Abstract
Reduction–oxidation (redox) signaling, the translation of an oxidative intracellular environment into a cellular response, is mediated by the reversible oxidation of specific cysteine thiols. The latter can result in disulfide formation between protein hetero- or homodimers that alter protein function until the local cellular redox environment has returned to the basal state. We have previously shown that this mechanism promotes the nuclear localization and activity of the Forkhead Box O4 (FOXO4) transcription factor. Aims: In this study, we sought to investigate whether redox signaling differentially controls the human FOXO3 and FOXO4 paralogs. Results: We present evidence that FOXO3 and FOXO4 have acquired paralog-specific cysteines throughout vertebrate evolution. Using a proteome-wide screen, we identified previously unknown redox-dependent FOXO3 interaction partners. The nuclear import receptors Importin-7 (IPO7) and Importin-8 (IPO8) form a disulfide-dependent heterodimer with FOXO3, which is required for its reactive oxygen species-induced nuclear translocation. FOXO4 does not interact with IPO7 or IPO8. Innovation and Conclusion: IPO7 and IPO8 control the nuclear import of FOXO3, but not FOXO4, in a redox-sensitive and disulfide-dependent manner. Our findings suggest that evolutionary acquisition of cysteines has contributed to regulatory divergence of FOXO paralogs, and that phylogenetic analysis can aid in the identification of cysteines involved in redox signaling. Antioxid. Redox Signal. 22, 15–28.
Introduction
Owing to their reactivity, cysteine residues catalyze reactions carried out by many classes of enzymes, including proteases, tyrosine phosphatases, and ubiquitin ligases. Cysteines also form structural disulfides in, for instance, membrane proteins. Due to these functions, catalytic and structural cysteines are highly conserved through evolution. At the same time, this reactivity probably drives selection against the evolution of nonfunctional cysteines at the surface of proteins. These properties give cysteines the most extreme conservation pattern of all amino acids; they are either highly or very poorly conserved (24). Indeed, the abundance of cysteines in proteins is far less than what would be expected by chance (cysteine is encoded by 2 out of the 61 amino acid coding DNA triplets; hence, 3.28% abundance is expected, but only ∼2.26% is found in the mammalian proteome) (11). Redox signaling is a form of cellular signal transduction that is initiated by or proceeds through reversible oxidation of cysteines in response to perturbations of the cellular redox state (16) and depends largely on surface-exposed cysteines (24). With the above in mind, we hypothesized that cysteines in intrinsically disordered protein domains that have been acquired and conserved throughout evolution could very well play a role in redox signaling.
Innovation.
To the best of our knowledge, the here presented data for FOXO are the first to show that evolutionary acquisition of reactive cysteines involved in redox signaling contributes to the differential regulation of proteins. It will be interesting to see if the cysteine-dependent regulatory divergence that we describe here for the FOXO family is maybe a more common feature of paralog divergence. Our observations also show that phylogenetic analysis of evolutionary acquired cysteines in exposed protein domains can aid to identify novel proteins involved in redox signaling. Furthermore, IPO7 and IPO8 were not previously described to be involved in redox signaling.
The mammalian family of FOXO transcription factors consists of FOXO1, 3, 4, and 6, all of which contain the highly conserved DNA-binding Forkhead (FKH) domain and the C-terminal transactivation domain (1). FOXO6 was discovered the latest and is structurally and functionally more distant from the other three proteins (20). Some paralog-specific functions have been described for FOXO1, 3, and 4, which could potentially be attributed to differences in the spatiotemporal expression patterns or different cofactor bindings. Nevertheless, the observation that a single allele of FOXO1, 3, or 4 prevents the formation of tumors in genetically engineered mice with inducible loss of these three FOXOs suggests that in adult mice they have largely overlapping functions (26). In general, FOXOs are thought to be tumor suppressor proteins that function to maintain cellular homeostasis under suboptimal (e.g., oxidative) conditions [reviewed in Eijkelenboom and Burgering (17)]. Previously, we have shown that redox signaling plays an important role in the regulation of the localization and activity of the FOXO4 transcription factor; cysteine-disulfide-dependent heterodimerization promotes the acetylation status (12), nuclear import by transportin-1 (TNPO1) and likely more aspects of FOXO4 signaling (27). The involvement of cysteine-dependent redox signaling in the regulation of the other FOXO family members has not been studied, and we wondered to what extent this is conserved. In this study, we report that FOXO1, FOXO3, and FOXO4 have acquired cysteines in different unstructured regions of the proteins during vertebrate evolution. We compared proteome-wide screens for redox-dependent binding partners of FOXO3 and FOXO4 (these paralogs both contain five cysteines) and found that paralog-specific cysteines are indeed involved in differential redox-dependent interactions. We demonstrate previously uncharacterized disulfide-dependent interactions of FOXO3 with IPO7 and IPO8 and found these interactions are required for efficient nuclear import of FOXO3 under oxidative conditions. Taken together, we conclude that evolutionary acquisition of redox-sensitive cysteines contributes to differences in post-translational regulation of the otherwise closely related proteins FOXO3 and FOXO4.
Results
FOXO1, 3, and 4 have acquired paralog-specific cysteines through evolution
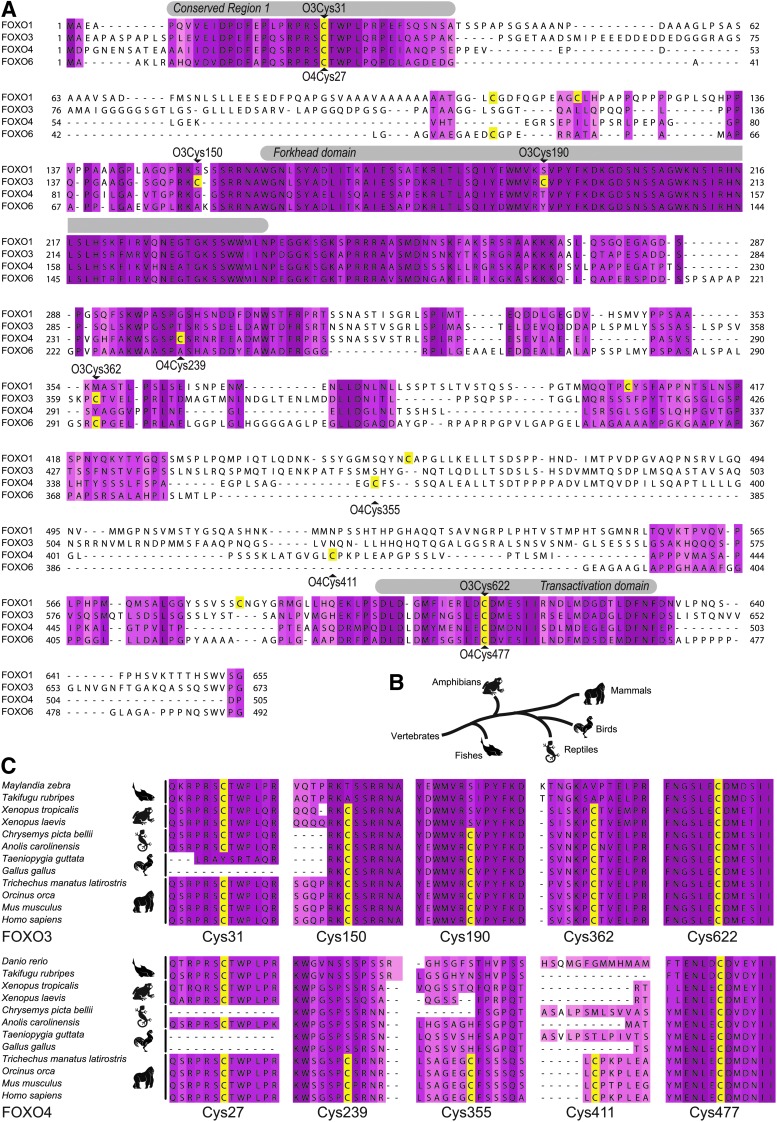
To determine whether FOXO paralogs contain divergent cysteines that might confer differential redox sensitivity, we analyzed a protein sequence alignment using JalView (7). We found that FOXO3 and FOXO4 contain five cysteines, whereas FOXO1 contains seven and FOXO6 contains four. We found two cysteines in annotated domains (in the conserved region 1 (CR1) near the N-terminus and the transactivation (TA) domain near the C-terminus) that were shared among all four paralogs (Fig. 1A), suggesting that these cysteines were present in the last common ancestral vertebrate homolog of human FOXOs. In contrast, there were differences in the distribution of the other cysteines, including Cys150 and Cys190 of FOXO3 and Cys239 of FOXO4, which are located in otherwise conserved parts of these proteins, suggesting that these cysteines occurred as the result of single base-pair mutations. Three other cysteines (Cys362 of FOXO3 and Cys355 and Cys411 of FOXO4) were in nonconserved regions of the proteins, suggesting that these could have occurred as the result of genetic deletions or insertions. To study the conservation and acquisition of cysteines during vertebrate evolution, we aligned the protein sequences of FOXO1, 3, and 4 of different vertebrate classes (2–4 species per class) (Supplementary Figs. S1–S3; Supplementary Data are available online at www.liebertpub.com/ars) and summarized our findings for FOXO3 and 4 in (indicated by the pictograms presenting the different vertebrate classes Fig. 1B) Figure 1C. The alignments show that the two cysteines shared between the FOXO paralogs near the N- and the C-terminus indeed are conserved throughout vertebrate evolution. For the cysteines in FOXO3, this analysis taught us that all three acquired cysteines were gained early in vertebrate evolution, whereas FOXO4 only acquired new cysteines just before or during mammalian evolution. Because only functional cysteines are thought to survive in evolution (24), the conservation that we observe for the newly acquired cysteines suggests that FOXOs could be regulated differentially by redox signaling.
