Abstract
Transforming growth factor beta2 (TGFβ2) is a multifunctional protein which is expressed in several embryonic and adult organs. TGFB2 mutations can cause Loeys Dietz syndrome, and its dysregulation is involved in cardiovascular, skeletal, ocular and neuromuscular diseases, osteoarthritis, tissue fibrosis, and various forms of cancer. TGFβ2 is involved in cell growth, apoptosis, cell migration, cell differentiation, cell-matrix remodeling, epithelial-mesenchymal transition, and wound healing in a highly context-dependent and tissue-specific manner. Tgfb2−/− mice die perinatally from congenital heart disease, precluding functional studies in adults. Here, we have generated mice harboring Tgfb2βgeo (knockout-first lacZ-tagged insertion) gene-trap allele and Tgfb2flox conditional allele. Tgfb2βgeo/βgeo or Tgfb2βgeo/− mice died at perinatal stage from the same congenital heart defects as Tgfb2−/− mice. β-galactosidase staining successfully detected Tgfb2 expression in the heterozygous Tgfb2βgeo fetal tissue sections. Tgfb2flox mice were produced by crossing the Tgfb2+/βgeo mice with the FLPeR mice. Tgfb2flox/− mice were viable. Tgfb2 conditional knockout (Tgfb2cko/−) fetuses were generated by crossing of Tgfb2flox/− mice with Tgfb2+/−;EIIaCre mice. Systemic Tgfb2cko/− embryos developed cardiac defects which resembled the Tgfb2βgeo/βgeo, Tgfb2βgeo/−, and Tgfb2−/− fetuses. In conclusion, Tgfb2βgeo and Tgfb2flox mice are novel mouse strains which will be useful for investigating the tissue specific expression and function of TGFβ2 in embryonic development, adult organs, and disease pathogenesis and cancer.
Keywords: transforming growth factor beta, Loeys Dietz syndrome, cardiovascular, cancer, fibrosis, lung, blood, vascular, craniofacial, eye, wound healing, neurological, epithelial mesenchymal transition
Transforming growth factor beta2 (TGFβ2) belongs to a family of multifunctional proteins, known as the TGFβ superfamily (Akhurst and Hata 2012;Doetschman et al. 2012a;Arthur and Bamforth 2011). The other two mammalian isoforms of this superfamily are TGFβ1 and TGFβ3 (Azhar et al. 2009;Doetschman et al. 2012b). TGFβs play critical autocrine and/or paracrine roles in embryonic tissue development and maintenance of tissue homeostasis (Conway and Kaartinen 2011;Horiguchi et al. 2012). TGFβs are immunoregulatory and profibrotic cytokines that regulate cell growth, apoptosis, cell migration, cell differentiation, cell-matrix remodeling, epithelial-mesenchymal transition, and wound healing in a highly context-dependent and tissue-specific manner (Sonnylal et al. 2007;Soderberg et al. 2009;Kulkarni et al. 2002). Activated TGFβs interacts with TGFβ receptor type II and type I, which propagate the TGFβ signal into the cell by phosphorylating TGFβ receptor-specific canonical Smads (i.e., Smad2 and Smad3) and non-Smad mediators (e.g., TAK1, ERK MAPK) (Iwata et al. 2012;Yumoto et al. 2013;Akhurst and Hata 2012). The dysregulation of the TGFβ pathway leads to a number of human diseases and disorders, including tissue fibrosis and cancer (Akhurst and Hata 2012), underscoring the essential roles the TGFβ isoforms have in vivo.
TGFB2 mutations have been identified in Loeys-Dietz syndrome (LDS) (OMIM# 614816613795, 610380) (Lindsay et al. 2012;Boileau et al. 2012;Renard et al. 2012). Loeys-Dietz syndrome is a connective tissue disorder, predisposing individuals to serious cardiovascular, craniofacial, cutaneous, ocular, and skeletal complications (Loeys et al. 2013). The cardiovascular complications of LDS patients include congenital heart defects, aortic aneurysm, cardiomyopathy, and heart valve complications (Maccarrick et al. 2014). TGFB2 signaling is associated with cardiovascular complications of Kawasaki disease (Shimizu et al. 2011). TGFB2 levels are elevated in the myocardial tissue of the patients of dilated cardiomyopathy (Pauschinger et al. 1999). Furthermore, TGFB2 is elevated in diseased mitral valves and aortas of Marfan syndrome patients, and mouse craniofacial defects, in which TGFβ signaling is also increased (Iwata, 2012 9286/id;Ng et al. 2004;Nataatmadja et al. 2006;Jain et al. 2009). Spatiotemporally restricted cardiac expression of Tgfb2 and its overlap with Tgfb1 or Tgfb3 in various cardiac cell lineages including endocardial, myocardial, cardiac neural crest, and vascular smooth muscle cells in embryonic hearts (Dickson et al. 1993;Azhar et al. 2003;Molin et al. 2003) suggest a critical cell type specific autocrine-paracrine and synergistic roles of TGFβ2 in regulation of TGFβ signaling during cardiovascular development and remodeling. Systemic knockout mice of Tgfb2 exhibit developmental defects in multiple organs and die at birth due to cardiac malformations, indicating that TGFβ2 is indispensable for embryonic tissue development (Sanford et al. 1997;Azhar et al. 2011;Bartram et al. 2001).
