Abstract
Genetically engineered mouse models (GEMMs) have dramatically improved our understanding of tumor evolution and therapeutic resistance. However, sequential genetic manipulation of gene expression and targeting of the host is almost impossible using conventional Cre-loxP–based models. We have developed an inducible dual-recombinase system by combining flippase-FRT (Flp-FRT) and Cre-loxP recombination technologies to improve GEMMs of pancreatic cancer. This enables investigation of multistep carcinogenesis, genetic manipulation of tumor subpopulations (such as cancer stem cells), selective targeting of the tumor microenvironment and genetic validation of therapeutic targets in autochthonous tumors on a genome-wide scale. As a proof of concept, we performed tumor cell–autonomous and nonautonomous targeting, recapitulated hallmarks of human multistep carcinogenesis, validated genetic therapy by 3-phosphoinositide-dependent protein kinase inactivation as well as cancer cell depletion and show that mast cells in the tumor microenvironment, which had been thought to be key oncogenic players, are dispensable for tumor formation.
Pancreatic ductal adenocarcinoma (PDAC) is a fearsome disease, with a mortality rate >95% that has remained unchanged for decades1.
GEMMs that faithfully recapitulate the histological, molecular, genetic and clinical hallmarks of human PDAC have been developed2–5. All are based on pancreatic expression of oncogenic KrasG12D or KrasG12V, which induce pancreatic intraepithelial neoplasias (PanINs) that progress to aggressive metastatic PDAC2–4,6–9. These GEMMs have elucidated the natural biology of pancreatic cancer, revealed potential diagnostic and therapeutic targets10,11 and highlighted the importance of the tumor stroma for PDAC maintenance, immune evasion and drug resistance5,12–14.
However, classical Cre-loxP–based GEMMs rely on a single Cre-mediated recombination step to activate mutant Kras expression and do not allow genetic modeling and manipulation of sequential multistep tumorigenesis and tumor heterogeneity, which are important hallmarks of the disease. A single-step approach does not enable genetic validation of possible targets by blocking PanIN progression or treating established PDAC, nor does it enable genetic investigation of resistance mechanisms or manipulation of the tumor microenvironment.
We addressed these limitations by generating a ‘next-generation’ mouse model that allows controlled independent or sequential manipulation of genes involved in the development and maintenance of PDAC.
RESULTS
Sequential genetic manipulation of the pancreas
To create a dual-recombinase system (DRS), we generated transgenic mice expressing Flprecombinase directed by the mouse Pdx1 promoter15 (Pdx1-Flp; Supplementary Fig. 1a,b). A Flp-activated alkaline phosphatase reporter allele (FSF-R26hpAP/+) revealed recombination in pancreatic islets, ducts and acini, and we observed extra-pancreatic recombination in bile duct, duodenum and stomach, paralleling established Pdx1-Cre lines2,15 (Supplementary Fig. 1c–f and Supplementary Table 1).
To manipulate Flp-recombined cells sequentially using Cre, we generated a latent tamoxifen-inducible allele (CreERT2) silenced by an FRT-stop-FRT (FSF) cassette under the control of the CAG promoter as a Rosa26 knock-in (FSF-R26CAG-CreERT2/+; Supplementary Fig. 2a, b). To activate CreERT2 expression and monitor tamoxifen-induced CreERT2-mediated recombination in the Flp-lineage, we used the Pdx1-Flp line together with a dual-fluorescent tdTomato-EGFP-Cre reporter (R26mT-mG) that switches expression from tdTomato to EGFP after Cre recombination (Supplementary Fig. 2c, d). Tamoxifen treatment induced EGFP expression in pancreatic acini, ducts and islets; stromal cells remained unrecombined (Supplementary Fig. 2d). Immunohistochemistry confirmed colocalization of EGFP with markers for acini (α-amylase), ducts (CK19) and islets (insulin); we observed sporadic extrapancreatic recombination in stomach, duodenum and bile duct (Supplementary Fig. 2c–f and data not shown). There was no evidence of recombination in the absence of tamoxifen, excluding leaky Cre activity in this system.
Tamoxifen-inducible genetic manipulation of the whole animal
Toxicity in non-neoplastic cells is a major limitation of many current drugs. It is thus important to evaluate the consequences of inactivating a specific therapeutic target or pathway in whole animals. To mimic drug treatment and reveal potential toxic side effects, we generated a CreERT2 deleter model, R26CAG-CreERT2, which ubiquitously expresses CreERT2 (Supplementary Fig. 3a).
High-dose tamoxifen treatment resulted in efficient Cre recombination of the R26mT-mG reporter in various tissues, at low dose in just a few cells, whereas untreated mice showed none (Supplementary Figs. 3b, c and 4).
Activation of oncogenic KrasG12D induces metastatic PDAC
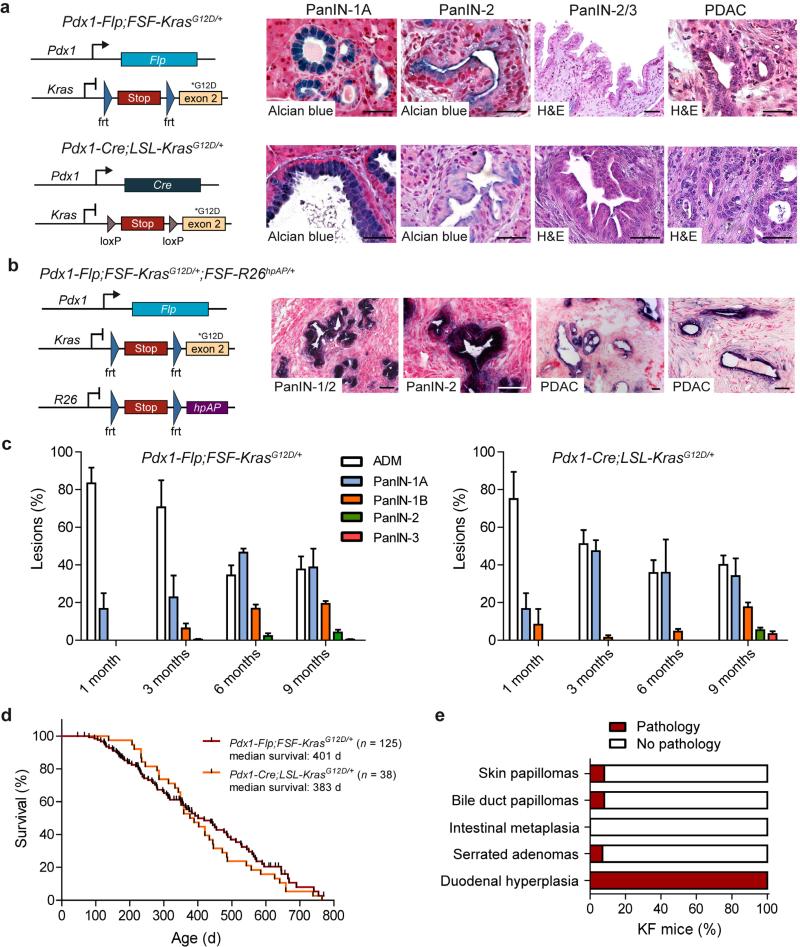
To achieve conditional activation of oncogenic Kras in the Pdx1-Flp lineage, we generated a FSF-silenced latent KrasG12D knock-in allele (FSF-KrasG12D/+; Supplementary Fig. 5a, b). KrasG12D expression from the endogenous promoter in Pdx1-Flp-FSF-KrasG12D/+ (termed KF) mice induced PanIN precursor lesions and PDAC originating from Pdx1-Flp–recombined cells, as confirmed by the FSF-R26hpAP reporter (Fig. 1a, b).
Figure 1.
Pdx1-Flp–activated expression of oncogenic KrasG12D induces premalignant PanIN and PDAC. (a) Left, genetic strategy to activate oncogenic Kras in the pancreas using the Flp-FRT (top) and the Cre-loxP (bottom) recombination system. Right, representative alcian blue and hematoxylin and eosin (H&E) stained sections of different grades (1-3) of pancreatic intraepithelial neoplasia (PanIN) and invasive PDAC of male and female Pdx1-Flp;FSFKrasG12D/+ (top) and Pdx1-Cre;LSL-KrasG12D/+ (bottom) mice. (b) Genetic strategy to induce oncogenic KrasG12D and human placental alkaline phosphatase (hpAP) reporter gene expression in the Pdx1-Flp lineage (left). Representative alkaline phosphatase staining (purple) of the indicated grades of PanIN lesions and PDAC cells in male and female Pdx1-Flp;FSF-KrasG12D/+;FSF-R26hpAP/+ mice (right). (c) Quantification of acinar to ductal metaplasia (ADM) and PanIN progression (from PanIN grade -1A (flat epithelial lesion), -1B (papillary lesion), -2 (lesion with nuclear abnormalities) to PanIN grade -3 (carcinoma in situ)) as a percentage of total lesions in Pdx1-Flp;FSF-KrasG12D/+ and Pdx1-Cre;LSLKrasG12D/+ mice (error bars, s.e.m.; n = 3 representative slides from three mice per time point and genotype). (d) Kaplan-Meier survival curves of Pdx1-Flp;FSF-KrasG12D/+ and Pdx1-Cre;LSL-KrasG12D/+ mice. (e) Occurrence of extrapancreatic tumors in KF mice. Scale bars, 50 μm.
