Abstract
Cells in the stromal microenvironment facilitate colorectal cancer (CRC) progression and “co-evolve” with the epithelial cancer cells. Genetic and epigenetic differences between normal colorectal mucosa fibroblasts (NF) and carcinoma-associated fibroblasts (CAF) are not known. The aim of this study is to identify differentially expressed genes and promoter methylation between NF and CAF in human CRC. RNA and DNA were extracted from cultured NF and CAF from CRC resections. Genome-wide gene expression and methylation analyses were performed using the Illumina Human HT-12 v4.0 Expression and Illumina Human Methylation 27 BeadChips. Gene expression values between NF and CAF were compared and correlated with methylation patterns. Data was analyzed using Partek Genomics Suite using one-way ANOVA and p<0.05 as significant. Ingenuity iReport™ was performed to identify potential differences in biological functions and pathways between the NF and CAF. Paired methylation and gene expression analyses from 11 NF and 10 CAF colorectal samples are reported. Unsupervised analysis of differentially expressed genes using iReport™ identified “Top Diseases” as “Cancer” and “Colorectal Cancer”. Previous genome wide studies have focused on the cancer cells. We have identified differentially expressed genes and differentially methylated promoter regions that are CAF-specific in CRC.
Keywords: carcinoma-associated fibroblasts, colorectal cancer, gene expression, normal fibroblasts, microarray, promoter methylation
INTRODUCTION
Gene expression profilings for colorectal cancer (CRC) samples have been performed using RNA extracted from whole tissue tumor samples. Since the tumor microenvironment is critical to the biological behavior of the cancer, elucidating the role of carcinoma-associated fibroblasts (CAF), the most abundant cell-type in the stroma, is crucial to understanding the pathogenesis of CRC [1]. A stroma-derived gene signature has been shown to correlate with prognosis, suggesting that tumor stroma contributes to the progression and metastatic potential of CRC [2–4].
Experimental data support the contention that fibroblasts associated with the normal colonic epithelium (NF) are phenotypically different from CAFs [5, 6]. Stable gene expression changes in CAF may be due to epigenetic changes [7] versus somatic mutations [8, 9]. It is now known that somatic mutations in the DNA sequence of CAF are rarely, if ever, encountered [10, 11], and thus, the acquisition of tumor-promoting activities by CAF, in part, are due to epigenetic alterations in the DNA [7, 12]. The most studied epigenetic modification is DNA methylation, which occurs on CpG islands within the gene promoter region. Genes are downregulated when promoter regions are heavily methylated, a process that entails the methyl donor S-adenosylmethionine transferring a methyl group to the 5’ carbon of cytosine.
The purpose of this study is to determine differences in the gene expression of resident fibroblasts in the normal colon mucosa (NF) versus CAF in human colorectal cancer and to determine which differentially expressed genes may be regulated by promoter methylation.
MATERIAL AND METHODS
Fibroblast isolation and culture
Under an IRB-approved discarded tissue protocol, human colon-derived fibroblasts were isolated from freshly resected operative specimens at the University of Texas Medical Branch, Galveston, TX. Surgical pathologists excised approximately 500 mm3 of tissue from grossly recognizable tumor and/or adjacent normal mucosa. In some cases, the normal mucosa from colectomies for diverticular disease or large adenomas was obtained for culture. Fibroblasts were derived from 10 normal colonic mucosa and 11 adenocarcinomas as previously described [13]. Primary CAF and NF cultures were routinely maintained in DMEM and 10% fetal bovine serum at 37°C in a humidified atmosphere containing 5% CO2. Relevant clinical and histopathologic information was extracted via retrospective chart reviews under a second IRB protocol (Table 1).
Table 1.
