Abstract
Enzymes are typically highly stereoselective catalysts that enforce a reactive conformation on their native substrates. We report here a rare example where the substrate controls the stereoselectivity of an enzyme-catalyzed Michael-type addition during the biosynthesis of lanthipeptides. These natural products contain thioether crosslinks formed by cysteine attack on dehydrated Ser and Thr residues. We demonstrate that several lanthionine synthetases catalyze highly selective anti additions in which the substrate (and not the enzyme) determines whether the addition occurs from the Re or Si face. A single point mutation in the peptide substrate completely inverted the stereochemical outcome of the enzymatic modification. Quantum mechanical calculations reproduced the experimentally observed selectivity and suggest that conformational restraints imposed by the amino acid sequence on the transition states determine the face selectivity of the Michael-type cyclization.
The stereochemical outcome of chemical reactions between chiral molecules in which one or more new stereocenters are generated is often governed by the stereochemistry of the substrate or reagent, resulting in substrate or reagent controlled processes1. For enzymatic reactions, the well-organized chiral environment of the enzyme active site typically results in reagent control for physiological processes. In this study, we discovered a rare case where the substrate controls the stereochemical outcome of an enzyme-catalyzed Michael-type addition during the biosynthesis of lanthipeptides.
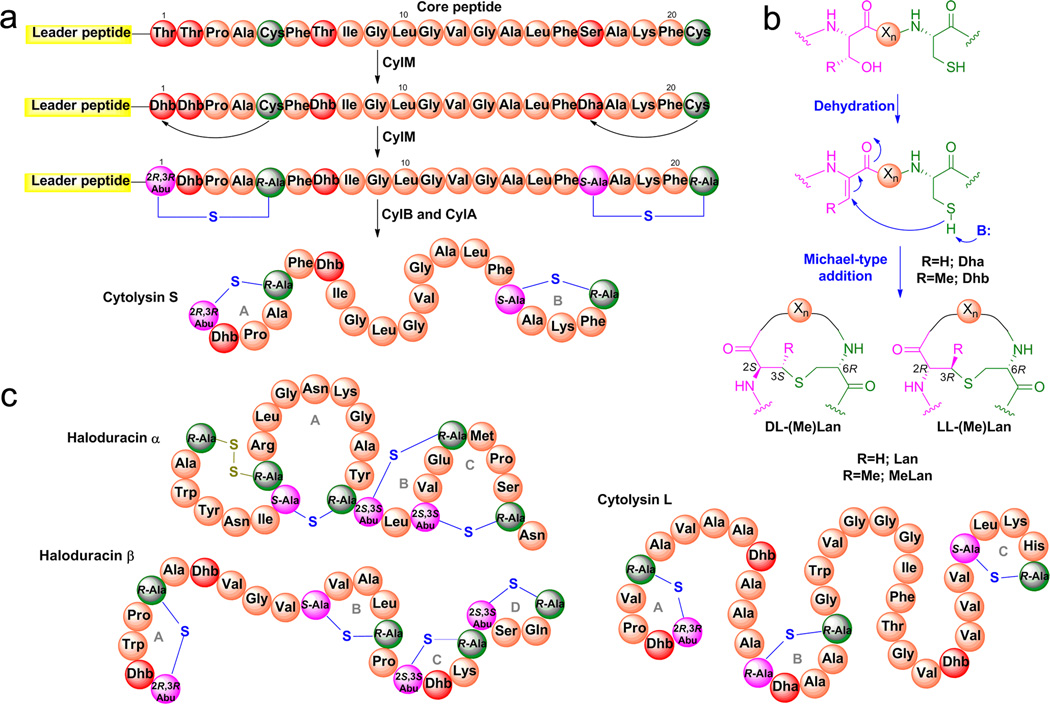
Lanthipeptides are a large family of ribosomally synthesized and post-translationally modified peptides (RiPPs), which constitute a growing class of natural products2. Their precursor peptides (called LanAs) consist of an N-terminal leader peptide that is essential for recognition by the enzymes that carry out the post-translational modifications3 (Fig. 1a). The latter take place in the C-terminal core region of the precursor peptide, and involve dehydration of serine and threonine residues to generate dehydroalanine (Dha) and dehydrobutyrine (Dhb), respectively (Fig. 1a and 1b). Subsequently, the thiols from cysteine residues are added to the newly formed double bonds in Dha and Dhb via a Michael-type addition. The resulting thioether bridges are termed lanthionine (Lan) and methyllanthionine (MeLan), respectively (Fig. 1b). In class II lanthipeptides, both the dehydration and cyclization reactions are catalyzed by one bi-functional enzyme that is generically termed LanM (e.g. CylM in Fig. 1a)4,5. Many lanthipeptides display antimicrobial activity and are designated lantibiotics6, with some members potently inhibiting the growth of a broad range of pathogens7.
Figure 1. Biosynthesis of class II lanthipeptides.
(a) Generic pathway of class II lanthipeptide biosynthesis with cytolysin S as an example. The CylLS peptide is synthesized by the ribosome as a linear precursor peptide with an N-terminal leader peptide and a C-terminal core peptide. The dehydration of Ser and Thr residues to Dha and Dhb residues as well as the Michael-type addition by Cys residues onto these α,β-unsaturated amino acids is catalyzed by the bi-functional lanthionine synthetase CylM. The leader peptide is then removed by the proteases CylB and CylA, affording mature cytolysin S. (b) Dehydration and cyclization reactions catalyzed by LanM enzymes that result in (Me)Lan residues with the canonical DL (i.e. D configuration at the α carbon originating from Ser or Thr and L configuration at the α carbon of the former Cys) and unusual LL (L configuration for both α carbons) stereochemistries. (c) The structures of haloduracin (Halα and Halβ) and cytolysin L. Abu, α-aminobutyric acid.