The KF and the classical Pdx1-Cre;LSL-KrasG12D/+ (termed KC)2 mice showed similar patterns of PanIN progression and PDAC formation (Fig. 1a, c, d). Tumor latency, morphology, survival time and rates of metastasis were nearly identical (Fig. 1d and Supplementary Fig 5c–e). PDAC in KF and KC mice were histopathologically indistinguishable, showing the full disease spectrum, from well-differentiated to undifferentiated tumors and typical liver and lung metastasis (Fig. 1a,b, Supplementary Fig. 5c–e and data not shown). Both KF and KC models thus faithfully recapitulate human PDAC2,12.
To accelerate PDAC formation, we generated KPF mice, in which the tumor suppressor p53 is inactivated through usage of an FRT-flanked Trp53 allele (Trp53frt) (Supplementary Fig. 6). We also crossed the KF model with a mouse line that lacks p53 in the whole body (Trp53Δ) and compared PDAC formation, metastasis rates and survival with the classical KPC (Pdx1-Cre;LSL-KrasG12D/+;LSL-Trp53R172H/+) model, which carries a p53R172H gain of function mutation 12,16. Again, tumors were histopathologically indistinguishable, revealing the full spectrum of human PDAC, from well- to poorly-differentiated and anaplastic tumors (Supplementary Fig. 6a–c). Median survival times and rates of metastasis were very similar (Supplementary Fig. 6a–e), and the number of metastatic lesions varied between one and several dozen in all models.
To assess extrapancreatic tumor development, we analyzed KF mice and observed sporadic noninvasive skin and bile duct papillomas and serrated duodenal adenomas in <10% of KF mice (Fig. 1e); by contrast, the incidence of skin papillomas is as high as 80% in KC mice2. Notably, we observed no extrapancreatic invasive carcinomas. Overall, the extrapancreatic pathologies of KF mice resembled those of the KC model2 but occurred at much lower frequencies. Inactivation of p53 did not affect the frequency or invasiveness of extrapancreatic tumors, probably owing to the rapidity of PDAC development (Supplementary Fig. 6a–c).
These results indicate that the KF and KPF models phenocopy the widely used KC and KPC models2,16 and may provide an improvement by reducing unwanted extrapancreatic tumor development.
Sequential genetic manipulation of PanIN lesions and PDAC
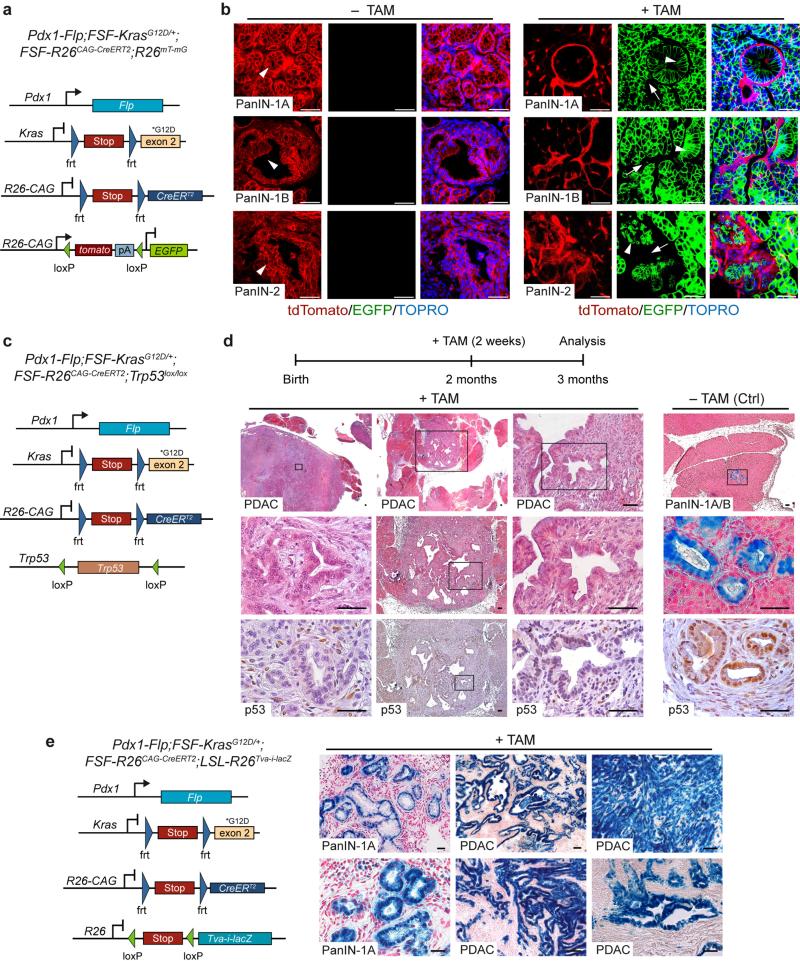
KF mice develop PanIN and PDAC. To target and manipulate these lesions at a chosen time point using Cre-loxP, we crossed the FSF-R26CAG-CreERT2 allele into KF mice and used the R26mT-mG-Cre reporter to visualize the secondary genetic alteration (Fig. 2a). Tamoxifen-activation of CreERT2 resulted in efficient excision of tdTomato and induction of EGFP in PanIN lesions in vivo (Fig. 2b). Tumor stroma showed no evidence of recombination. No recombination was detected in vehicle-treated mice (Fig. 2b and data not shown), excluding leaky Cre activity or spontaneous recombination events in this system.
Figure 2.
Secondary genetic manipulation of established KrasG12D-induced PanIN lesions and PDAC cells in the Pdx1-Flp lineage. (a) Genetic strategy to induce EGFP expression in the Flp lineage by tamoxifen-mediated activation of CreERT2. (b) Representative confocal microscopic images of tdTomato (red, non–Cre-recombined cells) and Cre-induced EGFP (green) expression in the pancreas and indicated PanIN lesions of tamoxifen- (+TAM) and vehicle-treated (–TAM) male and female mice. Arrowheads indicate highest-grade PanIN lesions; arrows indicate stromal cells. Nuclei were counterstained with TOPRO-3 (blue). (c) Genetic strategy to recapitulate human multistep carcinogenesis by time-specific p53 inactivation. (d) Top, schematic of tamoxifen treatment protocol. Bottom, representative alcian blue (blue) and H&E-stained pancreatic tissue sections of randomized tamoxifen-treated Pdx1-Flp;FSF-KrasG12D/+;FSF-R26CAG-CreERT2/+;Trp53lox/lox mice (+TAM) and untreated age- and sex-matched littermate controls (–TAM). n = 3 mice per group. Bottom, immunohistochemical p53 staining demonstrates loss of p53 expression in TAM-treated tumors but not in untreated controls. (e) Genetic strategy to induce lacZ expression by tamoxifen-mediated CreERT2 activation (left). Right, representative β-galactosidase staining (n = 3 representative slides from three mice) shows expression of lacZ in PanIN lesions and PDAC, but not in desmoplastic stroma of tamoxifen-treated male and female Pdx1-Flp;FSFKrasG12D/+;FSF-R26CAG-CreERT2;LSL-R26Tva-i-lacZ mice. Scale bars, 50 μm.
To investigate whether our model provides the opportunity to recapitulate crucial aspects of human multistep carcinogenesis, we uncoupled temporal activation of oncogenic Kras from elimination of p53. We activated CreERT2 by tamoxifen treatment in 2-month-old Pdx1-Flp;FSF-KrasG12D/+;FSF-R26CAG-CreERT2/+ mice with and without loxP-flanked (floxed) Trp53 (Fig. 2c,d) and analyzed tumor development 1 month later. Inactivation of p53 in a stage-specific fashion induced rapid formation of multifocal PDAC. Histopathology revealed tumors that were well-to-moderately differentiated or undifferentiated. Untreated Pdx1-Flp;FSF-KrasG12D/+;FSF-R26CAG-CreERT2/+;Trp53lox/lox littermate controls and tamoxifen-treated mice lacking the floxed Trp53 showed only PanIN lesions (Fig. 2d and Supplementary Fig. 7b).
To investigate whether gene expression can be modulated in a stage-specific manner, including in established PDAC, we used a Cre-activatable lacZ reporter line (LSL-R26Tva-i-lacZ)4. CreERT2 activation by tamoxifen resulted in lacZ expression in PanIN and PDAC in vivo (Fig. 2e and Supplementary Fig. 8a–c). Again, the tumor stroma showed no apparent recombination events. Importantly, we observed no difference in recombination efficacy between stroma-rich, well-differentiated tumors and undifferentiated PDAC lacking desmoplasia (Fig. 2e and Supplementary Fig. 8c).
To manipulate PDAC in vitro, we isolated and cultured tumor cells from tamoxifen-naive Pdx1-Flp;FSF-KrasG12D/+;FSF-R26CAG-CreERT2 mice carrying the R26mT-mG-Cre reporter. Administration of tamoxifen resulted in EGFP expression within 24 h (Supplementary Fig. 8d).