FIG. 1.
Alignment of FOXO transcription factors shows a selective gain of cysteine residues during recent evolution. (A) Human FOXO1, 3, and 4 were aligned and the cysteines are highlighted in yellow. (B) Phylogenetic tree showing the evolution of the classes used for the phylogenetic analysis shown in (C). (C) The evolution of FOXO cysteines was studied by aligning the coding sequences of FOXO3 and 4 of different classes of vertebrates (2–4 species per class). The regions surrounding the cysteines are depicted, the alignment of the complete genes (including FOXO1) are shown in Supplementary Figures S1–S3. The five mammalian cysteines in FOXO3 and 4 show different evolutionary patterns; cysteines in the conserved region (CR) and in the transactivation (TA) region were likely already present in the last common vertebrate ancestor and are conserved from fish to mammals in all FOXO paralogs, whereas the cysteines in the middle part of the proteins display a pattern of evolutionary acquisition. For instance, FOXO3-Cys150 is present in FOXO3 in amphibians, reptiles, birds, and mammals, suggesting that it was acquired somewhere after the fishes and before the amphibians branched off in vertebrate evolution. Note that no homologous cysteine is present in FOXO4 (A).
FOXO3 forms disulfide-dependent heterodimers with several proteins
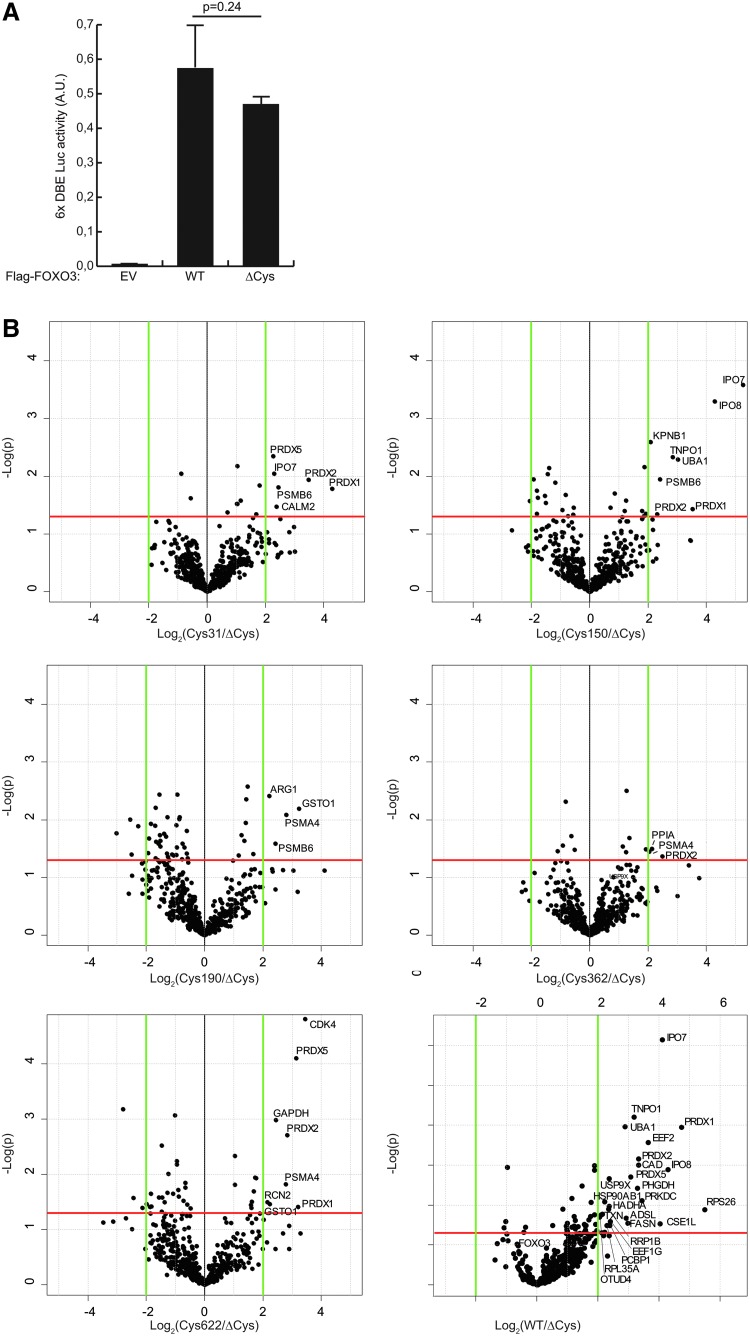
To see if the cysteines that have been acquired during vertebrate evolution by FOXO3 and 4 are differentially involved in redox signaling pathways, we compared the disulfide-dependent interactomes of the two proteins using a tandem mass spectrometry (MS/MS)-based screening method described before for FOXO4 (27). We generated Flag-tagged FOXO3 mutant constructs, in which either all cysteines were replaced with serines (FOXO3ΔCys) or in which four out of five cysteines were replaced with serines (e.g., FOXO3ΔCys-31C contains only the cysteine at position 31). It is to be expected that all these mutants are still functional, as the basal transcriptional activity of FOXO3ΔCys is equal to that of the wild-type (WT) FOXO3 (Fig. 2A).
FIG. 2.
Volcano plots of quantitative MS/MS results. (A) Volcano plots of the quantitative MS/MS data comparing FOXO3ΔCys with the different single cysteine-containing FOXO3 mutants or wild-type FOXO3 (containing all five cysteines). The log2 of the ratio of protein intensities of the pull-downs was plotted versus log10 transformed p-values of the t-test performed on biological quadruplicates. See also Supplementary Figure S4 for a schematic overview of the setup of the screen and Supplementary Table S1 for the raw data of the hits of the screen. (B) A luciferase assay using luciferase driven from an artificial promoter containing six FOXO-binding sites (6x DBE) shows that transcriptional activity of FOXO3ΔCys is equal to that of wild-type FOXO3. N=3, a representative experiment is shown. Error bars represent standard deviations of three technical replicates; p-values were calculated using a two-tailed Student's t-test.