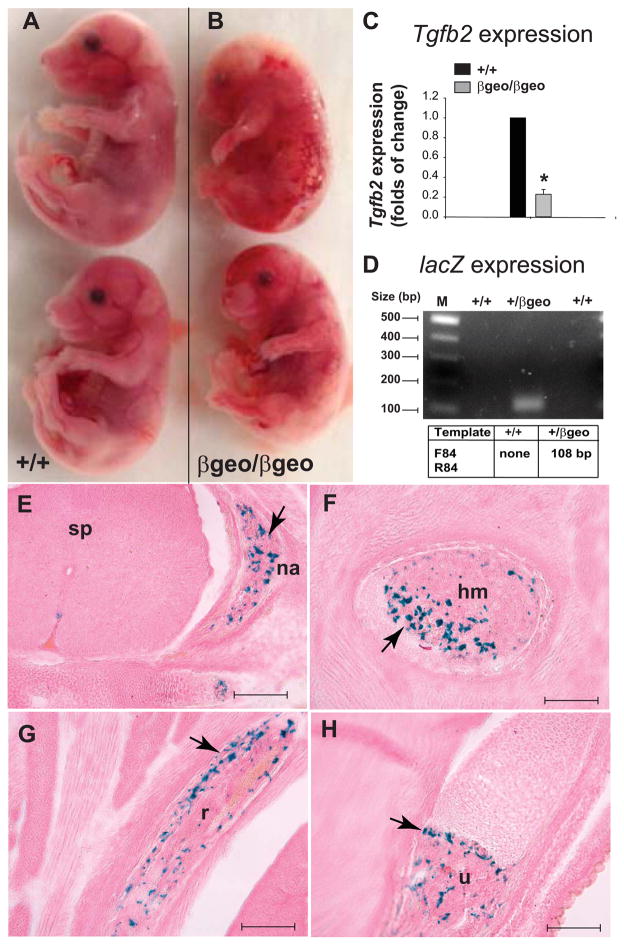
Here, we report on the generation and characterization of mice carrying a novel and flexible gene-trap knockout-first, lacZ tagged insertion allele of Tgfb2 (hereafter referred to as Tgfb2βgeo). Three independent lines of the correctly targeted Tgfb2βgeo ES cell clones were obtained from the European Conditional Mouse Mutagenesis Program (EUCOMM) for generating Tgfb2βgeo mice. These ES clones had passed all rigorous quality control tests of the EUCOMM (Skarnes et al. 2011). Tgfb2βgeo ES cell clones and Tgfb2βgeo mice were validated by an extensive 5′ and 3′ screening (Fig. 1A–B). In this targeting scheme, homologous recombination can result in a Tgfb2βgeo allele where gene function would be ablated by a polyadenylation (polyA) signal-mediated transcriptional stop at the end of the lacZ expression marker gene that is driven off the Tgfb2 promoter. Tgfb2+/βgeo mice were maintained on C57BL/6 genetic background. Genotyping analysis of the newborn offspring indicated that all Tgfb2βgeo/βgeo or Tgfb2βgeo/− mice died at the perinatal stage (Fig. 1C, 2A–B). Timed-pregnant heterozygous Tgfb2βgeo and Tgfb2+/− (C57BL/6) females that crossed to heterozygous Tgfb2βgeo males were used to produce embryos/fetuses for gross morphological and histological characterization. The data indicated that Tgfb2βgeo/βgeo and Tgfb2βgeo/− fetuses at E16.5–E18.5 appeared grossly abnormal and exhibited abnormal body vasculature (Fig. 2A–B). Next, Tgfb2 expression was measured in Tgfb2βgeo/βgeo hearts by real-time quantitative PCR (qPCR) via an intron spanning (exon 6–7) Universal ProbeLibrary assay. The data indicated that the amount of wild-type Tgfb2 transcript containing the exon 6–7 was significantly downregulated in Tgfb2βgeo/βgeo fetal hearts compared to the wild-type fetuses (Fig. 2C). This suggests that although Tgfb2 expression is abated, the polyA signal-mediated transcriptional stop at the end of the lacZ gene-trap cassette is not able to completely abolish the wild-type Tgfb2 expression. Since we expected the Tgfb2 promoter to drive the lacZ expression marker gene, the expression of lacZ was also analyzed by both RT-PCR, and β-galactosidase (X-gal) staining of fetal tissue cryo-sections. Limited data indicated remarkable Tgfb2 expression associated with ossification within cartilage primordium of neural arch (Fig. 2E), mid-shaft region of left humerus (Fig. 2F), rib (Fig. 2G), and distal part of shaft of right ulna (Fig. H) during late embryonic development. The data confirmed the presence of lacZ expression as an indicator of the endogenous Tgfb2 expression in Tgfb2+/βgeo fetuses. Overall, as reported previously in Tgfb2−/− fetuses (Sanford et al. 1997), the significant loss of wild-type Tgfb2 mRNA expression is consistent with the observed perinatal lethality of Tgfb2βgeo/βgeo or Tgfb2βgeo/− mice.
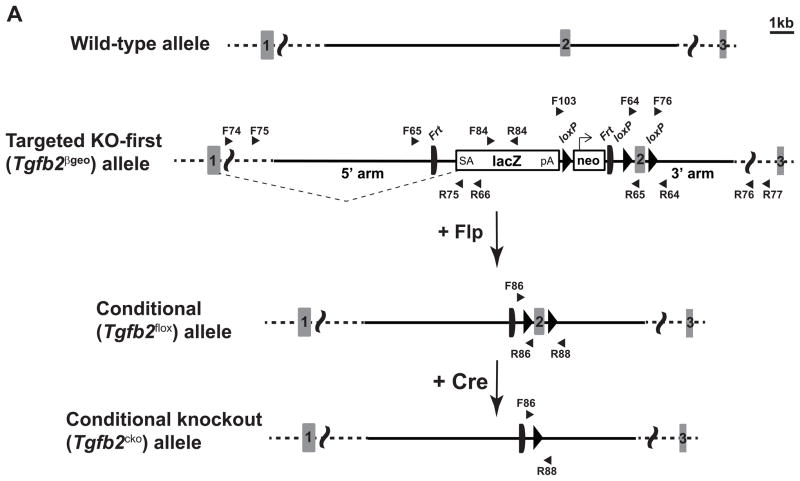
Figure 1. Gene targeting scheme for generating mice with a knockout-first, lacZ-tagged insertion and conditional allele of Tgfb2.
A: Schematic diagram of the Tgfb2 wild-type, targeted knockout-first and lacZ-tagged insertion (Tgfb2βgeo), conditional (Tgfb2flox), and conditional knockout (Tgfb2cko) allele. Boxes with numbers represent exons. Both left and right homology arms in the targeting vector are clearly indicated. The 5′ outside and 3′ outside regions of the Tgfb2 locus are shown in dotted lines. The targeting vector is designed to flank exon 2 with loxP to create Tgfb2 conditional deletion through Cre-mediated recombination. The targeted Tgfb2βgeo allele contains an IRES:lacZ trapping cassette and a floxed promoter-driven neomycin cassette inserted into the intron 1 of the Tgfb2. The presence of an Engrailed (En2) splice acceptor (SA) disrupts gene function, resulting in a lacZ fusion for studying gene expression localization. Splicing events are depicted in dotted lines. Flp recombinase can remove the FRT flanked gene trap cassette, convert the Tgfb2βgeo allele to a conditional Tgfb2flox allele and restore the TGFβ2 activity. Subsequent exposure to Cre recombinase can delete the floxed exon 2 of the Tgfb2flox allele resulting in a Tgfb2cko allele. All LR-PCR and genotyping screening primers are indicated by arrowheads. Drawn roughly to scale, the area, to the left and right of different alleles, beyond the vertical curved lines is not drawn to the scale.