Lanthipeptides have been extensively studied over the past 30 years, with structural information available for a subset of compounds8. Until recently, it was generally assumed that the cysteine addition always gave (2S, 6R)-Lan (hereafter called DL-Lan) and (2S, 3S, 6R)-MeLan (hereafter DL-MeLan) (Fig. 1b). This assumption was recently challenged by the discovery of (2R, 3R, 6R)-MeLan (LL-MeLan, Fig. 1b) in two class II lantibiotics - the cytolysin from Enterococcus faecalis and haloduracin produced by Bacillus halodurans9. The enterococcal cytolysin consists of two peptides called cytolysin L (CylLL”, Figure 1c) and cytolysin S (CylLS”, Figure 1a), which are generated from the precursor peptides CylLL and CylLS, respectively, by a single lanthionine synthetase CylM. The production of cytolysin enhances virulence in infection models of E. faecalis and is associated with acute patient mortality10–13. Haloduracin is an antimicrobial substance that also consists of two peptides (Fig. 1c)14. Based on sequence homology of the rings containing the LL-MeLan residues in cytolysin and haloduracin, we have suggested that a Dhb-Dhb-Xxx-Xxx-Cys motif (Xxx represents amino acids other than Dha, Dhb and Cys) (Fig. 1) induces the formation of the unusual MeLan stereochemistry9. In this model, the LL stereochemistry is formed by anti-addition of cysteine to the Re face of the alkene because of a conformational preference of the substrate that is unique to this motif9.
These findings have raised a number of intriguing questions. First, given that cytolysin and haloduracin contain both LL- and DL-(Me)Lan, how can one enzyme catalyze similar conjugate additions with different stereochemistries in a single polypeptide substrate? Second, does the substrate sequence indeed govern the diastereoselectivity? And third, is it possible to manipulate the stereochemical outcome by mutating the peptide substrate? In this study, we show that the formation of LL-MeLan from the Dhb-Dhb-Xxx-Xxx-Cys sequence is not specific to the enzymes involved in cytolysin or haloduracin biosynthesis. Furthermore, we demonstrate that a Dhb-to-Ala mutation at the second position of the motif results in MeLan formation with the canonical DL stereochemistry, supporting the substrate control hypothesis. Quantum mechanical simulations of dehydrated peptides confirm an intrinsic preference for Re face or Si face addition depending on the sequence of the substrate peptide.
Results
Generality of LL-MeLan formation from Dhb-Dhb-Xxx-Xxx-Cys
To test the hypothesis that the substrate peptide sequence containing the Dhb-Dhb-Xxx-Xxx-Cys motif determines the formation of LL stereochemistry independent of the identity of the lanthionine synthetase, we generated chimeric peptides with the CylLS core peptide connected to leader peptides from different class II lanthipeptides. These leader peptides were expected to be recognized by their cognate lanthionine synthetases that carry out the dehydration and cyclization reactions in the core peptide (Fig. 2a). The gene halA2-cylLS that encodes a chimeric peptide consisting of the leader peptide of the haloduracin β (Halβ) precursor peptide (HalA2) fused to the CylLS core peptide (Fig. 2a) was co-expressed in E. coli with halM2 encoding the Halβ synthetase HalM215. Modified HalA2-CylLS was purified and analysis by matrix-assisted laser desorption ionization time-of-flight (MALDI-TOF) mass spectrometry (MS) showed the desired four dehydrations (Supplementary Results, Supplementary Fig. S1). The modified chimeric peptide was hydrolyzed in 6 M HCl to individual amino acid residues, which were derivatized and analyzed by gas chromatography (GC) MS using a chiral GC column to reveal the stereochemistry of the (Me)Lan residues9,16. Signals corresponding to LL-MeLan and DL-Lan were observed as the predominant products by comparison with synthetic standards derivatized in the same way (Fig. 2b, 2c; Table 1 and Supplementary Fig. S2). Hence, HalM2 catalyzes the cyclization of the CylLS core peptide with the same stereochemical selectivity as CylM. This result was perhaps not too surprising since HalM2 converts a Dhb-Dhb-Trp-Pro-Cys sequence into an LL-MeLan cross-link in its native substrate HalA2 (Fig. 1c)9. We therefore turned to LanM enzymes that only form DL-(Me)Lan in their natural substrates. The lanthionine synthetase LtnM2 involved in the biosynthesis of the lantibiotic lacticin 3147 A217 was evaluated next. An LtnA2-CylLS peptide consisting of the leader peptide from the natural LtnM2 substrate (LtnA2) and the core peptide from CylLS was co-expressed with LtnM2 in E. coli. MALDI-TOF MS analysis illustrated the desired four dehydrations (Supplementary Fig. S3). Hydrolysis and derivatization of the modified LtnA2-CylLS peptide once again resulted in almost exclusive LL-MeLan and DL-Lan signals in GC-MS experiments (Fig. 2b, 2c; Table 1 and Supplementary Fig. S4), demonstrating that LtnM2 also preferentially forms the LL stereochemistry when guided by the substrate sequence. We then chose ProcM as our next LanM target. ProcM is a lanthionine synthetase with 29 natural substrates (ProcAs) that exhibits high substrate tolerance18. ProcM can generate many different ring topologies, and based on the currently structurally characterized ProcA products, they all have the DL-(Me)Lan stereochemistry (Supplementary Fig. S5)19. It has been suggested that it is the sequence of the ProcA peptides, rather than the ProcM enzyme, that primarily determines the cyclization patterns20. As such, ProcM would be the perfect candidate to test the hypothesis that the unusual LL-MeLan stereochemistry produced from the Dhb-Dhb-Xxx-Xxx-Cys motif is determined by the substrate. In this study, the ProcA3.2 leader peptide was chosen for a chimeric ProcA3.2-CylLS peptide and the latter was co-expressed with ProcM in E. coli. The modified ProcA3.2-CylLS peptide afforded several products that differed in the extent of dehydration with 2- and 4-fold dehydrated peptides as the major products (Supplementary Fig. S6). The modified peptides were hydrolyzed, and the resulting amino acids were derivatized and subjected to GC-MS analysis. In this case, we observed an LL-MeLan peak with a relatively low intensity as well as a DL-Lan signal (Fig. 2b, 2c; Table 1 and Supplementary Fig. S7). The low signal intensity of the derivatized MeLan compared with Lan could be the result of the observed incomplete dehydration of the non-cognate core peptide or because of partial cyclization by ProcM. Regardless, the MeLan that was generated clearly was formed with high selectivity for the LL stereochemistry. Collectively, these results demonstrate that class II LanM enzymes have a general preference for converting sequences containing the Dhb-Dhb-Xxx-Xxx-Cys motif into LL-MeLan (Table 1).