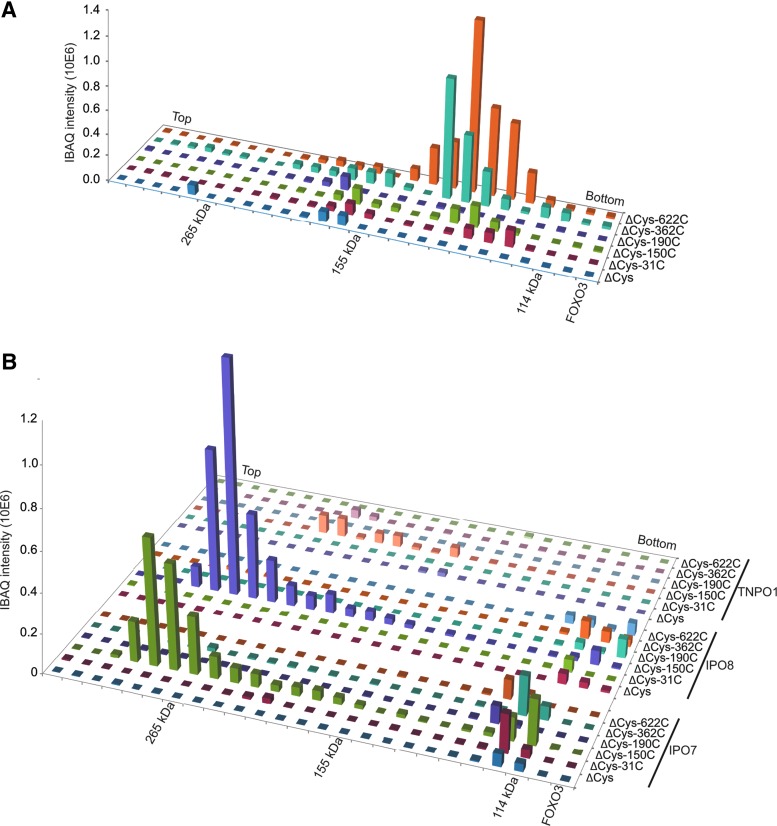
We used a label-free quantitative MS/MS method to identify cysteine-dependent binding partners (Supplementary Fig. S4A); the different mutants were immunoprecipitated (IP) from cells after addition of H2O2, MS/MS samples were prepared and measured, and the intensity of co-IP proteins was determined in a comparative manner using the iBAQ (intensity-based absolute quantification) algorithm (28). As we repeated the experiment four times, statistical analysis could be performed and we could identify the proteins enriched in the pull-downs for each single cysteine-containing mutant (Fig. 2B, right upper quadrants of the plots and Supplementary Table S1). Comparing WT-FOXO3 to FOXO3ΔCys (lower right plot) gave similar results in the sense that most cysteine-dependent binding partners identified with the single cysteine-containing FOXO3 mutants were found back. To assay the covalent complex formation with FOXO3, we performed a similar co-IP experiment, but now separated the samples by nonreducing (but denaturing) sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) (Supplementary Fig. S4B). The lanes of the gel were cut in millimeter strips and each strip was analyzed separately by quantitative MS/MS. In this way, the apparent MW of each of the cysteine-dependent binding partners (as visualized in Supplementary Fig. S4A) could be estimated. Proteins that migrate at a position in the gel with a shift of about the size of FOXO3 (∼85 kDa) were selected as candidate disulfide-dependent binding partners of FOXO3 (summarized in Table 1). Note that no evidence was found for heterodimerization of FOXO3 with other FOXO family members. Figure 3A shows the iBAQ quantification of the amount of the newly identified disulfide-dependent binding partner CDK4 (cyclin-dependent kinase-4) in each gel strip. It is clear from this analysis that the majority of CDK4 indeed migrates at the apparent combined MW of FOXO3 (85 kDa) and CDK4 (34 kDa): ∼120 kDa. Furthermore, the quantification also shows that CDK4 prefers FOXO3-Cys622 and to a lesser extent, FOXO3-Cys362 for binding. In Figure 3B, the amounts of three other identified disulfide-dependent binding partners (IPO7, IPO8, and TNPO1) in each gel strip are presented. It is clear that again the majority of these proteins pulled down with FOXO3 migrate at the apparent combined MW of FOXO3 and these proteins (∼220 kDa for IPO7 and IPO8, ∼190 kDa for TNPO1). Note that only IPO7 and IPO8 bound to FOXO3-Cys150 migrates at the size of the disulfide-dependent FOXO3-IPO7 complex. Residual binding to the other cysteine mutants migrates at the size of monomeric IPO7 and IPO8, respectively. The amount of IPO7 and IPO8 pulled down is much higher than the amount of TNPO1. For comparison, we presented our previously published data (27) on redox-dependent binding partners of FOXO4 in a similar manner for these three nuclear importers (Supplementary Fig. S5). In the case of FOXO4, TNPO1 binds much better than IPO7 and IPO8. There is some residual binding of IPO7 to FOXO4, but this largely migrates at the size of monomeric IPO7 and IPO8, and also FOXO4ΔCys shows this residual binding.
Table 1.
Identification of Cysteine-Disulfide-Dependent Binding Partners of FOXO3
| Protein names | Gene names | Uniprot | Unique peptide | PEP | p-value | Fold change | MW (kDa) | Shift |
|---|---|---|---|---|---|---|---|---|
| FOXO3-Cys31 | ||||||||
| Peroxiredoxin-1 | PRDX1 | Q06830 | 10 | 3.4979E-38 | 0.016366613 | 19.41407143 | 22.11 | Yes |
| Peroxiredoxin-2 | PRDX2 | P32119 | 6 | 1.9653E-22 | 0.011488354 | 11.1392564 | 21.892 | Yes |
| Proteasome subunit beta type-6 | PSMB6 | P28072 | 3 | 2.4438E-09 | 0.015472578 | 5.407060481 | 25.357 | Yes |
| Calmodulin | CALM2;CALM1 | P62158 | 4 | 1.5769E-56 | 0.033803563 | 5.198066127 | 20.762 | N.I. |
| Importin-7 | IPO7 | O95373 | 19 | 3.2462E-222 | 0.009029563 | 4.912392441 | 119.52 | Yes |
| Peroxiredoxin-5 | PRDX5 | P30044 | 7 | 5.636E-23 | 0.004519624 | 4.778972665 | 22.086 | N.D. |
| FOXO3-Cys150 | ||||||||
| Importin-7 | IPO7 | O95373 | 19 | 3.2462E-222 | 0.000259916 | 37.96989779 | 119.52 | Yes |
| Importin-8 | IPO8 | O15397 | 15 | 6.9487E-154 | 0.000508687 | 19.41086975 | 119.94 | Yes |
| Peroxiredoxin-1 | PRDX1 | Q06830 | 10 | 3.4979E-38 | 0.037154983 | 11.45135205 | 22.11 | Yes |
| Transportin-1 | TNPO1 | Q92973 | 9 | 4.0284E-43 | 0.005145949 | 8.134666145 | 102.35 | Yes |
| Ubiquitin-like modifier-activating enzyme 1 | UBA1 | P22314 | 11 | 3.4263E-99 | 0.004634444 | 7.181045658 | 117.85 | N.D. |
| Proteasome subunit beta type-6 | PSMB6 | P28072 | 3 | 2.4438E-09 | 0.01122133 | 5.314328148 | 25.357 | Yes |
| Peroxiredoxin-2 | PRDX2 | P32119 | 6 | 1.9653E-22 | 0.045877289 | 4.954137135 | 21.892 | Yes |
| Importin subunit beta-1 | KPNB1 | Q14974 | 5 | 3.6065E-25 | 0.002571971 | 4.249026831 | 97.169 | N.D. |
| FOXO3-Cys190 | ||||||||
| Glutathione S-transferase omega-1 | GSTO1 | P78417 | 9 | 2.3023E-28 | 0.006422468 | 9.394772677 | 27.566 | N.I. |
| Proteasome subunit alpha type-4 | PSMA4 | P25789 | 6 | 1.3338E-16 | 0.008221979 | 6.961045517 | 29.483 | N.D. |
| Proteasome subunit beta type-6 | PSMB6 | P28072 | 3 | 2.4438E-09 | 0.025890331 | 5.380638158 | 25.357 | Yes |
| Arginase-1 | ARG1 | P05089-2 | 4 | 5.8174E-08 | 0.003837175 | 4.647008858 | 35.664 | Yes |
| FOXO3-Cys362 | ||||||||
| Peroxiredoxin-2 | PRDX2 | P32119 | 6 | 1.9653E-22 | 0.043097546 | 5.60347922 | 21.892 | Yes |
| Proteasome subunit alpha type-4 | PSMA4 | P25789 | 6 | 1.3338E-16 | 0.031961578 | 4.377192513 | 29.483 | N.D. |
| Peptidyl-prolyl cis-trans isomerase A | PPIA | P62937 | 3 | 8.4601E-09 | 0.03539262 | 4.111145097 | 18.012 | N.D. |
| FOXO3-Cys622 | ||||||||
| Cyclin-dependent kinase 4 | CDK4 | P11802 | 6 | 5.4088E-22 | 1.54875E-05 | 10.81388506 | 33.729 | Yes |
| Peroxiredoxin-1 | PRDX1 | Q06830 | 10 | 3.4979E-38 | 0.039055561 | 9.240073572 | 22.11 | Yes |
| Peroxiredoxin-5 | PRDX5 | P30044 | 7 | 5.636E-23 | 7.90351E-05 | 8.809030507 | 22.086 | N.D. |
| Peroxiredoxin-2 | PRDX2 | P32119 | 6 | 1.9653E-22 | 0.001943848 | 7.126900789 | 21.892 | Yes |
| Proteasome subunit alpha type-4 | PSMA4 | P25789 | 6 | 1.3338E-16 | 0.015236304 | 6.8761906 | 29.483 | N.D. |
| Glyceraldehyde-3-phosphate dehydrogenase | GAPDH | P04406 | 14 | 1.1354E-191 | 0.001033415 | 5.413238551 | 36.053 | Yes |
| Glutathione S-transferase omega-1 | GSTO1 | P78417 | 9 | 2.3023E-28 | 0.034872177 | 4.706508784 | 27.566 | N.I. |
| Reticulocalbin-2 | RCN2 | F8WCY5 | 6 | 1.3415E-96 | 0.032031865 | 4.441045678 | 39.139 | N.I. |
N.I., not identified in secondary screen; N.D., mass-shift not observed in secondary screen.
FIG. 3.
Examples of key results from the secondary MS screen. (A) Representation of the FOXO3-CDK4 interaction as observed in the gel-based MS/MS experiment. CDK4 binds preferably to FOXO3-Cys622 and migrates at an apparent MW that represents a covalent complex. (B) Similar representations for nuclear import receptors that were identified to bind to FOXO3. Low amounts of TNPO1 were found, whereas substantial amounts of IPO7 and IPO8 were found to bind specifically to Cys150. See also Supplementary Figure S5 for a similar analysis of TNPO1, IPO7, and IPO8 binding to FOXO4.
FOXO3 and FOXO4 have overlapping and specific disulfide-dependent binding partners
Like we previously showed for FOXO4, each cysteine in FOXO3 is also involved in disulfide-dependent interactions with a limited set of proteins, which differs for each cysteine (Table 1). The latter indicates that there is specificity and strongly suggests that FOXO3 is also subject to cysteine-dependent redox signaling. When we compare the list of candidate disulfide-dependent binding partners of FOXO3 with that we previously published for FOXO4 (27), we find that both FOXOs bind extensively to peroxiredoxins, which could reflect the involvement of these proteins in disulfide formation, as suggested recently by Jarvis et al. (21). Otherwise, the overlap between the two lists is minor, indicating that there is a clear distinction in the disulfide-dependent binding partners of the two proteins and suggesting differential redox regulation of human FOXO3 and FOXO4.
