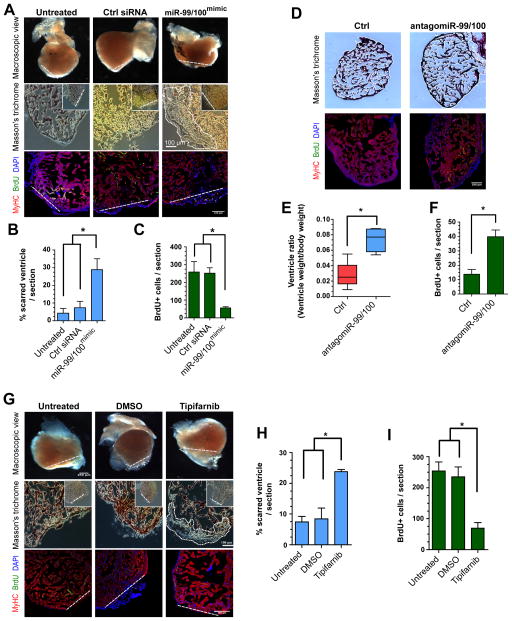
Figure 3. Heart regeneration is controlled by miR-99/100.
(A–C) Following adult zebrafish heart amputation, exogenous intra-cardiac administration of 0.2 μg miR-99/100 mimics every second day for a total of 14 days led to defective cardiac regeneration in amputated zebrafish (A, upper row), as determined by reduced BrdU incorporation (A, lower row and C). (D–F) miR-99/100 antagomiRs exerted the opposite effect in uninjured adult animals, inducing a significant increase in ventricle size (D, upper panels and E) and increasing cardiomyocyte proliferation in the absence of heart damage (D, lower panels and F). (G–I) Chemical inhibition of fnt activity with tipifarnib upon intraperitoneal injection (final concentration 0.02 mg/animal) every 2 days for 14 days dramatically reduced heart regeneration in amputated zebrafish as compared to control animals administered with the solvent DMSO control (G and H) and reducing cardiomyocyte proliferation as assessed by BrdU incorporation (G, lower panels and I). Dashed line: amputation plane. Boxed area: magnified detail. Data are represented as mean ± s.e.m. *p<0.05. n = 6 animals, three different sections per animal were used for quantitative analyses.
We next sought to investigate if mammals presented similarities with the adult zebrafish in terms of miR-99/100 and/or Let-7a/c expression and functionality. Therefore, we first evaluated miR-99/100, Let-7a/c and FNTβ/SMARCA5 expression at different stages during murine and human cardiomyocyte differentiation and heart development. qRT-PCR and immunofluorescence analyses demonstrated the progressive upregulation of miR-99/100 and Let-7a/c as heart development and maturation proceeds (Figures 4A–G and S4A–C). Interestingly, neonatal murine cardiomyocytes, which although still proliferative quickly lose those capabilities after birth, presented significantly higher levels of miR expression than embryonic cardiomyocytes, and no fntb neither smarca5 mRNAs could be detected (Figures 4C and 4D). Thus, suggesting that higher levels of miR-99/100 and let7 are required for cardiomyocyte maturartion in line with a recent report by Coppola et al (Coppola et al., 2014) or alternatively, that distinct mechanisms control neonatal and adult cardiomyocyte proliferation. By employing hESC-derived cardiomyocytes we were able to partially recapitulate human cardiomyocyte development in a dish, revealing that low levels of miR-99/100 and high levels of FNTβ/SMARCA5 are necessary at early stages for cardiac specification and are lost when cardiomyocytes mature (data not shown). Adult murine and human heart tissue demonstrated extremely high levels of miR-99/100, a point at which FNTβ/SMARCA5 expression was undetectable (Figures 4A–G). Interestingly, analyses of post-mortem injured human heart tissue suggested a failure to upregulate FNTβ/SMARCA5 expression, while both proteins were expressed in the developing embryonic human heart but absent in the adult (Figure S4D). To further demonstrate that this was not due to the temporal window analyzed, as adult human hearts can rarely be obtained from an individual right after an infarction has occurred, we additionally performed myocardial infarction experiments in adult mice. Our results indicated that murine hearts are unable to elicit a natural downregulation of miR-99/100 and Let-7a/c and presented levels comparable to those observed on sham-operated animals (Figures 4H, S4E and S4F). Collectively, these results suggested that adult mammals, contrary to zebrafish, might fail to regenerate their heart due to a failure in downregulating miR-99/100 and Let-7a/c, and inversely, upregulate the miR-99/100 targets, FNTβ/SMARCA5, after injury.
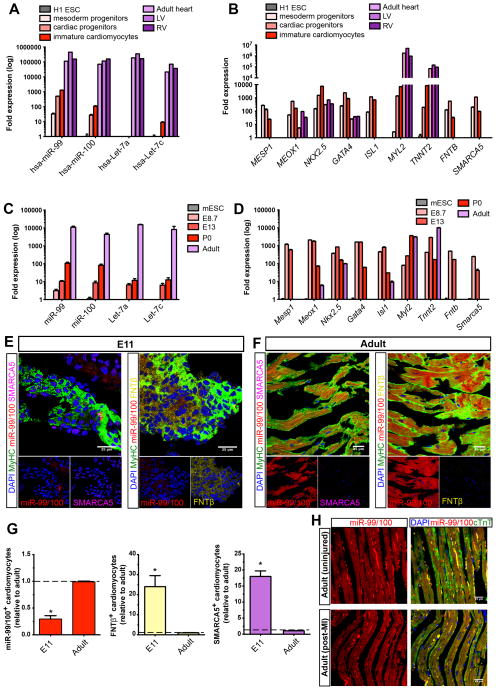
Figure 4. The miR-99/100-dependent heart regeneration pathway is developmentally conserved in mammals but fails to activate upon injury.
(A–B) qRT-PCR analysis showing the expression of miR-99/100 and Let-7a/c (A), their protein targets and key cardiac transcription factors (B) during human cardiac differentiation (n=5). Human embryonic stem cell line H1 (H1 ESC); left ventricle (LV and right ventricle (RV). (C–D) qRT-PCR analysis showing the expression of miR-99/100 and Let-7a/c (C), their protein targets and key cardiac transcription factors (D) during mouse development and adult stages (n=5). Mouse embryonic stem cells (mESC). (E–G) FISH/immunofluorescence (E, F) and quantitative analysis (G) demonstrating low levels of miR-99/100 and high levels of FNTβ and SMARCA5 in E11 murine embryonic hearts as opposed to adult hearts, which showed high levels of miR-99/100, and almost undetectable expression of FNTβ and SMARCA5. (H) FISH/immunofluorescence in adult mouse heart before (upper panels) and after myocardial infarction (lower panels) highlighted a failure to downregulate miR-99/100 upon injury in the murine heart. Data are represented as mean ± s.e.m. *p<0.05. (E, F): n = 6 animals; (H): n= 5 animals. In all cases, three different sections per animal were used for quantitative analyses (G). See also Figure S4.