Figure 2. GC-MS analysis of in vivo modified HalA2-CylLS, LtnA2-CylLS and ProcA3.2-CylLS chimeric peptides.
(a) HalA2-CylLS, LtnA2-CylLS and ProcA3.2-CylLS chimeras that were modified in E. coli by the lanthionine synthetases HalM2, LtnM2, and ProcM, respectively. A generic protein structure is shown for the three enzymes. GC-MS traces are shown for hydrolyzed and derivatized MeLan residues (b) and Lan residues (c) from modified HalA2-CylLS (blue), LtnA2-CylLS (red), and ProcA3.2-CylLS (magenta). All intensities were normalized with respect to the predominant product. For co-injection traces with synthetic (Me)Lan standards, see Supplementary Fig. S2, S4 and S7.
Table 1.
Stereochemical outcome of lanthipeptide precursor peptides modified by different lanthionine synthetases.
| Peptide substrate |
Lanthionine synthetase |
Configuration of (Me)Lan residue | Quantum mechanical prediction for A ring |
|
|---|---|---|---|---|
| A ringa | Other rings | |||
| CylLS | CylMb | LL | DL | LL |
| HalA2-CylLS | HalM2 | LL | DL | LL |
| LtnA2-CylLS | LtnM2 | LL | DL | LL |
| ProcA3.2-CylLS | ProcM | LL | DL | LL |
| HalA2 | HalM2b | LL | DL | LL |
| HalA2-T2A | HalM2 | DL | DL | DL |
The ring that contains the Dhb-Dhb-Xxx-Xxx-Cys motif.
Reported previously9.
Mutation of the Dhb-Dhb-Xxx-Xxx-Cys motif
To probe whether the two consecutive dehydro amino acids in the motif impart formation of the LL stereochemistry, Thr2 in HalA2 was mutated to Ala. The HalA2-T2A peptide was co-expressed with its synthetase HalM2 in E. coli, resulting in the desired elimination of six water molecules. After leader peptide removal via an engineered Factor Xa cleavage site15, the modified HalA2-T2A core peptide was subjected to tandem MS (MS/MS) analysis, which confirmed the formation of the expected ring pattern (Fig. 3a). The modified peptide was then hydrolyzed, and the resulting amino acids were derivatized and analyzed by GC-MS. A DL-MeLan signal was almost exclusively observed, compared with a 2:1 ratio of DL:LL MeLan signals observed for the wild-type HalA2 peptide after HalM2 modification (the LL-MeLan residue originating from the A ring and two DL-MeLan residues originating from the C and D rings) (Fig. 3b, Table 1 and Supplementary Fig. S8)9; as expected the DL-configuration of the Lan residue (ring B) remained unchanged in the mutant (Supplementary Fig. S8). To rule out the possibility that the A ring of the HalA2-T2A mutant was not properly cyclized, which would mean that the observed DL-MeLan signal originated solely from the C and D rings, we used trypsin to remove the leader peptide and analyzed the core peptide mass with liquid chromatography MS (LC-MS). If the A ring of the HalA2-T2A mutant was not properly cyclized, the core peptide would start with a Dhb, which would hydrolyze to a ketone after the removal of the leader peptide, giving a mass increase of 1 Da14. We chose trypsin over Factor Xa because trypsin exhibits minimal bias in terms of digestion efficiency when the cleavage site is located next to a dehydrated amino acid21. The observed mass for the HalA2-T2A core peptide was 2318.0476 Da (Fig. 3c), in agreement with the calculated mass of the cyclized peptide (2318.0469 Da). We did not observe any peak in the LC trace with a mass corresponding to the non-cyclized peptide (calculated 2319.0309 Da). Collectively, these results show that the A ring of the HalA2-T2A mutant was successfully cyclized and had the DL stereochemistry, strongly suggesting that the 2nd Dhb in the Dhb-Dhb-Trp-Pro-Cys sequence is critical for the formation of LL-MeLan by HalM2. By mutation of the 2nd Dhb to Ala, the stereochemistry of the MeLan residue in the A ring of HalA2 could be successfully changed from LL to DL.
Figure 3. Structural characterization of HalM2-modified HalA2-T2A peptide.
(a) MS/MS analysis of the HalA2-T2A core peptide modified by HalM2 in E. coli. Fragmentation is suppressed in the sequences spanning residues 1–5, 11–15, 17–20 and 21–24 as a result of ring formation. The b3 ion is attributed to in-ring fragmentation, which is rare but has also been observed for cytolysin, nisin and geobacillin9,15,34. (b) GC-MS traces for hydrolyzed and derivatized MeLan residues from HalM2-modified HalA2 peptide (blue) and HalM2-modified HalA2-T2A peptide (red). For co-injection traces of hydrolyzed and derivatized (Me)Lan residues obtained from modified HalA2-T2A peptide with synthetic (Me)Lan standards, see Supplementary Fig. S8. (c) Electrospray ionization (ESI) quadrupole TOF mass spectrum for modified HalA2-T2A core peptide. The [M+2H]2+ ion was observed with an m/z value of 1160.0316 (calculated m/z of cyclized HalA2-T2A core peptide with 2 protons: 1160.0313).
