Abstract
Polycomb/Trithorax response elements (PRE/TREs) can switch their function reversibly between silencing and activation, by mechanisms that are poorly understood. Here we show that a switch in forward and reverse noncoding transcription from the Drosophila vestigial (vg) PRE/TRE switches the status of the element between silencing (induced by the forward strand) and activation (induced by the reverse strand). In vitro, both ncRNAs inhibit PRC2 histone methyltransferase activity, but in vivo only the reverse strand binds PRC2. Over-expression of the reverse strand evicts PRC2 from chromatin and inhibits its enzymatic activity. We propose that interactions of RNAs with PRC2 are differentially regulated in vivo, allowing regulated inhibition of local PRC2 activity. Genome-wide analysis shows that strand switching of ncRNAs occurs at several hundred PcG binding sites in fly and vertebrate genomes. This work identifies a novel and potentially widespread class of PRE/TREs that switch function by switching the direction of ncRNA transcription.
INTRODUCTION
The Polycomb (PcG) and Trithorax Group (TrxG) proteins are essential for development and differentiation in flies and vertebrates1. In flies, these proteins provide epigenetic maintenance of repressed (PcG) or activated (TrxG) gene expression states that are initially determined by transcription factors2. PcG/TrxG proteins act antagonistically through Polycomb/Trithorax response elements (PRE/TREs)3. PRE/TREs are switchable regulatory DNA elements that can preserve a memory of the activated or silenced state of their associated genes over several cell generations2. How PRE/TREs switch between active and silent states and how this function is modulated in different developmental contexts is poorly understood. Many PcG/TrxG binding sites are transcribed into long noncoding (nc) RNAs4 and several ncRNAs are involved in PcG/TrxG function, though the mechanisms by which they act are currently the subject of debate5,6.
Here we show that the Drosophila vestigial (vg) PRE/TRE is transcribed in a bidirectional and developmentally regulated manner. Embryonic transcription of the PRE/TRE reverse strand correlates with vg gene activation, whereas larval transcription of the PRE/TRE forward strand correlates with vg gene repression. Ectopic transcription of each strand leads to trans-activation or trans-silencing at the endogenous vg locus. Thus, in vivo, two different strands transcribed from the same regulatory element have opposite effects on the properties of the element.
TrxG and PcG proteins have been shown to bind to several ncRNAs, and it has been proposed that these RNAs specifically recruit the TrxG and PcG proteins to their target genes7-12, reviewed in6. However, it has been shown that the Polycomb Repressive Complex 2 (PRC2) binds RNA in vitro with high affinity but no specificity13 challenging the hypothesis that PRC2 is recruited to its target genes via specific interactions with ncRNAs. In contrast to these findings, it has recently been reported that some RNAs show specificity in both binding and inhibition of the activity of PRC2 in vitro14. Thus, the specificity of PRC2- ncRNA interactions, in terms of both binding and inhibition is currently disputed.
Here we report that RNA promiscuously inhibits the enzymatic activity of PRC2 in vitro at equimolar RNA-PRC2 concentrations and with no specificity for the different RNAs tested, including those of the vg PRE/TRE. However, in vivo the reverse (but not the forward) vg PRE/TRE strand binds highly specifically to the PRC2 component Enhancer of Zeste (E(Z)). Over-expression of the reverse strand leads to both local inhibition of the catalytic activity of PRC2 and the eviction of E(Z) from chromatin. Thus, we show that despite nonspecific interaction in vitro, the interaction of different ncRNAs and PcG proteins in vivo is highly specific and has the potential to inhibit PRC2 at its site of action in a developmentally regulated manner. In summary, this work has broad implications for understanding the dynamic dual nature of PRE/TRE elements, and the role of specific ncRNAs in switching their function.
RESULTS
Opposite RNA strands of the vg PRE/TRE correlate inversely with vg mRNA
The Drosophila vg gene is essential for determining correct cell identity in wing and haltere15,16, in specific muscle lineages15,17,18, and is also expressed in the embryonic central nervous system16,19. The vg gene is a target of PcG regulation in embryos20,21 and in wing22-24. The locus contains two sites of PcG ChIP enrichment: one at the promoter, and one downstream of the gene (Fig. 1a)20,25. The latter site contains a PRE/TRE that has been characterized in several transgenic assays, and can maintain both silencing and activation of reporter gene activity in a PcG- and TrxG-dependent manner22,23,26.
Figure 1. Coding and noncoding transcription at the vg locus.