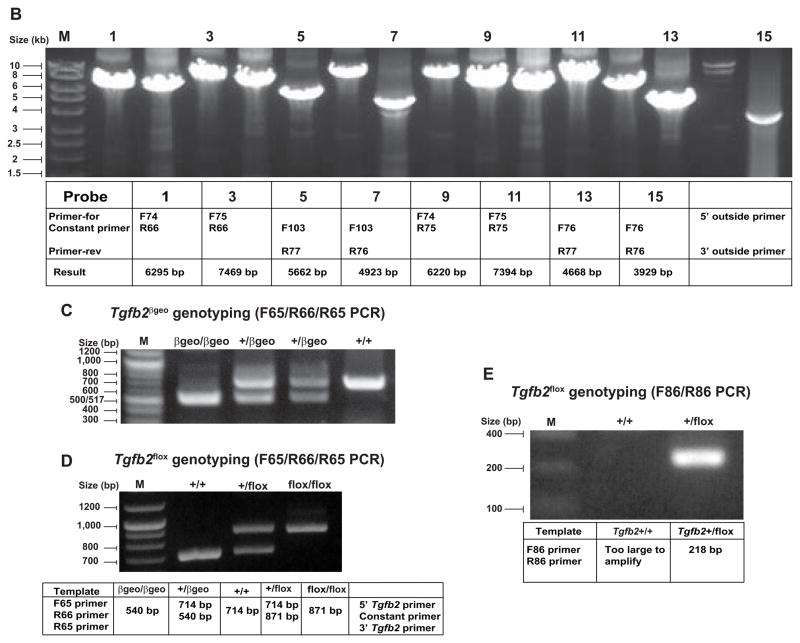
B: Long range PCR screening. LR-PCR screening of targeted ES cells is done using 8 different sets of 5′-outside or 3′-outside LR-PCR primers in combination with the constant cassette primers located within the targeting vector. Table indicates specific primer pairs and the observed large amplicon sizes, confirming the 5′ and 3′ end Tgfb2βgeo targeting. M, DNA marker.
C: PCR genotyping of fetuses with wild-type, heterozygous, and homozygous Tgfb2βgeo alleles. The primers used are: Tgfb2 intron 2 forward primer (F65), constant/cassette lacZ reverse primer (R66) and Tgfb2 exon 2 reverse primer (R65). The F65 and R66 primers produce a PCR product of 540 bp from the Tgfb2βgeo allele, whereas F65 and R65 primers give rise to a PCR product of 714 bp from the wild-type allele. Band size as measured by DNA size markers (M) is indicated.
D: PCR genotyping of Tgfb2flox mice. Tgfb2βgeo allele gives rise to Tgfb2flox allele (post Flp), which is specifically identified by 871 bp band in a three primer (F65, R66, R65) PCR reaction. Band size as measured by DNA size markers (M) is indicated.
E: PCR genotyping with a Tgfb2 forward primer (F86) and constant/cassette primer (R86) produces a unique PCR band of 218 bp from the Tgfb2flox allele but not wild-type or Tgfb2βgeo alleles.
Figure 2. Gross morphological and molecular analyses of Tgfb2βgeo/βgeo fetuses.
A–B: Gross morphological images of wild-type (A) and Tgfb2βgeo/βgeo (B) littermates at E17.5. Tgfb2βgeo/βgeo fetuses appear grossly abnormal with craniofacial features and excessive or abnormal blood vessels and profuse bleeding.
C: Real-time qPCR analysis of ‘wild-type’ Tgfb2 expression in control and Tgfb2βgeo/βgeo fetal hearts. Total RNA from hearts of E16.5 fetuses is used to prepare the cDNA. Intron spanning Universal ProbeLibrary assay with a mouse Tgfb2 exon 7 probe (#73) along with the Tgfb2 exon 6 (forward) and Tgfb2 exon 7 (reverse) primers are used for UPL qPCR analysis. Note that there is a significant loss of wild-type Tgfb2 transcript expression in Tgfb2βgeo/βgeo fetal hearts (*P = <0.005, n = 3 for wild-type and Tgfb2βgeo/βgeo). Expression levels are normalized to β-actin (via dual-color UPL real-time qPCR) and to the wild-type value.
D: Gel RT-PCR analysis indicates lacZ expression in Tgfb2+/βgeo fetal hearts. Note that lacZ PCR fails as the lacZ cassette is absent in wild-type sample.
E–H: lacZ staining of X-gal for cryo-section of Tgfb2+/βgeo fetues at (E16.5). The β-galactosidase staining indicated by blue color (arrow, E–H) is clearly visible in the ossified cartilage primordium of the neural arch (E), mid shaft region of left humerus (F), rib (G), and distal part of shaft of right ulna (H). Scale bar (E–H) = 200 μm. Abbreviations: na, neural arch; hm, humerus; r, rib; u, ulna.
