Abstract
Background: The automatic interpretation of electrocardiography (ECG) data can provide continuous analysis of heart activity, allowing the effective use of wireless devices such as the Holter monitor. Materials and Methods: We propose an intelligent heartbeat monitoring system to detect the possibility of arrhythmia in real time. We detected heartbeats and extracted features such as the QRS complex and P wave from ECG signals using the Pan–Tompkins algorithm, and the heartbeats were then classified into 16 types using a decision tree. Results: We tested the sensitivity, specificity, and accuracy of our system against data from the MIT-BIH Arrhythmia Database. Our system achieved an average accuracy of 97% in heartbeat detection and an average heartbeat classification accuracy of above 96%, which is comparable with the best competing schemes. Conclusions: This work provides a guide to the systematic design of an intelligent classification system for decision support in Holter ECG monitoring.
Key words: : heartbeat detection, heartbeat classification, decision tree, electrocardiography monitoring
Introduction
The symptoms of arrhythmia are diverse, ranging from minor chest palpitations, chest pain, and fainting (syncope) to sudden heart attack, depending on the type and severity of heart disease.1 Thus, even patients showing mild symptoms of arrhythmia should be diagnosed as early as possible.2 Many patients are unaware of their symptoms and consider it inconvenient to visit a hospital. Moreover, even those who seek diagnosis may show normal cardiac behavior during their visit. For these reasons, there has been a growing demand for remote electrocardiography (ECG) monitoring systems capable of functioning continuously at any location.3 Fortunately, the recently developed Holter monitoring device can be integrated with modern smartphones to serve precisely this function.4 It allows patients to continue their normal routines while long-term ECG information is collected.5
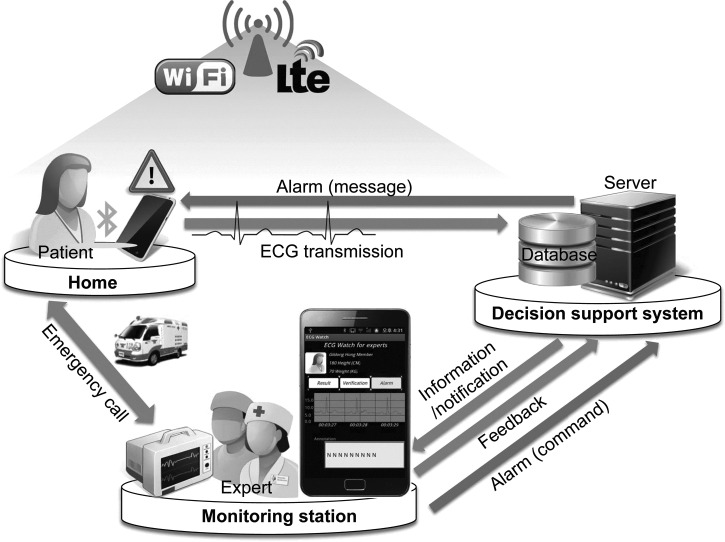
In this article, we propose an intelligent ECG monitoring system to detect the possibility of arrhythmia in real time by analyzing heartbeats in terms of the standard “PQRST” wave features. Figure 1 illustrates the system architecture and the operational scenario of the proposed system for intelligent ECG telemetry. A wearable ECG sensor continuously measures the activity of a patient's heart. The resulting digital stream is transmitted wirelessly to a mobile device (e.g., a smartphone) using a short-range wireless communication technology such as Bluetooth® (Bluetooth SIG, Kirkland, WA) or Zigbee® (Zigbee Alliance, San Ramon, CA).6 The mobile device forwards the data to a decision support system, which saves and analyzes the signal, using long-range wireless technologies such as IEEE 802.11 or long-term evolution (LTE). The results of this analysis are conveyed to medical staff, located at a remote monitoring station or carrying a mobile device, to support diagnosis in real time. If heartbeat features that require immediate attention are detected, the staff are notified immediately. They may advise a hospital visit, by sending a message back to the patient through the decision support system, or take immediate action.
Fig. 1.
System architecture and operational scenario for intelligent electrocardiography (ECG) monitoring. Lte, long-term evolution.
A patient's long-term ECG data are accumulated in a database at a decision support system to obtain his or her baseline ECG patterns. The decision support system uses these data to build a decision tree, based either on feedback from medical experts during an initial learning phase or on domain knowledge about heartbeat classification. This tree allows subsequent ECG data to be classified against the baseline by the decision support system. We use the Pan–Tompkins algorithm7 to extract heartbeat features, and the decision tree is then able to classify beats into 16 types on the basis of these features. The effectiveness of our system has been demonstrated by experiments on data from the MIT-BIH Arrhythmia Database.8
Materials and Methods
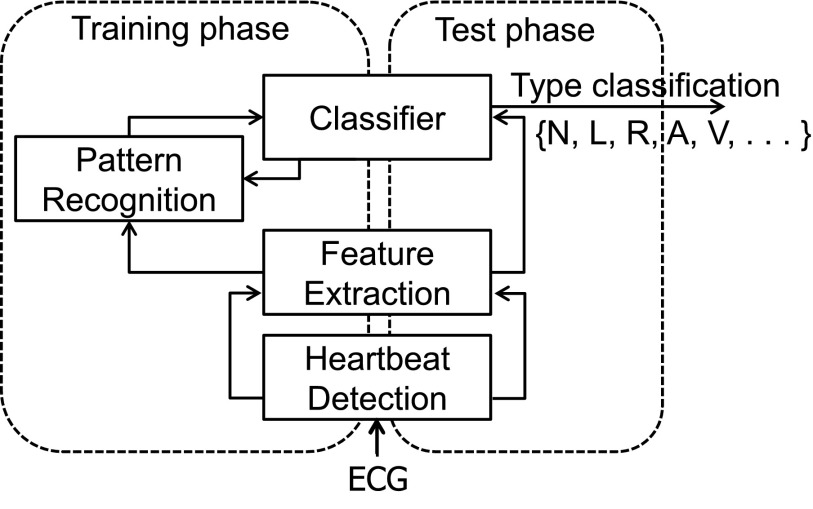
Our system is trained to classify a patient's ECG data by extracting individual heartbeats and their features from the waveform. The system considers both the QRS complex and the P wave, which serve as indicators of ventricular and atrial activity, respectively. Once trained, the system identifies the types of subsequent heartbeats on the basis of their features using a decision tree. An overview of the proposed heartbeat classification system is provided in Figure 2.