Experimental manipulation of conserved regenerative pathways allows for mammalian cardiomyocte proliferation by cell dedifferentiation
Adult zebrafish heart regeneration occurs by the spontaneous dedifferentiation of resident cardiomyocytes as a response to injury. Recently, dedifferentiation of somatic cells to a progenitor-like state with proliferative and re-differentiation potential has been described to occur as a consequence to injury in a number of different mammalian organs, including the gut and pancreas (Schwitalla et al., 2013; Zaret, 2008; Ziv et al., 2013), as an attempt to maintain tissue homeostasis. Neonatal murine heart regeneration by enhanced cardiomyocyte proliferation has been reported (Porrello et al, 2011). Yet, whether de novo proliferative cardiomyocytes were generated by dedifferentiation as opposed to boosting the inherent proliferative capacity of neonatal murine cardiomyocytes remains unknown. More interestingly, recent reports indicate that neonatal heart regeneration is accomplished by neovascularization and not necessarily cardiomyocyte dedifferentiation (Andersen et al., 2014). Regardless the case, cell dedifferentiation and repair does not naturally occur in the adult mammalian heart upon injury to any significant extent (Senyo et al, 2012). Therefore, we next wondered whether experimental downregulation of miR-99/100 and/or let-7a/c in vitro and ex vivo, or inversely, experimental upregulation of SMARCA5 and FNTβ, in mammalian cardiomyocytes, could lead to dedifferentiation in an analogous manner to that observed during zebrafish heart regeneration. Seven days after miR-99/100 and/or let-7a/c silencing by anti-miR delivery (Figures S5A and S5B), significant up-regulation of SMARCA5 and FNTβ was observed in cultured adult primary murine cardiomyocytes compared to the respective scrambled and mock vector controls (Figures 5A–C). Importantly, an increased amount of GATA4, a marker associated with dedifferentiated cardiomyocytes (Kikuchi et al., 2010), was also observed (Figures 5A, 5B and 5D). Accompanying dedifferentiation we observed PCNA re-expression (Figures 5A, 5B and 5D) and increased formation of human cardiomyocyte beating colonies (Movies S1, S2 and S3), suggesting the acquisition of a proliferative phenotype similar to that observed during zebrafish regeneration (Jopling et al., 2010; Kikuchi et al., 2010). In support of cardiomyocyte specificity, microRNA down-regulation did not affect proliferation neither FNTβ/SMARCA5 expression in human fibroblasts or vascular cells (Figures S5C and S5D). To provide further insights into the gene expression changes occurring during mammalian cardiomyocyte dedifferentiation, neonatal cardiomyocytes were transduced with the different anti-miRs and their respective negative control. Seven days after infection, GFP+ cells were sorted out and analyzed by RNA-seq (Figures S5E, S5F and Table S2). Transcriptomic analysis revealed differences in genes involved in epigenetic remodeling and demethylation, cardiac development, proliferation and unexpectedly, metabolic pathways (Figures 5E–H and S5G). Furthermore, and in an effort to offer a more comprehensive view of transcriptomic changes, we performed semi-quantitative mass spectrometry on regenerating zebrafish hearts, neonatal mouse hearts and ex vivo adult myocardial mouse tissue treated with the anti-miRs to find out changes at the translational level (Figure 5I and Table S3). A cross-species comparison of the differentially regulated proteins confirmed the transcriptomic findings and identified dozens of metabolic and mitochondrial processes as key actors, while also highlighting changes in the cytoskeleton and chromatin state in the regeneration-permissive state (Figure S5H and Table S3). Given the new-found importance of metabolic processes and considering that changes in metabolism have been ascribed to the dedifferentiation of somatic cells to induced Pluripotent Stem Cells (iPSCs) (Panopoulos et al., 2012; Shyh-Chang et al., 2013b) and regeneration (Shyh-Chang et al., 2013a), we decided to investigate whether similar processes underlined anti-miR elicited cardiomyocyte dedifferentiation. First, since Lin28 expression has been linked to an increase in oxidative phosphorylation, regeneration and Let-7a/c biogenesis, we evaluated Lin28 expression in primary adult murine cardiomyocytes subjected to anti-miR treatments. qPCR analyses demonstrated lack of Lin28 mRNA in any of the analyzed conditions (data not shown), in line with previous reports demonstrating that Lin28-mediated responses were context-specific and did not result in heart regeneration (Shyh-Chang et al., 2013a). Along the same line, seahorse analysis indicated an increase in the ratio between glycolysis and oxidative phosphorylation in line with the metabolic changes observed during dedifferentiation to iPSCs (Figure S6A). Interestingly, mitochondrial fusion has been recently described to accompany cardiomyocyte differentiation (Kasahara et al., 2013). In line with these observations, we observed a highly fused and organized mitochondrial network in differentiated primary cardiomyocytes whereas anti-miR delivery resulted in mitochondrial network fragmentation indicating cardiomyocyte dedifferentiation (Figures S6B). We then decided to employ a different experimental setting closer to in vivo conditions - i.e. ex vivo myocardial tissue cultures of adult murine hearts - to further understand the whole extend of the dedifferentiation process. Ex vivo tissue has the advantage of providing a microenvironment more closely resembling that present in the heart. Lentiviral-mediated silencing of miR-99/100 and/or miR-Let-7a/c to hypoxic and non-hypoxic murine heart organotypic slices (Brandenburger et al., 2012) (Figure 6A), resulted in sarcomeric disassembly (Figure 6B) and the downregulation of Connexin 43 (Cx43) and MyHC (Figures 6C, 6D and S6C). Accompanying cardiomyocyte dedifferentiation, H3 phosphorylation and re-expression of GATA4 were also observed (Figures 6C, 6D and S6C). Moreover, necrosis, which is readily observed in control organotypic slices subjected to hypoxia, was reduced after anti-miR delivery (Figure 6E). Taken together, our results demonstrate that experimental downregulation of miR-99/100 and Let-7a/c triggers the dedifferentiation of mammalian cardiomyocytes to a proliferative state able to reduce necrosis ex-vivo.