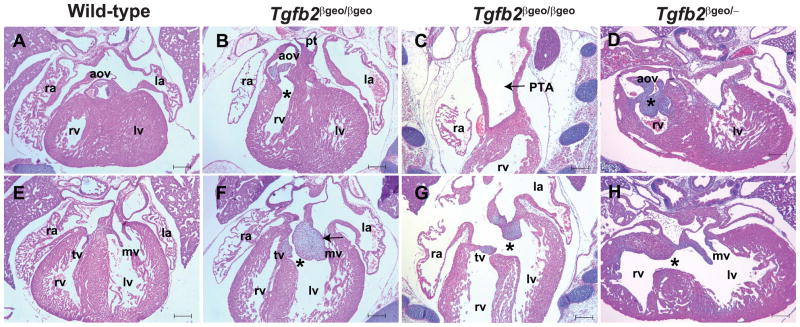
Histological examination of serial tissue sections indicated multiple cardiac structural defects in Tgfb2βgeo/βgeo and Tgfb2βgeo/− fetuses (Fig. 3A–H). Tgfb2βgeo/βgeo as well as Tgfb2βgeo/− fetuses developed similar cardiac malformations of both the outflow tract and inflow tract. The outflow tract malformations of the mutant fetuses included double-outlet right ventricle (DORV) (100% cases), persistent truncus arteriosus (PTA) (27.2% cases), and abnormal morphology and thickening of aortic and/or pulmonary valves (100% cases) (Fig. 3A–D). In addition, the mutant fetuses developed double-inlet left ventricle (DILV) and/or overriding of tricuspid valves orifice via a perimembranous inlet ventricular septal defect (VSD) (100% cases), and abnormal morphology and thickening of tricuspid and mitral valves (100%) (Fig. 3E–H). Malformations of myocardium, epicardium, and aortic arch arteries also found but were not carefully determined in Tgfb2βgeo/βgeo or Tgfb2βgeo/− fetuses. Notably, the overall penetrance of the observed cardiac valve and septal defects was significantly higher in Tgfb2βgeo/βgeo or Tgfb2βgeo/− fetuses compared to Tgfb2−/− fetuses. We attribute this difference in the phenotypic penetrance between the Tgfb2βgeo/βgeo or Tgfb2βgeo/− (C57BL/6) and Tgfb2−/− (129/BL-Swiss) (Azhar et al. 2011;Bartram et al. 2001) mice to different genetic background of the two strains. Collectively, our results show that Tgfb2βgeo/βgeo or Tgfb2βgeo/− fetuses exhibit high penetrance of similar cardiac phenotypes that are reported previously in Tgfb2−/− fetuses. Thus, Tgfb2βgeo allele represents a novel knockout-first, lacZ-tagged insertion and conditional-ready allele for Tgfb2.
Figure 3. Tgfb2βgeo/βgeo and Tgfb2βgeo/− fetuses develop similar cardiac structural defects.
A–H: Hematoxylin and eosin (H&E) staining of wild-type (A, E), Tgfb2βgeo/βgeo (B–C, F–G), and Tgfb2βgeo/− (D,H) fetuses at E17.5. Tgfb2βgeo/βgeo fetuses have double-outlet right ventricle (DORV) (asterisk, B) and thickened aortic valves (B), persistent truncus arteriosus (PTA) (arrow, C), double-inlet left ventricle (F, asterisk) and overriding tricuspid valve orifice via a perimembranous ventricular septal defect (VSD) (asterisk, G), and abnormally thickened mitral (arrow, F) and tricuspid valves (F,G). Tgfb2βgeo/− fetuses show many similar cardiac malformations including DORV, VSD, and thickening of semilunar and mitral valves (D,H). Scale bars =200 μm for A–H. Abbreviations: rv, right ventricle; lv, left ventricle; pt, pulmonary trunk; aov, aortic valves; tv, tricuspid valves; mv, mitral valves; ra, right atrium; la, left atrium
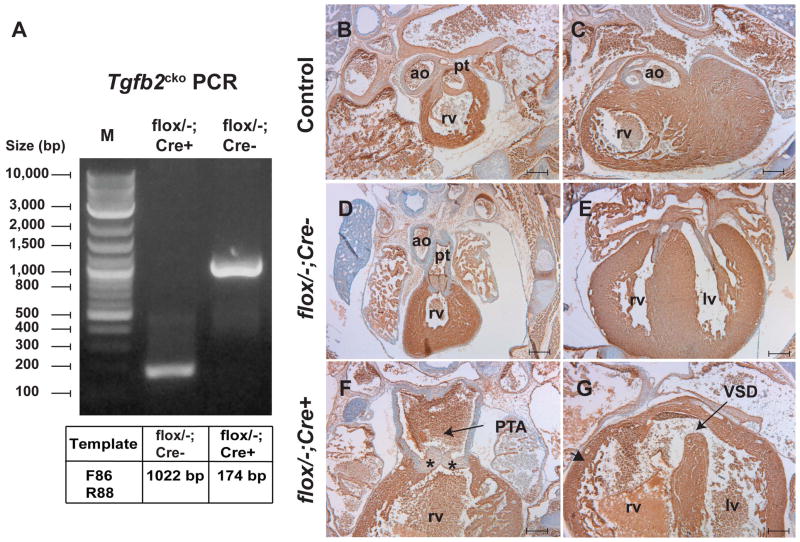
Mice with conditional (Tgfb2flox ) allele were produced by crossing the conditional-ready Tgfb2+/βgeo female mice with the Flp recombinase germline deleter (FLPeR) male mice (Farley et al. 2000), which removed the entire FRT-flanked lacZ-neomycin (βgeo) gene-trap cassette (Fig. 1A, C–E). Genomic PCR analysis confirmed that Flp recombinase resulted in mice harboring Tgfb2flox allele in which loxP sites flanked the exon 2 of Tgfb2 (Fig. 1D–E). Subsequently, Tgfb2flox/− mice were produced by intercrossing the Tgfb2+/flox and Tgfb2+/− (C57BL/6) mice. Tgfb2flox/− and Tgfb2flox/flox mice were viable and fertile, and the adult Tgfb2flox/− mice were normal and indistinguishable from the wild-type littermate mice (Fig. 1D). In a proof of principle experiment, embryos with a Tgfb2 conditional knockout (Tgfb2cko/−) allele were generated by crossing the Tgfb2flox/− mice with Tgfb2+/−;EIIaCre transgenic mice. EIIaCre mice have ubiquitous Cre activity and are known to generate germline or systemic knockout animals from the floxed animals (Holzenberger et al. 2000;Doetschman et al. 2012b). The data indicated that EIIaCre recombinase successfully excised the exon 2 of Tgfb2 in vivo (Fig. 4A). Histological and immunohistochemical analyses were done and the changes in cardiac structure and morphology were cataloged from the wild-type control, Tgfb2flox/−;EIIaCre−, and Tgfb2flox/−;EIIaCre+ (i.e., Tgfb2cko/−) fetuses at E16.5–E18.5. Cross comparison of cardiac phenotype indicated that Tgfb2cko/− fetuses developed a spectrum of heart defects which resembled the Tgfb2βgeo/βgeo, Tgfb2βgeo/−, and Tgfb2−/− fetuses (Fig. 4B–G). These data indicate that Tgfb2flox mice are fully capable of producing robust conditional Cre-mediated deletion of Tgfb2 in vivo.
Figure 4. Systemic Tgfb2 conditional knockout (Tgfb2cko) fetuses develop congenital heart defects.