Computational studies on the inherent cyclization stereoselectivity
Computational simulations have been successfully applied previously to correlate inherent substrate reactivity with observed enzymatic selectivities22,23. To investigate the possible origins of the experimentally observed sequence-dependent stereochemical preferences in this work, the cyclization reactions were studied computationally for the A rings of the CylLS and HalA2 core peptides. The high flexibility of the substrates confers the stereo-determining transition structures (TS) a potentially vast conformational diversity; therefore, a multilevel approach combining molecular dynamics (MD) and quantum mechanical (QM) calculations of optimized TS was employed to locate the minimum energy pathways for both Re and Si face additions for each model peptide.
The CylLS sequence leading to its A ring was very rigid in the MD simulations (Fig. 4a) as depicted by the Ramachandran plots of the residues involved in the macrocyclization (Fig. 4b–f). In the experimentally favored Re-face approach, the Dhb-Dhb and Pro-Ala sequences act synergistically to generate a stable secondary structure with characteristics of both α-helix (i+4→i hydrogen bonding) and 310-helix (i+3→i hydrogen bonding), as previously found for peptides containing α,β-dehydro amino acids24,25. The majority of the Cys population (64%) was also found in a helix-like secondary structure, whereas the non-reactive Dhb attained a distorted γ-turn-like arrangement in which the α,β-unsaturated carbonyl system deviates from planarity due to hydrogen bonding interactions and the influence of the neighboring proline. Well-defined hydrogen bonds between the carbonyl of the reactive Dhb and the amide hydrogens of Ala (i+3) and Cys (i+4) were observed along the entire MD trajectory. Additional hydrogen bonds between the carbonyl of the adjacent Dhb (i+1) and the amide hydrogens of Ala and Cys further stabilize this tightly folded motif. In contrast, the secondary structure of the CylLS A-ring sequence for Si face approach is more flexible. In order for the nucleophilic cysteine to approach the Si face of the alkene, the reactive Dhb changes from an α-helical (s-cis) to a PPII (s-trans) conformation (the latter having two mirror image populations, Figure 4b), forced by the rigidity of the proline backbone. This proline is in α-helix and PPII conformations in a 70:30 ratio, and the adjacent Dhb is also flexible, showing two main populations with γ-turn (distorted s-trans, 72%) or αL-helix-like (distorted s-cis, 23%) conformations. The four hydrogen bonds described above are frequently disrupted, which correlates with transitions between the described conformations (Supplementary Fig. S9). Overall, this higher flexibility and the disruption of the tight hydrogen bond network suggest that the Si face approach might be disfavored for the Dhb-Dhb-Pro-Ala-Cys sequence.
Figure 4. Secondary structure of the A ring of the CylLS core sequence derived from MD simulations.
(a) Overlay of 20 representative conformers obtained from the accelerated MD (aMD) trajectory restrained to the Re face approach. (b–f) Ramachandran plots obtained through the simulations for the five residues involved in the macrocyclization. The lowest energy QM optimized transition structures for both approximations are shown as blue squares (Re) and red circles (Si). The higher rigidity of the peptide backbone in the most stable Re face approach is apparent. Also, MD simulations show a change in the conformation of the reactive Dhb from α-helical (s-cis) to PPII (s-trans) in the Si face approach, while the remaining amino acids qualitatively preserve the same conformation.
These preliminary observations were confirmed by the large difference in the activation barriers (ΔΔG‡) calculated for the Michael-type additions to the Re and Si faces of Dhb, which favors the diastereoselective formation of LL-MeLan by 3.2 kcal mol−1 (>99:1 d.r. at 25 °C, Supplementary Table 2), in agreement with our experimental observations (Table 1 and Fig. 5). This preference for the Re face approach is rationalized in terms of the ability of the substrate to form a rigid oxyanion hole in this orientation (Fig. 5), allowing efficient stabilization of the incipient negative charge on the carbonyl oxygen by two amide hydrogens. Of note, the backbone amides stabilize the enolate sufficiently to make the nucleophilic attack slightly exergonic, which is not the normal trend for thio-Michael additions in solution26. Additionally, in the favored Re face approach, the enamide is in the more reactive s-cis conformation, which may further stabilize the corresponding TS compared to that of the Si face approach, in which the enamide is in a less reactive s-trans conformation26.
Figure 5. Lowest energy quantum mechanical transition structures for the Re and Si face Michael-type additions that generate the A ring in the CylLS core sequence.
Geometries were optimized at the IEF-PCM/ω-B97X-D/6-31G(d) level and the relative activation free energies (ΔΔG‡, in kcal mol−1) were calculated at the IEF-PCM/M06-2X/6-311+G(2d,p) level. The depicted kinetic diastereomeric percentage was derived from these relative activation energies. The presence of a tight hydrogen bond network in the Re face approach, which creates a rigid oxyanion hole to stabilize the developing negative charge on the carbonyl oxygen, stabilizes this transition state significantly and favors Re-face attack to occur almost exclusively.