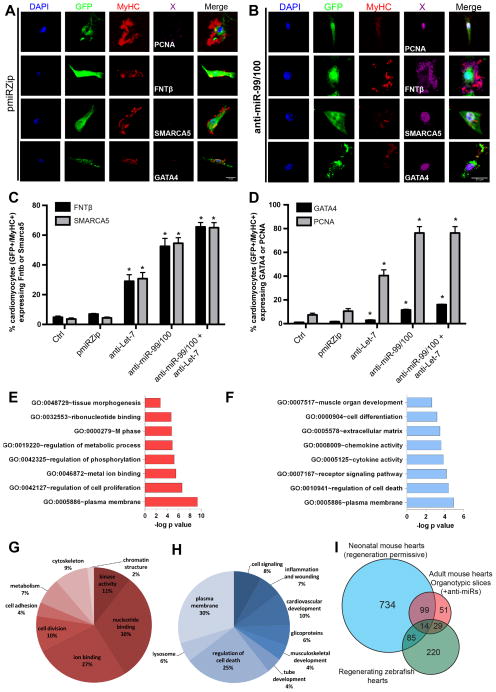
Figure 5. Forced down-regulation of miR-99/100 and Let-7a/c suffices to promote dedifferentiation and proliferation of adult murine cardiomyocytes in culture.
(A) Representative pictures showing that pmiRZip control vector-transduced adult murine cardiomyocytes spontaneously disorganized sarcomeric structures in vitro, but did not dedifferentiate to express FNTβ, SMARCA5, GATA4 or the proliferative marker, PCNA. (B) Upon lentiviral transduction with antimiR-99/100, adult cardiomyocytes dedifferentiate and express PCNA, GATA4 and both miR targets. (C, D) Quantitative analysis of FNTβ and SMARCA5 (C), or GATA4 and PCNA (D) positive adult cardiomyocytes after different anti-miR treatments. (E–F) Most relevant upregulated (E) and downregulated (F) functional gene ontology processes observed during in vitro cardiomyocyte dedifferentiation with antimiRs. (G–H) Clusters of genes indicating the most relevant upregulated (G) and downregulated (H) groups involved in cardiomyocyte dedifferentiation. As expected, epigenetic remodeling and developmental processes appeared as highly represented. (I) Comparative proteomic analysis of the heart during regenerative stages. Adult regenerating zebrafish hearts (uninjured, 3 dpa, 7dpa), neonatal regeneration-permissive mouse hearts (P0, P7, adult) and anti-miR treated adult myocardial mouse tissue (control, anti-miR-99/100, anti-Let-7a/c, anti-miR-99/100+anti-Let-7a/c) were collected and processed for semi-quantitative mass spectrometry to determine their translational profiles. The Venn diagram depicts differentially regulated protein candidates for each stage and species after a cross-comparison to find common protein effectors involved in the maintenance or transition to a regeneration-permissive state. Data are represented as mean +/− s.e.m. *p<0.05. n = 3 independent experiments. See also Figure S5 and Tables S2 and S3.
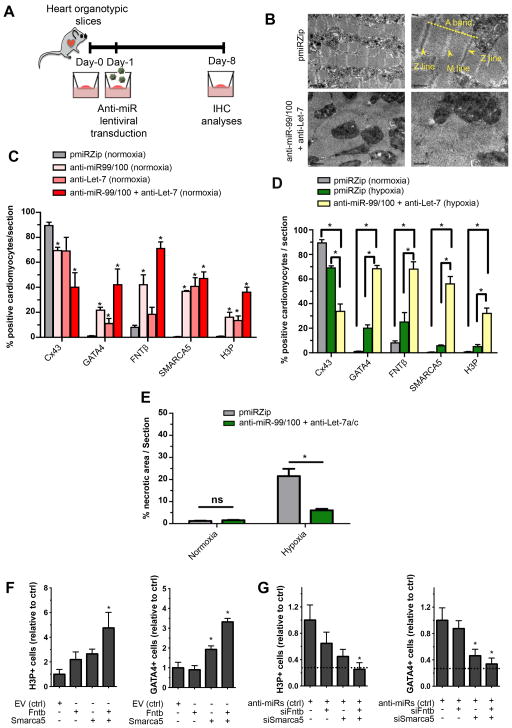
Figure 6. Myocardial tissue can be induced to a partially dedifferentiated proliferative state by anti-miR treatment.
(A) Diagram describing organotypic culture experiments: ventricles were obtained from wild-type mice, cut into slices and put in culture conditions. At 24 h, they were transduced with the antimiR constructs and evaluated by immunohistochemistry (IHC) analyses for cardiomyocte proliferation/dedifferentiation, after 7 days. (B) Representative pictures showing that sarcomeric disorganization was readily observed by electron microscopy after induced dedifferentiation in adult cardiac tissue. (C) Quantitative analyses of normoxic organotypic cultures demonstrating significant increases in FNTβ and SMARCA5, enhanced numbers of dedifferentiated cardiomyocytes -determined by Cx43 and GATA4 expression- and significantly increased numbers of proliferating cells, upon anti-miR treatment. (D) Quantitative evaluations of hypoxic organotypic cultures demonstrating cardiomyocyte dedifferentiation upon anti-miR treatment as indicated by GATA4 re-expression, reduced number of Cx43+ cells and increased Histone 3 phosphorylation (H3P). (E) Histomorphometric evaluation of the damaged myocardium in normoxic and hypoxic conditions after Masson’s trichrome staining. (F) Lentiviral overexpression of Fntb, Smarca5 or both is sufficient to impose a dedifferentiated profile in neonatal murine cardiomyocytes. (G) In primary neonatal murine cardiomyocytes, knock-down of miR 99/100 and Let-7a/c is insufficient to promote a dedifferentiated state in the absence of Fntb/Smarca5 (knocked-down with siRNAs), suggesting these two proteins are mostly responsible for the effects of the miRs. In G, dashed lines represent the baseline values in the untreated condition (absolute values being 8% H3P+ and 2.5% GATA4+ cells). Data are represented as mean +/− s.e.m. *p<0.05. n = 4 independent experiments/condition, 3 sections/experiment/condition were used for quantitative analyses. See also Figure S6.