Patient clinico-pathologic data of cultured fibroblasts.
| Sample | CRC Stage vs. Non- Malignant Disease |
Age | Sex | Pathology |
|---|---|---|---|---|
| CAF-1 | Stage 2 CRC | 68 | f | Moderately differentiated |
| CAF-2 | Stage 3 CRC | 59 | f | Poorly differentiated |
| CAF-3 | Stage 4 CRC | 62 | f | Moderately differentiated |
| CAF-4 | Stage 3 CRC | 54 | f | Moderately differentiated |
| CAF-5 | Stage 2 CRC | 52 | f | Poorly differentiated |
| CAF-6 | Stage 3 CRC | 56 | m | Moderately differentiated |
| CAF-7 | Stage 3 CRC | 77 | m | Moderately differentiated |
| CAF-8 | Stage 2 CRC | 55 | m | Moderately differentiated |
| CAF-9 | Stage 3 CRC | 76 | m | Well differentiated |
| CAF-10 | Stage 3 CRC | 67 | m | Moderately differentiated |
| NF-1 | Normal | 61 | f | Normal margin: diverticulitis |
| NF-2 | Normal | 38 | f | Normal margin: diverticulitis |
| NF-3 | Normal | 46 | m | Normal margin: diverticulitis |
| NF-4 | Normal | 75 | m | Vascular congestion |
| NF-5 | Normal | 77 | f | Normal margin: diverticulitis |
| NF-6 | Normal | 74 | m | Normal margin: cancer |
| NF-7 | Normal | 46 | m | Normal margin: cancer |
| NF-8 | Normal | 52 | f | Normal margin: in situ cancer |
| NF-9 | Tubovillous Adenoma | 68 | f | Normal Margin: Adenoma |
| NF-10 | Normal | 60 | f | Normal margin: cancer |
| NF-11 | Tubovillous Adenoma | 59 | m | Normal Margin: Adenoma |
Abbreviations: CAF: carcinoma-associated fibroblasts, NF: normal fibroblasts, CRC: colorectal carcinoma; f: female, m: male.
Microarray analyses
Total RNA was extracted using RNAqueous (Ambion) from NF and CAF early passage (P2-5) cultures. The purity and concentration of the RNA samples were determined using Agilent 2100 Bioanalyzer and NanoDrop ND-1000, respectively. Samples of cRNA were hybridized to Illumina Human HT-12 v4.0 Expression BeadChip, which covered the whole genome with over 47,000 probes. Genomic DNA was extracted from parallel cell cultures with lysis buffer (0.6% SDS, 10 mM EDTA, 10 mM Tris HCl, pH 7.5 and 100 µg/ml RNase-A). DNA underwent phenol and chloroform extraction, was precipitated with ethanol, and was rehydrated with sterile H20. Genomic DNA from each sample was modified by sodium bisulfite conversion and enzymatic fragmentation, and then hybridized to the Human Methylation27 BeadChip which interrogated 27,578 CpG loci, corresponding to 14,495 genes. Two probes for each CpG site are present, one corresponding to a methylated CpG locus and the other to the nonmethylated locus. Allele-specific primers are annealled and extended with DNP- and biotin-labeled ddNTPs. The array is fluorescently stained, scanned, and the intensities of the nonmethylated and methylated bead types are measured. The BeadStudio Software generated a DNA methylation level (β value) for each CpG site, which is determined by dividing the signal intensity for the methylated CpG by the sum of both the methylated and unmethylated CpGs. Beta values with a value of “0” indicate no methylation and “1” indicate a methylated promoter region. Quality control and interarray normalization was conducted according to manufacturer standards for each array. One sample failed the gene expression array quality control and another sample failed the methylation array; both were removed from the study, leaving a final N=11 NF and N=10 CAF samples for analyses.
Statistical analysis
Microarray data for both chips were analyzed using Partek Genomics Suite (Partek, Inc; St. Louis, MO). Data quality was assessed via PCA and the application of Partek’s quality control workflow. Differentially expressed candidate genes and differential β-values between comparison groups were identified by applying ANOVA, p<0.05. Thus, two filters, each with p- values <0.05, were applied to the final combined dataset, in order to decrease the false discovery rate. Ingenuity iReport™ for the microarray analysis (Qiagen, Valencia, CA) was performed to identify potential differences in biological functions and pathways between the NF and CAF.
All data used in this study are available at the NCBI Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/) under accession numbers GSE XX, and GSE YY.