(a) vg locus showing vg intron and exons, PRE/TRE (red) and enhancers (blue). BE, boundary enhancer38; QE, quadrant enhancer39. Sites of ChIP enrichment20,25 (Supplementary Fig. 2, Supplementary Fig. 10) are shown above the Figure (grey bars). qPCR primers used in Supplementary Figure 1 are shown below the Figure. (b) Noncoding transcripts at the vg PRE/TRE. See also Supplementary Figure 2 and Supplementary Figure 3. Tissue in which each transcript was detected, and transcript length are given. Colored bars indicate PRE/TRE DNA motifs. Grey box: 1.6 kb core PRE/TRE26 used for transgenes. (c-p) Double in situ hybridization on embryos (c-h) and 3rd instar larval wing discs (k-p) showing vg mRNA (red), PRE/TRE reverse (d,e,g,h) or forward (l,m,o,p) strand transcript (green) and DAPI (blue). Anterior is to the right, posterior to the left; embryos: ventral view. (c-e) 10h (stage 13) embryos. w, h, wing and haltere primordia; m, muscle precursors; CNS, central nervous system. (f-h) Single optical slice at high magnification of boxes in (c-e) showing muscle precursors with several nuclei. (g) Arrows indicate nuclear dots. (i,j) Diagram of multinucleate precursors outlined in (h), showing (i) founder colony (j) advanced colony. (k-p) Wing discs: single optical slices showing vg mRNA and PRE/TRE forward strand transcript are expressed in a complementary pattern. (n-p) high magnification of boxes in (k-m). (o) Arrows indicate nuclear dots. Embryo and wing disc in situ hybridizations were performed at least three times, and 5-10 discs or 10-20 embryos were analyzed.
We detected transcripts from the downstream vg PRE/TRE in tissues in which the vg mRNA is expressed (Supplementary Fig. 1). Mapping of 5' and 3' ends of PRE/TRE transcripts by 5' RACE, primer walking, and cDNA cloning in embryos, larval brains and wing discs identified several RNA Pol II transcripts arising from both strands of the PRE/TRE (Fig. 1b, Supplementary Fig. 2, Supplementary Fig. 3, Supplementary Table 1). Additional mapping showed that the forward strand transcripts are distinct from the vg mRNA itself, as no transcription was detected between the 3' end of vg and the 5' ends of the PRE/TRE transcripts. Interestingly, we observed a developmental switch in strand preference of the PRE/TRE transcripts: in embryos, both strands of the PRE/TRE were transcribed (Fig. 1b, Supplementary Fig. 2). In contrast, in larval wing discs and brains, only transcripts from the forward strand were detected (Fig. 1b, Supplementary Fig. 2). Thus, the PRE/TRE transcripts are highly developmentally regulated.
Double in situ hybridizations to each strand of the PRE/TRE and the vg mRNA showed that reverse strand transcripts of the PRE/TRE and the vg mRNA became visible at 10h of embryogenesis. Both of these transcripts were present in the somatic muscle lineage27 (Fig. 1c-j). The PRE/TRE transcript was detectable in muscle founder cells before the appearance of the vg mRNA, both in the shared cytoplasm of fused nuclei, and as nuclear dots (Fig. 1f-j, Supplementary Fig. 4). In later embryos, both the PRE/TRE reverse strand and the vg mRNA were expressed at high levels in all somatic muscles (Supplementary Fig. 4j-l). Thus, the reverse strand of the vg PRE/TRE is transcribed specifically in cells of the muscle lineage in which the vg mRNA later becomes activated.
In contrast, the spatial pattern of PRE/TRE transcription from the forward strand was reciprocal to that of the vg mRNA, with the highest levels of vg mRNA corresponding to the lowest levels of PRE/TRE transcripts, and vice versa. This reciprocal relationship was observed to some extent in late embryos (Supplementary Fig. 5), and most strikingly in 3rd instar larval wing discs (Fig. 1k-p) and brains (Supplementary Fig. 6). In summary, these results demonstrate that transcripts from opposite strands of the vg PRE/TRE correlate inversely with the vg mRNA.