Considering that microRNAs are pleiotropic molecules controlling expression of multiple downstream target genes, we next sought to investigate whether FNTβ/SMARCA5 upregulation alone could suffice for the induction of cardiomyocyte dedifferentiation and proliferation in mammalian cardiomyocytes. We speculated that if the main role of miR-99/100 during heart regeneration was to upregulate endogenous FNTβ/SMARCA5 expression, overexpression of these proteins might allow bypassing the requirement for microRNA downregulation altogether. Indeed, overexpression of either protein in primary murine cardiomyocytes cultured in vitro resulted in the appearance of dedifferentiated phenotypes, including GATA4 expression and ultimately, cell proliferation (Figure 6F). To further demonstrate specificity, we devised an experimental design conceptually similar to traditional rescue experiments. Contrary to our overexpression experiments, should FNTβ and SMARCA5 be the major players controlling mammalian cardiomyocyte dedifferentiation and heart regeneration downstream of miR-99/100, concomitant siRNA-mediated knockdown of FNTβ and SMARCA5 alongside delivery of anti-miR-99/100 should result in impaired cardiomyocyte dedifferentiation and proliferative potential. As expected, downregulation of the downstream effectors FNTβ and SMARCA5 in anti-miR-99/100 treated cells significantly abrogated mammalian cardiomyocyte dedifferentiation and proliferation (Figure 6G). Last, we wondered whether miR-99/100 and let-7 levels, and inversely FNTb and SMARCA5 levels, correlated not only with cardiomyocyte dedifferentiation but also reciprocally with cardiomyocyte specification. In line with recent findings (Coppola et al., 2014) reporting a positive effect of the miR-99/let-7 cluster during ESC-cardiomyocyte differentiation by enhancing mesoderm specification, our results further demonstrated that expression of miR-99/100 and let-7 mimics, or inversely knockdown of FNTb and SMARCA5, resulted in the maturation of ESC-derived human cardiomyocytes, Interestingly, we observed a significant upregulation of genes typically associated to adult mature cardiomyocytes including CX43 and MYL7 (Figure S6D). Together, these results highlight FNTβ and SMARCA5 as critical targets downstream of miR-99/100 for controlling mammalian cardiomyocyte dedifferentiation and inversely, maturation.
Induction of cardiomyocyte dedifferentiation facilitates mammalian heart regeneration in vivo
Last, we decided to test the in vivo regenerative potential of anti-miRs delivery in a murine model of myocardial infarction (MI). Following LAD artery ligation, we first focused on the use of lentiviral vectors encoding anti-miR-99/100 and anti-Let-7a/c and the respective mock controls. Intracardial delivery of lentiviruses resulted in significantly improved functional parameters including Ejection Fraction (EJ) and Fraction Shortening (FS) as compared to non-treated animals (Figure S7A). Indeed, the values achieved 15 days after treatment did not significantly differ from those obtained in sham operated animals. To evaluate the potential therapeutic use of anti-miR-99/100 and anti-Let-7a/c, we next decided to use adeno-associated viruses serotype 2/9 (AAV2/9) presenting cardiac tropism. AAVs encoding for anti-miR-99/100 and anti-Let-7a/c, or the respective scrambled controls, were administered by intracardiac injection in the periphery of the infarcted area. Importantly, functional heart parameters, as determined by echocardiography, including fractional shortening, ejection fraction and ventricular wall thickness significantly improved in the treated group after 14 and 90 days, as compared to scrambled controls, to levels closer to those observed in sham operated animals but significantly different (Figures 7A and S7B-D). In addition, a significant reduction in fibrotic scarring as well as in infarct size was observed when treated injured animals were compared to scrambled MI controls (Figures 7B, 7C, S7E and S7F). To determine if cardiomyocyte dedifferentation and proliferation were taking place, we performed histological analyses. Our results revealed increased numbers of cardiomyocytes positive for the miR-99/100 target genes FNTβ and SMARCA5 18 days after injury in anti-miRs treated animals (Figures 7D and 7E). Additionally, since knockdown of FNTb and SMARCA5 facilitated cardiomyocyte maturation, we wondered whether functional improvement could be due to the specification of resident cardiac precursors. Interestingly, re-expression of GATA4 and PCNA (Figures 7F, 7G, S7G and S7H) alongside increased H3 phosphorylation and BrdU incorporation (Figures 7H, 7I, S7I and S7J), demonstrated increased DNA synthesis, suggesting a process of cardiomyocyte dedifferentiation. However, whereas we fail to observe a substantial population of GATA4+ or PCNA+ cells in scrambled-treated nor in sham operated animals, we cannot totally rule out a contribution from cardiac precursors in this system. To rule out the possibility that DNA synthesis was occurring independently of cell division we next investigated the expression of aurora B kinase (ARK-2) and anillin, both indicative of cytokinesis. ARK-2 and anillin expression were readily observed in regenerating murine hearts (Figures 7J and S7K). In conclusion, our data demonstrate that failure to naturally downregulate miR-99/100 and Let-7a/c, and subsequent upregulation of their target genes FNTβ and SMARCA5, as a response to heart injury acts as a major roadblock preventing adult mammalian heart regeneration. More interestingly, barriers to mammalian heart regeneration could be experimentally overcome by anti-miR administration.
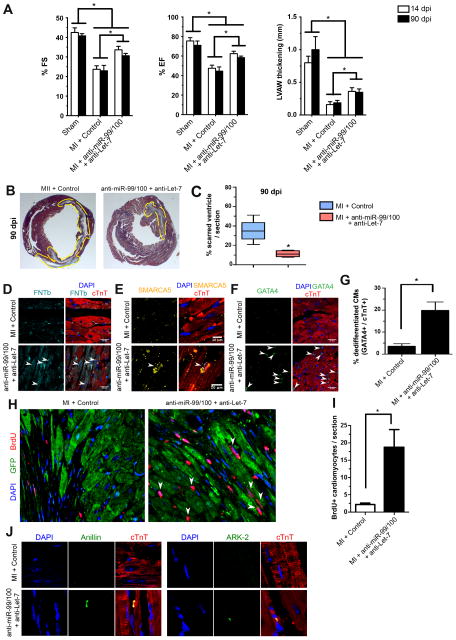
Figure 7. miR-99/100 and Let-7 silencing is sufficient to induce heart regeneration in a murine model of myocardial infarction.
(A) AAV2/9 (a serotype showing cardiomyocyte specificity) GFP-mediated anti-miR-99/100 and anti-Let-7 in vivo delivery in a mouse model of myocardial infarction (MI) resulted in the significant improvement of fractional shortening (FS, left panel), ejection fraction (EF, middle panel) and left ventricular anterolateral wall thickness (LVAW, right panel) at 14 and 90 days post-infarction (dpi) as compared to AAV2/9-GFP scrambled injected animals. (B, C) Reduced infarct size in anti-miR treated animals was confirmed by Masson’s trichromic staining at 90 dpi. Representative pictures are depicted in B, and the quantitative analysis is showed in C. (D–F) Immunofluorescent analysis of cardiac tissue at 18 dpi indicating that functional recovery was accompanied by re-expression of FNTβ (D) and SMARCA5 (E), as well as cardiomyocyte dedifferentiation as indicated by GATA4 re-expression (F). (G) Quantitative evaluation of cTnT+ cardiomyocytes (CMs) expressing GATA4 upon anti-miR treatment in vivo. (H–I) Qualitative (H) and quantitative (I) analysis demonstrated a significant increase in the number of BrdU positive cardiomyocytes upon anti-miR delivery. (J) Representative pictures demonstrating cytokinesis in cTnT+ cardiomyocytes from anti-miRs treated animals compared to control animals, as evaluated by anillin and aurora B kinase staining. Data are represented as mean ± s.e.m. *p<0.05. Arrowheads: cells of interest. n = 8 animals/group (14dpi); n = 7 animals/group (90dpi). In all cases, three different sections per animal were utilized for quantitative analyses. See also Figure S7.