A: Genomic PCR analysis of tail DNA samples indicating the Cre recombinase mediated in vivo deletion of loxP flanked exon 2 containing region of the Tgfb2 in Tgfb2flox/−;EIIaCre+ but not Tgfb2flox/−;EIIaCre− fetuses. Cre-mediated deletion of Tgfb2flox allele results in a 174 bp band.
B–G: Cardiac morphology of wild-type (B–C), Tgfb2flox/−;EIIaCre− (D–E), and Tgfb2flox/−;EIIaCre+ (F–G) is indicated by cardiac myosin heavy chain (MF20) immunohistochemistry at E17.5. Tissue sections are counterstained with hematoxylin. Tgfb2flox/−;EIIaCre− (D–E) hearts are normal. Interestingly, Tgfb2flox/−;EIIaCre+ fetuses develop severe malformations of the cardiac outflow tract (F) (arrow, PTA; asterisks, abnormally thickened trucal valves) and inflow tract (G) (arrow, VSD; arrowhead, abnormal myocardium). These cardiac malformations are similar to the ones that are seen in the Tgfb2βgeo/βgeo, Tgfb2βgeo/−, Tgfb2−/− fetuses. Scale bar =200 μm for B–G. Abbreviations: rv, right ventricle; lv, left ventricle; pt, pulmonary trunk; ao, aorta; PTA, persistent truncus arteriosus; VSD, ventricular septal defect
Systemic Tgfb2 deletion studies by very nature are limited in scope, and leave a fundamental gap in our understanding of the critical cell-source of TGFβ2 (endocardium, neural crest and/or myocardium, second heart field, epicardium) as well as its regulatory mechanisms (canonical and/or non-canonical) that mediate cardiovascular development and remodeling. TGFβ2 is involved in adult cardiovascular pathologies including aortic aneurysm, cardiac fibrosis and cardiomyopathy, mitral valve prolapse, and calcific aortic valve disease. In addition, TGFβ2 plays important role in muscular, craniofacial, ocular, chronic liver, kidney, neurodegenerative and autoimmune diseases, osteoarthritis, tissue fibrosis, and various forms of cancer. The expression of Tgfb2 in adult wild-type mouse cardiovascular tissues has not been determined yet. It is known that Tgfb2 expression increases in diseased tissues, and many other pathophysiological states and cancer (Iwata et al. 2012;Lindsay et al. 2012;Friess et al. 1993). Collectively, Tgfb2+/βgeo mice with lacZ-tagged insertion allele will be useful for localizing endogenous Tgfb2 expression in embryos and adults, changes in Tgfb2 expression and distribution in a longitudinal study in the pathogenesis of cardiovascular and other diseases, and in response to stress (i.e., Ang II, aortic coarctation, high fat diet) to induce cardiovascular disease states (e.g., aortic aneurysm, cardiac hypertrophy, atherosclerosis). Finally, Tgfb2flox mice open a new frontier, and have unlimited potential to advance the understanding of TGFβ2 function in embryonic development, tissue homeostasis in adults, pathogenesis of cardiovascular and other diseases, and various forms of cancer. In conclusion, the generation and characterization of Tgfb2βgeo and Tgfb2flox mice is a major first step towards defining the tissue-specific expression and function of Tgfb2 and TGFβ2 regulatory mechanisms in organ development, function, and disease.
Methods
Generation of Tgfb2βgeo and Tgfb2flox mice
All animal breeding and procedures are approved by the Institutional Animal Care and Use Committee (Indiana University School of Medicine). Tgfb2βgeo mice will be made available to other investigators consistent with the general guidelines, policies, and procedures of the Indiana University. Mouse ES cells with Tgfb2 knockout-first lacZ tagged insertion allele (ID:47128, Targeting Confirmed) are available to all non-profit non-commercial investigators through EUCOMM. Three independently targeted clones of Tgfb2βgeo ES cells were obtained from the EUCOMM. The ES cells were male and heterozygous for the Tgfb2βgeo allele. The specific details and the complete DNA sequence of the Tgfb2βgeo gene targeting construct (L1L2_Bact_P) are available in the GeneBank (Accession# JN955293). Tgfb2 is located on mouse chromosome 1 and has 7 exons (NCBI Gene ID# 21808). Tgfb2βgeo is a targeted trap allele which functions as a gene-trap knockout (Skarnes et al. 2011). The targeting vector contained an IRES-LacZ trapping cassette and a floxed promoter-neomycin cassette inserted into an intron 1 of the Tgfb2. The mutagenic cassette had an Engrailed (En2) splice acceptor sequence and poly-A transcription termination signals which was expected to disrupt the Tgfb2 function while expressing the lacZ gene under the control of the endogenous Tgfb2 promoter for studying its gene expression. Mycoplasma testing and chromosome counting was done by EUCOMM. All Tgfb2βgeo ES cell clones were mycoplasma negative. Also, chromosome counting had found no chromosomal abnormalities in Tgfb2βgeo ES cell clones.
We validated the Tgfb2βgeo ES cell clones by 3′ screen and 5′ screen for correct gene targeting using long range PCR (LR-PCR) method. These LR-PCR reactions amplified very large and specific PCR products, ranging from 3.9 kb to 7.4 kb in the Tgfb2βgeo targeted clones. The 5′ LR-PCR had confirmed correct integration of the Tgfb2 on the 5′ side by Tgfb2-specific 5′-outside forward primer (F74 or F75) and constant cassette specific 3′-reverse primer (R66 or R75). The subsequent 5′ LR-PCR product was sequenced with a primer to the upstream FRT to verify the Tgfb2 and FRT site. In addition, the 3′ LR-PCR had confirmed the correct integration of the Tgfb2 on the 3′ side by Tgfb2-specific 3′-outside reverse primer (R76 or R77) and constant cassette specific 5′-forward primer (F103, F76). The subsequent LR-PCR product was sequenced with a primer to the downstream loxP (Tgfb2-rev primer, R64) to verify the Tgfb2 and loxP site. Two or more sets of LR-PCR assays were used in both 5′ and 3′ screen with similar results. The following primers were used in the LR-PCR 5′ screen: CTCCTGATCTCCAGTGATCTTGTGTAAC (F74, forward 5′-outside primer), GTGATATGTGCAATGTCTGATGTACTC (F75, forward 5′-outside primer), CACAACGGGTTCTTCTGTTAGTCC (R66, reverse universal/cassette primer). The following primers were used in the LR-PCR 3′ screen: GCAATAGCATCACAAATTTCACAAATAAAGCA (F103, forward universal/cassette primer), GAGATGGCGCAACGCAATTAATG (F76, forward universal/cassette primer), CAACACACATGGTTCCAACACCACCGCCG (R76, reverse 3′-outside primer), CTCACTATCCTTAGAGAGCTAAGCAAGC (R77, reverse 3′-outside primer). The following PCR conditions were used: denaturation: 93°C/3 min; annealing and amplification: 92°C/15 sec, 65°C/30 sec, 65°C/8 min (−1°C/cycle) for 8x; 92°C/15 sec, 55°C/30 sec, 65°C/8 min (+20 sec/cycle) for 30x; 65°C/9 min, 4°C hold. Phusion® High-Fidelity PCR Master Mix with HF Buffer (New England Biolabs, Inc) was used for amplifying the large PCR products in the both LR-PCR screens.