The shift of the proline from the i+2 to the i+3 position with respect to the reacting Dhb, as found in the HalA2 sequence (Dhb-Dhb-Trp-Pro-Cys), completely changes the secondary structure in both reactive trajectories (Supplementary Fig. S10). In the Re face approach, the reacting Dhb still exhibits an α-helical conformation (planar s-cis), but the non-reacting Dhb and Pro adopt mainly an inverse PPII conformation (planar s-trans) due to the release of conformational constraints imposed in CylLS by the neighboring proline and the hydrogen bond network. The ability to attain a 310-helix is seriously hampered by the impossibility to establish i+3→i hydrogen bonds with the reacting Dhb, and therefore the intricate hydrogen bond pattern described above for the CylLS sequence is lost. The secondary structure for the Si face approach is similar, with no persistent hydrogen bonds along the simulation. Unlike in CylLS, the approach of the cysteine to the Si face of Dhb in HalA2 is achieved by maintaining the reactive Dhb in an α-helical type conformation and isomerizing the neighboring Dhb from a PPII (s-trans) to an α-helical (s-cis) conformation. However, the most stable TS for the Michael-type addition differ from those sampled preferentially in the restrained MD for the neutral cysteine (Supplementary Fig. S11). Notably, the lowest energy conformations of the two Dhb residues in the TS are completely reversed from those calculated for the CylLS sequence: s-trans for the reacting Dhb and s-cis for the adjacent Dhb in the Re face approach, and s-cis for both Dhb residues in the Si face approach. Nevertheless, very high stereoselectivity (96:4, see Supplementary Table 2) towards LL-MeLan formation was calculated for this sequence based upon the transition state energies, in agreement with the experiments. It should be noted that the HalA2 sequence does not allow the formation of the tight oxyanion hole observed for CylLS. Consequently, the thio-Michael addition shows the normal endergonic behavior yielding a higher energy enolate, with the overall transformation becoming favored only upon protonation. Interestingly, the HalA2 sequence confers the necessary conformational bias to override the suggested less reactive character of the s-trans enamide for the favored Re approach26.
With the intrinsic preference for the Re face addition established in the A rings of both cytolysin S and Halβ, we next investigated the reversal of stereochemistry in the HalA2-T2A mutant. The secondary structure of the A-ring sequence of the mutant is quite similar to that of HalA2 (Supplementary Fig. S10 & S12), but the mutation has a deep impact on the calculated stereoselectivity, favoring the Si face approach by 2 kcal mol−1 in line with the experimental observations. Intriguingly, the geometries of the lowest energy TS calculated for this sequence are virtually identical to those of the wild-type sequence, but the stereoselectivity is reversed (Table 1 and Supplementary Fig. S13). The presence of an sp3-hybridized alpha carbon in the Ala instead of the planar arrangement in Dhb perturbs the peptide backbone very slightly, but enough to alter the electrostatic environment of the reacting fragments. This change is reflected in the molecular dipole moments (µ) calculated for the lowest energy TS; whereas in HalA2, the TS for Re face approach is slightly more polar (µ = 12.6 D) than for the Si face approach (µ = 11.6 D), for HalA2-T2A both TS are equally polar (µ = 13.7 D and 13.6 D for Re and Si face approaches, respectively). In the absence of the strong structural driving forces observed for CylLS, more polar TS will be preferentially stabilized in moderately polar environments such as the continuum solvent used in the calculations and as found in protein active sites, and this is indeed observed for HalA2. However, when both TS are equally polar as in HalA2-T2A, the intrinsically more reactive character of the enamide s-cis conformation in the Si face approach appears to result in reversal of the stereoselectivity with respect to HalA2. Overall, these striking results reflect the complexity of the factors that govern the facial selectivity in the biosynthesis of lanthipeptides, since dissimilar sequences with significantly different three-dimensional arrangements can provide the same stereoselectivity, but a very subtle change in the sequence with almost no geometric effect completely reverts the face selectivity. These stereodetermining structural preferences were calculated to be intrinsic to the oligopeptide sequences consistent with the observation that the stereochemical outcome is independent of the lanthionine synthetases used.
Discussion
The discovery of LL-MeLan residues in the enterococcal cytolysin and haloduracin formed from a conserved Dhb-Dhb-Xxx-Xxx-Cys substrate motif revealed the first examples of enzymatic formation of non-canonical stereochemistry of MeLan residues. Very recently, LL stereochemistry was also reported for the lantibiotic carnolysin that also contains this motif27. We show in this work that the unusual stereochemistry is inherent to this sequence motif, because four different class II lanthionine synthetases with low sequence identities in their cyclase domains (<25% identity) all converted this motif to LL-MeLan products even though the enzymes install other rings in the same peptide with DL stereochemistry. Indeed, two of the tested lanthionine synthetases only give products with DL stereochemistry in their natural substrates, making it highly unlikely that they would have evolved features that favor LL-MeLan-containing products. Furthermore, the stereoselectivity was changed from LL to DL when the second Dhb in such a motif in HalA2 was mutated to Ala. Thus, these Michael-type additions are rare examples of natural enzymatic processes in which stereochemistry is substrate-controlled; we have not been able to find other such examples, but a related observation has been made in ketoreduction by iterative polyketides synthases where substrate length controlled the face selectivity of hydride transfer28. Of course, reversal of stereoselectivity of enzymatic reactions has been observed routinely with non-natural substrate analogs or mutant enzymes29–32.
The computational studies also support a model in which the observed stereochemistry is determined to a large extent by the sequence of the substrate peptide, even though the transition state structures can differ substantially depending on the location of a Pro residue. We suggest that for the formation of most (Me)Lan residues in lanthipeptides, the enzyme controls the face selectivity of attack and protonation, resulting in the predominantly observed DL stereochemistry. However, we hypothesize that two consecutive Dhb residues in the Dhb-Dhb-Xxx-Xxx-Cys motif result in a conformational preference that is unfavorable for the canonical interaction with the enzyme active site. The enzyme is likely to still activate the Cys residue given the very slow rates of non-enzymatic (Me)Lan formation33, but the face selectivity of attack is now governed by the substrate and not the enzyme. The role of the enzyme might be investigated in more detail if the cyclization reaction could be decoupled from the dehydration reaction such that the non-enzymatic reaction could be studied in isolation. Structural information of substrate bound to the enzyme will be required to determine whether all calculated transition states can be attained in the context of the protein; at present no structures of LanM enzymes are available.