Fig. 2.
The proposed heartbeat classification system. ECG, electrocardiography.
Heartbeat Detection
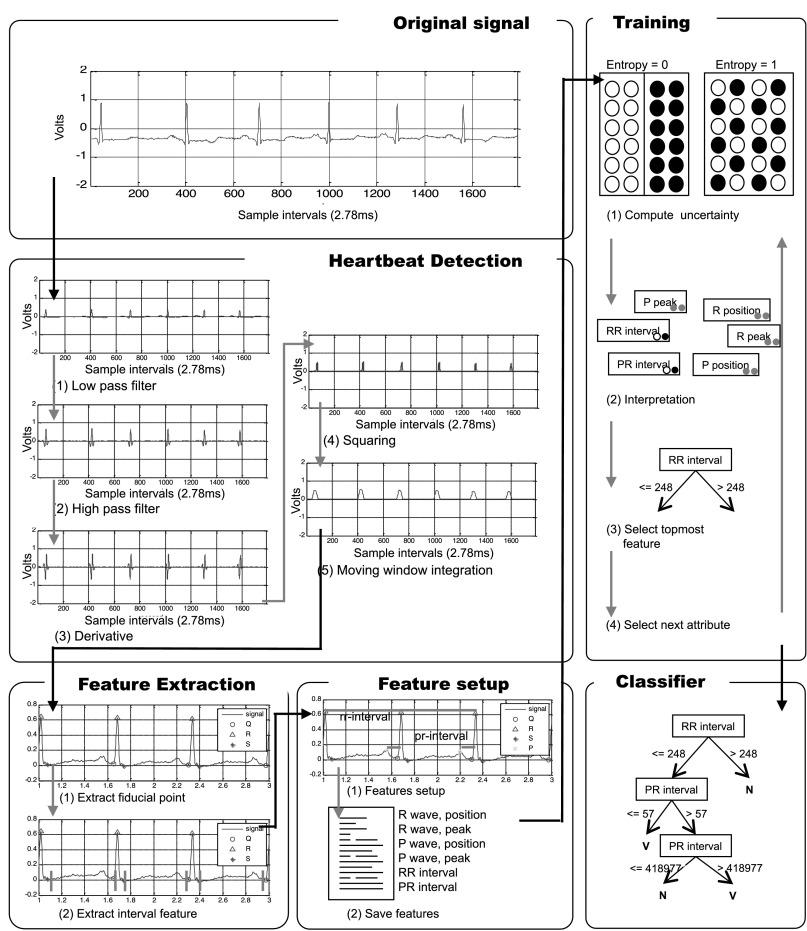
Our system looks for heartbeats in the ECG using the Pan–Tompkins algorithm,7 which performs band-pass filtering, differentiation, squaring, and integration over a moving window. The heartbeat detection panel in Figure 3 depicts the output of consecutive steps in the algorithm when processing record 100 from the MIT-BIH Arrhythmia Database.
Fig. 3.
Summary of proposed system for the classification of heartbeats. ecg/ECG, electrocardiography; MLII, modified limb lead II.
The band-pass filter reduces noise such as 60-Hz interference, baseline wander, muscle noise, and T-wave interference in the ECG signal. The difference equation of the low-pass filter is
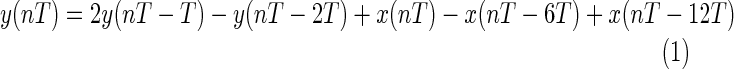 |
and the difference equation of the high-pass filter is
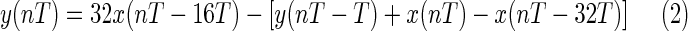 |
The low-pass filter has a cutoff frequency of 11 Hz, and the high-pass filter has a cutoff frequency of 5 Hz. After being filtered, the ECG signal is differentiated to provide slope information, using the following difference equation:
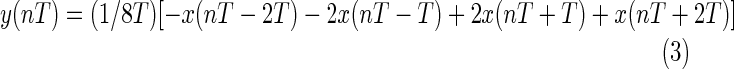 |
The slope approximates the ideal derivative for direct currents in a low frequency range of 0–30 Hz.
The next step is to square the signal, thus making all values positive, as well as emphasizing higher frequencies. The equation for squaring is
 |
An integrating moving window is then used to obtain waveform features, which are added to the slope of the R wave by the following equation:
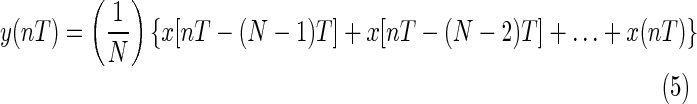 |
This produces repeated peaks.