Strain of origin of Tgfb2βgeo ES cell clones was C57BL/6N (Parental ES cell line: JM8) and the ES cells carried the genotype A/a (Agouti heterozygous). The dominant agouti coat color gene is restored in JM8 cells by targeted repair of the C57BL/6 nonagouti mutation (Pettitt et al. 2009). Tgfb2βgeo ES clones were thawed and expanded, and the blastocysts (C57BL/6) injection and embryo transfer were done by Transgenic & Knockout-Mouse Core Facility (Indiana University School of Medicine). Based on percent coat color (agouti/black), several male chimeras were identified from blastocyst injections of both ES cell clones. At 7 weeks of age the Tgfb2βgeo male chimeras were mated with C57BL/6 wild-type female mice, and a germ line transmission of Tgfb2βgeo allele was successfully established. For genotyping of Tgfb2βgeo mice, we used a Tgfb2 specific primer in the 5′ homology arm (F65) with the constant/cassette primer (R66) in the targeting cassette to detect the Tgfb2βgeo allele. Another Tgfb2 specific primer was designed in the 3′ homology arm (R65) in order to detect the wild-type allele with a PCR fragment between the two Tgfb2 specific primers. DNA sequence of some of the Tgfb2 specific primers that were used for the genotyping includes: CACCTTTTACCTACAGATGAAGTTGC (F65, forward primer), CTTAAGACCACACTGTGAGATAATCC (R65, reverse primer). The following PCR conditions were used: denaturation: 95°C/3 min; annealing and amplification 95°C/30 sec, 60°C/30 sec, 72°C/30 sec for 35x, 72°C/3 min; 4°C hold.
For generation of mice with a Tgfb2flox allele, Tgfb2+/βgeo female mice were crossed to FLPeR mice. PCR primers and PCR conditions for genotyping FLPeR transgenic mice were used as recommended (Farley et al. 2000). For initial screening of Tgfb2flox and Tgfb2βgeo allele, Tgfb2-5′ arm (F65), constant cassette (R66), Tgfb2-3′arm (R65) PCR primers were used. In addition, two specific sets of PCR primers which identified Tgfb2flox but not the Tgfb2βgeo allele were used for further confirmation. These two independent PCR genotyping reactions used gene-specific (F86) and constant cassette (R86), and gene-specific forward (F86) and reverse (R88) primers, respectively. Tgfb2flox female mice were crossed with EIIaCre deleter mice (Holzenberger et al. 2000). PCR genotyping for the EIIaCre transgenic pups were done as published (Doetschman et al. 2012b). Genomic PCR on tail DNA samples (F86 and R88) were used for detecting the Cre-mediated recombination in the Tgfb2flox/−;EIIaCre mice. PCR genotyping for detecting Tgfb2+/− allele was performed as described (Sanford et al. 1997). DNA sequence of the additional primers that were used for the Tgfb2flox or Tgfb2cko allele genotyping included: AAGGCGCATAACGATACCAC (F86, forward primer), CCGCCTACTGCGACTATAGAGA (R86, reverse primer), ACTGATGGCGAGCTCAGACC (R88, reverse primer). The following PCR conditions were used for genotyping Tgfb2flox or Tgfb2cko allele: denaturation: 94°C/3 min; annealing and amplification 94°C/30 sec, 58°C/30 sec, 72°C/45 sec for 35x, 72°C/5 min; 4°C hold.
Histological, immunohistochemical and X-gal staining
Wild-type control and various groups of experimental embryos collected between E13.5 and E18.5 were processed for histological and molecular analyses as described (Azhar et al. 2011). Embryos were genotyped using genomic DNA extracted from tail biopsies. Hematoxylin and eosin staining was performed on 7-μm-thick serial sections of heart for routine histological examination (n=11 for E13.5–E18.5). Cardiac structure and morphology was determined by immunohistochemistry using cardiac myosin heavy chain (MF20) antibody (Developmental Studies Hybridoma Bank, Iowa). Tissue collection, processing, and β-galactosidase staining on 14 μm thick frozen (O.C.T.) tissue sections were done according to the published protocol (Komatsu et al. 2014). X-gal-stained tissue sections were counterstained with nuclear fast red (Vector Lab, Burlingame, CA). Images of whole embryos were captured in a stereozoom microscope (Zeiss Stemi 2000-C). All sections were visualized under brightfield optics with a Zeiss AxioLab.A1 light microscope (Carl Zeiss Microimaging, Inc.), and the morphometric measurements on the captured images were done by AxioVision Zeiss imaging software and NIH Image J (Fiji).