PRE/TRE strand switching correlates with reporter switching
To gain further insight into the role of strand-specific PRE/TRE transcription, we used a transgenic reporter assay in which the central 1.6 kb of the downstream vg PRE/TRE was linked to a miniwhite (mw) reporter gene (Fig. 2a). This 1.6 kb vg PRE/TRE gives PcG-dependent repression and strong pairing sensitive silencing of the mw reporter in adult eyes26. ChIP-seq analysis of the endogenous vg locus in Drosophila cells25 and embryos (Supplementary Fig. 2b) showed substantial overlap of the 1.6 kb PRE/TRE with enrichments for Polycomb (PC), E(Z) and histone H3 lysine 27 trimethylation (H3K27me3). Most importantly, we chose to use this PRE/TRE subfragment because it contains a subset of embryonic and larval promoters, namely those driving transcripts through the core of the PRE/TRE (Fig. 1b, Supplementary Fig. 2a). PRE/TRE constructs and control constructs lacking the PRE/TRE were each independently integrated at two distinct genomic locations26 (Fig. 2a, b). Mapping of 5' and 3' ends of the transgenic transcripts was performed in early (0-5h) embryos, in which there was no background of endogenous PRE/TRE transcripts. At both integration sites, the transgene produced transcripts with the same 5' ends as the endogenous embryonic ones, confirming that the transgenic PRE/TRE uses its own promoters, and is not simply transcribed from promoters flanking the integration site (Fig. 2a, Supplementary Fig. 3). However the levels of the transgenic transcripts were higher than the endogenous levels, and were different at the two sites, probably due to the effects of flanking enhancers26,28(Fig. 2i,j,m,n,q,r). This enabled comparison of the levels of each PRE/TRE strand with reporter activity at both sites.
Figure 2. Switch in forward and reverse nc transcription correlates with a switch in the effect of the vg PRE/TRE on reporter gene expression.
(a) 1.6 kb vg PRE/TRE transgene. Black arrows: transgenic transcripts mapped in 0-5h embryos (see Supplementary Fig. 2 and Supplementary Fig. 3). (b) Positions of transgenes (red) relative to the endogenous vg locus (black) on chromosome 2R. CEN, centromere; TEL, telomere. (c-t) At each of sites 1 and 2, two transgenic lines were generated: a PRE/TRE transgene, as in (a, pKC27vg.fwd), and a control transgene lacking the PRE/TRE (pKC27mw). Levels of PRE/TRE transcript on each strand, and of the mw reporter were assessed in embryos, wing discs and larval brains by qPCR on transgenic animals homozygous for pKC27vg.fwd and pKC27mw transgenes. (c-f) Adult eye color of male homozygous flies carrying reporter constructs as indicated. Scale bar = 200 μm. (g,h) Quantitation of eye pigment in (c-f) 26. Error bars show max and min. of 2 independent measurements for each line. (i,j) Strand-specific RT-qPCR analysis of PRE/TRE transcription in 3rd instar larval eye discs, shown as % TBP (TATA Binding Protein). (k,l) RT-qPCR of mw reporter transcript, and (m,n) PRE/TRE transcription on each strand in embryos. (o,p) RT-qPCR of mw reporter transcript, and (q,r) PRE/TRE transcription on each strand in 3rd instar larval brains. For all RT-qPCR analyses, mean and s.e.m. of 4 independent cDNAs prepared from two independent RNAs are shown. (s) For each transgenic line and tissue, the relative proportion of PRE/TRE transcripts from each strand is shown on the y-axis as (rev strand/ fwd strand). x-axis: fold activation of mw reporter, calculated as (mw in PRE/TRE transgene/mw level in control transgene). (t) Left: ratio between transgenic forward and reverse strand transcripts in 3rd instar larval eye discs (data from i,j) at sites 1 and 2. Right: pairing sensitive silencing (PSS) score of transgenes at sites 1 and 2 (from26). PSS score was calculated from adult eye pigment levels as heterozygote/homozygote for each transgenic line, normalized to the same ratio calculated for the ‘no PRE/TRE’ control line at the same landing site (see Fig. 3d in 26).
In adult eyes, as shown previously26, the PRE/TRE gave strong repression of the mw reporter. Repression was stronger at site 2 (48.5 fold) than at site 1 (13.4 fold; Fig. 2c-h)26. Interestingly, in eye imaginal discs of 3rd instar larvae, the levels of forward strand transcripts correlated with the extent of repression at each site (Fig. 2g-j). Remarkably, in embryos, both the regulatory properties and the strand preference of the PRE/TRE element were reversed. At both sites, we observed robust activation of the reporter, accompanied by higher levels of reverse PRE/TRE strand than forward strand (Fig. 2k-n). Finally, in larval brains, the transgene at site 1 showed higher forward than reverse strand PRE/TRE transcript levels, accompanied by repression of the reporter (Fig. 2o,q). Conversely, at site 2, the reverse strand of the PRE/TRE was present at higher levels than the forward strand, and the reporter was activated (Fig. 2p,r). In summary, this analysis shows that those lines and tissues in which the forward PRE/TRE strand is present at higher levels than the reverse strand give repression of the mw reporter, whereas those with higher levels of the reverse strand give activation (summarized in Fig. 2s). Thus, these data document a remarkable correlation between the switch in forward and reverse nc transcription and the switch in the effect of the vg PRE/TRE on reporter gene expression. We name this element a “GEAR box” (Gene Expression Alternating RNA) element.