A heartbeat spans one peak, from which the system can locate the QRS complex: Q is the starting point, S is the ending point, and R is the summit of the peak. The system then finds the P wave from the QRS complex. The P wave is located between the S point of the current heartbeat and the Q point of the next heartbeat. The system divides the duration of the P wave into two and uses the peak in the second interval as the P point.
Feature Extraction
The position and amplitude of Q, R, S, and P [Fig. 3, feature extraction stage (1)] are calculated for each beat. Ventricular activity is related to the QRS complex, and atrial activity, to the P wave.
Six interval features are calculated [Fig. 3, feature extraction stage (2)]. The RR interval stretches from the beginning of the P wave to the beginning of the QRS complex. The other five features are extracted from consecutive R waves. The RR interval is the time that elapses between two consecutive R waves. The Pre-RR interval is the time between the previous R wave and the current R wave, and the Post-RR interval is the time between the current R wave and the following R wave. The Average-RR interval is the mean length of RR intervals over 60 cycles, and the Local-RR interval is the mean RR interval over 10 cycles. The Variable-RR interval is the average of the Pre-RR and Post-RR intervals.
The Average-RR and Local-RR intervals have had other definitions in previous work. For example, in de Chazal et al.,9 the Local-RR interval was defined as the mean RR interval over 10 heartbeats, centered on the current beat, and the Average-RR interval was defined as the mean of all RR intervals in the same ECG recording. These definitions require data about heartbeats later than the current beat and are therefore unsuitable for real-time applications. We have thus redefined the mean RR intervals in terms of past heartbeats.
Heartbeat Classification Using the Decision Tree
Decision trees built by algorithms such as ID3 (Iterative Dichotomiser 3)10 and C4.511 are commonly used for heartbeat classification. ID3 is a statistical algorithm and is thus less sensitive to errors in training data. However, it is not suitable for our application because the decision tree that it creates has discrete attributes. Instead, we use the C4.5 algorithm, which allows the use of numerical attributes.
The C4.5 algorithm constructs the decision tree by determining the uncertainty of features in the training data and selecting those that are most reliable. The iteration continues until the system has classified all of the heartbeats in the training data. Let T be a set of training data, and let F be a set of 14 extracted features represented by 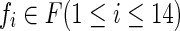 Uncertainty is expressed as an entropy value E(T), as follows:
Uncertainty is expressed as an entropy value E(T), as follows:
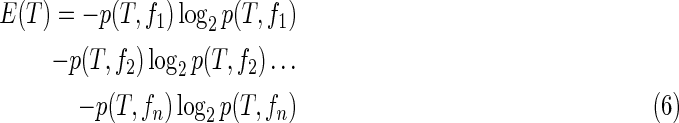 |
If all of the heartbeats exhibiting a particular feature are in the same class, then the entropy of that feature is 0; if the numbers of positive and negative heartbeats are equal, then the entropy of the feature is 1, as shown in the first training stage of Figure 3. We use information gain as a metric of uncertainty. The information gain Gain (T, F) of a feature F from training data T is defined as follows:
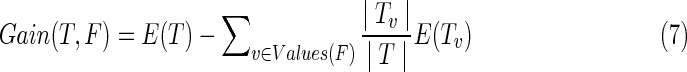 |
where Values(F) is the set of all possible values for feature F and Tv is the subset of T for which feature F has value v. The features are interpreted in terms of information gain. The C4.5 algorithm recursively places the feature with the highest gains as the attribute at the root of the tree.
Results
We used the MIT-BIH Arrhythmia Database for training and testing the proposed decision tree–based classifier. The database contains 48 half-hour recordings, each containing two ECG lead signals (denoted as lead A and lead B). In 45 of the recordings, lead A is a modified limb lead II (MLII), and lead B is a modified lead V1. In the other recordings, lead A is V5, and lead B is V2 or MLII. The lead signals were band-pass-filtered to the range of 0.1–100 Hz and were digitized at 360 Hz.
The database contains approximately 109,000 heartbeats. Each beat is labeled with its type and the location of the QRS complex. These labels can be used to determine the accuracy of classification, and our system takes account of 16 of these labels. These labels are listed in Table 1, which also gives totals for each type of beat in the database. Table 2 relates these heartbeat types to individual records.
Table 1.
Heartbeat Types
| HEARTBEAT TYPE | ANNOTATION | NUMBER OF HEARTBEATS |
|---|---|---|
| Normal beat (NOR) | N | 75,016 |
| Left bundle branch block beat (LBBB) | L | 8,075 |
| Right bundle branch block beat (RBBB) | R | 7,259 |
| Atrial premature beat (APC) | A | 2,546 |
| Aberrated atrial premature beat (AP) | a | 150 |
| Nodal (junctional) premature beat (NP) | J | 83 |
| Supraventricular premature beat (SP) | S | 2 |
| Premature ventricular contraction (PVC) | V | 7,130 |
| Fusion of ventricular and normal beat (VFN) | F | 803 |
| Ventricular flutter wave (VF) | ! | 472 |
| Atrial escape beat (AE) | e | 16 |
| Nodal (junctional) escape beat (NE) | j | 229 |
| Ventricular escape beat (VE) | E | 106 |
| Paced beat (PACE) | / | 7,028 |
| Fusion of paced and normal beat (FPN) | f | 982 |
| Unclassifiable beat (UN) | Q | 33 |
Table 2.