Methods
General methods
All polymerase chain reactions (PCR) were carried out on a C1000™ thermal cycler (Bio-Rad). DNA sequencing was performed by ACGT, Inc. MALDI-TOF MS was carried out on a Bruker Daltonics UltrafleXtreme MALDI TOF/TOF instrument (Bruker). LC-ESI-Q/TOF MS and MS/MS analyses were conducted using a Synapt G1 instrument with an Acquity UPLC system (Waters), which was equipped with a Jupiter Proteo C12 column (5 µm; 90 Å; 100 × 1.0 mm) (Phenomenex). GC-MS analysis was performed in the Mass Spectrometry Laboratory (School of Chemical Sciences, UIUC) on an Agilent 7890 gas chromatograph (Agilent) with a CP-Chirasil-L-Val fused silica column (25 m × 0.25 mm × 0.15 µm) (Agilent). Solid phase extraction was performed with Strata-X polymeric reverse phase columns (Phenomenex).
Materials
All oligonucleotides were obtained from Integrated DNA Technologies and used as received. Restriction endonucleases, DNA polymerases, and T4 DNA ligase were purchased from New England Biolabs. Media components were obtained from Difco Laboratories. Trypsin was purchased from Worthington Biochemical Corporation; Factor Xa was obtained from New England Biolabs and other endoproteinases were ordered from Roche Biosciences.
Strains and plasmids
E. coli DH5α and E. coli BL21 (DE3) cells were used as host for cloning and plasmid propagation, and host for protein expression, respectively. Co-expression vector pRSFDuet-1 was obtained from Novagen.
Construction of plasmids and expression of peptides
See the method section in the Supplementary Information.
GC-MS analysis
The synthesis of Lan and MeLan standards and the preparation of samples for GC-MS analysis were carried out following a reported procedure published elsewhere16,19. The modified full length peptides with their leader peptides attached were hydrolyzed and the resulting solutions were dried and directly used for derivatization. The derivatized samples were analyzed by GC-MS using an Agilent 7890 gas chromatograph equipped with a CP-Chirasil-L-Val fused silica column (25 m × 0.25 mm × 0.15 µm). Samples were dissolved in methanol and introduced to the instrument via a split (1:20) injection at a flow rate of 1.7 mL/min helium gas. The temperature method used was 160 °C for 2 min, 160 °C to 180 °C at 3 °C/min, and 180 °C for 2 min. The selected-ion monitoring (SIM) was achieved by extracting the total ion spectra for peaks containing the characteristic fragment masses of 365 Da for Lan and 379 Da for MeLan residues.
Protease cleavage of the leader peptides
Modified HalA2-CylLS, ProcA3.2-CylLS and HalA2-T2A peptides were dissolved in H2O to a concentration of 3 mg/mL. To a total 200 µL reaction with a final peptide concentration of 200 µM, 20 µL of 500 mM HEPES buffer (pH 7.5) was added, followed by 10 µL of 0.02 mg/mL AspN protease (for HalA2-CylLS), 1 mg/mL GluC protease (for ProcA3.2-CylLS), and 1 mg/mL Factor Xa or 5 mg/mL trypsin (for HalA2-T2A). The protease cleavage reactions were incubated at 25 °C for 6 to 16 h.
LC-ESI-Q/TOF MS and MS/MS analysis
LC-ESI-Q/TOF MS and MS/MS analysis were performed on a Waters Synapt G1 mass spectrometer. For detailed instrument settings, experimental conditions, and data analysis, see the method section in the Supplementary Information.
MD simulations
Constrained MD simulations in water were performed using the AMBER 12 package (Case, D. A. et al., UCSF, 2012). Parameters for non-natural dehydro amino acids were generated using the general AMBER force field (gaff)35, with partial charges set to fit the electrostatic potential generated at the HF/6-31G(d) level by the RESP model36. Water molecules were treated with the SHAKE algorithm. Long-range electrostatic effects were modeled using the particle-mesh-Ewald method37. An 8 Å cutoff was applied to Lennard-Jones and electrostatic interactions. The Langevin equilibration scheme was used to control and equalize the temperature with a 2 fs time step at a constant pressure of 1 atm and temperature of 300 K. Production trajectories were run for 100 ns using McCammon’s accelerated MD (aMD) algorithm38,39.
DFT calculations
All calculations were carried out with the Gaussian 09 package (Frisch, M. J. et al., Gaussian, Inc., 2009). Full geometry optimizations and transition state searches were carried out with the ω-B97X-D range-separated hybrid functional40 and 6-31G(d) basis set. Frequency analyses were carried out at the same level used in the geometry optimizations. Single point energies were calculated with the M06-2X hybrid functional41 on the optimized geometries. Bulk electrostatic effects were considered implicitly during optimization and energy evaluation through the IEF-PCM polarizable continuum model42. Cartesian coordinates, electronic energies, entropies, enthalpies, Gibbs free energies and lowest frequencies of the low energy conformations are available in Supplementary Table 2. For details of the computational studies, see the method section in the Supplementary Information.
Supplementary Material
Acknowledgments
We thank Dr. M. Carmen Martínez-Cuesta for providing the pBAC105 plasmid encoding LtnM2 from Lactococcus lactis IFPL105 and Dr. Leigh Anne Furgerson for constructing the pET15b-HalA2-T2A plasmid. We thank Furong Sun for assistance with GC-MS analysis. This work was supported by the National Institutes of Health (GM 58822 to W.v.d.D. and GM 075962 to K.N.H.). Calculations were performed on the Hoffman2 cluster at UCLA and the Extreme Science and Engineering Discovery Environment (XSEDE), which is supported by the National Science Foundation (OCI-1053575).