RESULTS
Establishment of primary fibroblast cell lines
The fibroblast cell lines are derived from either male or female patients and used without regard to sex. The purity of the fibroblast cultures were verified as previously described using anti-vimentin (V6630 1:40, Sigma), anti-α-smooth muscle actin (αSMA) (A5228; 1:200, Sigma), and anti-pancytokeratin antibodies (AE1/3 1:200 Santa Cruz) (data not shown) [13]. The antibody to CD31 was used in several cultures to demonstrate lack of endothelial cell contamination (data not shown). The clinico-pathologic data from the 11 NF and 10 CAF samples are shown in Table 1. Then NF was derived from the normal margin of colectomies for either cancer or benign lesions such as diverticulitis; 2 samples were derived from tubovillous adenomas. The CAFs were obtained from mainly Stage II and III operable colorectal cancers. One sample was from a patient who was found to have distant metastatic disease intraoperatively.
Differential gene expression in CAF vs. NF
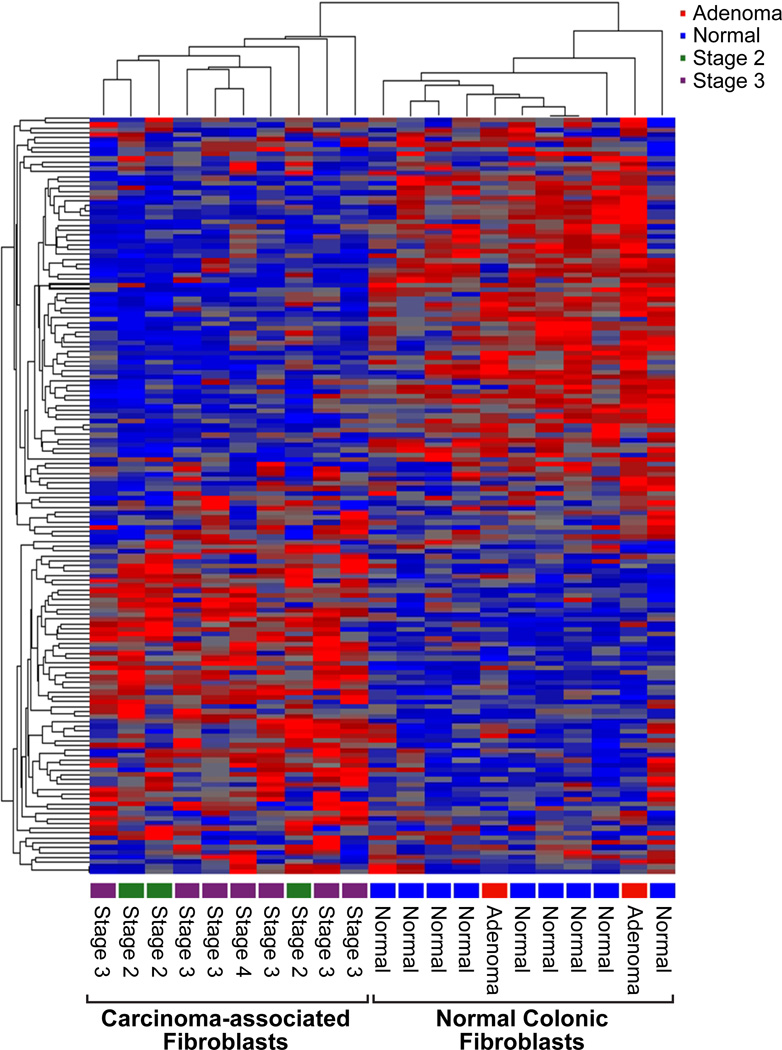
Unsupervised cluster analysis (Partek Genomics Suite) of gene expression profiles generated using the Human HT-12 v4.0 Expression BeadChip array comparing NF and CAF identified 2,472 differentially expressed genes with p-values <0.05. Compared to NF, CAF overexpressed 1,168 (47.2%) gene transcripts. In contrast, NF over-expressed 1,304 genes (52.8%) compared to CAF. If the p-value was set at p<0.001, only 84 genes were significant between the two groups (Fig. 1). Fifty genes (60%) were over-expressed in NF relative to CAF, whereas 34 genes (40%) were over-expressed in CAF relative to NF.
Figure 1.
Heat Map showing differential gene expression of colorectal derived carcinoma-associated fibroblasts (CAF) versus normal mucosal fibroblasts (NF), ANOVA, p<0.001.