Occurrence of Different Types of Heartbeats in MIT-BIH Records
| RECORDING NUMBER | N | L | R | A | a | J | S | V | F | ! | e | j | E | / | f | Q |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 100 | 2,239 | — | — | 33 | — | — | — | 1 | — | — | — | — | — | — | — | — |
| 101 | 1,860 | — | — | 3 | — | — | — | — | — | — | — | — | — | — | — | 2 |
| 102 | 99 | — | — | — | — | — | — | 4 | — | — | — | — | — | 2,028 | 56 | — |
| 103 | 2,082 | — | — | 2 | — | — | — | — | — | — | — | — | — | — | — | — |
| 104 | 163 | — | — | — | — | — | — | 2 | — | — | — | — | — | 1,380 | 666 | 18 |
| 105 | 2,526 | — | — | — | — | — | — | 41 | — | — | — | — | — | — | — | 5 |
| 106 | 1,507 | — | — | — | — | — | — | 520 | — | — | — | — | — | — | — | — |
| 107 | — | — | — | — | — | — | — | 59 | — | — | — | — | — | 2,078 | — | — |
| 108 | 1,739 | — | — | 4 | — | — | — | 17 | 2 | — | — | 1 | — | — | — | — |
| 109 | — | 2,492 | — | — | — | — | — | 38 | 2 | — | — | — | — | — | — | — |
| 111 | — | 2,123 | — | — | — | — | — | 1 | — | — | — | — | — | — | — | — |
| 112 | 2,537 | — | — | 2 | — | — | — | — | — | — | — | — | — | — | — | — |
| 113 | 1,789 | — | — | — | 6 | — | — | — | — | — | — | — | — | — | — | — |
| 114 | 1,820 | — | — | 10 | — | 2 | — | 43 | 4 | — | — | — | — | — | — | — |
| 115 | 1,953 | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — |
| 116 | 2,302 | — | — | 1 | — | — | — | 109 | — | — | — | — | — | — | — | — |
| 117 | 1,534 | — | — | 1 | — | — | — | — | — | — | — | — | — | — | — | — |
| 118 | — | — | 2,166 | 96 | — | — | — | 16 | — | — | — | — | — | — | — | — |
| 119 | 1,543 | — | — | — | — | — | — | 444 | — | — | — | — | — | — | — | — |
| 121 | 1,861 | — | — | 1 | — | — | — | 1 | — | — | — | — | — | — | — | — |
| 122 | 2,476 | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — |
| 123 | 1,515 | — | — | — | — | — | — | 3 | — | — | — | — | — | — | — | — |
| 124 | — | — | 1,531 | 2 | — | 29 | — | 47 | 5 | — | — | 5 | — | — | — | — |
| 200 | 1,743 | — | — | 30 | — | — | — | 826 | 2 | — | — | — | — | — | — | — |
| 201 | 1,625 | — | — | 30 | 97 | 1 | — | 198 | 2 | — | — | 10 | — | — | — | — |
| 202 | 2,061 | — | — | 36 | 19 | — | — | 19 | 1 | — | — | — | — | — | — | — |
| 203 | 2,529 | — | — | — | 2 | — | — | 444 | 1 | — | — | — | — | — | — | 4 |
| 205 | 2,571 | — | — | 3 | — | — | — | 71 | 11 | — | — | — | — | — | — | — |
| 207 | — | 1,457 | 86 | 107 | — | — | — | 105 | — | 472 | — | — | 105 | — | — | — |
| 208 | 1,586 | — | — | — | — | — | 2 | 992 | 373 | — | — | — | — | — | — | 2 |
| 209 | 2,621 | — | — | 383 | — | — | — | 1 | — | — | — | — | — | — | — | — |
| 210 | 2,423 | — | — | — | 22 | — | — | 194 | 10 | — | — | — | 1 | — | — | — |
| 212 | 923 | — | 1,825 | — | — | — | — | — | — | — | — | — | — | — | — | — |
| 213 | 2,641 | — | — | 25 | 3 | — | — | 220 | 362 | — | — | — | — | — | — | — |
| 214 | — | 2,003 | — | — | — | — | — | 256 | 1 | — | — | — | — | — | — | 2 |
| 215 | 3,159 | — | — | 3 | — | — | — | 164 | 1 | — | — | — | — | — | — | — |
| 217 | 244 | — | — | — | — | — | — | 162 | — | — | — | — | — | 1,542 | 260 | — |
| 219 | 2,082 | — | — | 7 | — | — | — | 64 | 1 | — | — | — | — | — | — | — |
| 220 | 1,954 | — | — | 94 | — | — | — | — | — | — | — | — | — | — | — | — |
| 221 | 2,031 | — | — | — | — | — | — | 396 | — | — | — | — | — | — | — | — |
| 222 | 2,062 | — | — | 208 | — | 1 | — | — | — | — | — | 212 | — | — | — | — |
| 223 | 2,029 | — | — | 72 | 1 | — | — | 473 | 14 | — | 16 | — | — | — | — | — |
| 228 | 1,688 | — | — | 3 | — | — | — | 362 | — | — | — | — | — | — | — | — |
| 230 | 2,255 | — | — | — | — | — | — | 1 | — | — | — | — | — | — | — | — |
| 231 | 314 | — | 1,254 | 1 | — | — | — | 2 | — | — | — | — | — | — | — | — |
| 232 | — | — | 397 | 1,382 | — | — | — | — | — | — | — | 1 | — | — | — | — |
| 233 | 2,230 | — | — | 7 | — | — | — | 831 | 11 | — | — | — | — | — | — | — |
| 234 | 2,700 | — | — | — | — | 50 | — | 3 | — | — | — | — | — | — | — | — |
!, ventricular flutter wave; /, paced beat; a, aberrated atrial premature beat; A, atrial premature beat; e, atrial escape beat; E, ventricular escape beat; f, fusion of paced and normal beat; F, fusion of ventricular and normal beat; j, nodal (junctional) escape beat; J, nodal (junctional) premature beat; L, left bundle branch block beat; N, normal beat; Q, unclassifiable beat; R, right bundle branch block beat; S, supraventricular premature beat; V, premature ventricular contraction.