DISCUSSION
Despite the fact that yet unknown mechanisms prevent adult mammals to naturally mount a robust and efficient regenerative response upon injury (Senyo et al., 2012), our results and those from others indicate that heart regeneration can be activated in mammals under certain circumstances (Porrello et al., 2011b, 2013; Senyo et al., 2012). Porrello et al. first demonstrated that an efficient regenerative response occurs in the neonatal murine heart during the days immediately after birth. This regenerative response is completely absent from day ~8 onwards and in adult animals. Interestingly, neonatal murine cardiomyocytes differ from adult cardiomyocytes in that they are able to naturally proliferate. Therefore, it remains unclear whether neonatal murine heart regeneration is due to enhanced cardiomyocyte proliferation or to a regenerative response mounted de novo. Most strikingly, neonatal murine heart regeneration has been recently ascribed to neo-angiogenesis as opposed to a major contribution due to newly generated myocardial mass (Andersen et al., 2014). Regardless of the precise mechanisms, these observations collectively suggest that developmental mechanisms are still present in the neonatal murine heart and that they might contribute to tissue repair by promoting cardiomyocyte proliferation. Indeed, our previous work identified microRNAs boosting cardiomyocyte proliferation in neonatal murine cardiomyocytes that sufficed for the partial healing of the mammalian heart (Eulalio et al., 2012), albeit the mechanisms at work remain obscure.
Contrary to neonate murine models, the adult heart is largely comprised of post-mitotic cardiomyocytes. Indeed, natural regeneration is preceded by cardiomyocyte dedifferentiation and proliferation in organisms able to regenerate their adult heart (Jopling et al., 2010; Kikuchi et al., 2010; Witman et al., 2011). These dedifferentiated cardiomyocytes are able to re-enter the cell cycle and fully repair the injured adult heart as a response to injury (Jopling et al., 2010; Raya et al., 2003; Wang et al., 2011). Interestingly, similar pro-regenerative phenomena, based on cell dedifferentiation, have been recently described to underlie mammalian tissue repair and homeostasis as a response to injury in the pancreas and intestine (Schwitalla et al., 2013; Zaret, 2008). However, mammalian hearts do not regenerate, nor do adult mammalian cardiomyocytes naturally dedifferentiate to a proliferative state upon injury (Jopling et al., 2011). Here we sought to identify the molecular mechanisms regulating cardiomyocyte dedifferentiation in the zebrafish heart and speculated that, if conserved, they might provide a platform for the development of in vivo regenerative strategies for the healing of the mammalian heart.
By systematic screening of molecules differentially regulated during zebrafish heart regeneration, we report on the identification of the molecular effectors controlling zebrafish heart regeneration and their application for the induction of regeneration in the injured adult mammalian heart. miR-99/100 expression was high in the uninjured zebrafish heart and underwent rapid down-regulation upon injury. Concomitant to miR-99/100 downregulation, expression of multiple downstream target genes was observed. Among all different target genes, we identified Fntβ and Smarca5 as critical regulators of zebrafish heart development and regeneration. Interestingly, similar patterns of expression were observed during mammalian heart development. Mature non-proliferative cardiomyocytes in the zebrafish, human and mouse demonstrated maximal levels of microRNA expression accompanied by absent GATA4, FNTβ and SMARCA5 expression. Noticeably, whereas the adult zebrafish efficiently downregulated miR-99/100 expression to undergo regeneration as a response to injury, mammals seemingly failed to endogenously activate such mechanisms in spite of being present and playing the exact same role. Experimental downregulation of miR99-100 in human, neonatal and adult murine cardiomyocytes resulted in the acquisition of a dedifferentiated phenotype, as best exemplified by GATA4 re-expression, expression of proliferative markers and sarcomeric disassembly. Noticeably, dedifferentiation seemed limited to mononucleated cardiomyocyte populations, raising the interesting question of whether mononucleated cardiomyocytes are more responsive to the activation of this signaling pathway or alternatively, that polynucleated/polyploidy cardiomyocytes can somehow revert to a mononucleated state. Genetic lineage tracing approaches might be able shed some light on this issue in the future.
Among all different target genes regulated by miR-99/100, we unveiled two major proteins, FNTβ and SMARCA5, as critical regulators of mammalian cardiomyocyte dedifferentiation and regeneration. Overexpression of either protein resulted in mammalian cardiomyocyte dedifferentiation and induction of proliferation even when miR-99/100 expression was not experimentally downregulated, a phenomenon enhanced when both proteins were simultaneously overexpressed. Conversely, downregulation of miR-99/100 by anti-miR delivery in the presence of siRNAs targeting FNTβ andSMARCA5 abrogated mammalian cardiomyocyte dedifferentiation and proliferation. Instead, miR-99/100 and let7 overexpression, or inversely, FNTb and SMARCA5 knockdown, facilitated cardiomyocyte maturation.
Interestingly, metabolic reprogramming as a response to Lin28 upregulation, a regulator of let-7 biogenesis, has been shown to elicit a regenerative response in mammalian organs other than the heart (Shyh-Chang et al., 2013a). Thus, adult mammalian heart regeneration might be subjected to different mechanisms in a context-dependent manner. Indeed, our results indicate that treatment with Let-7a/c alone did not suffice for mounting and efficient regenerative response, as observed by incomplete sarcomeric disassembly, whereas blocking miR-99/100 efficiently led to sarcomeric disassembly and GATA4 expression, indicating cell dedifferentiation. More robust results in inducing cardiomyocyte dedifferentiation and adult heart regeneration were obtained when both miR99-100 and Let7c were inhibited by their respective antimiRs, therefore, suggesting that sarcomeric disassembly represents a barrier to the dedifferentiation effect of Let-7. Ultimately, modulation of the identified regenerative machinery in adult murine models of myocardial infarction resulted in the significant improvement of heart functionality. Induced mammalian heart regeneration seem to be accomplished by a process of cardiomyocyte dedifferentiation resembling what naturally occurrs in the zebrafish as a response to injury (Jopling et al., 2011; Laflamme and Murry, 2011).
Our results represent a proof-of-concept on how the identification of conserved regenerative effectors in animal models naturally able regenerate their tissues may serve for the development of strategies towards inducing in vivo reprogramming to a dedifferentiated state in mammals. Altogether, in vivo activation of conserved cardiac regenerative mechanisms may help to circumvent concerns associated with heart cell transplantation as well as those associated with other reprogramming technologies (Abad et al., 2013; Aguirre et al., 2013; Qian et al., 2012; Song et al., 2012; Vierbuchen and Wernig, 2011), adding an additional tool to the clinical armamentarium of regenerative medicine towards the treatment of human heart disease.