Ingenuity pathway analyses of differentially expressed genes
We performed an unbiased analysis using iReport™, which offers pathway, process and disease associations without the input of key words. Unlike the Ingenuity Pathway Analysis which requires input of filters and key words, Ingenuity iReport™ identified “Cancer”, “Metastatic Colorectal Cancer” and “Colorectal Cancer” as “Top Diseases”. For example, tumor suppressor gene Deleted in Colon Cancer was decreased in the CAF compared to NF and microRNAs specific to colon cancer (MIR653 and MIR601) were upregulated in CAF. Cell Migration and Invasion were identified as “Top Processes”. Finally, “Top Pathways” identified the Wnt/βcatenin signaling pathway, and the FXR/RXR nuclear receptors which are conditional (ligand-activated) transcription factors that either have been implicated in or associated with colon cancer progression [14–16].
Specific genes upregulated in CAF which may promote tumor cell invasion by increasing cell adhesion and/or migration and invasion include: COL4A6, ITGB1BP1, CEACAM20, NCAM1, NEDD9, CD34, EXOC2, SMAGP, GLT25D2, TIAM2, LIM2, RAB11FIP1, and PRKD2. Genes that may enhance CAF-mediated cell-cell interaction signaling (PCDHGB6, PCDHGA9, IL12RB1, CD83, IL1RAP, IL18RAP, TAC1, LYVE-1), and promote extracellular region degradation (HYAL3, ADAMTS1, ADAMTS7, MMP16, GPR65, PCSK6, USP22) were over-expressed in CAF compared to NF. Other differentially overexpressed genes in CAFs include growth factors (HGF, CDNF, and CSH1), transcriptional regulators (IRF5, KLF5, ELK4, SSX4B, and ATF3), cell survival/proliferation-associated genes (AKT1, CDKL2, CNNM1, CCNE1, NOL3, GRB2, and PRAMEF19), members of the Wnt signaling pathway (SNAI3), and other intracellular signaling cascades (PLCB3, ERBB3, and BMX). Several genes encode secreted proteins (NRG4, TNF9, PCSK6, CMTM4, BOLA2, and TNSF8) that may also contribute to tumor progression (see GSEXX).
Differential methylation between CAF vs. NF
A total of 1,772 differential methylation of cytosine residues at CpG dinucleotides were identified between NF and CAF: 1,057 (60%) were hypomethylated and 715 (40%) were hypermethylated in CAF compared to NF. To correlate differentially expressed transcripts, which may be regulated epigenetically by promoter methylation, we matched the gene expression data with differentially methylated CpG loci between NF and CAF. Compared to NF, CAF overexpressed 26 genes corresponding with promoter hypomethylation, and CAF cells had decreased expression in 33 genes corresponding with promoter hypermethylation (Table 2). Among the 26 overexpressed genes in CAF, 6 (23%) genes are located on chromosome 6, and among the 33 hypermethylated or under-expressed genes, 4 (12%) were located on chromosome 9.
Table 2.
Genes regulated by promoter regulation: A. 26 genes overexpressed in CAF compared to NF.