Real-time quantitative PCR Analysis
Real-time qPCR analysis (via intron spanning assay) was done using Universal ProbeLibrary (UPL) assay according to the manufacturer’s protocols (Roche Inc, Indianapolis, IN). Expression of lacZ was detected using RT-PCR. Whole hearts from wild-type or Tgfb2 gene-trap knockout or conditional knockout fetuses at E16.5 were collected under a stereozoom microscope (Zeiss Stemi 2000-C). Total RNA was isolated by RNeasy Mini kit (Qiagen, Valencia, CA). Three different samples of wild-type and experimental fetal hearts were assessed by real-time qPCR in LightCycler 480 (Roche Inc, Indianapolis, IN). ProbeFinder Assay Design Software (Roche Inc, Indianapolis, IN) was used to select the target-specific primer sequences and the matching Mouse Universal ProbeLibrary probe. Each reaction was performed in triplicate. The relative amount of target mRNA normalized to β-actin and to the wild-type was calculated. Specific primers and probes that were used in the qPCR or RT-PCR assays included: ATCACGACGCGCTGTATC (lacZ forward primer), ACATCGGGCAAATAATATCG (lacZ reverse primer), TGGAGTTCAGACACTCAACACA (Tgfb2 exon 6-specific forward primer), AAGCTTCGGGATTTATGGTGT (Tgfb2 exon 7-specific reverse primer), TCCTCAGC (Tgfb2 exon 7-specific UPL probe #73). The UPL Reference Gene Assay for β-actin (#05046190001, Roche, Inc) was used for quantification of gene expression using dual-color real-time qPCR. The following conditions were used for lacZ PCR: denaturation- 94°C/5 min; annealing and amplification- 94°C/30 sec, 58°C/30 sec, 72°C/45 sec for 35x, 72°C/5 min; 12°C hold. For UPL qPCR, the following conditions were used: Pre-incubation: 95°C/4 min; Amplification: 95°C/10 sec, 60°C/30 sec, 72°C/01 sec for 45x; Cooling: 40°C/30 sec.
Statistical Analysis
All experiments were done on three or more embryos per genotype per developmental stage with similar results. Microsoft Excel was used for managing the raw data. Statistics was performed using pair-wise comparisons between the groups, utilizing analysis of variance and unpaired two-tailed t-test (SigmaPlot, Systat Software, Inc., CA). Data was reported as means ± SE of the mean. Probability values <0.05 were considered as significant.
Acknowledgments
Supported, in part, by the Indiana University Department of Pediatrics (Neonatal-Perinatal Medicine), Riley Children’s Foundation, and Simmons Clinical Fund, Biomedical Research Grant (IU School of Medicine), and the Indiana CTSI pilot fund (via Grant # TR000006 from the National Institutes of Health, National Center for Advancing Translational Sciences, Clinical and Translational Sciences Award) grants (M.A.).
We thank the Transgenic & Knockout-Mouse Core Facility at the Indiana University School of Medicine and Mr. Bill Carter for generating the chimeric Tgfb2βgeo mice.
LITERATURE CITED
- Akhurst RJ, Hata A. Targeting the TGFbeta signalling pathway in disease. Nat Rev Drug Discov. 2012;11:790–811. doi: 10.1038/nrd3810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arthur HM, Bamforth SD. TGFbeta signaling and congenital heart disease: Insights from mouse studies. Birth Defects Res A Clin Mol Teratol. 2011;91:423–434. doi: 10.1002/bdra.20794. [DOI] [PubMed] [Google Scholar]
- Azhar M, Brown K, Gard C, Chen H, Rajan S, Elliott DA, Stevens MV, Camenisch TD, Conway SJ, Doetschman T. Transforming growth factor Beta2 is required for valve remodeling during heart development. Dev Dyn. 2011;240:2127–2141. doi: 10.1002/dvdy.22702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azhar M, Schultz JE, Grupp I, Dorn GW, Meneton P, Molin DG, Gittenberger-de Groot AC, Doetschman T. Transforming growth factor beta in cardiovascular development and function. Cytokine Growth Factor Rev. 2003;14:391–407. doi: 10.1016/s1359-6101(03)00044-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azhar M, Yin M, Bommireddy R, Duffy JJ, Yang J, Pawlowski SA, Boivin GP, Engle SJ, Sanford LP, Grisham C, Singh RR, Babcock GF, Doetschman T. Generation of mice with a conditional allele for transforming growth factor beta 1 gene. Genesis. 2009;47:423–431. doi: 10.1002/dvg.20516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartram U, Molin DG, Wisse LJ, Mohamad A, Sanford LP, Doetschman T, Speer CP, Poelmann RE, Gittenberger-de GA. Double-outlet right ventricle and overriding tricuspid valve reflect disturbances of looping, myocardialization, endocardial cushion differentiation, and apoptosis in Tgfb2 knockout mice. Circulation. 2001;103:2745–2752. doi: 10.1161/01.cir.103.22.2745. [DOI] [PubMed] [Google Scholar]
- Boileau C, Guo DC, Hanna N, Regalado ES, Detaint D, Gong L, Varret M, Prakash SK, Li AH, d’Indy H, Braverman AC, Grandchamp B, Kwartler CS, Gouya L, Santos-Cortez RL, Abifadel M, Leal SM, Muti C, Shendure J, Gross MS, Rieder MJ, Vahanian A, Nickerson DA, Michel JB, Jondeau G, Milewicz DM. TGFB2 mutations cause familial thoracic aortic aneurysms and dissections associated with mild systemic features of Marfan syndrome. Nat Genet. 2012;44:916–921. doi: 10.1038/ng.2348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conway SJ, Kaartinen V. TGFbeta superfamily signaling in the neural crest lineage. Cell Adh Migr. 2011;5:232–236. doi: 10.4161/cam.5.3.15498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickson MC, Slager HG, Duffie E, Mummery CL, Akhurst RJ. RNA and protein localisations of TGF beta 2 in the early mouse embryo suggest an involvement in cardiac development. Development. 1993;117:625–639. doi: 10.1242/dev.117.2.625. [DOI] [PubMed] [Google Scholar]
- Doetschman T, Barnett JV, Runyan RB, Camenisch TD, Heimark RL, Granzier HL, Conway SJ, Azhar M. Transforming growth factor beta signaling in adult cardiovascular diseases and repair. Cell Tissue Res. 2012a;347:203–223. doi: 10.1007/s00441-011-1241-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doetschman T, Georgieva T, Li H, Reed TD, Grisham C, Friel J, Estabrook MA, Gard C, Sanford LP, Azhar M. Generation of mice with a conditional allele for the transforming growth factor beta3 gene. Genesis. 2012b;50:59–66. doi: 10.1002/dvg.20789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farley FW, Soriano P, Steffen LS, Dymecki SM. Widespread recombinase expression using FLPeR (flipper) mice. Genesis. 2000;28:106–110. [PubMed] [Google Scholar]