The DRS enables validation of therapeutic targets
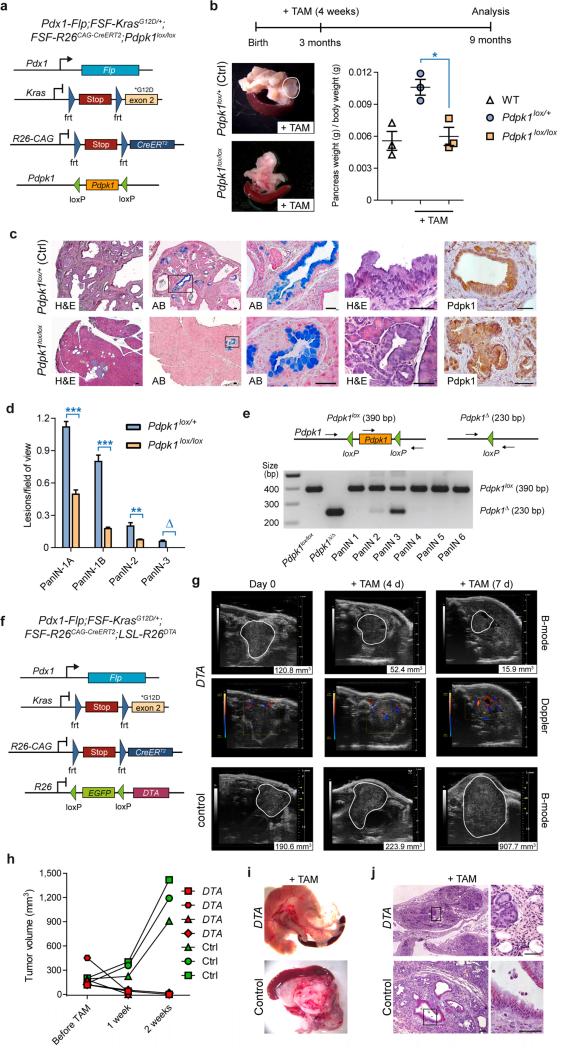
To test whether the DRS can be used to define and validate therapeutic targets in vivo, we crossed Pdx1-Flp;FSF-KrasG12D/+;FSF-R26CAG-CreERT2/+ mice with conditional floxed Pdpk1 (encoding 3-phosphoinositide-dependent protein kinase 1 (Pdpk1)) knockout mice17. This enables tamoxifen-mediated deletion of Pdpk1 in KrasG12D-induced PanIN (Fig. 3a,b). Pdpk1 is an important downstream effector of phosphoinositide 3-kinase and essential for KrasG12D-induced PDAC initiation17. However, it remains unclear whether Pdpk1 is also necessary for PDAC progression. To inactivate floxed Pdpk1 (Pdpk1lox/lox) in a time- and stage-controlled fashion after PanIN formation, we activated CreERT2 in 3-month-old Pdx1-Flp;FSFKrasG12D/+;FSF-R26CAG-CreERT2;Pdpk1lox/lox conditional knockout animals and Pdx1-Flp;FSFKrasG12D/+;FSF-R26CAG-CreERT2;Pdpk1lox/+ controls and analyzed pancreata in mice at age 9 months. Inactivation of both Pdpk1 alleles resulted in macroscopically normal pancreata with normal size and weight, whereas tamoxifen-treated heterozygous Pdpk1lox/+ controls showed increased pancreatic weight and size and macroscopic signs of tumor development (Fig. 3b). Pdpk1-deleted mice retained normal pancreatic tissue architecture and showed only sporadic PanINs, whereas pancreata from control mice were nearly completely replaced by tumorous tissue owing to induction of low- and high-grade PanIN lesions (Fig. 3c,d). Thus, Pdpk1 ablation in PanIN-bearing animals blocked tumor progression almost completely. Notably, the few PanINs that were present in tamoxifen-treated Pdpk1lox/lox mice expressed Pdpk1 (Fig. 3c), suggesting incomplete recombination of the Pdpk1 locus. This was confirmed by laser-capture microdissection and genotyping PCR, which showed the intact floxed Pdpk1 band (Fig. 3e). Thus, KrasG12D-driven PDAC progression depends on intact Pdpk1 expression.
Figure 3.
Validation of therapeutic targets in vivo by Cre-induced time-specific Pdpk1 inactivation or DTA-mediated tumor cell depletion. (a) Genetic strategy to delete Pdpk1 in established PanIN lesions by time-specific tamoxifen-mediated CreERT2 activation. (b) Top, schematic of tamoxifen (TAM) treatment protocol. Bottom, representative macroscopic view (left) and weight (right) of pancreata from TAM-treated homozygous conditional Pdpk1-knockout mice (Pdpk1lox/lox) and age- and sex-matched TAM-treated heterozygous controls (Pdpk1lox/+). Visible tumors are outlined in white. WT, wild type. Data represent mean ± s.e.m; n = 3 female mice per genotype; *P = 0.0148, Student's t test. (c) Representative pancreatic sections of TAM-treated Pdpk1lox/+ control (top) and Pdpk1lox/lox (bottom) mice stained with standard H&E or alcian blue (AB) to visualize mucinous PanIN lesions stained in blue and immunohistochemical Pdpk1 staining. (d) Absolute quantification of different grades of PanIN lesions (grade −1A, flat epithelial lesion; grade −1B, papillary lesion; grade −2, lesion with nuclear abnormalities; grade −3, carcinoma in situ) in 9-month-old sex-matched TAM-treated mice with genotypes as indicated (mean and s.e.m.; n = 3 female mice per genotype; 3 representative slides per mouse; ***P < 0.001, **P < 0.01, Student's t test; ∆P = 0.05, Fisher's exact test). (e) Top, genotyping strategy to detect Pdpk1 alleles. Bottom, PCR analysis of nonrecombined Pdpk1lox/lox DNA from a mouse without Cre expression, recombined pancreatic tissue from a Ptf1aCre/+;LSL-KrasG12D/+;Pdpk1lox/lox (Pdpk1Δ/Δ;17) mouse (positive control for recombined allele) and microdissected PanIN lesions from TAM-treated Pdpk1lox/lox mice. (f) Genetic strategy to induce DTA expression in established PDAC by tamoxifen-mediated CreERT2 activation. (g) Tumor growth was monitored by high-resolution ultrasound in sex-matched DTA and control mice with mean tumor diameters >5 mm before (day 0) and 4 and 7 d after treatment with tamoxifen (+TAM). Macroscopic tumor perfusion was evaluated by Doppler ultrasound. Visible lesions are outlined in white; numbers (bottom right corner) indicate tumor burden as determined by automated three-dimensional B-mode imaging. (h) Quantification of tumor volume in TAM-treated DTA (n = 4) and control (Ctrl, n = 3) mice. (i) Representative macroscopic view of pancreas from TAM-treated DTA (top) and control (bottom) mouse. (j) Representative H&E stains of TAM-treated DTA (top) and control (bottom) mouse. Scale bars, 50 μm.
To investigate whether the DRS can be used to validate therapeutic targets in established invasive PDAC in vitro and in vivo, we crossed Pdx1-Flp;FSF-KrasG12D/+;FSFR26CAG-CreERT2 mice with conditional LSL-R26DTA/+ mice, which carry a latent DTA allele (encoding diphtheria toxin A) (Fig. 3f). This enables depletion of all Flp-recombined cells as a proof of concept.
Tamoxifen-mediated Cre activation and DTA expression in cultured PDAC cells led to extensive cell death and depletion of PDAC cells in vitro (Supplementary Fig. 9a–c). To investigate the feasibility of PDAC depletion in vivo, we monitored PDAC formation in Pdx1-Flp;FSF-KrasG12D/+;FSF-R26CAG-CreERT2 mice with (DTA) or without (control) the LSLR26DTA allele by high-resolution ultrasound. Mice with established tumors of comparable size were then enrolled and treated with tamoxifen to activate Cre and express DTA. Tamoxifen-mediated DTA induction led to rapid regression of tumor volume, whereas control mice showed rapid disease progression (Fig. 3f–h and Supplementary Fig. 9d).
After 7–14 d tamoxifen treatment, mice in the conditional DTA cohort showed tumor regression, and pancreatic masses were rarely detectable by ultrasound, macroscopic pathological examination or histopathology (Fig. 3g–j and Supplementary Fig. 9d–f). Other pancreata of the DTA cohort displayed some residual small tumor nodules, with histopathological signs of intact tumor tissue (Supplementary Fig. 9e,f). As we expected, tamoxifen treatment resulted in pancreatitis and pancreatic atrophy due to DTA-mediated cell depletion of both neoplastic and normal pancreatic tissue (Fig. 3i,j and Supplementary Fig. 9e,f). Histopathological analysis and abnormally elevated serum lipase concentrations of tamoxifen-treated Pdx1-Flp;FSF-R26CAG-CreERT2;LSL-R26DTA mice without the oncogenic KrasG12D allele confirmed pancreatitis induction by DTA expression. Tamoxifen-treated control mice showed normal pancreatic histology, no signs of inflammation or atrophy and normal serum lipase levels (Supplementary Fig. 9g,h).
To investigate whether tamoxifen or activation of CreERT2 itself affects PDAC growth, we also treated Pdx1-Flp;FSF-KrasG12D/+;FSF-R26CAG-CreERT2 tumor cells and mice with the FSF-R26CAG-CreERT2 allele but not the latent DTA allele with tamoxifen. These cells and tumors continued to grow after treatment (Fig. 3g,h and Supplementary Fig. 9b,c). The effects thus depend on DTA expression and not Cre recombinase toxicity18.