The vg PRE/TRE forward strand represses endogenous vg mRNA
To ask whether forward and reverse GEAR box transcripts themselves play a role in regulating gene expression, we examined the effects of ectopically over-expressing each strand from the transgene on the endogenous vg locus. PRE/TREs have been shown to mediate long-range interactions29. We observed that the relative levels of vg PRE/TRE forward strand in eye discs correlated with the extent of pairing-sensitive silencing (PSS) at each site (Fig. 2t;26), suggesting that the forward strand may be involved in mediating or stabilizing pairing between homologous PRE/TREs, and may thus facilitate PSS. To determine whether the transgenic vg PRE/TRE interacts physically with the endogenous vg locus upon induced transcription of either the forward or the reverse ncRNA strands, we performed 3D DNA FISH (Fig. 3a,b). We used the GAL4-UAS system in combination with transgenes designed to over-express either the reverse or forward strand, integrated at site 1 (Fig. 2a,b, 3b). Engrailed GAL4 (enGAL4) was used to drive elevated transcription of the transgenic PRE/TRE forward or reverse strand in the posterior compartment of the wing disc, while the anterior compartment of the same disc lacking enGAL4, served as a negative control (see Fig. 3g,l for enGAL4 expression domain).
Figure 3. Forward strand PRE/TRE transcription induces long-range interactions and repression of endogenous vg mRNA.
(a,b) 3D FISH analysis in 3rd instar larval wing discs. (a) Representative images showing DNA FISH signal for endogenous vg locus (red) and transgenic locus (green). Note that each locus shows only one spot due to somatic pairing of homologs. (b) Boxplots showing distribution of 3D distances between the transgenic locus at site 1 and the endogenous vg locus. Median, first and third quartiles are shown. Whiskers extend to 1.5 IQR. Transgenic animals carrying a control construct (pKC27mw, left) expressing the reverse strand (pKC27vg.rev', middle) or the forward strand (pKC27vg.fwd', right) of the vg PRE/TRE, were crossed to the enGAL4 driver, which is expressed in the posterior half of the wing disc (P). The anterior half of the wing disc (A) serves as a control (see g, l, for expression patterns). Combined normalised data from three discs for each genotype are shown (data for each individual disc are shown in Supplementary Fig. 7, for normalization procedures, see Methods). Y-axis shows 3D distance between the transgenic locus and the endogenous vg locus. Average nuclear radius: 3.6μm. Induced transcription of the forward strand but not the reverse strand leads to a significant decrease in distances between the transgene and the endogenous locus in the posterior compartment compared to the anterior (n.s., p-value = 0.1 or larger, Wilcoxon rank-sum test). (c-n) Double in situ hybridization to detect the reverse strand (c,g,k) or the forward strand (d,h,l) of the vg PRE/TRE, and the vg mRNA (e,I,m) in wing discs of transgenic 3rd instar larvae carrying the reporters and control constructs integrated at site 1, when crossed to the enGAL4 driver. Posterior is to the left, anterior is to the right. Scale bar = 100μm. (c-f) control transgene without PRE/TRE (pKC27mw). (c) and (d) show different discs, (d,e) same disc. (f) merge of (d,e). (g-j) Transgene over-expressing PRE/TRE reverse strand (pKC27vg.rev'). (g) and (h) show different discs, (g,i) same disc. (j) merge of (g,i). (k-n) Transgene over-expressing the forward strand (pKC27vg.fwd'). (k) and (l) show different discs, (l,m) same disc. (n) merge of (l,m). Blue: DAPI. (m,n) Arrows indicate borders of domain of down-regulation of vg mRNA, see also Supplementary movie 1. Each in situ hybridization was performed at least twice and 4 to 10 discs were analyzed for each line. (o) For each disc the domain of enGAL4 expression was masked in 3D using the ncRNA signal (P), and the remaining volume was separately masked (A). (p) vg mRNA signal intensity was quantified in 3D within A and P domains for individual discs, and the ratio of P/A was calculated. Mean and s.e.m. of the P/A ratio for 4 discs for each line are shown. p-value: Student's t-test, one tailed.
Induced transcription of the control transgene lacking the PRE/TRE, or induction of the PRE/TRE reverse strand transcript, had no significant effect on the distance distribution between the endogenous vg locus and the transgene (compare A and P in Fig. 3b, Supplementary Fig. 7), showing that transcription per se has little effect on the localization of the transgene relative to the endogenous vg locus. In contrast, induction of the PRE/TRE forward strand led to a significant increase in proximity of the transgene to the endogenous locus (Fig. 3b) in two of the three discs examined (Supplementary Fig. 7). This effect is transcription-dependent and strand-specific, and suggests that the forward strand, but not the reverse strand, may induce or stabilize long-range interactions between two copies of the PRE/TRE.