- Friess H, Yamanaka Y, Buchler M, Ebert M, Beger HG, Gold LI, Korc M. Enhanced expression of transforming growth factor beta isoforms in pancreatic cancer correlates with decreased survival. Gastroenterology. 1993;105:1846–1856. doi: 10.1016/0016-5085(93)91084-u. [DOI] [PubMed] [Google Scholar]
- Holzenberger M, Lenzner C, Leneuve P, Zaoui R, Hamard G, Vaulont S, Bouc YL. Cre-mediated germline mosaicism: a method allowing rapid generation of several alleles of a target gene. Nucleic Acids Res. 2000;28:E92. doi: 10.1093/nar/28.21.e92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horiguchi M, Ota M, Rifkin DB. Matrix control of transforming growth factor-beta function. J Biochem. 2012;152:321–329. doi: 10.1093/jb/mvs089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwata J, Hacia JG, Suzuki A, Sanchez-Lara PA, Urata M, Chai Y. Modulation of noncanonical TGF-beta signaling prevents cleft palate in Tgfbr2 mutant mice. J Clin Invest. 2012;122:873–885. doi: 10.1172/JCI61498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komatsu Y, Kishigami S, Mishina Y. In situ hybridization methods for mouse whole mounts and tissue sections with and without additional beta-galactosidase staining. Methods Mol Biol. 2014;1092:1–15. doi: 10.1007/978-1-60327-292-6_1.:1-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulkarni AB, Thyagarajan T, Letterio JJ. Function of cytokines within the TGF-beta superfamily as determined from transgenic and gene knockout studies in mice. Curr Mol Med. 2002;2:303–327. doi: 10.2174/1566524024605699. [DOI] [PubMed] [Google Scholar]
- Lindsay ME, Schepers D, Bolar NA, Doyle JJ, Gallo E, Fert-Bober J, Kempers MJ, Fishman EK, Chen Y, Myers L, Bjeda D, Oswald G, Elias AF, Levy HP, Anderlid BM, Yang MH, Bongers EM, Timmermans J, Braverman AC, Canham N, Mortier GR, Brunner HG, Byers PH, Van EJ, van LL, Dietz HC, Loeys BL. Loss-of-function mutations in TGFB2 cause a syndromic presentation of thoracic aortic aneurysm. Nat Genet. 2012;44:922–927. doi: 10.1038/ng.2349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loeys BL, Mortier G, Dietz HC. Bone lessons from Marfan syndrome and related disorders: fibrillin, TGF-B and BMP at the balance of too long and too short. Pediatr Endocrinol Rev. 2013;10(Suppl 2):417–23. 417–423. [PubMed] [Google Scholar]
- Maccarrick G, Black JH, III, Bowdin S, El-Hamamsy I, Frischmeyer-Guerrerio PA, Guerrerio AL, Sponseller PD, Loeys B, Dietz HC., III Loeys-Dietz syndrome: a primer for diagnosis and management. Genet Med10. 2014 doi: 10.1038/gim.2014.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molin DG, Bartram U, Van der HK, Van Iperen L, Speer CP, Hierck BP, Poelmann RE, Gittenberger-de-Groot AC. Expression patterns of Tgfbeta1-3 associate with myocardialisation of the outflow tract and the development of the epicardium and the fibrous heart skeleton. Dev Dyn. 2003;227:431–444. doi: 10.1002/dvdy.10314. [DOI] [PubMed] [Google Scholar]
- Pettitt SJ, Liang Q, Rairdan XY, Moran JL, Prosser HM, Beier DR, Lloyd KC, Bradley A, Skarnes WC. Agouti C57BL/6N embryonic stem cells for mouse genetic resources. Nat Methods. 2009;6:493–495. doi: 10.1038/nmeth.1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renard M, Callewaert B, Malfait F, Campens L, Sharif S, Del CM, Valenzuela I, McWilliam C, Coucke P, De PA, De BJ. Thoracic aortic-aneurysm and dissection in association with significant mitral valve disease caused by mutations in TGFB2. Int J Cardiol. 2012;10 doi: 10.1016/j.ijcard.2012.09.029. [DOI] [PubMed] [Google Scholar]
- Sanford LP, Ormsby I, Gittenberger-de GA, Sariola H, Friedman R, Boivin GP, Cardell EL, Doetschman T. TGFbeta2 knockout mice have multiple developmental defects that are non- overlapping with other TGFbeta knockout phenotypes. Development. 1997;124:2659–2670. doi: 10.1242/dev.124.13.2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu C, Jain S, Davila S, Hibberd ML, Lin KO, Molkara D, Frazer JR, Sun S, Baker AL, Newburger JW, Rowley AH, Shulman ST, Davila S, Burgner D, Breunis WB, Kuijpers TW, Wright VJ, Levin M, Eleftherohorinou H, Coin L, Popper SJ, Relman DA, Fury W, Lin C, Mellis S, Tremoulet AH, Burns JC. Transforming Growth Factor-{beta} Signaling Pathway in Patients With Kawasaki Disease. Circ Cardiovasc Genet. 2011;4:16–25. doi: 10.1161/CIRCGENETICS.110.940858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skarnes WC, Rosen B, West AP, Koutsourakis M, Bushell W, Iyer V, Mujica AO, Thomas M, Harrow J, Cox T, Jackson D, Severin J, Biggs P, Fu J, Nefedov M, de Jong PJ, Stewart AF, Bradley A. A conditional knockout resource for the genome-wide study of mouse gene function. Nature. 2011;474:337–342. doi: 10.1038/nature10163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soderberg SS, Karlsson G, Karlsson S. Complex and context dependent regulation of hematopoiesis by TGF-beta superfamily signaling. Ann N Y Acad Sci. 2009;1176:55–69. doi: 10.1111/j.1749-6632.2009.04569.x.:55-69. [DOI] [PubMed] [Google Scholar]
- Sonnylal S, Denton CP, Zheng B, Keene DR, He R, Adams HP, Vanpelt CS, Geng YJ, Deng JM, Behringer RR, de CB. Postnatal induction of transforming growth factor beta signaling in fibroblasts of mice recapitulates clinical, histologic, and biochemical features of scleroderma. Arthritis Rheum. 2007;56:334–344. doi: 10.1002/art.22328. [DOI] [PubMed] [Google Scholar]
- Yumoto K, Thomas PS, Lane J, Matsuzaki K, Inagaki M, Ninomiya-Tsuji J, Scott GJ, Ray MK, Ishii M, Maxson R, Mishina Y, Kaartinen V. TGF-beta-activated kinase 1 (Tak1) mediates agonist-induced Smad activation and linker region phosphorylation in embryonic craniofacial neural crest-derived cells. J Biol Chem. 2013;288:13467–13480. doi: 10.1074/jbc.M112.431775. [DOI] [PMC free article] [PubMed] [Google Scholar]