| Function | Probeset | Gene name | Gene Symbol |
Fold Change |
P value |
|---|---|---|---|---|---|
| Metabolism/Transport | cg13077930 | Albumin | ALB | 1.71E+18 | 0.000272652 |
| cg26984624 | Ankrin 1, erythrocytic | ANK1 | 3.48E+07 | 0.00736581 | |
| cg20857947 | Arginisuccinate lyase | ASL | 121184 | 0.027152 | |
| cg22063653 | 2,3 bisphosphoglycerate mutase | BPGM | 28981.8 | 0.0218035 | |
| cg09425611 | Carboxylesterase 1 | CES1 | 431.142 | 0.0358082 | |
| cg00901704 | Solute carrier family 25 (mitochondrial thiamine pyrophosphate carrier), member 19 | SLC25A19 | 1.83E+27 | 0.0170674 | |
| cg08724474 | Solute carrier family 36 (proton/amino acid symporter), member 1 | SLC36A1 | 1.77E+36 | 0.0304509 | |
| cg14265670 | Uridine phosphorylase 1 | UPP1 | 2.91E+07 | 0.0365465 | |
| cg18412613 | YKT6-v-SNARE homolog | YKT6 | 478304 | 0.0461634 | |
| cg14493899 | Transporter 2, ATP-binding cassette, subfamily B (MDR/TAP) | TAP2 | 2.17E+14 | 0.0232357 | |
| Adhesion/Signaling | cg01288598 | CD83 molecule | CD83 | 68474.7 | 0.0271523 |
| cg05649009 | Cholinergic receptor, nicotinic, alpha1 (muscle) | CHRNA1 | 4.72E+27 | 0.049423 | |
| cg11552293 | Collagen, type IV, alpha 6 | COL4A6 | 1067.49 | 0.030782 | |
| cg11393453 | GRB2 associated regulator of MAPK1 | FAM59A | 1.61E+10 | 0.0329179 | |
| cg13060405 | Myosin XVIIIA | MYO18A | 1.43E+27 | 0.0304239 | |
| cg25187533 | Pregnancy specific beta-1-glycoprotein 2 | PSG2 | 1.13E+32 | 0.000976575 | |
| cg00950418 | SRSF protein kinase 2 | SRPK2 | 1.41E+10 | 0.0240908 | |
| Cell cycle | cg25522312 | MAD2 mitotic arrest deficient-like 1 (yeast) | MAD2L1 | 3.81E+07 | 0.0084277 |
| cg08062469 | Sperm associated antigen 5 | SPAG5 | 4.13E+09 | 0.0369162 | |
| cg19738333 | Receptor accessory protein 4 | REEP4 | 6.08E+12 | 0.018247 | |
| Transcription | cg20150743 | Zinc finger protein 3 | ZNF3 | 9578.43 | 0.0128924 |
| Autophagy | cg10732834 | Ectopic P-granules autophagy protein 5 homolog (C. elegans) | KIAA1632 | 5.03E+33 | 0.0311191 |
| Unknown | cg22010317 | chromosome X open reading frame 36 | Cxorf36 | 43048.4 | 0.0126714 |
| cg17178888 | FLJ14166 | 7.39E+07 | 0.0392013 | ||
| cg02890926 | Transmembrane protein 53 | TMEM53 | 1.40E+06 | 0.02627 | |
| cg25861458 | FL23356 | 130539 | 0.00114061 |
| B. 33 Genes under-expressed in CAF compared to NF | |||||
|---|---|---|---|---|---|
| Function | Probeset | Gene name | Gene Symbol |
Fold Change |
P value |
| Signaling | cg22930187 | Artemin | ARTN | −4008.21 | 0.0195053 |
| cg04929736 | Clusterin | CLU | −1.21E+14 | 0.0126985 | |
| cg03278643 | Ellis van Creveld syndrome 2 | EVC2 | −9.16E+06 | 0.022855 | |
| cg24888049 | Feline sarcoma oncogene | FES | −1.31E+29 | 0.005809 | |
| cg18977436 | Fibroblast growth factor 14 | FGF14 | −68717.7 | 0.025556 | |
| cg24222324 | Tumor necrosis factor superfamily, member 11 | TNFSF11 | −2227.31 | 0.008463 | |
| cg11072113 | von Willebrand factor C and EGF domains | VWCE | −1.21E+18 | 0.049167 | |
| Cell-cell; cytoskeletal | cg02512860 | Claudin 15 | CLDN15 | −37932.3 | 0.025022 |
| cg06799664 | Dynein, axonemal, heavy chain like 1 | DNAHL1 | −6381.26 | 0.044435 | |
| cg07126559 | Sarcoglycan, gamma (35kDa dystrophinassociated glycoprotein) | SGCG | −6.45E+29 | 0.03913 | |
| Enzyme | cg22333888 | ADAM metallopeptidase domain 33 | ADAM33 | −1.33E+29 | 0.0146697 |
| cg23026995 | X-prolyl aminopeptidase (aminopeptidase P) 2, membrane-bound | XPNPEP2 | −2.29E+23 | 0.030756 | |
| Transcription factors | cg19556572 | AT-hook transcription factor | AKNA | −5.29E+35 | 0.007186 |
| cg10003443 | forkhead box A2 | FOXA2 | −7451.05 | 0.012987 | |
| cg08789630 | K(lysine) acetyltransferase 6B | MYST4 | −1.74E+07 | 0.029483 | |
| cg16722536 | T-box 1 | TBX1 | −17684 | 0.020172 | |
| cg12788467 | HNF1 homeobox B | TCF2 | −2542.91 | 0.037723 | |
| cg19352038 | Paired box 3 | PAX3 | −27179.6 | 0.013469 | |