Functional analysis of cell populations within the tumor microenvironment — mast cells are dispensable for PDAC initiation.]
Mast cells link the innate and adaptive immune systems and have been implicated in tumor initiation, progression and metastasis of various tumor entities19. Our data show mast cell infiltration in human PDAC and in KC mice (Supplementary Fig. 10a–e). However, the pro- or antitumorigenic function of mast cells is controversial20–22.
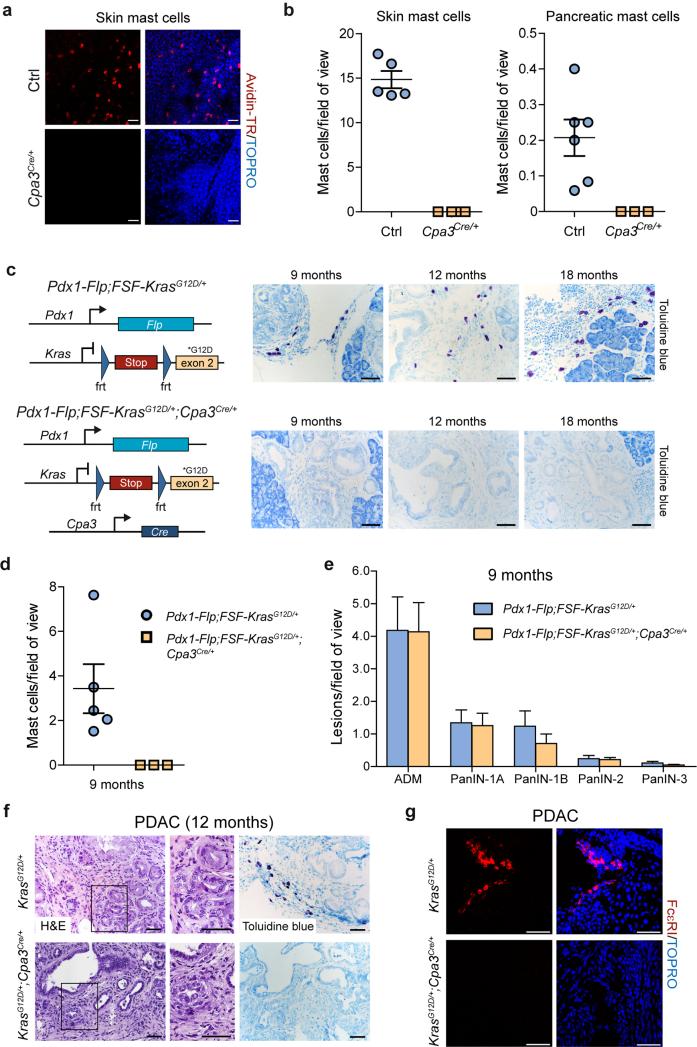
We used the DRS method to evaluate the contribution of mast cells to pancreatic carcinogenesis. To deplete mast cells in vivo, we used the mast cell–deficient Cpa3Cre/+ line, which is not based on Kit hypomorphism23. This line lacks both connective tissue and mucosal mast cells owing to Cre-mediated eradication of the entire mast cell lineage. In addition to mast cells, carboxypeptidase A3 (Cpa3) is weakly expressed in hematopoietic progenitors and splenic basophils23.
We verified mast cell deficiency in skin and pancreas of Cpa3Cre/+, as compared to wild-type mice (Fig. 4a,b). We then analyzed mast cell infiltration and PanIN/PDAC formation in KF mice with and without Cpa3Cre/+ (Fig. 4c-g). Staining with toluidine blue and immunofluorescent staining of the high affinity IgE receptor FcεRI revealed the absence of mast cells in PanIN and PDAC lesions of Pdx1-Flp;FSF-KrasG12D/+;Cpa3Cre mice, whereas KF controls frequently showed mast cell infiltration (Fig. 4c,d,f,g). To investigate the role of mast cells in PDAC initiation, we analyzed mice deficient and proficient in mast cells at 9 months old, when all grades of PanIN are present in the KF model (Figs. 1c and 4e). The number and grade of PanIN were the same, regardless of the presence of mast cells (Fig. 4e). We also analyzed some mice at age 12 months. These showed sporadic occurrence of invasive PDAC, again regardless of whether mast cells were present (Fig. 4f,g).
Figure 4.
Mast cells are dispensable for PDAC development. (a) Representative confocal microscopic images of skin whole-mounts of male Cpa3Cre/+ and littermate control (Ctrl) mice on a C57BL/6 genetic background. Mast cells were stained with avidin-TexasRed (red) and nuclei were counterstained with TOPRO-3 (blue). (b) Quantification of skin (left) and pancreatic (right) mast cells in male and female Cpa3Cre/+ and control mice (mean, ± s.e.m.; 10–15 fields of view per animal; each dot in the graph represents one mouse). (c) Genetic strategy to eradicate mast cells in the KF model by Cre recombinase expression using Cpa3Cre/+ mice (left). Representative images of toluidine blue–stained metachromatic mast cells (purple) in PanIN-bearing pancreata of male and female KF (top) and mast cell-depleted Pdx1-Flp;FSF-KrasG12D/+;Cpa3Cre/+ (bottom) mice (n = 3 animals per genotype). (d) Quantification of tumor-infiltrating mast cells in 9-month-old PanIN-bearing male and female mice with genotypes as indicated (mean ± s.e.m.; 15 fields of view per animal). (e) Absolute quantification of ADM and different grades of PanIN lesions in 9-month-old male and female Pdx1-Flp;FSF-KrasG12D/+;Cpa3Cre/+ (n = 3) and Pdx1-Flp;FSF-KrasG12D/+ (n = 4) mice (mean and s.e.m.; three representative slides per mouse). (f) Representative H&E and toluidine blue staining of tissue sections from 12-month-old male Pdx1-Flp;FSF-KrasG12D/+ (top) and Pdx1-Flp;FSF-KrasG12D/+;Cpa3Cre/+ mouse with established PDAC (three representative slides per mouse). (g) Representative immunofluorescence staining of FcεRI (red) in mast cells in PDAC-bearing male Pdx1-Flp;FSF-KrasG12D/+ (top) and Pdx1-Flp;FSF-KrasG12D/+;Cpa3Cre/+ (bottom) mouse (three representative slides per mouse). Nuclei are counterstained with TOPRO-3 (blue). Scale bars, 50 μm.
Mast cells are therefore dispensable for PDAC formation. Previous reports have, however, described dramatically altered tumor development and growth in mast cell–deficient hypomorphic Kit-mutant mice (e.g. KitW-sh), or mice treated with cromolyn, a mast cell stabilizer20,24,25. We investigated expression of the Kit proto-oncogene during pancreatic carcinogenesis and frequently observed Kit expression in PanIN and PDAC cells by immunohistochemistry (Supplementary Fig. 10f,g). Kit expression in pancreas was also analyzed by lineage tracing with a tamoxifen-inducible KitCreERT2 knock-in mouse line26, revealing that Kit marks a distinct pancreatic cell population (Supplementary Fig. 10h). The R26mT-mG-Cre reporter indicated no leaky Cre activity in the absence of tamoxifen.
These data indicate that hypomorphic Kit mutations may contribute to the altered tumor phenotype of mast cell–deficient models in a mast cell–independent manner23.
To gain insight into gene expression changes associated with mast cell deficiency, we performed mRNA expression profiling of pancreatic tumors and obtained a statistically significant gene expression signature of mast cell deficiency in Pdx1-Flp;FSF KrasG12D/+;Cpa3Cre/+ mice using “gene set enrichment analysis” (GSEA) (Normalized Enrichment Score: -1.5837048; Nominal P-value: 0.008152174; False Discovery Rate (FDR) q-value: 0.0096; see also Supplementary Results). mRNA expression analysis of genes relevant to PDAC revealed no significant (P < 0.05) changes due to mast cell ablation, except for C-X-C motif chemokine 13 (Cxcl13), fibroblast growth factor 21 (Fgf21) and interleukin 2 receptor gamma (Il2rg) (Supplementary Fig. 11a,b and data not shown). Only 155 probe sets (0.44%), corresponding to 108 annotated genes, showed significantly (P < 0.05) different changes (>2-fold) in mRNA expression between mast cell deficient and proficient pancreatic tumors (Supplementary Fig. 11c). In addition to the aforementioned three gene products, cathepsin E and the mucin Muc5ac have been linked to pancreatic disease (Supplementary Fig. 11d). Thus, cancer-relevant pathways are not broadly affected by mast cell deficiency, corroborating our finding that mast cells do not influence PDAC development.
DISCUSSION
Breakthroughs in cancer research, such as genomic analysis and high-throughput drug and genetic screening, are providing a wealth of resources and large, complex data sets. Understanding and interpreting the biological and clinical relevance of these resources is a significant challenge but holds the promise of breakthrough treatments for particularly difficult conditions, such as PDAC.