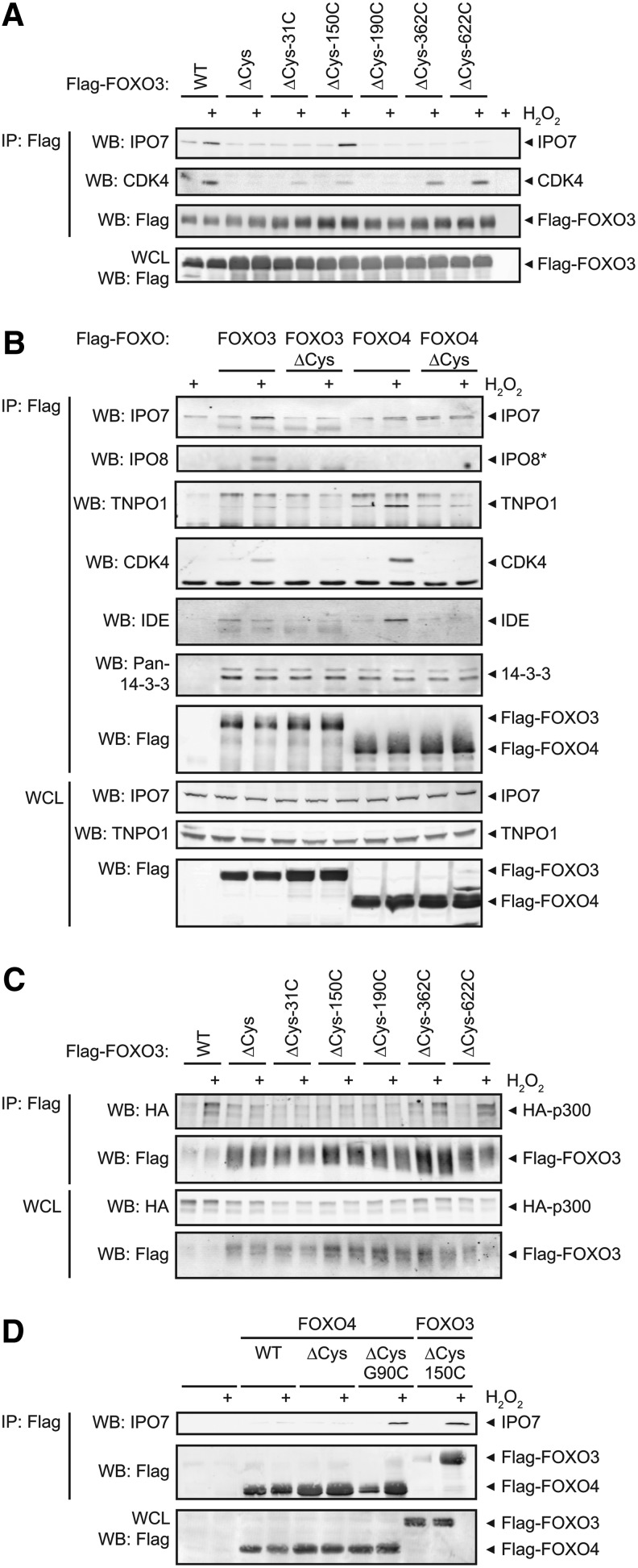
As with every screening method, this method also has its limitations and we next set out to validate some of the newly found redox-dependent FOXO3 interaction partners, their preference for a certain FOXO3 cysteine, and to what extent they are selective for the FOXO3 or FOXO4 paralog. Co-IP experiments followed by Western blot analysis for some of the hits were performed (Fig. 4A). Indeed, IPO7 and CDK4 interact with FOXO3 in a H2O2-induced and cysteine-dependent manner and prefer binding to Cys150 and Cys622, respectively, as was also observed in our MS/MS screen. We next tested whether these proteins could also bind FOXO4 and whether previously identified redox-dependent binding partners of FOXO4 [like insulin degrading enzyme (IDE) (27)] and p300 (12) also bound to FOXO3 (Fig. 4B, C). We found that CDK4 and TNPO1 bind both FOXO3 and FOXO4 in a redox-dependent manner, whereas IPO7 and IPO8 preferentially bind FOXO3. IDE preferentially binds FOXO4. A quantification of the Western blots of the co-IP experiments for IPO7 and TNPO1 to FOXO3 and FOXO4, amalgamated from multiple experiments, can be found in Supplementary Figure S6. Although in our screen the histone acetyl transferase (HAT) p300 was not identified, this protein binds FOXO3 in a cysteine-dependent manner with a preference for Cys622 in the TA domain (Fig. 4C). The binding of p300 to FOXO4 was also shown to be dependent on the cysteine in its TA domain (Cys477 in FOXO4) (12). Taken together, these data indicate that whether a redox-dependent binding partner prefers FOXO3 (IPO7 and IPO8) or FOXO4 (IDE and to a lesser extent TNPO1) or has no preference for FOXO3 or FOXO4 (CDK4 and p300) correlates largely to whether the cysteine it preferentially binds to is conserved between FOXO3 and FOXO4. We therefore propose that evolutionary acquisition of paralog-specific cysteines in human FOXO proteins determines differential redox signaling and could contribute to the divergence of paralog-specific regulation. In agreement with this, we can make IPO7 bind to FOXO4 in a redox-dependent manner (with comparable stoichiometry as with FOXO3) by introduction of a single cysteine residue in FOXO4ΔCys at the position homologous to FOXO3-Cys150 (FOXO4G90C) (Fig. 4D).
FIG. 4.
Disulfide-dependent binding partners differ between FOXO3 and FOXO4 and this correlates with the conservation of specific FOXO cysteines. (A) Verification of two of the hits of our MS/MS screen. Flag-IPs were analyzed on reducing sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) and subsequent Western blot stained with antibodies specific for IPO7 and CDK4. Both proteins show cysteine-dependent binding to FOXO3 in a ROS-induced and cysteine-specific manner, confirming the MS/MS results. Blots represent N=2. (B) Comparison of FOXO3 and FOXO4 cysteine-dependent binding partners. TNPO1 and IDE were previously identified cysteine-dependent FOXO4 interaction partners. IDE binding was not found in our screen for FOXO3 interaction partners or in this assay. As previously reported, TNPO1 does bind to FOXO3 in a cysteine-dependent manner, but to a lesser extent than to FOXO4. CDK4, identified to preferably bind to the cysteine in the TA-domain (FOXO3-Cys622) binds both FOXO3 and FOXO4. IPO7 and IPO8 bind to FOXO3 but not FOXO4. 14-3-3-binding was used as a positive control for co-IP. Blots represent N=4 for IPO7 and TNPO1 and N=3 for IDE, CDK4, and 14-3-3, a representative figure is shown. Quantification of all IPO7 and TNPO1 co-IPs can be found in Supplementary Figure S6A, (B) * IPO8 co-IP was performed in a different experiment than the one that is presented here for the other interaction partners. The complete figure of this experiment is shown in Supplementary Figure S6C. (C) HA-p300 preferably binds to FOXO3-Cys622. This is similar to p300-binding to FOXO4, which was shown to prefer binding the TA-domain cysteine as well (12). (D) A mutation in FOXO4ΔCys that introduces a cysteine at the position homologous to FOXO3-Cys150 (FOXO4ΔCys-G90C) allows the formation of a ROS-induced FOXO4-IPO7 complex.