| cg07403255 | Paired box 8 | PAX8 | −6.64E+06 | 0.022095 | |
| Apoptosis | cg06493930 | Growth arrest-specific 2 | GAS2 | −699.226 | 0.021412 |
| cg05924583 | Tumor protein p73 | TP73 | −211.453 | 0.027257 | |
| Transport | cg27461196 | FXYD domain containing ion transport regulator 1 | FXYD1 | −1.94E+21 | 0.048906 |
| cg04502814 | Selenoprotein P, plasma, 1 | SEPP1 | −1.24E+13 | 0.033794 | |
| cg16509045 | transient receptor potential cation channel, subfamily M, member 6 | TRPM6 | −600.168 | 0.030977 | |
| Metabolism | cg10432859 | UDP glucuronosyltransferase 1 family, polypeptide A7 | UGT1A7 | −11322.6 | 0.017452 |
| Unknown | cg01813965 | Coiled-coil domain containing 135 | C16orf50 | −9979.22 | 0.009274 |
| cg18752880 | C1q and tumor necrosis factor related protein 3 | C1QTNF3 | −1.03E+06 | 0.028981 | |
| cg24691461 | cerebral cavernous malformation 2-like, CCM2L | C20orf160 | −4898.6 | 0.00563 | |
| cg02654291 | chromosome 9 open reading frame 64 | C9orf64 | −254582 | 0.016801 | |
| cg21489873 | MIR600 host gene (non-protein coding) | C9orf45 | −2.41E+19 | 0.049695 | |
| cg15679095 | GLTSCR1-like | KIAA0240 | −6.85E+31 | 0.034122 | |
| cg02077702 | immunoglobulin superfamily containing leucine-rich repeat | ISLR | −1.13E+23 | 0.030866 | |
| cg19170321 | Calpain 3, p94 | CAPN3 | −4.66E+13 | 0.0233799 | |
DISCUSSION
We identified differentially expressed genes in CAF primarily cultured from human colorectal cancer specimens compared to NF from normal/non-malignant colorectal tissue obtained from surgical resections. Although primary cultures from tissue explants may be biased by clonal selection [10], it assures higher purity of the tissue population (fibroblasts) studied. Laser capture microdissection will allow for isolating stromal tissue components in situ, but introduces confounders, such as the inclusion of other cells from the stroma, e.g., endothelial cells and immune cells.
Importantly, the tumor-promoting phenotype persists when early passage CAF are cultured in absence of carcinoma cells [17, 18], suggesting hereditable changes in CAF. However, genetic mutations in CAF have been found to be largely the result of experimental artifact [11]. Both DNA methylation and miRNA-mediated epigenetic changes have been identified in CAF [7, 12], but our understanding of genotypic differences between colorectal CAF versus NF are limited and require further study.
Chang et al. [19] cultured fibroblasts in 10% fetal bovine serum from ten different anatomical sites and identified 512 genes, which he named “the fibroblast core serum response (CSR) genes”. The CSR genes resemble a wound response signature. The authors showed that the CSR signature was present in cancer tissues from breast, lung, prostate, gastric and hepatocellular origin, but not in corresponding normal tissue [19]. However, Troester and colleagues [20] showed that the CSR signature was prominent in “normal” breast tissue from the margin of breast cancers, but not in tissue from reduction mammoplasties. Thus, if applied colorectal cancers, the specificity of a CSR gene set may not accurately distinguish NF and CAF. Furthermore, the CSR gene set is not specific for CRC, since multiple solid tumors were found to have the CSR gene signature. Analysis of our list of differentially expressed genes between CAF and NF using Ingenuity iReport™ demonstrated that CAF are involved in cell migration, invasion and Wnt/β-catenin signaling, as may be predicted based on the known functions of fibroblasts, similar to the functions of the genes on the CSR list. Additionally, unsupervised analyses of our gene list by iReport™ identified “Top Diseases” to be “Cancer”, and specifically “Colorectal Cancer”/“Metastatic Colorectal Cancer”. Importantly, our analysis identified differentially expressed genes that demonstrate both tissue specificity (colorectum) and also distinguishes between NF versus CAF.