To evaluate the performance of our system, we used three standard metrics, sensitivity, specificity, and accuracy. Sensitivity is the ability of the system to identify heartbeats of a particular type correctly, expressed as follows:
 |
where TP is the number of true-positives and FN is the number of false-negatives. Specificity is the ability of the system not to identify heartbeats incorrectly if they are not of a particular type, expressed as follows:
 |
where TN is the number of true-negatives and FP is the number of false-positives. Accuracy is an overall measure of the system's ability to classify heartbeats, which combines sensitivity and specificity as follows:
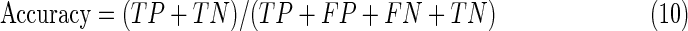 |
We evaluated the performance of our system in two stages. We first considered heartbeat detection, which affects classification performance. Table 3 summarizes the detection of heartbeats from the MIT-BIH data. Overall, our system achieved a mean sensitivity of 97.22% and a mean specificity of 96.89%. The most problematic recording was 107, which had many abnormal beats with unusually large P waves exhibiting atypical peaks, resulting in the lowest specificity of 50.05%. Performance on this recording might be improved through deeper analysis of the P wave, but this is beyond the scope of this study.
Table 3.
Performance Evaluation of Heartbeat Detection
| RECORDING NUMBER | SENSITIVITY (%) | SPECIFICITY (%) | ACCURACY (%) |
|---|---|---|---|
| 100 | 99.96 | 100.00 | 99.98 |
| 101 | 99.84 | 99.82 | 99.83 |
| 102 | 99.82 | 98.91 | 99.37 |
| 103 | 99.67 | 100.00 | 99.84 |
| 104 | 96.58 | 75.30 | 85.94 |
| 105 | 96.17 | 92.82 | 94.50 |
| 106 | 93.99 | 99.80 | 96.90 |
| 107 | 99.86 | 50.05 | 74.96 |
| 108 | 96.38 | 84.03 | 90.21 |
| 109 | 99.64 | 99.84 | 99.74 |
| 111 | 99.58 | 99.62 | 99.60 |
| 112 | 99.60 | 99.53 | 99.57 |
| 113 | 99.94 | 100.00 | 99.97 |
| 114 | 99.52 | 99.16 | 99.34 |
| 115 | 99.59 | 100.00 | 99.80 |
| 116 | 98.80 | 99.67 | 99.24 |
| 117 | 99.80 | 99.74 | 99.77 |
| 118 | 99.09 | 97.19 | 98.14 |
| 119 | 94.94 | 99.50 | 97.22 |
| 121 | 99.31 | 99.15 | 99.23 |
| 122 | 99.96 | 100.00 | 99.98 |
| 123 | 100.00 | 100.00 | 100.00 |
| 124 | 99.14 | 99.63 | 99.39 |
| 200 | 93.19 | 95.80 | 94.50 |
| 201 | 95.14 | 99.38 | 97.26 |
| 202 | 99.30 | 99.72 | 99.51 |
| 203 | 95.53 | 95.37 | 95.45 |
| 205 | 99.37 | 99.25 | 99.31 |
| 207 | 86.83 | 99.90 | 93.37 |
| 208 | 94.11 | 96.23 | 95.17 |
| 209 | 98.59 | 99.90 | 99.25 |
| 210 | 97.58 | 98.42 | 98.00 |
| 212 | 99.53 | 99.85 | 99.69 |
| 213 | 98.48 | 94.52 | 96.50 |
| 214 | 98.22 | 99.30 | 98.76 |
| 215 | 98.88 | 99.88 | 99.38 |
| 217 | 96.75 | 96.08 | 96.42 |
| 219 | 93.12 | 100.00 | 96.56 |
| 220 | 99.03 | 100.00 | 99.52 |
| 221 | 98.21 | 99.67 | 98.94 |
| 222 | 94.31 | 98.26 | 96.29 |
| 223 | 98.34 | 100.00 | 99.17 |
| 228 | 96.26 | 87.15 | 91.71 |
| 230 | 91.57 | 100.00 | 95.79 |
| 231 | 78.17 | 100.00 | 89.09 |
| 232 | 98.13 | 99.22 | 98.68 |
| 233 | 97.30 | 99.22 | 98.26 |
| 234 | 99.46 | 100.00 | 99.73 |
| Average | 97.22 | 96.89 | 97.06 |
The second part of the performance evaluation was heartbeat classification. We used two methods to evaluate the performance of our decision tree classifier. In the first method, we applied the 10-fold cross-validation procedure.12 All of the heartbeats in the MIT-BIH data were distributed into 10 subsets of approximately equal size. Then, the classifier was trained 10 times, leaving out one of the subsets each time. The omitted subset was used to compute the prediction error of the classifier on the training set. The data for each heartbeat have two signals derived from leads A and B. When using the training data obtained from lead A, the accuracy was 94.75%, and it was 92.7% when using the data from lead B, as shown in Table 4. When we applied our classifier to the training data from both leads A and B, the accuracy rose to 96%.
Table 4.