IPO7 is a cysteine-disulfide-dependent binding partner of FOXO3
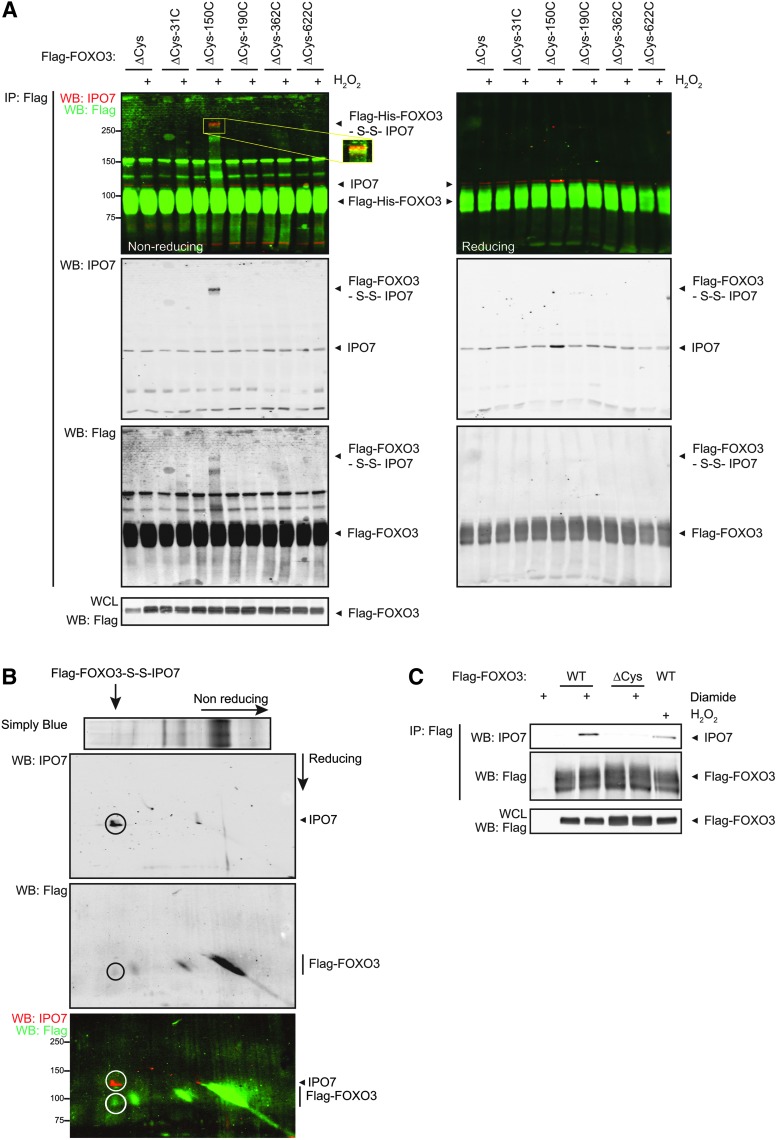
The striking preference of IPO7 and IPO8 for binding FOXO3 over FOXO4 (Supplementary Figs S3B, S4B, and S5) and the requirement of FOXO3-Cys150 herein (Figs. 3B and 4A) attracted our attention and we decided to look further into the molecular details of this interaction. The interaction is indeed disulfide dependent as the two proteins form a covalent complex under nonreducing (but denaturing SDS-PAGE) conditions (Fig. 5A, B). IP followed by parallel reducing and nonreducing Western blot analysis for FOXO3 and IPO7 shows that FOXOΔCys-150C and IPO7 migrate at the same apparent molecular weight under nonreducing conditions, whereas they migrate at their own molecular weight under reducing conditions (Fig. 5A). Note that all IPO7 that is bound to FOXO3-Cys150 migrates in the shifted band under nonreducing conditions, suggesting that the interaction is fully dependent on disulfide formation. Diagonal SDS-PAGE confirms these observations (Fig. 5B). Finally, treatment of cells with the disulfide bond inducing agent diamide instead of H2O2 before lysis induces an even stronger FOXO3-IPO7 interaction when compared to H2O2-treated samples (Fig. 5C). Taken together, the redox-dependent FOXO3-Cys150-IPO7 heterodimer depends on a disulfide.
FIG. 5.
IPO7 is a cysteine-disulfide-dependent binding partner of FOXO3. (A) IPs of the different cysteine mutants of Flag-FOXO3 were analyzed in parallel by nonreducing (left) and reducing (right) SDS-PAGE followed by Western blot for IPO7 and FOXO3. The proteins migrate in one complex under nonreducing conditions (yellow band in enlarged inset), whereas they run in separate bands under reducing conditions. (B) A diagonal SDS-PAGE experiment confirms that the interaction between FOXO3 and IPO7 is direct and disulfide dependent. Flag-FOXO3 overexpressing cells were incubated in H2O2 before lysis, a Flag-IP was performed, and the sample was analyzed by diagonal SDS-PAGE gel electrophoresis followed by Western blot analysis for IPO7 (red) and Flag (green). FOXO3 and IPO7 migrate in a vertical line below the diagonal in the second dimension, indicating that they were migrating in one disulfide-mediated complex before reduction. An explanation and schematic overview of a diagonal SDS-PAGE experiment can be found in Supplementary Figure S9. (C) The FOXO3-IPO7 complex is induced by the disulfide-inducing reagent diamide. Note that the induction of binding is even more efficient than with H2O2.
IPO7 is required for efficient reactive oxygen species-induced nuclear translocation of FOXO3
IPO7 is a nuclear import receptor belonging to the β-karyopherin superfamily of proteins, which are responsible for nucleocytoplasmic transport of macromolecules like proteins, RNA and DNA. IPO7 is described to be involved in the nuclear import of several transcription factors (4–6), ribosomal subunits (19), Histone1 (2), exogenous DNA (15), and kinases (6).
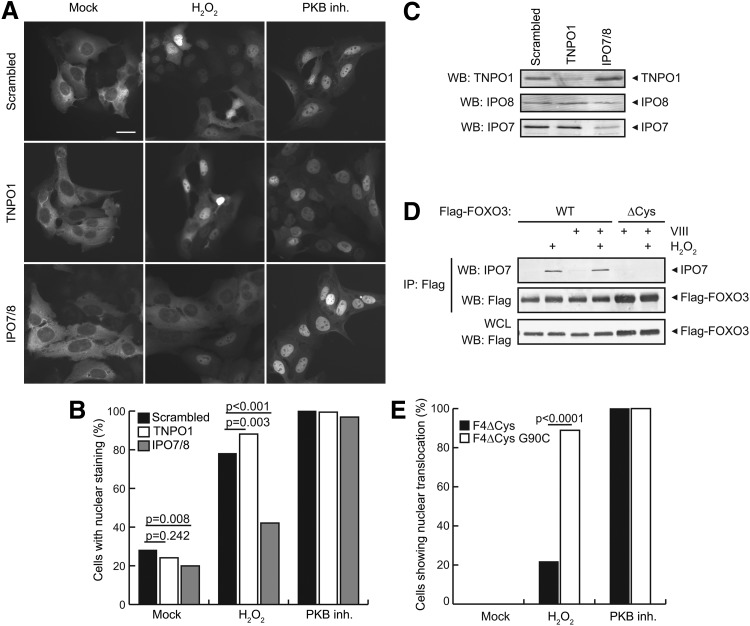
FOXO3 has been described to accumulate in the nucleus and become activated in response to reactive oxygen species (ROS) (3). To investigate whether IPO7 plays a role in FOXO3 nuclear accumulation in response to ROS, we studied the nucleocytoplasmic shuttling upon ipo7, ipo8, and tnpo1 knockdown. IPO7 and IPO8, but not TNPO1, are required for efficient nuclear accumulation of FOXO3 upon H2O2 treatment (Fig. 6A–C and Supplementary Fig. S7). Note that the combined knockdown of ipo7 and ipo8 has more effect than the knockdown of either ipo7 or ipo8, suggesting that these proteins can compensate for each other. In line with this, the primary sequence of human TNPO1 is also very different from that of human IPO7 and IPO8. The ClustalW analysis (PMID 17846036) (21a) shows that IPO7 and IPO8 share 64.64% amino acid sequence identity, whereas TNPO1 shares only 15.2% identity with IPO7 and 16.23% with IPO8. Nuclear translocation upon inhibition of the insulin signaling pathway (PKB inhibition) was not changed upon ipo7, ipo8, or tnpo1 knockdown (Fig. 6A–C and Supplementary Fig. S7), suggesting that ROS-dependent and loss-of-insulin-signaling-dependent nuclear accumulation of FOXO3 is mediated by different import proteins. In agreement with this, induction of nuclear accumulation of FOXO3 by inhibition of PKB does not result in the FOXO3-IPO7 complex formation (Supplementary Fig. S6D). A GFP-tagged version of the above-described FOXO4ΔCys-G90C mutant, which mimics the evolutionary acquisition of FOXO3-Cys150 and hence IPO7 (and IPO8) binding (Fig. 4D), indeed translocates more efficiently in response to ROS as compared to GFP-tagged FOXO4ΔCys (Fig. 6E).
FIG. 6.
IPO7 and IPO8 are required for ROS-induced nuclear translocation of FOXO3. (A) Inducible GFP-FOXO3 expression in U2OS cells transfected with negative control (Scrambled), TNPO1, or IPO7 and IPO8 siRNAs and incubated with H2O2 (500 μM) or PKB inhibitor VIII (10 nM). The scale bar represents 20 μm. (B) Quantification of experiment shown in (A). Localization of GFP-FOXO3 was assessed in >300 cells (divided over two technical replicates) and p-values were calculated in a Fischer's exact test using a 2×2 contingency table (Supplementary Table S2 for the raw data). The percentage of cells showing predominant nuclear FOXO3 localization is plotted. N=3, a representative experiment is shown. A similar experiment using independent siRNAs is shown in Supplementary Figure S7. (C) Western blot for TNPO1, IPO8, and IPO7 on lysates from U2OS cells transfected with the indicated siRNAs. Data are representative of N=2 blots. (D) Treatment with the PKB inhibitor VIII is known to induce nuclear accumulation of FOXO3. However, this does not result in FOXO3-IPO7 complex formation. This indicates that the nuclear import upon PKB inhibition is mediated by a different and currently unknown nuclear importer. (E) FOXO4ΔCys-G90C shows a more efficient nuclear translocation compared to its cysteine-less counterpart FOXO4ΔCys. Localization of GFP-FOXO4ΔCys and GFP-FOXO4ΔCys-G90C was followed over time by time-lapse imaging. Nuclear translocation was scored for all cells that showed cytoplasmic localization at the start of the movie. The percentage of cells showing nuclear translocation during the movie is plotted. Data are representative of N=4 experiments. p-values were calculated in a Fischer's exact test using a 2×2 contingency table (Supplementary Table S2 for the raw data).