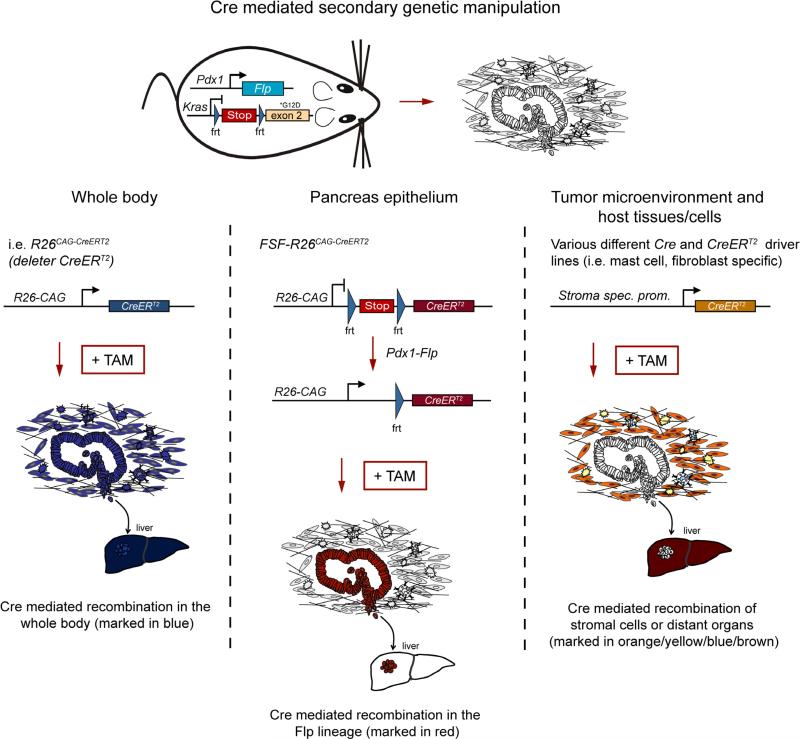
What has so far made it difficult to take full advantage of these developments is a means of realistically modeling and manipulating the developing tumor entity and its microenvironment. This is vital for the identification and assessment of candidate therapeutic targets. We have generated, characterized and validated a next-generation DRS that enables sequential genetic manipulation of PanIN and PDAC cells and the host on a genome-wide scale in vivo. Combining the Flp-FRT recombination system for tumor initiation with the ‘universal’ Cre-loxP system for secondary genetic manipulation provides researchers with new opportunities to model, manipulate and investigate diverse aspects of malignant tumors in whole animals. This includes rigorous genetic analysis of (i) genetic alterations in multistep carcinogenesis, (ii) tumor cell subpopulations such as cancer stem cells, (iii) the tumor microenvironment, (iv) immune cell subpopulations, (v) the metastatic niche of host organs, (vi) therapeutic targets and (vii) resistance mechanisms (Fig. 5).
Figure 5.
Applications of DRS. Stroma-rich PDAC tumors can be induced by Pdx1-Flp– mediated activation of an FRT-stop-FRT (FSF) silenced oncogenic KrasG12D allele in the pancreas. Middle, secondary genetic manipulation of tumor cells in the Flp lineage can be achieved by a FSF-silenced latent CreERT2 allele (FSF-R26CAG-CreERT2). CreERT2 can be activated by tamoxifen treatment at any time point during tumor formation and progression to recombine floxed sequences. In this way it is possible to model and analyze multistep carcinogenesis, manipulate tumor subpopulations, validate therapeutic targets in established tumors and analyze resistance mechanisms by genetic approaches. The use of various Flp driver lines or a Flp-expressing virus allows expression of oncogenic KrasG12D or any other Flp-FRT-regulated oncogene or tumor suppressor in various different tissue and cell types. Left, use of a CreERT2 deleter allele, which is expressed from the Rosa26 locus under the control of the CAG promoter in all cells of the body, allows whole-organism, time-specific deletion of a target or pathway, making it possible to mimic drug treatment and thus evaluate its value as a drug target for therapy of established PDAC. Right, Cre driver lines and expression models can be used to target subpopulations of the tumor microenvironment, such as fibroblasts (orange), stellate cells (yellow), endothelial cells, pericytes, adipocytes and mast cells (blue), as well as other immune cells. In addition, metastasis target organs and the metastatic niche and other host factors, tissues and cells can be genetically manipulated.
The PDAC stroma supports tumor growth and mediates drug resistance and is clearly relevant to future therapies12,14. It is therefore important to understand the biology of the different stromal subcomponents. To demonstrate the utility of the DRS-based PDAC model as a means of targeting the tumor microenvironment, we investigated the role of mast cells and showed that they are dispensable for PDAC initiation and progression (Supplementary Results).
Besides evaluating stromal subpopulations, the DRS-based PDAC model enables sequential mutagenesis within tumor-initiating cells or precursor PanIN lesions, as previously used to study p53 loss during sarcomagenesis27. It also allows rigorous genome-wide testing of tumor cell–intrinsic therapeutic targets. Synthetic lethal interaction partners of oncogenic Kras28 and multiple novel candidate genes and pathways identified via genome-wide small hairpin RNA– and transposon-based forward genetic screens and next-generation sequencing28–30 demonstrate impressively that potential treatment targets are available. These await rigorous genetic validation in vivo.
We demonstrated secondary genetic targeting, thus mimicking therapeutic interventions, using a floxed Pdpk1 mouse line and a latent DTA allele as proof of concept. Our model thus provides an ideal complement for genome-wide mouse embryonic stem cell repositories and mutant lines with Cre-inducible somatic inactivation of every gene, enabling genome-wide target validation31. The synergistic power of these developments will spur the field.
Two groups have generated transgenic GEMMs that allow tetracycline (TET)-inducible expression of oncogenic Kras in pancreas and its time-specific inactivation32,33. These showed that continuous oncogenic Kras activity is essential for tumor maintenance32,33. Combined with Cre-loxP, Flp-FRT and other recombination systems such as Dre-rox, these TET-Kras mice, as well as novel embryonic stem cell–based PDAC models34, will further increase the possibilities of modeling and manipulating PDAC cells and their microenvironment.
In summary, we developed a next-generation Flp-FRT–based spontaneous model of Kras-driven stroma-rich PDAC that recapitulates major hallmarks of human PDAC with minimal extrapancreatic disease. The use of secondary modifications allows genetic dissection of the native biology of PDAC tumors and their associated stroma and rigorous genetic validation of candidate therapeutic targets. The DRS model can thus contribute directly to the development of new therapeutic strategies. Generation of tissue-specific Flp driver lines or viral Flp delivery27,35 will allow spatial and temporal control of gene expression in various cancer models. The DRS approach is highly versatile with wide applicability and has the potential to significantly advance cancer research.
ONLINE METHODS
Materials
All cell culture reagents were obtained from Invitrogen (Groningen, The Netherlands). Primers were made by MWG (Ebersberg, Germany) and restriction endonucleases obtained from New England Biolabs (Mannheim, Germany). The E. coli strains TOP10 and Stbl3 (Invitrogen, Groningen, The Netherlands) were used for transformation and plasmid amplification. 4-Hydroxytamoxifen (H6278) and peanut oil (P2144) were purchased from Sigma (Deisenhofen, Germany).
Mouse strains and tumor models
LSL-KrasG12D/+2, Pdx1-Cre2, LSL-Trp53R172H/+16, Trp53frt/+35, Trp53lox/+36, Trp53Δ/+37, LSL-R26Tva-i-lacZ/+4, R26mT-mG/+38, FSF-R26hpAP/+39, Pdpk1lox/+40, Cpa3Cre/+23, KitCreERT2/+26, R26Flpo/+41 and LSL-R26DTA/+42 have been described previously. Unless otherwise stated, animals were on a mixed C57Bl/6;129S6/SvEv genetic background. The strains were interbred to obtain mice with activation of oncogenic KrasG12D in the pancreas as previously described10. Age- and sex-matched randomized animals were used as indicated and no animals were excluded from analyses. All animal studies were conducted in compliance with European guidelines for the care and use of laboratory animals and were approved by the Institutional Animal Care and Use Committees (IACUC) of Technische Universität München, Regierung von Oberbayern and UK Home Office.
Construction of the Pdx1-Flp transgenic line
To generate the Pdx1-Flp construct, a 6.3 kb fragment containing the Pdx1 promoter from pKSpdx-1SalI (#571, a kind gift from C. Wright; see Supplementary Figure 1a) was ligated to the codon optimized Flp-o cDNA (a kind gift from P. Soriano; Addgene plasmid # 13792;41). The start codon of the Flp-o cDNA was directly fused to the ATG start codon of the Pdx1 gene. The human beta globin polyadenylation signal sequence was ligated 3’ of the Flp-o expression construct. The 8.1 kb insert was released from pBluescript by SalI-PmeI digestion and was used for pronuclear injection into C57BL/6 zygotes (Polygene, Rümlach, Switzerland).
Construction of targeting vectors and generation of FSF-R26CAG-CreERT2 and FSF KrasG12D knock-in mouse lines
Rosa26 targeting by a knock-in strategy was performed on the basis of plasmid pROSA26–1 as previously described4. A targeting vector containing a CAG promoter and a FRT-flanked transcriptional and translational stop cassette (FRT-stop-FRT; FSF) with a neomycin resistance gene 5’ of the CreERT2 expression cassette was generated by standard cloning procedures (Supplementary Figure 2a). The targeting vector was linearized and electroporated into 129S6/SvEv embryonic stem (ES) cells. Cells were selected with 200 μg ml–1 Geneticin, and correctly targeted homologous recombined clones identified by PCR as described4. Correct recombination and single copy insertion was verified by long range and quantitative PCR, respectively. Germ-line transmission was achieved in 2/2 clones harboring the targeted allele. A three-primer PCR strategy (Supplementary Figure 2b) was used to genotype FSF-R26CAG-CreERT2 animals as described4.