Results of Classification Using the First Method
| LEAD | SENSITIVITY (%) | SPECIFICITY (%) | ACCURACY (%) |
|---|---|---|---|
| A | 94.90 | 94.60 | 94.75 |
| B | 92.80 | 92.60 | 92.70 |
| A+B | 95.60 | 95.50 | 96.00 |
In the second method, both the training and the test data were extracted from a single record using the 10-fold cross-validation procedure. Each record was divided into 10 subsets, and the classifier was trained using nine of these subsets, whereas the remaining subset was used for testing. This process was repeated for each of the 10 subsets. Because the MIT-BIH Arrhythmia Database is not large (its data are taken from 30-min ECG recordings of about 48 patients), we increased the experimental dataset by replicating each heartbeat and then carried out 10-fold cross-validation. In this case, the system achieved an average sensitivity of 99.5%, specificity of 89.97%, and accuracy of 96.73%, as shown in Table 5. Although there may be some learning bias helping the system to analyze the heartbeat of an individual, this result suggests the potential of our Holter ECG monitoring system in providing reliable and intelligent personal ECG telemetry using the proposed decision tree–based decision support system.
Table 5.
Results of Classification Using the Second Method
| RECORDING NUMBER | SENSITIVITY (%) | SPECIFICITY (%) | ACCURACY (%) |
|---|---|---|---|
| 100 | 99.90 | 95.70 | 97.80 |
| 101 | 99.80 | 80.10 | 89.95 |
| 102 | 99.50 | 95.90 | 97.70 |
| 103 | 99.90 | 50.00 | 74.95 |
| 104 | 97.30 | 98.30 | 97.80 |
| 105 | 99.70 | 92.50 | 96.10 |
| 106 | 99.90 | 99.70 | 99.80 |
| 107 | 100.00 | 98.40 | 99.20 |
| 108 | 99.50 | 78.10 | 88.80 |
| 109 | 99.80 | 91.20 | 95.50 |
| 111 | 100.00 | 0.00 | 50.00 |
| 112 | 100.00 | 100.00 | 100.00 |
| 113 | 100.00 | 100.00 | 100.00 |
| 114 | 99.70 | 96.70 | 98.20 |
| 115 | 100.00 | 100.00 | 100.00 |
| 116 | 99.80 | 99.10 | 99.45 |
| 117 | 99.90 | 0.10 | 50.00 |
| 118 | 99.80 | 98.70 | 99.25 |
| 119 | 100.00 | 100.00 | 100.00 |
| 121 | 99.80 | 100.00 | 99.90 |
| 122 | 100.00 | 100.00 | 100.00 |
| 123 | 100.00 | 100.00 | 100.00 |
| 124 | 99.40 | 95.20 | 97.30 |
| 200 | 99.00 | 99.10 | 99.05 |
| 201 | 98.90 | 97.70 | 98.30 |
| 202 | 99.60 | 94.00 | 96.80 |
| 203 | 98.70 | 96.80 | 97.75 |
| 205 | 99.60 | 94.90 | 97.25 |
| 207 | 99.30 | 99.70 | 99.50 |
| 208 | 98.30 | 98.90 | 98.60 |
| 209 | 99.60 | 98.70 | 99.15 |
| 210 | 99.20 | 96.60 | 97.90 |
| 212 | 99.60 | 99.40 | 99.50 |
| 213 | 98.80 | 99.20 | 99.00 |
| 214 | 99.60 | 98.40 | 99.00 |
| 215 | 99.90 | 98.30 | 99.10 |
| 217 | 98.40 | 98.10 | 98.25 |
| 219 | 99.70 | 93.30 | 96.50 |
| 220 | 99.90 | 98.00 | 98.95 |
| 221 | 99.80 | 99.10 | 99.45 |
| 222 | 97.30 | 92.40 | 94.85 |
| 223 | 98.30 | 98.10 | 98.20 |
| 228 | 99.70 | 99.20 | 99.45 |
| 230 | 100.00 | 0.00 | 50.00 |
| 231 | 99.60 | 99.20 | 99.40 |
| 232 | 99.90 | 99.80 | 99.85 |
| 233 | 99.70 | 99.80 | 99.75 |
| 234 | 100.00 | 100.00 | 100.00 |
| Average | 99.50 | 89.97 | 96.73 |
Discussion
The original Holter monitor simply records aspects of the electrical activity of a patient's cardiovascular system, with the aim of subsequent diagnosis of abnormal cardiac rhythms, especially those that only occur occasionally, and abnormal cardiac impulse conduction. Holter monitoring has usually been performed in a hospital, where a patient must stay for a couple of days at a time, but abnormalities that occur very infrequently may nevertheless remain undetected. Automatic heartbeat classification for real-time decision support has not been a part of established Holter monitoring processes. Our system allows the patient much more freedom of movement, thus permitting longer-term monitoring, as well as providing the automatic heartbeat classification needed for decision support.
Several other studies have proposed ECG detection systems based on the QRS complex.13–23 These include the classification of ECG heartbeat characteristics using wavelet features13–17 or waveform features.9,18–21,24 Karimifard et al.22 used Hermitian coefficients, and Osowski et al.23 used higher-order statistics. Other methods of heartbeat characterization are based on P wave detection using local search25–27 or ventricular and atrial source separation.28,29
Other automatic methods of heartbeat classification have been based on machine learning.9,13–24 Arif et al.13 and Karimifard et al.22 used the k-nearest neighbor method with good results. Ye et al.14 and Osowski et al.23 used a support vector machine, which is known to be a good tool for classification.30,31 De Oliveira et al.20 investigated a Bayesian network, and de Lannoy et al.24 used weighted conditional random fields. Yet other studies have involved neural networks15,16,21 or linear discriminates.9,18 However, none of these techniques classifies heartbeats automatically using a decision tree.