Finally, our findings suggest a differential regulation of FOXO3 and FOXO4 nuclear import under oxidative conditions. Indeed, when we compare translocation dynamics of GFP-tagged versions of FOXO3 and FOXO4 under similar circumstances, nuclear accumulation of FOXO4 upon H2O2 treatment seems more rapid compared with FOXO3, whereas basal localization, export, and insulin-signaling pathway-dependent dynamics are comparable (Supplementary Fig. S8). It remains to be explored what the consequences of the here described redox-signaling-dependent regulatory divergence of FOXO3 and FOXO4 will be in a physiological context.
Discussion
In this study, we compared disulfide-dependent heterodimerization of two functionally closely related transcription factors FOXO3 and FOXO4. Intermolecular disulfide formation as a mode of redox signaling has not been studied much and, until recently, only about 30 such interactions were described (29). However, using the MS/MS-based approach described here for FOXO3 and previously for FOXO4, we find several functional intermolecular disulfides, suggesting that this could be a common mechanism in redox signaling. It is currently not known whether disulfide-dependent heterodimerization in response to ROS is a common mechanism in TNPO1-, IPO7-, and IPO8-dependent nuclear transport.
Our data show that FOXO3, like FOXO4, is being shuttled into the nucleus in response to ROS through formation of a disulfide with a nuclear import protein. However, whereas we previously showed that FOXO4 dimerizes with TNPO1 (27), we now find that FOXO3 dimerizes preferentially with IPO7 and IPO8. This implies that not only a differential spatiotemporal expression of FOXO3 and 4 but also a hypothetical differential spatiotemporal expression of IPO7, IPO8, or TNPO1 during, for example, development, differentiation, or tumorigenesis could, in principle, lead to differential responses of FOXO3 and 4. It remains to be explored how the here observed regulatory divergence of FOXO3 and FOXO4 translates to a functional divergence. This is not trivial as FOXO3ΔCys and FOXO4ΔCys eventually still accumulate in the nucleus in response to ROS, which might be due to JNK-dependent phosphorylation (18), mono-ubiquitinylation (32), inhibition of Crm-1-dependent nuclear export (30), or a combination of these. The delay in nuclear accumulation of the FOXOΔCys mutants is clearly measurable, but relatively short as compared to the establishment of a measurable FOXO transcriptional program-dependent phenotype. We will therefore aim to address the functional consequence of our observations in future studies.
The disulfide bond formation between IPO7 (or IPO8) and FOXO3 is specific for Cys150 in FOXO3, and this is different from what we have seen previously for disulfide-dependent binding of p300 and TNPO1 to FOXO4; p300 binds preferably to FOXO4-Cys477, however, when a mutant is used that lacks this cysteine but has FOXO4-Cys411 available, it can form a disulfide with FOXO4-Cys411 (12). Likewise, TNPO1 forms a disulfide preferably with FOXO4-Cys239, but once this is mutated to a serine, FOXO4-Cys355 is able to mediate the interaction (27). One explanation for this adaptability could lie in the unstructured nature of FOXO proteins; only the DNA binding domain is structured and the remainder is largely intrinsically disordered. This could allow for flexibility in cysteine presence in disulfide formation. It is believed that disulfide formation occurs in a two-step manner (11) Once a cysteine becomes oxidized to sulfenic acid or by formation of a disulfide with peroxiredoxin, it becomes highly reactive and will form a disulfide with a neighboring thiol. If it is the cysteine thiol in a FOXO-binding partner that becomes oxidized first, it will seek any thiol in FOXO in its vicinity to form a disulfide bond with. Indeed, sulfenic acid formation has been observed on both IPO7 and TNPO1 (22).This model could potentially also explain why TNPO1 can still interact with FOXO3 (Figs. 3B and 4B), although the preferred cysteine it binds to in FOXO4 (Cys239) is not present in FOXO3. The observation that IPO7 and IPO8 strictly require FOXO3Cys150 could potentially be due to the fact that this cysteine lies close to the structured FKH domain (Fig. 1A).
We have previously shown that stress-induced nuclear accumulation of the Caenorhabditis elegans FOXO homolog DAF-16 depends on the TNPO1 homolog IMB-2 (27). In light of our findings, we wondered whether an IPO7 or IPO8 homolog could also function in C. elegans to transport DAF-16 to the nucleus. However, a BLAST search revealed that there are no proteins in C. elegans that resemble human IPO7 or IPO8.
Gene duplications are believed to be one of the most important factors in evolution; due to the resulting redundancy, they allow for the release of negative evolutionary pressure and thus for an increased mutation rate. Not all gene duplicates are maintained in the genome; functional divergence is often required to prevent the loss of two completely redundant genes (8, 31). The mammalian FOXO transcription factors FOXO1, 3, 4, and 6 are the result of gene duplications during evolution. When comparing the sequences of FOXO1, 3, and 4, the diversification becomes easily clear (Fig. 1A); although the CR1, FKH, and TA domains have an overlap of approximately 80% (1), the other regions are quite diverse. However, although the protein sequences clearly show differences, several of the upstream regulators are shared between the paralogs, and some even originate from before the genome duplications. While functional divergence has been appreciated for almost a decade, not many thorough comparisons have been made between the upstream regulators of the different paralogs up to now (e.g., in (34)).We show here that cysteines that are conserved between FOXO paralogs indeed can bind the same proteins (e.g., CDK4, p300) in a disulfide-dependent manner, whereas paralog-specific cysteines can give rise to paralog-specific interaction partners (e.g., IPO7 and IPO8 for FOXO3 and IDE and to a lesser extent TNPO1 for FOXO4). We mimicked the evolutionary acquisition of FOXO3-Cys150 by introducing a cysteine in the homologous position in FOXO4ΔCys (FOXO4ΔCys-G90C). This mutant, which has no other cysteines but the one that mimics FOXO3-Cys150, was now indeed able to bind IPO7 in a redox-dependent manner and translocates rapidly to the nucleus in response to ROS.
Materials and Methods
Alignment of FOXO proteins
The amino acid sequences of the different human FOXO paralogs and vertebrate FOXO homologs were obtained through the NCBI database. In some cases, an mRNA sequence gave better coverage of a certain FOXO homolog and in that case, the mRNA was translated and the corresponding amino acid sequence was used. The GI-numbers of the used sequences are listed in the alignment figures for each sequence. To analyze the homologs of each vertebrate FOXO isoform, we used the sequences of two species of fish, two species of amphibians, two species of reptiles, two species of birds, and four species of mammals. We used the same species for the different FOXO isoforms, except for FOXO4, where the genome of Danio rerio had a better coverage than Maylandia zebra. The used species are listed in the corresponding figures. Alignments were performed using the T-Coffee algorithm with default settings (25). We used Jalview software for the representation (35). The human FOXO paralog alignment was colored by conservation (without threshold). The FOXO ortholog alignments were colored by conservation (threshold 15). Darker purple means more conservation or consensus. Cysteine residues were colored Yellow in Adobe Illustrator.
Cell culture
All cells were kept under standard conditions (bicarbonate-buffered Dulbecco's modified Eagle's medium (10% fetal bovine serum), 37°C, 6% CO2 atmosphere). All overexpression experiments were carried out in HEK293T cells. Transfections were performed using polyethylenimine (Sigma Aldrich), and experiments were performed 48 h post-transfection. RNAi transfections were performed according to protocol using HiPerfect (Qiagen).
For localization studies, U2OS cells stably expressing tetracycline (Tet)-repressor and doxycycline-inducible GFP-Flag-FOXO3, GFP-Flag-FOXO4, FOXO4ΔCys, or FOXO4ΔCys-G90C were used. These cells were generated by the transient transfection of a stable Tet-repressor expressing U2OS cell line (U2OS TRs C2.1) (13) with pTON-bEGFP-FOXOX. Upon selection, a monoclonal cell line was obtained and used for experiments using WT FOXO3 and 4, whereas polyclonal cell lines were used for the experiments assessing localization of FOXO4ΔCys and FOXO4ΔCys-G90C. Expression of GFP-Flag-FOXO was induced with doxycycline for 3 days before starting experiments to ensure reproducible low and cytoplasmic expression of GFP-Flag-FOXO.
Plasmids and reagents
Flag-His-FOXO3 and doxycycline-inducible GFP-FOXO3 constructs (pTON-bEGFP-FOXO3) were created by Gateway cloning (Invitrogen). An HA-FOXO3 construct was used as a template for creating a FOXO3 entry clone. These were subsequently swapped in the donor vector according to the standard Gateway protocol. Cysteine mutant constructs were created by point mutation PCR (using the following respective forward primers: C31: 5′-CCGCGATCCTCTACGTGGCCC-3′, C150: 5′-CCGAGGAAATCTTCGTCGCGG-3′, C190: 5′-ATGGTGCGTTCCGTGCCCTAC-3′, C362: 5′-AGCAAGCCGTCCACGGTGGAA-3′, and C622: 5′-AGCTTGGAATCTGACATGGAG-3′) and verified by sequencing. For doxycycline-inducible GFP-FOXO3 constructs, a modified version of pcDNA4-TO (Invitrogen) carrying a biotinylation-tag, a Flag-tag, and a GFP (referred to as (pTON-bEGFP) was converted to a Gateway donor vector.