Footnotes
Author Contributions
W.T. and W.A.v.d.D. designed the study. W.T. performed all experiments. G.J.O. performed all the theoretical studies. W.T., G.J.O., K.N.H. and W.A.v.d.D. analyzed the data and wrote the manuscript.
Competing financial interests
The authors declare no competing financial interests.
References
- 1.Masamune S, Choy W, Petersen JS, Sita LR. Double asymmetric synthesis and a new strategy for stereochemical control in organic synthesis. Angew. Chem. Int. Ed. Engl. 1985;24:1–30. [Google Scholar]
- 2.Arnison PG, et al. Ribosomally synthesized and post-translationally modified peptide natural products: overview and recommendations for a universal nomenclature. Nat. Prod. Rep. 2013;30:108–160. doi: 10.1039/c2np20085f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yang X, van der Donk WA. Ribosomally synthesized and post-translationally modified peptide natural products: new insights into the role of leader and core peptides during biosynthesis. Chem.- Eur. J. 2013;19:7662–7677. doi: 10.1002/chem.201300401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Siezen RJ, Kuipers OP, de Vos WM. Comparison of lantibiotic gene clusters and encoded proteins. Antonie van Leeuwenhoek. 1996;69:171–184. doi: 10.1007/BF00399422. [DOI] [PubMed] [Google Scholar]
- 5.Xie L, et al. Lacticin 481: in vitro reconstitution of lantibiotic synthetase activity. Science. 2004;303:679–681. doi: 10.1126/science.1092600. [DOI] [PubMed] [Google Scholar]
- 6.Bierbaum G, Sahl HG. Lantibiotics: mode of action, biosynthesis and bioengineering. Curr. Pharm. Biotechnol. 2009;10:2–18. doi: 10.2174/138920109787048616. [DOI] [PubMed] [Google Scholar]
- 7.Cotter PD, Hill C, Ross RP. Bacterial lantibiotics: strategies to improve therapeutic potential. Curr. Protein Pept. Sci. 2005;6:61–75. doi: 10.2174/1389203053027584. [DOI] [PubMed] [Google Scholar]
- 8.Chatterjee C, Paul M, Xie L, van der Donk WA. Biosynthesis and mode of action of lantibiotics. Chem. Rev. 2005;105:633–684. doi: 10.1021/cr030105v. [DOI] [PubMed] [Google Scholar]
- 9.Tang W, van der Donk WA. The sequence of the enterococcal cytolysin imparts unusual lanthionine stereochemistry. Nat. Chem. Biol. 2013;9:157–159. doi: 10.1038/nchembio.1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cox CR, Coburn PS, Gilmore MS. Enterococcal cytolysin: a novel two component peptide system that serves as a bacterial defense against eukaryotic and prokaryotic cells. Curr. Prot. Pept. Sci. 2005;6:77–84. doi: 10.2174/1389203053027557. [DOI] [PubMed] [Google Scholar]
- 11.Huycke MM, Spiegel CA, Gilmore MS. Bacteremia caused by hemolytic, high-level gentamicin-resistant Enterococcus faecalis. Antimicrob. Agents Chemother. 1991;35:1626–1634. doi: 10.1128/aac.35.8.1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Libertin CR, Dumitru R, Stein DS. The hemolysin/bacteriocin produced by enterococci is a marker of pathogenicity. Diagn. Microbiol. Infect. Dis. 1992;15:115–120. doi: 10.1016/0732-8893(92)90033-p. [DOI] [PubMed] [Google Scholar]
- 13.Chow JW, et al. Plasmid-associated hemolysin and aggregation substance production contribute to virulence in experimental enterococcal endocarditis. Antimicrob. Agents Chemother. 1993;37:2474–2477. doi: 10.1128/aac.37.11.2474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McClerren AL, et al. Discovery and in vitro biosynthesis of haloduracin, a two-component lantibiotic. Proc. Natl. Acad. Sci. U.S.A. 2006;103:17243–17248. doi: 10.1073/pnas.0606088103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shi Y, Yang X, Garg N, van der Donk WA. Production of lantipeptides in Escherichia coli. J. Am. Chem. Soc. 2011;133:2338–2341. doi: 10.1021/ja109044r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu W, Chan AS, Liu H, Cochrane SA, Vederas JC. Solid supported chemical syntheses of both components of the lantibiotic lacticin 3147. J. Am. Chem. Soc. 2011;133:14216–14219. doi: 10.1021/ja206017p. [DOI] [PubMed] [Google Scholar]
- 17.Suda S, Cotter PD, Hill C, Ross RP. Lacticin 3147- biosynthesis, molecular analysis, immunity, bioengineering and applications. Curr. Protein Pept. Sci. 2012;13:193–204. doi: 10.2174/138920312800785021. [DOI] [PubMed] [Google Scholar]
- 18.Li B, et al. Catalytic promiscuity in the biosynthesis of cyclic peptide secondary metabolites in planktonic marine cyanobacteria. Proc. Natl. Acad. Sci. U.S.A. 2010;107:10430–10435. doi: 10.1073/pnas.0913677107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tang W, van der Donk WA. Structural characterization of four prochlorosins: a novel class of lantipeptides produced by planktonic marine cyanobacteria. Biochemistry. 