To ascertain whether transgenic PRE/TRE transcription also affects regulation of the endogenous vg mRNA, we used enGAL4 to drive either the forward or reverse strand from site 1, and analyzed the levels of vg mRNA (Fig. 3c-p). In control lines in which enGAL4 was used to drive the UAS mw transgene lacking the PRE/TRE (Fig. 3c-f) or to drive the reverse PRE/TRE strand (Fig. 3g-j), the vg mRNA pattern was identical to wild type (Fig. 1k). In contrast, enGAL4 driven over-expression of the PRE/TRE forward strand from the transgene caused substantial down-regulation of vg mRNA within the enGAL4 expression domain (Fig. 3k-p, see also Supplementary movie 1). Analysis of enGAL4 expression levels using vg RNAi as a sensor revealed that this domain corresponds to the highest level of enGAL4 expression (Supplementary Fig. 8). Taken together, these results demonstrate that ectopic vg PRE/TRE forward strand expression causes both long-range interactions and repression of the endogenous vg locus.
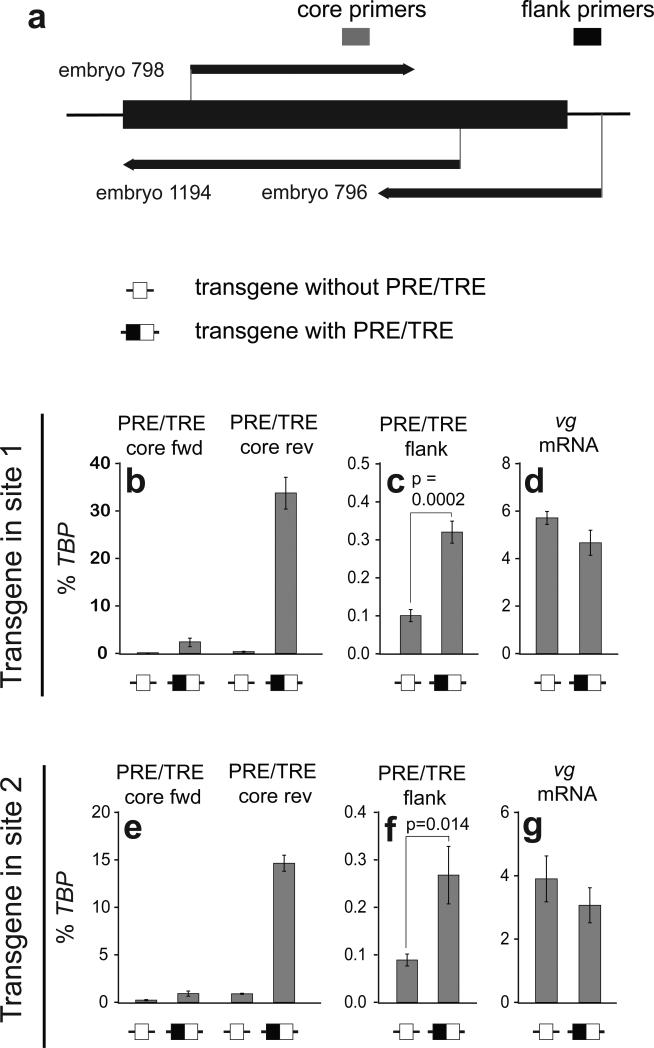
The PRE/TRE reverse strand activates the endogenous PRE/TRE
The above experiments demonstrate a role for the forward strand in vg regulation, but the reverse strand had little effect in these assays. Since the reverse strand is not normally expressed in larval tissues (Fig. 1b), we reasoned that its principal function may be in embryos, where it is normally present. To address this, we examined the effects of transgenic transcripts on the endogenous vg locus in embryos (Fig. 4). In early embryos (0-5h), neither the endogenous vg mRNA nor the PRE/TRE transcripts are detectable (Supplementary Fig. 1c), allowing unambiguous identification of induced transcripts from the transgene and the endogenous locus. We took advantage of the fact that the transgenic reverse strand promoter is active at this early stage without the need for GAL4-induced transcription (Fig. 4a,b,e). To distinguish between transgenic and endogenous PRE/TRE transcripts, we designed PCR primers to specifically detect the 5' end of the “embryo 796” transcript (Fig. 1b, 4a). This sequence is not present on the transgene and is therefore unique to the endogenous locus (Fig. 4a: “flank primers”). In early (0-5h) embryos carrying control transgenes lacking the PRE/TRE, endogenous PRE/TRE transcription was barely detectable (Fig. 4b,e). In contrast, the PRE/TRE transgene was highly transcribed on the reverse strand (Fig. 4b,e 4th column). Remarkably, this ectopic reverse strand transcription was accompanied by a three-fold up-regulation of transcripts from the endogenous flanking site (Fig. 4c,f), although the vg mRNA itself was unaffected (Fig. 4d,g). Additional mapping by primer walking confirmed that these ectopically induced endogenous transcripts correspond to the endogenous reverse strand transcript “embryo 796” (Fig. 4a, Supplementary Fig. 3). In summary, these results show that early embryonic transcription of the reverse strand of the transgenic vg PRE/TRE activates reverse strand transcription at the endogenous vg PRE/TRE.