Flag-FOXO4 wild-type and cysteine mutant constructs were described before (12,33). Flag-FOXO4-G90C mutant was created by point mutation PCR using the following primer: Fwd: 5′-GTCCTCGGAAGTGTGGCTCCCGCCG GAATG. HA-p300 was described before (12).
TNPO1 (L-011308), IPO7 (L-012255, smartpool and singles -06 and -07), and IPO8 (L-012256, smartpool and single -19) knockdown was performed using smartpool siRNAs from Dharmacon and used in a concentration of 20 nM. PKB inhibitor VIII (Merck Chemicals) was used in a concentration of 10 nM for the indicated time. Leptomycin B (Sigma-Aldrich) was used in a concentration of 20 ng/ml for the indicated time (similar to H2O2). Thirty-seven percent H2O2 was purchased from Merck and freshly diluted to a stock of 100 mM in H2O for every experiment. H2O2 treatments were 200 μM for 10 min for IPs and 500 μM for 45 min in localization studies. Diamide (Sigma Aldrich) was used in a concentration of 0.1 mM.
Co-IPs and Western blotting
For co-IPs, cells were treated as indicated and subsequently lysed in the lysis buffer (50 mM Tris-HCl pH 7.5, 1% TX100, 1.5 mM MgCl2, 5 mM EDTA, 100 mM NaCl, NaF, Leupeptin, Aprotinin) containing 100 mM iodoacetamide to prevent postlysis oxidation and to prevent reduction events. Lysates were incubated with Flag-M2 beads (Sigma Aldrich) while shaking for approximately 2 h and washed in the lysis buffer containing 1 M NaCl. Samples were either used for Western blot analysis (reducing or nonreducing conditions) or eluted of the beads in 0.1 M glycine (pH 2). For parallel reducing and nonreducing SDS-PAGE analysis, IPs were washed as described above and divided in two after the last washing.
SDS-PAGE and Western blotting were performed according to standard protocols. Nonreducing SDS-PAGE of Flag-FOXO3 was performed on 6% polyacrylamide gel to obtain maximum separation. (For an explanation of diagonal SDS-PAGE, see the cartoon in Supplementary Fig. S9).
Antibodies that were used are Flag M2 (Sigma, F1804), IPO7, IPO8, and TNPO1 (Abcam; ab99273, ab72109, and ab10303, respectively)], HA (12CA5, monoclonal from hybridoma cell lines), CDK4 and Pan-14-3-3 (Santa Cruz; SC-260 and SC-629, respectively), and IDE (ITK diagnostics; A301-906A). Detection of fluorescent secondary antibodies was performed using the LI-COR Biosciences Odyssey Infrared Imaging System; detection of secondary HRP antibodies was performed on the FUJIFILM Luminescent Image Analyzer LAS-4000 mini.
Luciferase assay
The activity of Flag-FOXO3 and Flag-FOXO3ΔCys was measured in HEK293T cells, using firefly luciferase driven from an artificial promoter containing six FOXO-binding sites (6×DBE) and was described before (33). The luciferase activity was normalized to the activity of Renilla luciferase expressed from a thymidine kinase promoter. Both luciferase activities were measured according to the manufacturer's instructions using the Dual-Luciferase reporter Assay system (Promega; product E1960) on a Berthold Techonolgies MicroLumatPlus LB 96V luminometer.
Mass spectroscopy and MS/MS data analysis
MS/MS sample preparation, measurements, and data analysis were described extensively before (27). In short, raw files were analyzed with the Maxquant software version 1.3.0.5 (10) with deamidation of glutamine and asparagine, oxidation of methionine, and cysteine alkylation with iodoacetamide as well as N-Ethylmaleimide set as variable modification. For identification, the human Uniprot database was searched with both the peptide as well as the protein false discovery rate set to 1%. Proteins identified with two or more unique peptides were filtered for reverse hits and standard contaminants.
For the first part of the screen, the label-free quantification (LFQ) algorithm (9) was used in combination with the match between runs tool (option set at 2 min), all of which are integral parts of the Maxquant software (23). LFQ intensities, which are based on the extracted ion current normalized on total peptide abundance, were log2 transformed, after which identified proteins were filtered for at least three valid values in at least one sample group. Missing values were replaced by imputation based on a normal distribution (using a width of 0.3 and a downshift of 1.8 (14)), to simulate a more realistic value for the noise. p-values were calculated in a Student's t-test and plotted against the difference of the mean log2 transformed LFQ values using the program R (www.R-project.org). For the hits of our screen, the raw data used for this analysis are shown in Supplementary Table S1.
For the second branch of the screen, we used the Intensity-Based Absolute Quantification algorithm (iBAQ) (28), to compare protein abundance within and between lanes of a nonreducing SDS-PAGE gel. All MS runs were treated as separate experiments and adjacent gel pieces (in the same lane or at the same height in the other lanes) were used for the match between runs option. When using iBAQ, intensities are not normalized to the total peptide abundance, which allows us to compare the intensities of the different gel pieces within a lane.
Localization studies
Doxycycline-inducible GFP-Flag-FOXO3 or GFP-Flag-FOXO4 cells were seeded on glass, transfected with the indicated siRNAs, and GFP-FOXO expression was induced with doxycycline, both for 72 h. Cells were treated as indicated, fixed for 1 h in 3.7% formaldehyde, DNA was stained with DAPI (4′,6-diamidino-2-phenylindole; Sigma-Aldrich), and the slides were mounted on glass. Analysis was performed on a Zeiss Axioskop 2 microscope equipped with a Zeiss Axiocam CCD camera. Localization of GFP-FOXO3 was scored to be either predominantly nuclear or cytoplasmic for >300 cells per condition (divided over two technical replicates). Statistical analysis of the data was performed in Graphpad Prism using a 2×2 contingency table; p-values were calculated in a two-tailed Fischer's exact test (see Supplementary Table S2 for the raw data). Knockdown of ipo7 was checked by Western blot.
Time-lapse imaging
Time-lapse imaging was performed as described before (27). Doxycycline-inducible GFP-Flag-FOXO4ΔCys and FOXO4ΔCys-G90C cells were seeded in 24-well plates, and FOXO expression was induced with doxycycline for 72 h. Time-lapse imaging was performed on an Olympus IX-81 microscope equipped with a 20x/0.5NA UPLFLN objective and an ORCA-ER camera (Hamamatsu Photonics) in a permanently heated chamber (37°C, 5% CO2). Images were acquired every 2 min for 30 min before treatments, as indicated, and followed in time until translocation had occurred. Five hundred micromolar H2O2 was used to induce translocation. Resulting movies were analyzed using ImageJ. Only cells with basal cytoplasmic localization of FOXO4 at the beginning of the movie (GFP-intensity cytoplasm>GFP-intensity nucleus) were taken into account. Cells undergoing mitosis or apoptosis during the movie were excluded from analysis. Statistical analysis of the data was performed in Graphpad Prism using 2×2 contingency tables; p-values were calculated in a two-tailed Fischer's exact test (Supplementary Table S2 for the raw data).
Supplementary Material
Abbreviations Used
- CR
conserved region
- FKH
Forkhead
- IDE
insulin-degrading enzyme
- IP
immunoprecipitation
- LFQ
label-free quantification
- ROS
reactive oxygen species
- SDS-PAGE
sodium dodecyl sulfate–polyacrylamide gel electrophoresis
- TA
transactivation
- WT
wild type
Acknowledgments
This research was sponsored by the following grants: ECHO, which is financed by the Netherlands Organization for Scientific Research (NWO) (M.V. and T.D.), Dutch Cancer Society (KWF Kankerbestrijding) (T.D., M.V.), Center for Biomedical Genetics (B.B.), and CancerGenomiCs.nl (B.B.).
Authors' Contributions
M.P., H.R.V., and T.B.D. designed the study and analyzed the data. T.B.D. and B.S. performed phylogenetic analysis. M.P., H.R.V., K.D., and H.R. performed the biochemical experiments. M.P. and A.G.D. performed the immunofluorescence experiments. H.R.V. performed the MS/MS analysis. M.V., B.S., and B.M.T.B. provided essential reagents, facilities, and input. M.P. and T.B.D. wrote the article. All authors discussed the results and commented on the article.
Data and Materials Availability
The mass spectrometry proteomics data have been deposited at the ProteomeXchange Consortium via the PRIDE partner repository (www.ebi.ac.uk/pride) with the dataset identifiers PXD001018, PXD001040, and PXD001041. For further details, see the corresponding parameter.txt files. Please contact the corresponding author (T.B.D.) for material requests.
Author Disclosure Statement
The authors have no competing interests.