Some issues remain in achieving practical use of the ECG monitoring system that we propose. First, the patient-worn ECG sensor module needs to be small and lightweight and to operate with low power, to ensure portability. This is the reason why we considered Bluetooth as the technology for communication between the sensor and the patient's mobile device. However, this requires that the patient does not move out of Bluetooth range of his or her own smartphone, which is not always practical. The alternative is to integrate a communication module for wide-area networking into the ECG sensor, but this increases its weight, power, and size.
Second, medical telemetry applications typically require only a moderate data rate; the primary requirements that they place on a wireless system are wide coverage, high transmission reliability, and low cost. Infrastructure-oriented wireless technologies, including 3GPP LTE, provided by cellular operators, and versions of IEEE 802.11 (Wi-Fi), have been recommended as means of supporting ECG monitoring in diverse environments. Wi-Fi was not, of course, originally designed for medical telemetry, but has been considered for this application because of its wide deployment and low cost.32 However, Wi-Fi suffers from limited coverage and variable latency.33 Conversely, LTE technologies, while more expensive, support high-mobility devices and have broad coverage.34 If monitoring systems can reuse existing LTE infrastructure, then the initial cost of deployment is greatly reduced.
A compromise solution is offered by machine-type communications (MTC)35 technologies, which operate over LTE networks. MTC has recently received a lot of attention because it has many advantages, such as low cost and low power, while inheriting the wide coverage of existing LTE networks. An ECG sensor that communicate through MTC could be small and light, but nevertheless transmit directly to a decision support system without the need for an intermediate mobile device.
Finally, we observe that telemetry applications require high transmission reliability and low service latency, as their effectiveness is sure to be compromised by the loss, corruption, or late delivery of data. However, guaranteeing those requirements is challenging because the data rate of a wireless channel is variable, and such channels are subject to errors. Designing a protocol for proper error control, which can also be combined with an appropriate diversity scheme, is therefore essential.36,37
Conclusions
We have proposed and evaluated an intelligent system for the wireless monitoring of cardiac activity. This system classifies an ECG waveform in terms of features using a decision tree. Our system achieved a high level of accuracy when evaluating data from the MIT-BIH Arrhythmia Database. This form of test is appropriate for a project at an early stage. In the future, we intend to implement and evaluate our system on data obtained directly from a patient, by adding a “live mode” to our classification system, which will allow it to be combined with Holter monitoring. We also plan to improve the accuracy of classification by reformulating the method of P wave detection.
Acknowledgments
This research was supported by the Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Science, ICT & Future Planning (NRF-2013R1A1A1059188).
Disclosure Statement
No competing financial interests exist.
References
- 1.Belgacem N, Bereksi-Reguig F. Bluetooth portable device for ECG and patient motion monitoring. Nat Technol 2011;4:19–23 [Google Scholar]
- 2.Ko S-Y, Wang K-M, Lian W-C, Kao C-H. A portable ECG recorder. In: Proceedings of the 2nd International Conference on Consumer Electronics, Communications and Networks Piscataway, NJ: IEEE, 2012;3063–3067 [Google Scholar]
- 3.Tello PJP, Manjarres O, Quijano M, Blanco AU. Remote monitoring system of ECG and temperature signals using bluetooth. In: Proceedings of the 4th International Symposium on Information Technology in Medicine and Education Piscataway, NJ: IEEE, 2012;860–863 [Google Scholar]
- 4.Lee Y-G, Jeong WS, Yoon G. Smartphone-based mobile health monitoring. Telemed J E Health 2012;18:585–590 [DOI] [PubMed] [Google Scholar]
- 5.D'Angelo LT, Tarita E, Zywietz TK, Lueth TC. A system for intelligent home care ECG upload and priorisation. In: Proceedings of the 32nd Annual International Conference of the IEEE Engineering Medicine and Biology Society Piscataway, NJ: IEEE, 2010;2188–2191 [DOI] [PubMed] [Google Scholar]
- 6.Lee J-S, Su Y-W, Shen C-C. A comparative study of wireless protocols: Bluetooth, UWB, ZigBee, and Wi-Fi. In: Proceedings of the 33rd Annual Conference of the IEEE Industrial Electronics Society Piscataway, NJ: IEEE, 2007;46–51 [Google Scholar]
- 7.Pan J, Tompkins WJ. A real-time QRS detection algorithm. IEEE Trans Biomed Eng 1985;32:230–236 [DOI] [PubMed] [Google Scholar]
- 8.Mark R, Moody G. MIT-BIH arrhythmia database. Available at http://ecg.mit.edu/dbinfo.html (last accessed July27, 2014) [DOI] [PubMed]
- 9.de Chazal P, Reilly RB. A patient-adapting heartbeat classifier using ECG morphology and heartbeat interval features. IEEE Trans Biomed Eng 2006;53:2535–2543 [DOI] [PubMed] [Google Scholar]
- 10.Quinlan JR. Induction of decision trees. Machine Learning 1986;1:81–106 [Google Scholar]