To generate a novel tamoxifen-inducible CreERT2 deleter mouse line (termed R26CAG-CreERT2), which ubiquitously expresses CreERT2, we deleted the FRT-flanked transcriptional stop cassette in FSF-R26CAG-CreERT2 animals using the R26Flpo/+41 deleter line.
To construct the FSF-KrasG12D targeting vector, we used a Kras genomic clone, amplified the long and short arm of the Kras targeting vector by PCR and introduced a codon 12 aspartic acid mutation into exon 2 by site-directed mutagenesis. Finally, a FSF cassette with a splice acceptor 3’ of the first FRT recombination site and a promoterless neomycin resistance cassette was introduced into the first intron of the Kras targeting vector (Supplementary Figure 5a). All constructs were verified by sequencing. We electroporated the PacI linearized targeting vector into 129S6/SvEv ES cells and selected appropriately targeted clones with 200 μg ml–1 Geneticin. Homologous recombined clones were identified by PCR and correct recombination and single copy insertion was verified by long range PCR and quantitative PCR, respectively. Two clones were injected into blastocysts (Polygene, Rümlach, Switzerland). Germ-line transmission was achieved in 2/2 clones harboring the targeted allele. A three-primer PCR strategy (Supplementary Figure 5b) was used to genotype animals.
Fluorescence stereomicroscopy and quantification of metastasis frequency
Macroscopic analysis of mouse tissues, PDAC and metastases was performed using a Zeiss Stemi 11 fluorescence stereomicroscope. At necropsy, all abdominal organs and the lungs were investigated macroscopically for metastases as described17,43. For microscopic quantification of metastases at least three series of sections (100 μm between the different series) of paraffin embedded lungs and livers of 20 mice per genotype with PDAC were prepared, H&E stained and investigated for the presence of metastases by an examiner blinded to the genotype of the animals.
Fluorescence in situ hybridization (FISH)
Metaphase spreads were prepared from splenocytes using standard protocols and fluorescence in situ hybridization was performed as described44. Probes specific for the Flp transgene were amplified using GenomePlex® Complete Whole Genome Amplification (WGA) kit (WGA2, Sigma) and labeled directly with ChromaTide Texas Red dUTP (Invitrogen) using GenomePlex® WGA Reamplification Kit (WGA3, Sigma) and custom-made dNTP mixture. Probes were then denatured for 10 min at 65 °C and preannealed for approximately 30 – 45 min at 37 °C. Slides were prepared one week in advance, to age metaphases at RT with a desiccant. Slides were then pretreated in prewarmed 2 × SSC and pepsin (75 μl of 1% pepsin in 50 ml of 0.01 M HCl) at 37 °C for 5 min each. Slides were washed 3 times in 2 × SSC for 3 min, and then treated for 10 min in formaldehyde fixative (1.25 ml of formaldehyde, 40% w/v and 2.5 ml of 1 M MgCl2, 50 mM MgCl2 in PBS) at RT. Slides were washed 3 times in 2 × SSC for 3 min, passed through an ethanol series and air dried. Slides were then denatured in 70% formamide, 2 × SSC for 1 min 30 s, at 63 °C and passed again through an ethanol series and air dried. Denatured preannealed probes were added to the slides, covered with coverslips, sealed with Fixogum rubber cement (Marabu) and incubated overnight at 37 °C. To remove unbound probes, slides were subjected to post-hybridization washes and mounted in SlowFade (Invitrogen) with DAPI (4,6 diamidino-2-phenylindole), overlayed with coverslip and sealed with nail varnish. Metaphases were scanned using an epifluorescence microscope equipped with a CCD camera and narrow band-pass filters. Images were captured using the SmartCapture software (Digital Scientific, UK) and metaphases were karyotyped using the SmartType Karyotyper software (Digital Scientific).
Generation and culture of primary mouse PDAC cell lines
Primary dispersed mouse pancreatic cancer cells were established and cultivated as previously described43. PDAC cells were cultured in DMEM supplemented with 10% FCS and 0.1 or 0.5 μM 4-hydroxytamoxifen or vehicle (ethanol) to delete loxP flanked sequences in vitro. Cell proliferation was quantified by cell counting on consecutive days. All cell lines were authenticated by genotyping and tested for mycoplasma contamination.
Tamoxifen treatment of mice
Mice were fed with tamoxifen-containing chow (400 mg tamoxifen citrate kg–1 chow; CreActive TAM400, LASvendi, Soest, Germany) for 2 or 4 weeks to activate CreERT2.
High-resolution ultrasound
Mice were screened for tumors by abdominal palpation. Palpable tumors were verified by high-resolution ultrasound (Vevo 2100, VisualSonics). Animals with average tumor diameters greater than 5 mm were enrolled in the treatment study as described17. Tumor volumes were determined by automated three-dimensional (3D) B-mode imaging along the entire length of the tumor (VisualSonics). Reconstructed three-dimensional ultrasound imaging data sets were analyzed and tumor volumes were quantified using the integrated Vevo 2100 software package (VisualSonics) by an examiner blinded to the genotype of the animals. Macroscopic tumor perfusion was analyzed using the Doppler mode of the Vevo 2100 system.
Quantitative reverse-transcriptase PCR
Total RNA was isolated from tissues using the RNeasy kit (Qiagen, Hilden, Germany) following the manufacturer's instructions and reverse transcribed (Invitrogen). Quantitative mRNA analyses were performed using real-time PCR analysis (TaqMan, PE Applied Biosystems, Norwalk, CT) and QuantiTect Primer Assays (Qiagen) as previously described45.
Laser capture microdissection and DNA extraction
PanIN lesions and normal acini from 8 μm thick, dewaxed, hematoxylin and eosin stained formalin fixed paraffin embedded tissue sections were microdissected using a P.A.L.M. laser capture microdissection system (Zeiss, Göttingen, Germany) as described previously46. DNA was isolated from the microdissected cells using the QIAamp DNA Micro Kit (Qiagen, Hilden, Germany).
Histochemistry and immunohistochemistry
For histopathology, mouse tissue specimens were fixed in 4% buffered formalin overnight, embedded in paraffin and sectioned (2.5 μm thick). Quantification and grading of mouse ADM and PanIN lesions was performed on three sections per mouse and 3 mice per time point according to the established nomenclature for the grading of PanIN lesions in mice47. The examiner was blinded to the genotype of the animals. Alcian blue staining was performed on paraffin embedded tissue sections using aqueous alcian blue solution (pH 2.5). Sections were counterstained with nuclear fast red, as previously described17. Toluidine blue staining of mast cells was performed using an aqueous toluidine blue staining solution (pH 2.0) for 10 min (all staining solutions from Sigma). For immunodetection, formalin-fixed paraffin-embedded tissue sections were dewaxed, rehydrated and placed in a microwave (10 min, 600 watt) to recover antigens. Sections were incubated with primary c-Kit (C-19; sc-168; 1:50; Santa Cruz Biotechnology, Santa Cruz, CA), p53 (NCL-p53-CM5p; rabbit, 1:400; Novocastra/Leica Mikrosysteme, Wetzlar, Germany) and Pdpk1 (Pdk1, #3061; rabbit, 1:50; Cell Signaling Technology, Danvers, MA) antibodies followed by a secondary antibody conjugated to biotin (Vector Laboratories, Burlingame, CA).
β-galactosidase staining
β-galactosidase staining of cryosections was performed as described previously4. Counterstaining was carried out with eosin or nuclear fast red.
Alkaline phosphatase (AP) staining
Cryosections were post-fixed in 4% paraformaldehyde in PBS for 10 min at 4 °C. After rinsing with cold PBS, sections were incubated at 70 °C in preheated PBS for 30 min, followed by incubation in AP detection buffer (100 mM NaCl, 100 mM Tris-HCl pH 9.5, 50 mM MgCl2) for 30 min. Sections were subsequently placed in AP staining solution (AP detection buffer with 0.8 mg ml–1 nitroblue tetrazolium (NBT), 0.1 mg ml–1 5-bromo-4-chloro-3-indolyl phosphate dimethylformamide (BCIP), 0.01% sodium deoxycholate, 0.02% NP-40, and 0.5 mM levamisole) at room temperature overnight. Once color development was complete, sections were counterstained with nuclear fast red or eosin.