EXPERIMENTAL PROCEDURES
Animals
All protocols were previously approved and performed under institutional guidelines. Wild-type (AB) and cmlc2:GFP zebrafish were maintained at 28.5 °C following standard methods. CD1 and C57BL mice were housed and maintained at the Salk Institute and ICGEB animal facilities.
Zebrafish heart amputation
Adult fish were anaesthetized in 0.4% Tricaine and secured, ventral side uppermost, in a slotted sponge. Watchmaker forceps were used to remove the surface scales and penetrate the skin, muscle and pericardial sac. Once exposed, the ventricle was gently pulled at the apex and cut with iridectomy scissors. After surgery, fish were immediately returned to system water.
Myocardial infarction
Myocardial infarction was induced in CD1 mice (8–12 weeks old) by permanent left anterior descending (LAD) coronary artery ligation as described elsewhere (Eulalio et al., 2012). Briefly, mice were anesthetized with an injection of ketamine and xylazine, intubated and placed on a rodent ventilator. Body temperature was maintained at 37°C. After removing the pericardium, a descending branch of the LAD coronary artery was visualized with a stereomicroscope and occluded with a nylon suture. Ligation was confirmed by the whitening of a region of the left ventricle. Recombinant AAV vectors or lentiviruses, at a dose of 1011 viral genome particles per animal, were injected immediately after LAD ligation into the myocardium bordering the infarct zone (single injection), using an insulin syringe with incorporated 30-gauge needle. Several groups of animals were studied: receiving control lentiviruses (coding for an empty vector) or antimiRs-99/100 and Let-7a/c coding lentiviruses or AAV9-control (shRNA-Luc) or AAV9-antimiR-99/100 + AAV9-anti-Let-7a/c. Sham operated groups and non-infarcted but injected groups were additionally included as control groups for AAV and lentivirus injected groups, respectively. The chest was closed, and the animals moved to a prone position until the occurrence of spontaneous breathing. Echocardiography analysis was performed at day 15 (lentiviral injections) or at days 14 and 90 (AAV-injected animals) after surgery, and hearts were collected at 18 and 90 days after infarction. All groups were analyzed in a blinded and randomized manner by a minimum of 2 experimenters.
Lentiviral and AAV constructs
Anti-miR constructs, miRZip-99/100 and miRZip-let7 (SBI), were used according to the manufacturer instructions. As respective controls, the anti-miRs were removed from the parent vector by digesting with BamH1 and EcoR1, end filled and re-ligated. Lentiviruses were packaged by transfecting in 293T cells followed by spinfection in the respective mouse or human ES derived cardiomyocytes. AAVs were generated as described before (Eulalio et al, 2012). Briefly, the antimiR constructs contained in the miRZip vectors were excised and ligated into pZacf-U6-luc-ZsGreen. Serotype 9 AAVs were packaged by transfection of 293T cells with the appropriate plasmids.
Organotypic heart slice culture
Mice ventricles (C57BL) were washed in cold Modified Tyrode’s Solution, embedded in 4% low melting point agarose and immediately cut into 300 μm slices using a vibratome (Leica). Heart slices were then maintained in complete IMDM 5 %, 1 % Pent/Strep in 12-well plates at the medium-air interface using 0.4 μm membrane transwells (Corning) at 37 C in a 5 %CO 2 incubator. For experimental hypoxia-like conditions, slices were kept in a hypoxia chamber incubator for 4 hours at 37 C, 5% O 2. Lentiviral transduction was performed by immersion of the slices in virus-containing medium for 24 h.
Confocal microscopy
Samples were imaged using a Zeiss L710 confocal microscope. For every sample, at least two different fields were examined at two different magnifications (using a 20x objective and a 63x oil-immersion objective). Z-stacks were obtained for further analysis and 3D reconstruction. For intensity comparison purposes, images were taken under the exact same conditions (pinhole size, laser intensity, etc). Automatic cell counting was performed with ImageJ and Metamorph software.
Statistical analysis
Results are expressed as mean ± s.e.m. Results are representative of at least 3 independent experiments except when otherwise indicated. Statistical analysis was carried out using Prism Software (GraphPad). For statistical comparison of two groups, unpaired, two-tailed Student’s t-test was used; for the comparison of three or more groups, one-way ANOVA followed by Tukey’s post-hoc test was used. A value of P<0.05 was considered significant.
Supplementary Material
Acknowledgments
We thank M. Schwarz, P. Schwarz for administrative support. We are grateful to Thai B. Nguyen, Yun Xia, Chris Benner, Daiji Okamura, Manching Ku, James Kasubowski, Merce Marti and the Waitt Advanced Biophotonicis Core at the Salk Institute for excellent technical assistance and useful discussions. We would also like to thank Louise C. Laurent and Robert Morey for their help with bioinformatics analyses. A.A. was partially supported by the Ipsen Foundation. L.K. was partially supported by the California Institute for Regenerative Medicine. I.S.M. was partially supported by a Nomis Foundation postdoctoral fellowship. Work in the laboratory of J.C.I.B. was supported by grants from the NHLBI (U01 HL107442-04), the G. Harold and Leila Y. Mathers Charitable Foundation and The Leona M. and Harry B. Helmsley Charitable Trust (2012-PG-MED002).
Footnotes
AUTHOR CONTRIBUTIONS
A.A. and J.C.I.B. designed all experiments. A.A., I.S.M., E.N. and J.C.I.B. wrote and revised the manuscript. A.A. performed and analyzed all zebrafish experiments. A.A., L.K. and M.N.K. performed all in vitro cardiomyocyte cell culture experiments. S.Z., M.G. and N.M. performed all murine in vivo experiments. T.H., E.V. and L.K. constructed all the different vectors. A.A., E.N. and C.R. performed immunofluorescence studies. S.K. analyzed genome-wide array analysis. A.O. performed metabolic studies. J.J.M. and J.R.Y. performed all proteomics analysis.