Figure 4. The reverse strand PRE/TRE transcript activates the endogenous vg PRE/TRE.
(a) Map of endogenous vg PRE/TRE locus showing position of sequence present in transgenes (thick bar), and PCR primers used in b-g. (b-g) RT-qPCR as indicated, on cDNA from 0-5h embryos. Error bars show s.e.m. of two to six independent cDNAs prepared from at least two independent RNAs. p-values: one tailed Student's t-test.
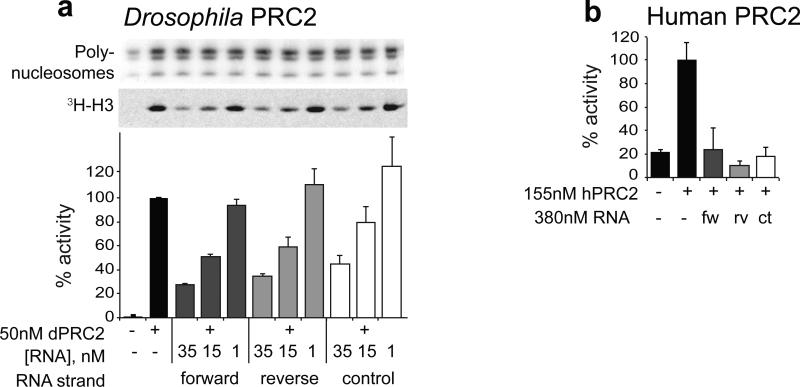
RNA promiscuously inhibits PRC2 HMTase activity
Our in vivo data show that the forward and reverse transcripts of the vg PRE/TRE have different properties. In vitro binding assays with recombinant vertebrate and fly PRC2 proteins (EZH2-EED or EZH2 and E(Z) alone) have reported specific binding to some RNAs and not to others8,12, and specific inhibition by some RNAs14. However, a recent study reports that recombinant human PRC2 binds RNA in vitro with high affinity and with no specificity for different RNAs13. To determine whether there is any difference in the interaction of the different strands of the vg PRE/TRE with PRC2 in vitro, we asked whether the forward and reverse strands might inhibit or stimulate the enzymatic activity of recombinant PRC2 to different extents. To this end, we assayed the in vitro H3K27 methyltransferase (HMTase) activity of recombinant Drosophila and human PRC2 in the presence of RNAs. We tested vg PRE/TRE forward or reverse strand, or control RNAs (of the same length but derived from bacterial plasmid sequences) (Fig. 5, Supplementary Fig. 9). This analysis revealed that all RNAs tested caused inhibition of both Drosophila and human PRC2 HMTase activity to equal extents. A similar effect was also observed for DNA (Supplementary Fig. 9). Robust inhibition occurred at approximately equimolar concentrations of PRC2 and RNA. This suggests that there is no inherent difference in the ability of different RNAs to inhibit PRC2 HMTase activity. These results are consistent with the observed lack of RNA binding specificity for human PRC213, and the proposed inhibition of PRC2 by RNA14,30, and raise the question of whether the two strands interact differently with PRC2 in vitro and in vivo.
Figure 5. RNAs inhibit PRC2 HMTase activity in vitro.
(a) In vitro HMTase assay. Drosophila PRC2 was incubated with HeLa polynucleosomes, 3H-labelled S-adenosylmethionine and with a 1.6 kb RNA in vitro transcribed from the 1.6 kb PRE/TRE shown in (4a), in forward or reverse orientation, or a bacterial control RNA of the same length. The experiment was performed three times. Mean and SD of two quantifications of a representative experiment are shown. (b) Human PRC2 was incubated with an H3 tail peptide (residues 21-40) and RNAs as in (a), detection of H3K27me3 was performed using DELFIA. The experiment was performed three times. Mean and s.e.m. of three technical replicates within a representative experiment are shown. All RNAs show similar inhibition of Drosophila and human PRC2 HMTase activity.