- 11.Quinlan JR. C4.5: Programs for machine learning. San Francisco: Morgan Kaufmann Publishers, 1993 [Google Scholar]
- 12.Kohavi R. A study of cross-validation and bootstrap for accuracy estimation and model selection. In: Proceedings of the 14th International Joint Conference on Artificial Intelligence, Vol. 2 1995;1137–1143 [Google Scholar]
- 13.Arif M, Akram MU, Afsar FA. Arrhythmia beat classification using pruned fuzzy k-nearest neighbor classifier. In: Proceedings of the 1st International Conference of Soft Computing and Pattern Recognition Piscataway, NJ: IEEE, 2009;37–42 [Google Scholar]
- 14.Ye C, Kumar BV, Coimbra MT. Heartbeat classification using morphological and dynamic features of ECG signals. IEEE Trans Biomed Eng 2012;59:2930–2941 [DOI] [PubMed] [Google Scholar]
- 15.Prasad GK, Sahambi JS. Classification of ECG arrhythmia using multi-resolution analysis and neural networks. In: Proceedings of the 18th IEEE Conference on Convergent Technologies for Asia-Pacific Region, Vol. 1 Piscataway, NJ: IEEE, 2003;227–231 [Google Scholar]
- 16.Ceylan R, Ozbay Y, Karlik B. A novel approach for classification of ECG arrhythmias: Type 2 fuzzy clustering neural network. Expert Syst Applic 2009;36:6721–6726 [DOI] [PubMed] [Google Scholar]
- 17.Zheng H, Wu J. A real-time QRS detector based on discrete wavelet transform and cubic spline interpolation. Telemed J E Health 2008;14:809–815 [DOI] [PubMed] [Google Scholar]
- 18.de Chazal P, O'Dwyer M, Reilly RB. Automatic classification of heartbeats using ECG morphology and heartbeat interval features. IEEE Trans Biomed Eng 2004;51:1196–1206 [DOI] [PubMed] [Google Scholar]
- 19.Llamedo M, Martinez JP. Heartbeat classification using feature selection driven by database generalization criteria. IEEE Trans Biomed Eng 2011;58:616–625 [DOI] [PubMed] [Google Scholar]
- 20.de Oliveira LSC, Andreão RV, Sarcinelli-Filho M. Premature ventricular beat classification using a dynamic Bayesian network. In: Proceedings of the 33rd Annual International Conference of the IEEE Engineering Medicine and Biology Society Piscataway, NJ: IEEE, 2011;4984–4987 [DOI] [PubMed] [Google Scholar]
- 21.Yeap TH, Johnson F, Rachniowski M. ECG beat classification by a neural network. In: Proceedings of the 12th Annual International Conference of the IEEE Engineering Medicine and Biology Society Piscataway, NJ: IEEE, 1990;1457–1458 [Google Scholar]
- 22.Karimifard S, Ahmadian A, Khoshnevisan M, Nambakhsh MS. Morphological heart arrhythmia detection using Hermitian basis functions and knn classifier. In: Proceedings of the 29th Annual International Conference of the IEEE Engineering Medicine and Biology Society Piscataway, NJ: IEEE, 2006;1367–1370 [DOI] [PubMed] [Google Scholar]
- 23.Osowski S, Hoai LT, Markiewicz T. Support vector machine-based expert system for reliable heartbeat recognition. IEEE Trans Biomed Eng 2004;51:582–589 [DOI] [PubMed] [Google Scholar]
- 24.de Lannoy G, Francois D, Delbeke J, Verleysen M. Weighted conditional random fields for supervised interpatient heartbeat classification. IEEE Trans Biomed Eng 2012;59:241–247 [DOI] [PubMed] [Google Scholar]
- 25.Almeida R, Martinez JP, Olmos S, Rocha AP, Laguna P. Automatic delineation of T and P wave using a wavelet-based multiscale approach. In: Proceedings of the 1st International Congress on Computational Bioengineering 2003;243–247 [Google Scholar]
- 26.Dotsinky I. Atrial wave detection algorithm for discovery of some rhythm abnormalities. Physiol Meas 2007;29:595–610 [DOI] [PubMed] [Google Scholar]
- 27.Graja S, Boucher J. Hidden Markov tree model applied to ECG delineation. IEEE Trans Instrum Meas 2005;54:2163–2168 [Google Scholar]
- 28.Lemay M, Jacquemet V, Forchaz A, Vesin JM, Kappenberger L. Spatiotemporal QRST cancellation method using separate QRS and T-waves templates. Comput Cardiol 2005;32:611–614 [Google Scholar]
- 29.Vaya C, Rieta JJ, Sanchez C, Moratal D. Convolutive blind source separation algorithms applied to the electrocardiogram of atrial fibrillation: Study of performance. IEEE Trans Biomed Eng 2007;54:1530–1533 [DOI] [PubMed] [Google Scholar]
- 30.Mitchell TM. Machine learning. New York: McGraw-Hill, 1997 [Google Scholar]
- 31.Witten H, Frank E. Practical machine learning tools and techniques. San Francisco: Morgan Kaufmann Publishers, 2005 [Google Scholar]
- 32.Cypher D, Chevrollier N, Montavont N, Golmie N. Prevailing over wires in healthcare environments: Benefits and challenges. IEEE Commun Mag 2006;44:56–63 [Google Scholar]
- 33.Zhu H, Li M, Chlamtac I, Prabhakaran B. A survey of quality of service in IEEE 802.11 networks. IEEE Wireless Commun 2004;11:6–14 [Google Scholar]
- 34.Ghosh A, Ratasuk R, Mondal B, Mangalvedhe N. LTE-advanced: Next-generation wireless broadband technology. IEEE Wireless Commun 2010;17:10–22 [Google Scholar]
- 35.Taleb T, Kunz A. Machine type communications in 3GPP networks: Potential, challenges, and solutions. IEEE Commun Mag 2012;50:178–184 [Google Scholar]
- 36.Kang K, Park K-J, Song J-J, Yoon C-H, Sha L. A medical-grade wireless architecture for remote electrocardiography. IEEE Trans Inform Technol Biomed 2011;15:260–267 [DOI] [PubMed] [Google Scholar]
- 37.Kang K, Ryu J, Hur J, Sha L. Design and QoS of a wireless system for real-time remote electrocardiography. IEEE J Biomed Health Inform 2013;17:745–755 [DOI] [PubMed] [Google Scholar]