Immunofluorescence staining
Mouse tissues were fixed in 4% buffered formalin for 2 h, dehydrated in sucrose solution at 4 °C (15% sucrose in PBS for 4 h; 30% sucrose in PBS overnight) and embedded in Tissue-Tek® (Sakura, Torrance, CA) before being rapidly frozen in liquid nitrogen. 20 μm thick frozen sections were post-fixed for 1 min in 4% buffered formalin, washed twice in PBS and incubated for 1 h in PBS with 3% (w/v) bovine serum albumin (BSA), 1% (w/v) Saponin and 1% (v/v) Triton-X 100. Subsequently, slides were incubated for 48 h at 4 °C in the dark with primary antibodies: Kit (M14; goat, 1:100; sc-1494, Santa Cruz Biotechnology, Santa Cruz, CA), alpha-amylase (A8273; rabbit, 1:100; Sigma-Aldrich), insulin (C27C9; rabbit, 1:100; #3014, Cell Signaling Technology), CK19 (ab52625; rabbit, 1:75; abcam, Cambridge, UK), or PE labeled FcεRI alpha (#12-5898-82; hamster, 1:100; eBioscience, Frankfurt, Germany) as described26. Kit, α-amylase, insulin and CK19 antibody stained slides were incubated in the dark with secondary antibody Alexa Fluor® 488 donkey anti-goat (1:100, Invitrogen), DyLight® 680 goat anti-rabbit IgG (H+L) (1:100; #5366), or DyLight® 800 goat anti-rabbit IgG (H+L) (1:100; #5151, both from Cell Signaling Technology). Nuclei were counterstained in selected sections with TOPRO®-3-iodide (1:1000, Invitrogen) for 2 h at RT. Selected sections were additionally counterstained with Alexa Fluor® 594 labeled phalloidin (1:250, Invitrogen) to visualize actin filaments and thus cell morphology. After three rinses in PBS, slides were mounted with Vectashield Mounting Medium (Vector Laboratories, Burlingame, CA).
Confocal laser-scanning microscopy
Cryosections were examined by confocal laser scanning confocal microscopy (LSM) with a Zeiss LSM 510 Axiovert 100 microscope (Zeiss, Oberkochen, Germany) with a ×20/0.5 air and a ×40/1.3 oil-immersion objective (optical section thickness 4.4 mm) as described26. Images (single optical sections and z-stacks, z-step size 0.5 mm) with a frame size of 1,024 × 1,024 pixels and an image size of 225 × 225 mm (×40/1.3 objective, Zeiss), or 450 × 450 mm (×20/0.5 air objective, Zeiss) were collected. Images were merged and converted with Zeiss LSM 510 software.
Staining and counting of skin mast cells
Staining of skin mast cells was performed as described26. In brief, ears of the respective animals were separated into a ventral and dorsal half and fixed in 1% buffered formalin overnight at 4 °C. After incubation for 1 h at RT in 1% BSA in PBS, mast cells were stained with avidin-Texas Red (1:500, Invitrogen) as described26. Nuclei were counterstained with TOPRO®-3-iodide. For mast cell-counting, the ears of at least 3 mice per genotype and 10 – 15 fields of view of each animal were analysed by LSM using the Zeiss LSM 510 microscope with a × 20/0.5 air objective. The examiner was blinded to the genotype of the animals.
RNA expression analyses
RNA extraction and microarray expression analyses were performed as recently described43,48. In brief, total RNA was prepared using RNeasy Kit (Qiagen) and processed with the Ambion WT expression kit (Applied Biosystems). Purified single-stranded cRNA was fragmented and labeled by using the GeneChip WT terminal labeling kit (Affymetrix, Santa Clara, CA) and hybridized onto Affymetrix GeneChip Mouse Gene 1.0 ST Array. Calculation of expression values was done by taking the mean of all perfect match probes for each probeset. The expression dataset was quantile normalized and each probeset was assessed for differential expression with standard Student's t test. Resulting P-values are corrected for the number of tests (48107) with the method by Benjamini & Hochberg49. Gene set enrichment analysis (GSEA) was performed with GSEA software (Broad Institute, MIT, MA; www.broadinstitute.org)50. Genes down-regulated > 2 fold in mast cell-deficient tissues (peritoneum and ear)23 were defined as gene set “Mast_cell_deficiency”. P-values < 0.05 with a false discovery rate (FDR) q-value < 0.05 (5%) and a family-wise error rate (FWER) P-value < 0.05 were considered to be statistically significant. Heatmaps for the expression levels of selected genes were assembled with the GenePattern software package (Broad Institute). Microarray data are available in the EMBLEBI ArrayExpress database (www.ebi.ac.uk/arrayexpress) under accession number E-MTAB-2551.
Data analysis
Unless otherwise indicated, all data were determined from at least three independent experiments and expressed as mean values ± s.e.m. Sample size was calculated using GraphPad StatMateSoftware 2.00 (GraphPad Software, Inc.; La Jolla, CA). Statistical comparisons between data sets were made with analysis of normality and variance, followed by a two-sided unpaired Student's t test. A Bonferroni correction of the P values was performed for pair wise multiple testing. Fisher's exact test was performed by using STATXACT 4.0.1 software (Cytel Software, Cambridge, MA) as described51. Values of P < 0.05 were considered to be statistically significant.
Supplementary Material
ACKNOWLEDGMENTS
The authors thank A. Berns (Netherlands Cancer Institute), S. Dymecki (Harvard Medical School), T. Jacks (Massachusetts Institute of Technology), L. Luo (Stanford University), J. Martinez-Barbera (University College London) and D. Tuveson (Cold Spring Harbor Laboratory) for providing transgenic animals, C. Wright (Vanderbilt University) for the mouse Pdx1 promoter construct, P. Soriano (Mount Sinai School of Medicine) for the Flp-o expression vector and the R26 targeting vector, T. Schmidt and M. Bewerunge-Hudler (DKFZ Microarray Core Facility) for mRNA analyses, and J. Götzfried, U. Götz and S. Jaeckel for technical assistance. This work was supported by funding from Deutsche Forschungsgemeinschaft (DFG SA 1374/4-1 to D.S. and SFB824, TP C9 to G.S. and D.S.), the Helmholtz Alliance Preclinical Comprehensive Cancer Center (to H.R.R., R.R., R.M.S. and D.S.), the German Cancer Consortium (DKTK) (to R.R., R.M.S. and D.S.), the Wilhelm-Sander Foundation (2012.084.1 to G.S.), the Spanish Ministerio de Economía y Competitividad subprograma Ramón y Cajal (I.V.), the European Union (ERC Advanced Grant No.233074 to H.R.R.), and the National Cancer Institute USA (R01 CA138265 to D.G.K. and CA155620 to A.M.L.).
Footnotes
AUTHOR CONTRIBUTIONS
B.S. and D.S. designed research; N.S., B.S., K.S., C.V., C.S., M.Z., S.E., M.C.P., P.E., S.K., R.B., F.Y., A.S., I.V., R.R., G.S., and D.S., performed research; T.B.F., A.M.L., C.L.L, E.J.M, D.G.K., A.S., D.R.A., I.V., A.B., A.K., A.E.S., H.R.R., R.R. and R.M.S. contributed new reagents/analytic tools; N.S., B.S., K.S., C.V., C.S., M.Z., S.E., M.C.P., P.E., S.K., R.B., F.T.Y., I.V., R.R., G.S. and D.S. analyzed data; and B.S. and D.S. wrote the paper. N.S., B.S., K.S., and C.V. contributed equally to this manuscript.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
Accession codes. Microarray data are available in the EMBL-EBI ArrayExpress database under accession number E-MTAB-2551.