2012;51:4271–4279. doi: 10.1021/bi300255s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang Q, Yu Y, Velásquez JE, van der Donk WA. Evolution of lanthipeptide synthetases. Proc. Natl. Acad. Sci. U. S. A. 2012;109:18361–18366. doi: 10.1073/pnas.1210393109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bindman NA, van der Donk WA. A general method for fluorescent labeling of the N-termini of lanthipeptides and its application to visualize their cellular localization. J. Am. Chem. Soc. 2013;135:10362–10371. doi: 10.1021/ja4010706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Michelson AZ, et al. Gas-phase studies of substrates for the DNA mismatch repair enzyme MutY. J. Am. Chem. Soc. 2012;134:19839–19850. doi: 10.1021/ja309082k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zu L, et al. Effect of isotopically sensitive branching on product distribution for pentalenene synthase: support for a mechanism predicted by quantum chemistry. J. Am. Chem. Soc. 2012;134:11369–11371. doi: 10.1021/ja3043245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zanuy D, Casanovas J, Alemán C. The conformation of dehydroalanine in short homopeptides: molecular dynamics simulations of a 6-residue chain. Biophys. Chem. 2002;98:301–312. doi: 10.1016/s0301-4622(02)00108-4. [DOI] [PubMed] [Google Scholar]
- 25.Mathur P, Ramakumar S, Chauhan VS. Peptide design using alpha,beta-dehydro amino acids: from beta-turns to helical hairpins. Biopolymers. 2004;76:150–161. doi: 10.1002/bip.10571. [DOI] [PubMed] [Google Scholar]
- 26.Krenske EH, Petter RC, Zhu Z, Houk KN. Transition states and energetics of nucleophilic additions of thiols to substituted alpha,beta-unsaturated ketones: substituent effects involve enone stabilization, product branching, and solvation. J. Org. Chem. 2011;76:5074–5081. doi: 10.1021/jo200761w. [DOI] [PubMed] [Google Scholar]
- 27.Lohans CT, Li JL, Vederas JC. Structure and biosynthesis of carnolysin, a homologue of enterococcal cytolysin with D-amino acids. J. Am. Chem. Soc. 2014;136:13150–13153. doi: 10.1021/ja5070813. [DOI] [PubMed] [Google Scholar]
- 28.Zhou H, et al. A fungal ketoreductase domain that displays substrate-dependent stereospecificity. Nat. Chem. Biol. 2012;8:331–333. doi: 10.1038/nchembio.912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.May O, Nguyen PT, Arnold FH. Inverting enantioselectivity by directed evolution of hydantoinase for improved production of L-methionine. Nat. Biotechnol. 2000;18:317–320. doi: 10.1038/73773. [DOI] [PubMed] [Google Scholar]
- 30.Turner NJ. Controlling chirality. Curr. Opin. Biotechnol. 2003;14:401–406. doi: 10.1016/s0958-1669(03)00093-4. [DOI] [PubMed] [Google Scholar]
- 31.Mugford PF, Wagner UG, Jiang Y, Faber K, Kazlauskas RJ. Enantiocomplementary enzymes: classification, molecular basis for their enantiopreference, and prospects for mirror-image biotransformations. Angew. Chem. Int. Ed. 2008;47:8782–8793. doi: 10.1002/anie.200705159. [DOI] [PubMed] [Google Scholar]
- 32.Reetz MT. Laboratory evolution of stereoselective enzymes: a prolific source of catalysts for asymmetric reactions. Angew. Chem. Int. Ed. 2011;50:138–174. doi: 10.1002/anie.201000826. [DOI] [PubMed] [Google Scholar]
- 33.Mukherjee S, van der Donk WA. Mechanistic studies on the substrate-tolerant lanthipeptide synthetase ProcM. J. Am. Chem. Soc. 2014;136:10450–10459. doi: 10.1021/ja504692v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Garg N, Tang W, Goto Y, Nair SK, van der Donk WA. Lantibiotics from Geobacillus thermodenitrificans. Proc. Natl. Acad. Sci. U.S.A. 2012;109:5241–5246. doi: 10.1073/pnas.1116815109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang J, Wolf RM, Caldwell JW, Kollman PA, Case DA. Development and testing of a general amber force field. J. Comput. Chem. 2004;25:1157–1174. doi: 10.1002/jcc.20035. [DOI] [PubMed] [Google Scholar]
- 36.Bayly CI, Cieplak P, Cornell WD, Kollman PA. A well-behaved electrostatic potential based method using charge restraints for deriving atomic charges: the RESP model. J. Phys. Chem. 1993;97:10269–10280. [Google Scholar]
- 37.Darden T, York D, Pedersen L. Particle mesh Ewald: an N-log(N) method for Ewald sums in large systems. J. Chem. Phys. 1993;98:10089–10092. [Google Scholar]
- 38.Hamelberg D, Mongan J, McCammon JA. Accelerated molecular dynamics: a promising and efficient simulation method for biomolecules. J. Chem. Phys. 2004;120:11919–11929. doi: 10.1063/1.1755656. [DOI] [PubMed] [Google Scholar]
- 39.Pierce LC, Salomon-Ferrer RF, de Oliveira CA, McCammon JA, Walker RC. Routine access to millisecond time scale events with accelerated molecular dynamics. J. Chem. Theory Comput. 2012;8:2997–3002. doi: 10.1021/ct300284c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chai JD, Head-Gordon M. Long-range corrected hybrid density functionals with damped atom-atom dispersion corrections. Phys. Chem. Chem. Phys. 2008;10:6615–6620. doi: 10.1039/b810189b. [DOI] [PubMed] [Google Scholar]
- 41.Zhao Y, Truhlar DG. The M06 suite of density functionals for main group thermochemistry, thermochemical kinetics, noncovalent interactions, excited states, and transition elements: two new functionals and systematic testing of four M06-class functionals and 12 other functionals. Theor. Chem. Acc. 2008;120:215–241. [Google Scholar]
- 42.Scalmani G, Frisch MJ. Continuous surface charge polarizable continuum models of solvation. I. General formalism. J. Chem. Phys. 2010;132:114110. doi: 10.1063/1.3359469. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.