CONCLUSIONS
The study of gene expression and its regulation in colorectal CAF is critical to the understanding of how the cancer microenvironment facilitates carcinogenesis and promotes cancer metastasis. Using the Illumina Human Methylation27 BeadChip, we further identified CAF genes that may be regulated by promoter regulation. One limitation of our study is that other mechanisms of epigenetic regulation of gene expression were not studied (for example, histone modification). Additionally, followup studies will be needed for independent technical validations of gene expression and the methylation of CpG sites, and verification of the gene expression-promoter methylation relationships in an independent set of colorectal NF and CAF. Future studies to identify specific epigenetic step-wise changes in the transition from NF to CAF in colorectal cancer are needed to develop therapies that target CAF in the cancer microenvironment [21].
Supplementary Material
ACKNOWLEDGEMENTS
This work was supported by grants from the National Institutes of Health (K08CA125209 [CC] and T32DK007639 [AM, FJB; PI: MRH]) and Society of Surgical Oncology Clinical Investigator Award (CC).
We thank the Mind Research Network; Neurogenetics Core Lab (Albuquerque, NM) for their assistance with the Illumina arrays. We are grateful to Drs. Akaash Gajar, Dennis Gore, and Guillermo Gomez in the Department of Surgery for their assistance in identifying discarded colorectal cancer tissue after surgical resection, and Eileen Figueroa and Steve Schuenke for their assistance with manuscript preparation.
LIST OF ABBREVIATIONS
- CRC
colorectal cancer
- CAF
carcinoma-associated fibroblasts
Footnotes
CONFLICTS OF INTEREST
The authors have no conflicts of interest.
Author contributions:
Conception and design (CC, MRH JRC, TW), analysis and interpretation (CC, JRC, AM), data collection (CC, JRC, ME, FJB, AM) writing the article (CC, AM), critical revision of the article (n/a) and obtaining funding (CC, MRH).
REFERENCES
- 1.De Wever O, Demetter P, Mareel M, Bracke M. Stromal myofibroblasts are drivers of invasive cancer growth. Int J Cancer. 2008;123(10):2229–2238. doi: 10.1002/ijc.23925. [DOI] [PubMed] [Google Scholar]
- 2.Huijbers A, Tollenaar RA, v Pelt GW, et al. The proportion of tumor-stroma as a strong prognosticator for stage II and III colon cancer patients: validation in the VICTOR trial. Ann Oncol. 2013;24(1):179–185. doi: 10.1093/annonc/mds246. [DOI] [PubMed] [Google Scholar]
- 3.Ngan CY, Yamamoto H, Seshimo I, et al. Quantitative evaluation of vimentin expression in tumour stroma of colorectal cancer. Br J Cancer. 2007;96(6):986–992. doi: 10.1038/sj.bjc.6603651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wikberg ML, Edin S, Lundberg IV, et al. High intratumoral expression of fibroblast activation protein (FAP) in colon cancer is associated with poorer patient prognosis. Tumour Biol. 2013;34(2):1013–1020. doi: 10.1007/s13277-012-0638-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Olumi AF, Grossfeld GD, Hayward SW, Carroll PR, Tlsty TD, Cunha GR. Carcinoma-associated fibroblasts direct tumor progression of initiated human prostatic epithelium. Cancer Res. 1999;59(19):5002–5011. doi: 10.1186/bcr138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kuperwasser C, Chavarria T, Wu M, et al. Reconstruction of functionally normal and malignant human breast tissues in mice. Proc Natl Acad Sci U S A. 2004;101(14):4966–4971. doi: 10.1073/pnas.0401064101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hu M, Yao J, Cai L, et al. Distinct epigenetic changes in the stromal cells of breast cancers. Nat Genet. 2005;37(8):899–905. doi: 10.1038/ng1596. [DOI] [PubMed] [Google Scholar]