References
- 1.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2013. CA Cancer J. Clin. 2013;63:11–30. doi: 10.3322/caac.21166. [DOI] [PubMed] [Google Scholar]
- 2.Hingorani SR, et al. Preinvasive and invasive ductal pancreatic cancer and its early detection in the mouse. Cancer Cell. 2003;4:437–450. doi: 10.1016/s1535-6108(03)00309-x. [DOI] [PubMed] [Google Scholar]
- 3.Guerra C, et al. Chronic pancreatitis is essential for induction of pancreatic ductal adenocarcinoma by K-Ras oncogenes in adult mice. Cancer Cell. 2007;11:291–302. doi: 10.1016/j.ccr.2007.01.012. [DOI] [PubMed] [Google Scholar]
- 4.Seidler B, et al. A Cre-loxP-based mouse model for conditional somatic gene expression and knockdown in vivo by using avian retroviral vectors. Proc. Natl. Acad. Sci. USA. 2008;105:10137–10142. doi: 10.1073/pnas.0800487105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pylayeva-Gupta Y, Grabocka E, Bar-Sagi D. RAS oncogenes: weaving a tumorigenic web. Nat. Rev. Cancer. 2011;11:761–774. doi: 10.1038/nrc3106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carrière C, Seeley ES, Goetze T, Longnecker DS, Korc M. The Nestin progenitor lineage is the compartment of origin for pancreatic intraepithelial neoplasia. Proc. Natl. Acad. Sci. USA. 2007;104:4437–4442. doi: 10.1073/pnas.0701117104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Habbe N, et al. Spontaneous induction of murine pancreatic intraepithelial neoplasia (mPanIN) by acinar cell targeting of oncogenic Kras in adult mice. Proc. Natl. Acad. Sci. USA. 2008;105:18913–18918. doi: 10.1073/pnas.0810097105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gidekel Friedlander SY, et al. Context-dependent transformation of adult pancreatic cells by oncogenic K-Ras. Cancer Cell. 2009;16:379–389. doi: 10.1016/j.ccr.2009.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Morris J.P.t., Wang SC, Hebrok M. KRAS, Hedgehog, Wnt and the twisted developmental biology of pancreatic ductal adenocarcinoma. Nat. Rev. Cancer. 2010;10:683–695. doi: 10.1038/nrc2899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eser S, et al. In vivo diagnosis of murine pancreatic intraepithelial neoplasia and early-stage pancreatic cancer by molecular imaging. Proc. Natl. Acad. Sci. USA. 2011;108:9945–9950. doi: 10.1073/pnas.1100890108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mazur PK, Siveke JT. Genetically engineered mouse models of pancreatic cancer: unravelling tumour biology and progressing translational oncology. Gut. 2012;61:1488–1500. doi: 10.1136/gutjnl-2011-300756. [DOI] [PubMed] [Google Scholar]
- 12.Olive KP, et al. Inhibition of Hedgehog signaling enhances delivery of chemotherapy in a mouse model of pancreatic cancer. Science. 2009;324:1457–1461. doi: 10.1126/science.1171362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pylayeva-Gupta Y, Lee KE, Hajdu CH, Miller G, Bar-Sagi D. Oncogenic Kras-induced GM-CSF production promotes the development of pancreatic neoplasia. Cancer Cell. 2012;21:836–847. doi: 10.1016/j.ccr.2012.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Provenzano PP, et al. Enzymatic targeting of the stroma ablates physical barriers to treatment of pancreatic ductal adenocarcinoma. Cancer Cell. 2012;21:418–429. doi: 10.1016/j.ccr.2012.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gannon M, Herrera PL, Wright CV. Mosaic Cre-mediated recombination in pancreas using the pdx-1 enhancer/promoter. Genesis. 2000;26:143–144. doi: 10.1002/(sici)1526-968x(200002)26:2<143::aid-gene13>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- 16.Hingorani SR, et al. Trp53R172H and KrasG12D cooperate to promote chromosomal instability and widely metastatic pancreatic ductal adenocarcinoma in mice. Cancer Cell. 2005;7:469–483. doi: 10.1016/j.ccr.2005.04.023. [DOI] [PubMed] [Google Scholar]
- 17.Eser S, et al. Selective requirement of PI3K/PDK1 signaling for Kras oncogene-driven pancreatic cell plasticity and cancer. Cancer Cell. 2013;23:406–420. doi: 10.1016/j.ccr.2013.01.023. [DOI] [PubMed] [Google Scholar]
- 18.Schmidt-Supprian M, Rajewsky K. Vagaries of conditional gene targeting. Nat. Immunol. 2007;8:665–668. doi: 10.1038/ni0707-665. [DOI] [PubMed] [Google Scholar]
- 19.Theoharides TC, Conti P. Mast cells: the Jekyll and Hyde of tumor growth. Trends Immunol. 2004;25:235–241. doi: 10.1016/j.it.2004.02.013. [DOI] [PubMed] [Google Scholar]
- 20.Theoharides TC. Mast cells and pancreatic cancer. N. Engl. J. Med. 2008;358:1860–1861. doi: 10.1056/NEJMcibr0801519. [DOI] [PubMed] [Google Scholar]
- 21.Nielsen HJ, et al. Independent prognostic value of eosinophil and mast cell infiltration in colorectal cancer tissue. J. Pathol. 1999;189:487–495. doi: 10.1002/(SICI)1096-9896(199912)189:4<487::AID-PATH484>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- 22.Rajput AB, et al. Stromal mast cells in invasive breast cancer are a marker of favourable prognosis: a study of 4,444 cases. Breast Cancer Res. Treat. 2008;107:249–257. doi: 10.1007/s10549-007-9546-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Feyerabend TB, et al. Cre-mediated cell ablation contests mast cell contribution in models of antibody- and T cell-mediated autoimmunity. Immunity. 2011;35:832–844. doi: 10.1016/j.immuni.2011.09.015. [DOI] [PubMed] [Google Scholar]
- 24.Soucek L, et al. Mast cells are required for angiogenesis and macroscopic expansion of Myc-induced pancreatic islet tumors. Nat. Med. 2007;13:1211–1218. doi: 10.1038/nm1649. [DOI] [PubMed] [Google Scholar]
- 25.Chang DZ, et al. Mast cells in tumor microenvironment promotes the in vivo growth of pancreatic ductal adenocarcinoma. Clin. Cancer Res. 2011;17:7015–7023. doi: 10.1158/1078-0432.CCR-11-0607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Klein S, et al. Interstitial cells of Cajal integrate excitatory and inhibitory neurotransmission with intestinal slow-wave activity. Nat. Commun. 2013;4:1630. doi: 10.1038/ncomms2626. [DOI] [PubMed] [Google Scholar]
- 27.Young NP, Crowley D, Jacks T. Uncoupling cancer mutations reveals critical timing of p53 loss in sarcomagenesis. Cancer Res. 2011;71:4040–4047. doi: 10.1158/0008-5472.CAN-10-4563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Luo J, et al. A genome-wide RNAi screen identifies multiple synthetic lethal interactions with the Ras oncogene. Cell. 2009;137:835–848. doi: 10.1016/j.cell.2009.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Biankin AV, et al. Pancreatic cancer genomes reveal aberrations in axon guidance pathway genes. Nature. 2012;491:399–405. doi: 10.1038/nature11547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pérez-Mancera PA, et al. The deubiquitinase USP9X suppresses pancreatic ductal adenocarcinoma. Nature. 2012;486:266–270. doi: 10.1038/nature11114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Skarnes WC, et al. A conditional knockout resource for the genome-wide study of mouse gene function. Nature. 2011;474:337–342. doi: 10.1038/nature10163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ying H, et al. Oncogenic Kras maintains pancreatic tumors through regulation of anabolic glucose metabolism. Cell. 2012;149:656–670. doi: 10.1016/j.cell.2012.01.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Collins MA, et al. Oncogenic Kras is required for both the initiation and maintenance of pancreatic cancer in mice. J. Clin. Invest. 2012;122:639–653. doi: 10.1172/JCI59227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Saborowski M, et al. A modular and flexible ESC-based mouse model of pancreatic cancer. Genes Dev. 2014;28:85–97. doi: 10.1101/gad.232082.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee C-L, et al. Generation of primary tumors with Flp recombinase in FRT-flanked p53 mice. Dis. Model. Mech. 2012;5:397–402. doi: 10.1242/dmm.009084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jonkers J, et al. Synergistic tumor suppressor activity of BRCA2 and p53 in a conditional mouse model for breast cancer. Nat. Genet. 2001;29:418–425. doi: 10.1038/ng747. [DOI] [PubMed] [Google Scholar]
- 37.Olive KP, et al. Mutant p53 gain of function in two mouse models of Li-Fraumeni syndrome. Cell. 2004;119:847–860. doi: 10.1016/j.cell.2004.11.004. [DOI] [PubMed] [Google Scholar]
- 38.Muzumdar MD, Tasic B, Miyamichi K, Li L, Luo L. A global double-fluorescent Cre reporter mouse. Genesis. 2007;45:593–605. doi: 10.1002/dvg.20335. [DOI] [PubMed] [Google Scholar]
- 39.Awatramani R, Soriano P, Mai JJ, Dymecki S. An Flp indicator mouse expressing alkaline phosphatase from the ROSA26 locus. Nat. Genet. 2001;29:257–259. doi: 10.1038/ng1101-257. [DOI] [PubMed] [Google Scholar]
- 40.Lawlor MA, et al. Essential role of PDK1 in regulating cell size and development in mice. EMBO J. 2002;21:3728–3738. doi: 10.1093/emboj/cdf387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Raymond CS, Soriano P. High-efficiency FLP and PhiC31 site-specific recombination in mammalian cells. PLoS ONE. 2007;2:e162. doi: 10.1371/journal.pone.0000162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ivanova A, et al. In vivo genetic ablation by Cre-mediated expression of diphtheria toxin fragment A. Genesis. 2005;43:129–135. doi: 10.1002/gene.20162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.von Burstin J, et al. E-cadherin regulates metastasis of pancreatic cancer in vivo and is suppressed by a SNAIL/HDAC1/HDAC2 repressor complex. Gastroenterology. 2009;137:361–371. 371, e361–365. doi: 10.1053/j.gastro.2009.04.004. [DOI] [PubMed] [Google Scholar]
- 44.Rad R, et al. PiggyBac transposon mutagenesis: a tool for cancer gene discovery in mice. Science. 2010;330:1104–1107. doi: 10.1126/science.1193004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Saur D, et al. CXCR4 expression increases liver and lung metastasis in a mouse model of pancreatic cancer. Gastroenterology. 2005;129:1237–1250. doi: 10.1053/j.gastro.2005.06.056. [DOI] [PubMed] [Google Scholar]
- 46.Flisikowska T, et al. A porcine model of familial adenomatous polyposis. Gastroenterology. 2012;143:1173–1175.e1171-1177. doi: 10.1053/j.gastro.2012.07.110. [DOI] [PubMed] [Google Scholar]
- 47.Hruban RH, et al. Pathology of genetically engineered mouse models of pancreatic exocrine cancer: consensus report and recommendations. Cancer Res. 2006;66:95–106. doi: 10.1158/0008-5472.CAN-05-2168. [DOI] [PubMed] [Google Scholar]
- 48.Diersch S, et al. Efemp1 and p27(Kip1) modulate responsiveness of pancreatic cancer cells towards a dual PI3K/mTOR inhibitor in preclinical models. Oncotarget. 2013;4:277–288. doi: 10.18632/oncotarget.859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Benjamini Y, Hochberg Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc., B. 1995;57:289–300. [Google Scholar]
- 50.Subramanian A, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Saur D, et al. Single-nucleotide promoter polymorphism alters transcription of neuronal nitric oxide synthase exon 1c in infantile hypertrophic pyloric stenosis. Proc. Natl. Acad. Sci. USA. 2004;101:1662–1667. doi: 10.1073/pnas.0305473101. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.