E(Z) binds to the reverse strand of the vg PRE/TRE in vivo
Experiments in which the interaction of different RNAs with PRC2 components was assayed by incubation with nuclear extracts have shown a high specificity for one strand in preference to its antisense counterpart for several ncRNAs7,8,12. To evaluate whether the two strands of the vg PRE/TRE bind differentially to PRC2 components in living cells, we performed RNA immunoprecipitation (RNA IP) from Drosophila S2 cells. Cells were cotransfected with GFP or GFP::E(Z) and a plasmid encoding either the forward or the reverse PRE/TRE strand (Fig. 6a,b). Surprisingly, this analysis revealed a highly strand-specific interaction between E(Z) and the reverse RNA strand of the PRE/TRE (Fig. 6c-f). Thus, although PRC2 does not interact specifically with different RNAs in vitro (Fig. 5, Supplementary Fig. 9)13, the PRC2 component E(Z) binds highly specifically to only one strand of the PRE/TRE in vivo (Fig. 6e,f). These results indicate that other factors present in the cell, rather than an intrinsic property of PRC2 itself, mediate the specificity of binding by facilitating or preventing interactions between PRC2 and specific RNAs.
Figure 6. The reverse strand PRE/TRE transcript binds specifically to E(Z) in vivo and removes E(Z) from the locus from which it is transcribed.
(a-f) The PcG protein E(Z) binds specifically to the PRE/TRE reverse strand in S2 cells. (a) Constructs containing the PRE/TRE in each orientation under control of the hsp70 promoter (pKC27vg.fwd or pKC27vg.rev) were transfected separately into Drosophila S2 cells. (b) Plasmids expressing either GFP::EZ or GFP were cotransfected separately with each of the constructs in (a). GFP- trap was used to IP the protein and associated RNAs from whole cell extracts without crosslinking. (c) Strand-specific cDNA synthesis and RT-qPCR shows that each of the transfected plasmids predominantly transcribes one strand (data normalised to highest expressed strand, typically between 200 and 1000 % of TBP). The forward strand was transcribed typically 3-20 fold higher in the forward plasmid transfection than the reverse strand in the reverse plasmid transfection (data not shown). Mean and s.e.m. of at least three independent IP experiments are shown. (d) Western blot using αGFP and αTubulin, showing GFP (top) and GFP::E(Z) (bottom) present in input (I), not in unbound (U) and enriched after GFP trap (IP). (e) Enrichment of PRE/TRE RNA after GFP trap on cells transfected with either plasmid as indicated, and GFP or GFP::E(Z). IP levels were normalised to input and further to GFP mRNA levels, and were controlled to exclude DNA contamination. Mean and s.e.m. of at least three independent IP experiments are shown. cDNA synthesis was performed by random priming. Whereas RNA did not coIP with GFP::E(Z) after transfection of the forward construct, GFP::E(Z) cotransfected with the reverse plasmid, which transcribes predominantly reverse strand (c), shows PRE/TRE RNA enrichment of over 100 fold after IP (4th column). (f) This IP material was subjected to strand-specific cDNA analysis, showing that only the reverse strand is detectable. Mean and SD of two experiments is shown; scale shows IP as % of TBP in input. (g) ChIP-seq analysis of E(Z) and H3K27me3 in 0-16h embryos over-expressing forward (top; pKC27vg.fwd') or reverse (bottom; pKC27vg.rev') strand ncRNAs. Left: transgenic locus. Black arrow indicates position and orientation of 1.6 kb PRE/TRE. Grey box: 1.6 kb PRE/TRE sequence identical between transgene and endogenous vg locus. White area: sequence unique to transgenic locus, corresponding to the ampicillin resistance gene of pKC27. Asterisk and arrowhead indicate differences between forward and reverse over-expression. Data are shown as ChIP/input to correct for different copy numbers of the PRE/TRE and the flank. Right: no difference was detectable at the endogenous vg locus (Chr 2R: 8,791,079- 8,794,193; see also Supplementary Fig. 10) or at the Fab-7 PRE/TRE (Chr 3R: 12,723,848- 12,728,057). Thick lines indicate position of PRE/TRE. RPM, reads per million. The experiment was repeated three times for H3K27me3 on pKC27vg.fwd’, once for H3K27me3 on pKC27vg.rev’ and once for E(Z) on both pKC27vg.fwd’ and pKC27vg.rev’. Tracks from one representative ChIP experiment are shown.