Supplementary Information is available in the online version of the paper.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abad M, Mosteiro L, Pantoja C, Cañamero M, Rayon T, Ors I, Graña O, Megías D, Domínguez O, Martínez D, et al. Reprogramming in vivo produces teratomas and iPS cells with totipotency features. Nature. 2013;502:340–345. doi: 10.1038/nature12586. [DOI] [PubMed] [Google Scholar]
- Addis RC, Epstein Ja. Induced regeneration-the progress and promise of direct reprogramming for heart repair. Nat Med. 2013;19:829–836. doi: 10.1038/nm.3225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguirre A, Sancho-Martinez I, Izpisua Belmonte JC. Reprogramming toward heart regeneration: stem cells and beyond. Cell Stem Cell. 2013;12:275–284. doi: 10.1016/j.stem.2013.02.008. [DOI] [PubMed] [Google Scholar]
- Andersen DC, Ganesalingam S, Jensen CH, Sheikh SP. Do Neonatal Mouse Hearts Regenerate following Heart Apex Resection? Stem Cell Reports. 2014;2:406–413. doi: 10.1016/j.stemcr.2014.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bersell K, Arab S, Haring B, Kühn B. Neuregulin1/ErbB4 signaling induces cardiomyocyte proliferation and repair of heart injury. Cell. 2009;138:257–270. doi: 10.1016/j.cell.2009.04.060. [DOI] [PubMed] [Google Scholar]
- Brandenburger M, Wenzel J, Bogdan R, Richardt D, Nguemo F, Reppel M, Hescheler J, Terlau H, Dendorfer A. Organotypic slice culture from human adult ventricular myocardium. Cardiovasc Res. 2012;93:50–59. doi: 10.1093/cvr/cvr259. [DOI] [PubMed] [Google Scholar]
- Brenner JL, Kemp BJ, Abbott AL. The mir-51 family of microRNAs functions in diverse regulatory pathways in Caenorhabditis elegans. PLoS One. 2012;7:e37185. doi: 10.1371/journal.pone.0037185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D, Chen Z, Jin Y, Dragas D, Zhang L, Adjei BS, Wang A, Dai Y, Zhou X. MicroRNA-99 family members suppress Homeobox A1 expression in epithelial cells. PLoS One. 2013;8:e80625. doi: 10.1371/journal.pone.0080625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brockes JP, Kumar A. Comparative aspects of animal regeneration. Annu Rev Cell Dev Biol. 2008;24:525–549. doi: 10.1146/annurev.cellbio.24.110707.175336. [DOI] [PubMed] [Google Scholar]
- Collins N, Poot Ra, Kukimoto I, García-Jiménez C, Dellaire G, Varga-Weisz PD. An ACF1-ISWI chromatin-remodeling complex is required for DNA replication through heterochromatin. Nat Genet. 2002;32:627–632. doi: 10.1038/ng1046. [DOI] [PubMed] [Google Scholar]
- Coppola A, Romito A, Borel C, Gehrig C, Gagnebin M, Falconnet E, Izzo A, Altucci L, Banfi S, Antonarakis SE, et al. Cardiomyogenesis is controlled by the miR-99a/let-7c cluster and epigenetic modifications. Stem Cell Res. 2014;12:323–337. doi: 10.1016/j.scr.2013.11.008. [DOI] [PubMed] [Google Scholar]
- Eulalio A, Mano M, Ferro MD, Zentilin L, Sinagra G, Zacchigna S, Giacca M. Functional screening identifies miRNAs inducing cardiac regeneration. Nature. 2012;492:376–381. doi: 10.1038/nature11739. [DOI] [PubMed] [Google Scholar]
- Han P, Hang CT, Yang J, Chang CP. Chromatin remodeling in cardiovascular development and physiology. Circ Res. 2011;108:378–396. doi: 10.1161/CIRCRESAHA.110.224287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivey KN, Srivastava D. MicroRNAs as regulators of differentiation and cell fate decisions. Cell Stem Cell. 2010;7:36–41. doi: 10.1016/j.stem.2010.06.012. [DOI] [PubMed] [Google Scholar]
- Jopling C, Sleep E, Raya M, Martí M, Raya A, Izpisúa Belmonte JC. Zebrafish heart regeneration occurs by cardiomyocyte dedifferentiation and proliferation. Nature. 2010;464:606–609. doi: 10.1038/nature08899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jopling C, Boue S, Izpisua Belmonte JC. Dedifferentiation, transdifferentiation and reprogramming: three routes to regeneration. Nat Rev Mol Cell Biol. 2011;12:79–89. doi: 10.1038/nrm3043. [DOI] [PubMed] [Google Scholar]
- Kasahara A, Cipolat S, Chen Y, Dorn GW, Scorrano L. Mitochondrial Fusion Directs Cardiomyocyte Differentiation via Calcineurin and Notch Signaling. Science. 2013;342:734–737. doi: 10.1126/science.1241359. [DOI] [PubMed] [Google Scholar]
- Kikuchi K, Holdway JE, Werdich Aa, Anderson RM, Fang Y, Egnaczyk GF, Evans T, Macrae Ca, Stainier DYR, Poss KD. Primary contribution to zebrafish heart regeneration by gata4(+) cardiomyocytes. Nature. 2010;464:601–605. doi: 10.1038/nature08804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kragl M, Knapp D, Nacu E, Khattak S, Maden M, Epperlein HH, Tanaka EM. Cells keep a memory of their tissue origin during axolotl limb regeneration. Nature. 2009;460:60–65. doi: 10.1038/nature08152. [DOI] [PubMed] [Google Scholar]
- Kwon SH, Workman JL. The changing faces of HP1: From heterochromatin formation and gene silencing to euchromatic gene expression: HP1 acts as a positive regulator of transcription. Bioessays. 2011;33:280–289. doi: 10.1002/bies.201000138. [DOI] [PubMed] [Google Scholar]
- Laflamme Ma, Murry CE. Heart regeneration. Nature. 2011;473:326–335. doi: 10.1038/nature10147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehoczky Ja, Robert B, Tabin CJ. Mouse digit tip regeneration is mediated by fate-restricted progenitor cells. Proc Natl Acad Sci U S A. 2011;108:20609–20614. doi: 10.1073/pnas.1118017108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang G, Malmuthuge N, McFadden TB, Bao H, Griebel PJ, Stothard P, Guan LL. Potential regulatory role of microRNAs in the development of bovine gastrointestinal tract during early life. PLoS One. 2014;9:e92592. doi: 10.1371/journal.pone.0092592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller aC, Sun D, Dutta a. The miR-99 family regulates the DNA damage response through its target SNF2H. Oncogene. 2012:1–9. doi: 10.1038/onc.2012.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paige SL, Thomas S, Stoick-Cooper CL, Wang H, Maves L, Sandstrom R, Pabon L, Reinecke H, Pratt G, Keller G, et al. A Temporal Chromatin Signature in Human Embryonic Stem Cells Identifies Regulators of Cardiac Development. Cell. 2012:1–12. doi: 10.1016/j.cell.2012.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panopoulos AD, Yanes O, Ruiz S, Kida YS, Diep D, Tautenhahn R, Herrerías A, Batchelder EM, Plongthongkum N, Lutz M, et al. The metabolome of induced pluripotent stem cells reveals metabolic changes occurring in somatic cell reprogramming. Cell Res. 2012;22:168–177. doi: 10.1038/cr.2011.