References
- 1.Anderson MJ, Viars CS, Czekay S, Cavenee WK, and Arden KC. Cloning and characterization of three human forkhead genes that comprise an FKHR-like gene subfamily. Genomics 47: 187–199, 1998 [DOI] [PubMed] [Google Scholar]
- 2.Bauerle M, Doenecke D, and Albig W. The requirement of H1 histones for a heterodimeric nuclear import receptor. J Biol Chem 277: 32480–32489, 2002 [DOI] [PubMed] [Google Scholar]
- 3.Brunet A, Sweeney LB, Sturgill JF, Chua KF, Greer PL, Lin Y, Tran H, Ross SE, Mostoslavsky R, Cohen HY, Hu LS, Cheng HL, Jedrychowski MP, Gygi SP, Sinclair DA, Alt FW, and Greenberg ME. Stress-dependent regulation of FOXO transcription factors by the SIRT1 deacetylase. Science 303: 2011–2015, 2004 [DOI] [PubMed] [Google Scholar]
- 4.Chachami G, Paraskeva E, Mingot JM, Braliou GG, Gorlich D, and Simos G. Transport of hypoxia-inducible factor HIF-1alpha into the nucleus involves importins 4 and 7. Biochem Biophys Res Commun 390: 235–240, 2009 [DOI] [PubMed] [Google Scholar]
- 5.Chen J, Liu MY, Parish CR, Chong BH, and Khachigian L. Nuclear import of early growth response-1 involves importin-7 and the novel nuclear localization signal serine-proline-serine. Int J Biochem Cell Biol 43: 905–912, 2011 [DOI] [PubMed] [Google Scholar]
- 6.Chuderland D, Konson A, and Seger R. Identification and characterization of a general nuclear translocation signal in signaling proteins. Mol Cell 31: 850–861, 2008 [DOI] [PubMed] [Google Scholar]
- 7.Clamp M, Cuff J, Searle SM, and Barton GJ. The Jalview Java alignment editor. Bioinformatics 20: 426–427, 2004 [DOI] [PubMed] [Google Scholar]
- 8.Conant GC. and Wolfe KH. Turning a hobby into a job: how duplicated genes find new functions. Nat Rev Genet 9: 938–950, 2008 [DOI] [PubMed] [Google Scholar]
- 9.Cox J, Hein MY, Luber CA, Paron I, Nagaraj N, and Mann M. MaxLFQ allows accurate proteome-wide label-free quantification by delayed normalization and maximal peptide ratio extraction. Mol Cell Proteomics 2014[Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cox J. and Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol 26: 1367–1372, 2008 [DOI] [PubMed] [Google Scholar]
- 11.Cremers CM. and Jakob U. Oxidant sensing by reversible disulfide bond formation. J Biol Chem 288: 26489–26496, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dansen TB, Smits LM, van Triest MH, de Keizer PL, van Leenen D, Koerkamp MG, Szypowska A, Meppelink A, Brenkman AB, Yodoi J, Holstege FC, and Burgering BM. Redox-sensitive cysteines bridge p300/CBP-mediated acetylation and FoxO4 activity. Nat Chem Biol 5: 664–672, 2009 [DOI] [PubMed] [Google Scholar]
- 13.de Graaf P, Mousson F, Geverts B, Scheer E, Tora L, Houtsmuller AB, and Timmers HT. Chromatin interaction of TATA-binding protein is dynamically regulated in human cells. J Cell Sci 123: 2663–2671, 2010 [DOI] [PubMed] [Google Scholar]
- 14.Deeb SJ, D'Souza RC, Cox J, Schmidt-Supprian M, and Mann M. Super-SILAC allows classification of diffuse large B-cell lymphoma subtypes by their protein expression profiles. Mol Cell Proteomics 11: 77–89, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dhanoya A, Wang T, Keshavarz-Moore E, Fassati A, and Chain BM. Importin-7 mediates nuclear trafficking of DNA in mammalian cells. Traffic 14: 165–175, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dickinson BC. and Chang CJ. Chemistry and biology of reactive oxygen species in signaling or stress responses. Nat Chem Biol 7: 504–511, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Eijkelenboom A. and Burgering BM. FOXOs: signalling integrators for homeostasis maintenance. Nat Rev Mol Cell Biol 14: 83–97, 2013 [DOI] [PubMed] [Google Scholar]
- 18.Essers MA, Weijzen S, de Vries-Smits AM, Saarloos I, de Ruiter ND, Bos JL. and Burgering BM. FOXO transcription factor activation by oxidative stress mediated by the small GTPase Ral and JNK. EMBO J 23: 4802–4812, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Golomb L, Bublik DR, Wilder S, Nevo R, Kiss V, Grabusic K, Volarevic S, and Oren M. Importin 7 and exportin 1 link c-Myc and p53 to regulation of ribosomal biogenesis. Mol Cell 45: 222–232, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jacobs FM, van der Heide LP, Wijchers PJ, Burbach JP, Hoekman MF, and Smidt MP. FoxO6, a novel member of the FoxO class of transcription factors with distinct shuttling dynamics. J Biol Chem 278: 35959–35967, 2003 [DOI] [PubMed] [Google Scholar]
- 21.Jarvis RM, Hughes SM, and Ledgerwood EC. Peroxiredoxin 1 functions as a signal peroxidase to receive, transduce, and transmit peroxide signals in mammalian cells. Free Radic Biol Med 53: 1522–1530, 2012 [DOI] [PubMed] [Google Scholar]
- 21a.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, and Higgins DG. Clustal W and Clustal X version 2.0. Bioinformatics 23: 2947-2948, 2007 [DOI] [PubMed] [Google Scholar]
- 22.Leonard SE, Reddie KG, and Carroll KS. Mining the thiol proteome for sulfenic acid modifications reveals new targets for oxidation in cells. ACS Chem Biol 4: 783–799, 2009 [DOI] [PubMed] [Google Scholar]
- 23.Luber CA, Cox J, Lauterbach H, Fancke B, Selbach M, Tschopp J, Akira S, Wiegand M, Hochrein H, O'Keeffe M, and Mann M. Quantitative proteomics reveals subset-specific viral recognition in dendritic cells. Immunity 32: 279–289, 2010 [DOI] [PubMed] [Google Scholar]
- 24.Marino SM. and Gladyshev VN. Cysteine function governs its conservation and degeneration and restricts its utilization on protein surfaces. J Mol Biol 404: 902–916, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Notredame C, Higgins DG, and Heringa J. T-Coffee: A novel method for fast and accurate multiple sequence alignment. J Mol Biol 302: 205–217, 2000 [DOI] [PubMed] [Google Scholar]
- 26.Paik JH, Kollipara R, Chu G, Ji H, Xiao Y, Ding Z, Miao L, Tothova Z, Horner JW, Carrasco DR, Jiang S, Gilliland DG, Chin L, Wong WH, Castrillon DH, and DePinho RA. FoxOs are lineage-restricted redundant tumor suppressors and regulate endothelial cell homeostasis. Cell 128: 309–323, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Putker M, Madl T, Vos HR, de Ruiter H, Visscher M, van den Berg MC, Kaplan M, Korswagen HC, Boelens R, Vermeulen M, Burgering BM, and Dansen TB. Redox-dependent control of FOXO/DAF-16 by transportin-1. Mol Cell 49: 730–742, 2013 [DOI] [PubMed] [Google Scholar]
- 28.Schwanhausser B, Busse D, Li N, Dittmar G, Schuchhardt J, Wolf J, Chen W, and Selbach M. Global quantification of mammalian gene expression control. Nature 473: 337–342, 2011 [DOI] [PubMed] [Google Scholar]
- 29.Sun MA, Wang Y, Cheng H, Zhang Q, Ge W, and Guo D. RedoxDB—a curated database for experimentally verified protein oxidative modification. Bioinformatics 28: 2551–2552, 2012 [DOI] [PubMed] [Google Scholar]
- 30.Sunayama J, Tsuruta F, Masuyama N, and Gotoh Y. JNK antagonizes Akt-mediated survival signals by phosphorylating 14-3-3. J Cell Biol 170: 295–304, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Taylor JS. and Raes J. Duplication and divergence: the evolution of new genes and old ideas. Annu Rev Genet 38: 615–643, 2004 [DOI] [PubMed] [Google Scholar]
- 32.van der Horst A, de Vries-Smits AM, Brenkman AB, van Triest MH, van den Broek N, Colland F, Maurice MM, and Burgering BM. FOXO4 transcriptional activity is regulated by monoubiquitination and USP7/HAUSP. Nat Cell Biol 8: 1064–1073, 2006 [DOI] [PubMed] [Google Scholar]
- 33.van der Horst A, Tertoolen LG, de Vries-Smits LM, Frye RA, Medema RH, and Burgering BM. FOXO4 is acetylated upon peroxide stress and deacetylated by the longevity protein hSir2(SIRT1). J Biol Chem 279: 28873–28879, 2004 [DOI] [PubMed] [Google Scholar]
- 34.van der Vos KE, and Coffer PJ. FOXO-binding partners: it takes two to tango. Oncogene 27: 2289–2299, 2008 [DOI] [PubMed] [Google Scholar]
- 35.Waterhouse AM, Procter JB, Martin DM, Clamp M, and Barton GJ. Jalview Version 2—a multiple sequence alignment editor and analysis workbench. Bioinformatics 25: 1189–1191, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.