- 8.Kurose K, Gilley K, Matsumoto S, Watson PH, Zhou XP, Eng C. Frequent somatic mutations in PTEN and TP53 are mutually exclusive in the stroma of breast carcinomas. Nat Genet. 2002;32(3):355–357. doi: 10.1038/ng1013. [DOI] [PubMed] [Google Scholar]
- 9.Weber F, Shen L, Fukino K, et al. Total-genome analysis of BRCA1/2-related invasive carcinomas of the breast identifies tumor stroma as potential landscaper for neoplastic initiation. Am J Hum Genet. 2006;78(6):961–972. doi: 10.1086/504090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jiang L, Gonda TA, Gamble MV, et al. Global hypomethylation of genomic DNA in cancer-associated myofibroblasts. Cancer Res. 2008;68(23):9900–9908. doi: 10.1158/0008-5472.CAN-08-1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Campbell I, Polyak K, Haviv I. Clonal mutations in the cancer-associated fibroblasts: the case against genetic coevolution. Cancer Res. 2009;69(17):6765–6769. doi: 10.1158/0008-5472.CAN-08-4253. [DOI] [PubMed] [Google Scholar]
- 12.Mitra AK, Zillhardt M, Hua Y, et al. MicroRNAs reprogram normal fibroblasts into cancer-associated fibroblasts in ovarian cancer. Cancer Discov. 2012;2(12):1100–1108. doi: 10.1158/2159-8290.CD-12-0206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chao C, Tallman ML, Ives KL, Townsend CM, Jr, Hellmich MR. Gastrointestinal hormone receptors in primary human colorectal carcinomas. J Surg Res. 2005;129(2):313–321. doi: 10.1016/j.jss.2005.04.038. [DOI] [PubMed] [Google Scholar]
- 14.Lax S, Schauer G, Prein K, et al. Expression of the nuclear bile acid receptor/farnesoid X receptor is reduced in human colon carcinoma compared to nonneoplastic mucosa independent from site and may be associated with adverse prognosis. Int J Cancer. 2012;130(10):2232–2239. doi: 10.1002/ijc.26293. [DOI] [PubMed] [Google Scholar]
- 15.Modica S, Murzilli S, Salvatore L, Schmidt DR, Moschetta A. Nuclear bile acid receptor FXR protects against intestinal tumorigenesis. Cancer Res. 2008;68(23):9589–9594. doi: 10.1158/0008-5472.CAN-08-1791. [DOI] [PubMed] [Google Scholar]
- 16.Jacobs ET, Martinez ME, Campbell PT, et al. Genetic variation in the retinoid X receptor and calcium-sensing receptor and risk of colorectal cancer in the Colon Cancer Family Registry. Carcinogenesis. 2010;31(8):1412–1416. doi: 10.1093/carcin/bgq127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Orimo A, Gupta PB, Sgroi DC, et al. Stromal fibroblasts present in invasive human breast carcinomas promote tumor growth and angiogenesis through elevated SDF-1/CXCL12 secretion. Cell. 2005;121(3):335–348. doi: 10.1016/j.cell.2005.02.034. [DOI] [PubMed] [Google Scholar]
- 18.Ishii G, Hashimoto H, Asada K, et al. Fibroblasts associated with cancer cells keep enhanced migration activity after separation from cancer cells: a novel character of tumor educated fibroblasts. Int J Oncol. 2010;37(2):317–325. doi: 10.3892/ijo_00000680. [DOI] [PubMed] [Google Scholar]
- 19.Chang HY, Sneddon JB, Alizadeh AA, et al. Gene expression signature of fibroblast serum response predicts human cancer progression: Similarities between tumors and wounds. Plos Biol. 2004;2(2):206–214. doi: 10.1371/journal.pbio.0020007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Troester MA, Lee MH, Carter M, et al. Activation of host wound responses in breast cancer microenvironment. Clin Cancer Res. 2009;15(22):7020–7028. doi: 10.1158/1078-0432.CCR-09-1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Takebe N, Ivy P, Timmer W, Khan N, Schulz T, Harris PJ. Review of cancer - associated fibroblasts and therapies that interfere with their activity. Tumor Microenvironment and Therapy. 2013;1:19–36. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.