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porrello ER, Mahmoud AI, Simpson E, Hill Ja, Richardson Ja, Olson EN, Sadek Ha. Transient regenerative potential of the neonatal mouse heart. Science. 2011a;331:1078–1080. doi: 10.1126/science.1200708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porrello ER, Mahmoud AI, Simpson E, Hill Ja, Richardson Ja, Olson EN, Sadek Ha. Transient regenerative potential of the neonatal mouse heart. Science. 2011b;331:1078–1080. doi: 10.1126/science.1200708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porrello ER, Mahmoud AI, Simpson E, Johnson BA, Grinsfelder D, Canseco D, Mammen PP, Rothermel BA, Olson EN, Sadek HA. Regulation of neonatal and adult mammalian heart regeneration by the miR-15 family. Proc Natl Acad Sci U S A. 2013;110:187–192. doi: 10.1073/pnas.1208863110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poss KD. Advances in understanding tissue regenerative capacity and mechanisms in animals. Nat Rev Genet. 2010;11:710–721. doi: 10.1038/nrg2879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poss KD, Wilson LG, Keating MT. Heart regeneration in zebrafish. Science. 2002;298:2188–2190. doi: 10.1126/science.1077857. [DOI] [PubMed] [Google Scholar]
- Qian L, Huang Y, Spencer CI, Foley A, Vedantham V, Liu L, Conway SJ, Fu J, Srivastava D. In vivo reprogramming of murine cardiac fibroblasts into induced cardiomyocytes. Nature. 2012 doi: 10.1038/nature11044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raya A, Koth CM, Büscher D, Kawakami Y, Itoh T, Raya RM, Sternik G, Tsai H-J, RodríguezEsteban C, Izpisúa-Belmonte JC. Activation of Notch signaling pathway precedes heart regeneration in zebrafish. Proc Natl Acad Sci U S A. 2003;100(Suppl):11889–11895. doi: 10.1073/pnas.1834204100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwitalla S, Fingerle AA, Cammareri P, Nebelsiek T, Göktuna SI, Ziegler PK, Canli O, Heijmans J, Huels DJ, Moreaux G, et al. Intestinal tumorigenesis initiated by dedifferentiation and acquisition of stem-cell-like properties. Cell. 2013;152:25–38. doi: 10.1016/j.cell.2012.12.012. [DOI] [PubMed] [Google Scholar]
- Seifert AW, Kiama SG, Seifert MG, Goheen JR, Palmer TM, Maden M. Skin shedding and tissue regeneration in African spiny mice (Acomys) Nature. 2012;489:561–565. doi: 10.1038/nature11499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senyo SE, Steinhauser ML, Pizzimenti CL, Yang VK, Cai L, Wang M, Wu T-D, Guerquin-Kern J-L, Lechene CP, Lee RT. Mammalian heart renewal by pre-existing cardiomyocytes. Nature. 2012:2–6. doi: 10.1038/nature11682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shyh-Chang N, Zhu H, Yvanka de Soysa T, Shinoda G, Seligson MT, Tsanov KM, Nguyen L, Asara JM, Cantley LC, Daley GQ. Lin28 Enhances Tissue Repair by Reprogramming Cellular Metabolism. Cell. 2013a;155:778–792. doi: 10.1016/j.cell.2013.09.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shyh-Chang N, Daley GQ, Cantley LC. Stem cell metabolism in tissue development and aging. Development. 2013b;140:2535–2547. doi: 10.1242/dev.091777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song K, Nam YJ, Luo X, Qi X, Tan W, Huang GN, Acharya A, Smith CL, Tallquist MD, Neilson EG, et al. Heart repair by reprogramming non-myocytes with cardiac transcription factors. Nature. 2012;485:599–604. doi: 10.1038/nature11139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart S, Tsun ZY, Izpisua Belmonte JC. A histone demethylase is necessary for regeneration in zebrafish. Proc Natl Acad Sci U S A. 2009;106:19889–19894. doi: 10.1073/pnas.0904132106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun D, Lee YS, Malhotra A, Kim HK, Matecic M, Evans C, Jensen RV, Moskaluk Ca, Dutta A. miR-99 family of MicroRNAs suppresses the expression of prostate-specific antigen and prostate cancer cell proliferation. Cancer Res. 2011;71:1313–1324. doi: 10.1158/0008-5472.CAN-10-1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi JK, Lou X, Alexander JM, Sugizaki H, Delgado-Olguín P, Holloway AK, Mori AD, Wylie JN, Munson C, Zhu Y, et al. Chromatin remodelling complex dosage modulates transcription factor function in heart development. Nat Commun. 2011;2:187. doi: 10.1038/ncomms1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vierbuchen T, Wernig M. Direct lineage conversions: unnatural but useful? Nat Biotechnol. 2011;29:892–907. doi: 10.1038/nbt.1946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Panáková D, Kikuchi K, Holdway JE, Gemberling M, Burris JS, Singh SP, Dickson AL, Lin YF, Sabeh MK, et al. The regenerative capacity of zebrafish reverses cardiac failure caused by genetic cardiomyocyte depletion. Development. 2011;3430:3421–3430. doi: 10.1242/dev.068601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witman N, Murtuza B, Davis B, Arner A, Morrison JI. Recapitulation of developmental cardiogenesis governs the morphological and functional regeneration of adult newt hearts following injury. Dev Biol. 2011;354:67–76. doi: 10.1016/j.ydbio.2011.03.021. [DOI] [PubMed] [Google Scholar]
- Xin M, Kim Y, Sutherland LB, Murakami M, Qi X, McAnally J, Porrello ER, Mahmoud AI, Tan W, Shelton JM, et al. Hippo pathway effector Yap promotes cardiac regeneration. Proc Natl Acad Sci U S A. 2013a;110:13839–13844. doi: 10.1073/pnas.1313192110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin M, Olson EN, Bassel-Duby R. Mending broken hearts: cardiac development as a basis for adult heart regeneration and repair. Nat Rev Mol Cell Biol. 2013b;14:529–541. doi: 10.1038/nrm3619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaret KS. Genetic programming of liver and pancreas progenitors: lessons for stem-cell differentiation. Nat Rev Genet. 2008;9:329–340. doi: 10.1038/nrg2318. [DOI] [PubMed] [Google Scholar]
- Zhang R, Han P, Yang H, Ouyang K, Lee D, Lin Y-F, Ocorr K, Kang G, Chen J, Stainier DYR, et al. In vivo cardiac reprogramming contributes to zebrafish heart regeneration. Nature. 2013 doi: 10.1038/nature12322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziv O, Glaser B, Dor Y. The plastic pancreas. Dev Cell. 2013;26:3–7. doi: 10.1016/j.devcel.2013.06.013. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.