The reverse strand inhibits and evicts E(Z) from chromatin
The fact that both strands can inhibit E(Z) activity in vitro, but only the reverse strand can bind in vivo, suggests that the reverse strand may activate the PRE/TRE in vivo by inhibiting local E(Z) HMTase activity, whilst the forward strand is unable to bind and has no inhibitory effect. To test this idea we performed ChIP on embryos over-expressing either the forward or reverse strand of the vg PRE/TRE from transgenes integrated at site 1, under the control of the ubiquitous daughterless GAL4 (daGAL4) driver. We examined H3K27me3 and E(Z) levels at both the transgenic and the endogenous vg loci (Fig. 6g). The transgene contains sequences that are also found elsewhere in the genome (the 1.6 kb PRE/TRE itself, and the markers white and yellow). Any effects occurring within the 1.6 kb PRE/TRE will not be discernable because the transgenic and endogenous PRE/TRE sequences are identical, but the sequences unique to each locus will be informative. The sequence that is unique to the transgene is the downstream sequence immediately flanking the PRE/TRE, which is bacterially derived (Fig. 6g, left, unique region shown on white background, 1.6 kb PRE/TRE on grey). The sequences that are unique to the endogenous vg locus are those flanking the 1.6 kb PRE/TRE (Fig. 6g, middle).
For the transgenic locus, embryos over-expressing the forward strand showed similar enrichment profiles across the transgene flank for both E(Z) and H3K27me3 (Fig. 6g, top). In contrast, embryos over-expressing the reverse strand showed two striking effects in this flanking region (Fig. 6g, bottom). Distal to the PRE/TRE, both E(Z) and H3K27me3 signals were reduced compared to the forward strand over-expressing line (arrowhead, Fig. 6g). This suggests that the reverse strand may titrate E(Z) away from the locus. In addition, the flank directly adjacent to the PRE/TRE displayed high E(Z) enrichment, but low H3K27me3 (asterisk, Fig. 6g). This is consistent with inhibition of E(Z) activity by the reverse strand (however, it may also reflect removal of nucleosomes from this site). There was no detectable difference between the over-expression of forward and reverse strands at the endogenous vg locus (Fig. 6g, middle, Supplementary Fig. 10). This is consistent with the lack of vg mRNA up-regulation observed in early embryos expressing the reverse strand (Fig. 4d,g). We also observed no detectable difference at several other targets of E(Z) regulation (Fig. 6g, right, and data not shown). In summary, this analysis suggests that the forward strand does not inhibit the enzymatic activity of E(Z) in vivo, whereas the reverse strand may act both by inhibiting activity, and by removing E(Z) from the locus from which it is transcribed.
GEAR box elements are potentially widespread
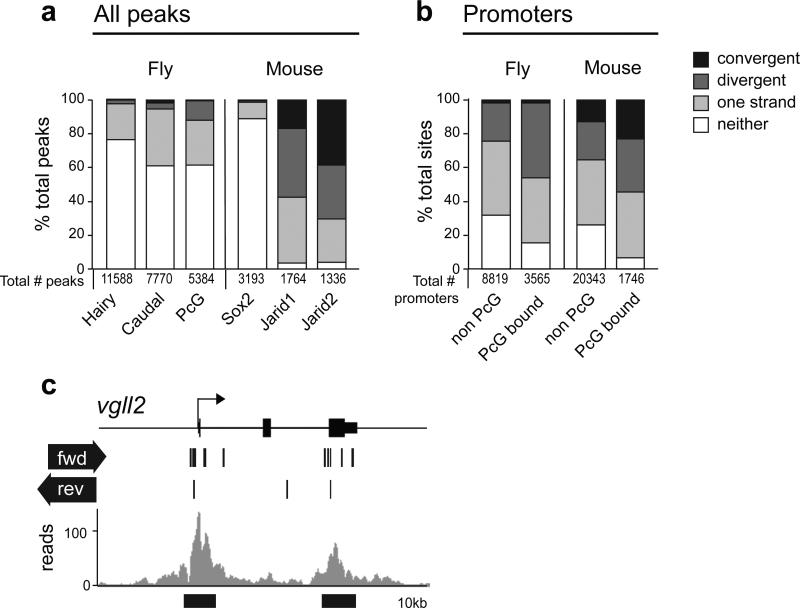
To determine whether further GEAR box elements may exist in the fly and mouse genomes, we mined genome-wide data sets20,21,31-34 (Fig. 7, Supplementary Table 2, Supplementary Table 3). This analysis identified several hundred PcG binding sites in the fly and mouse genomes, that show transcription from both strands (either convergent or divergent) passing through the PcG binding site (Fig. 7a,b; see Supplementary Table 2 and Supplementary Table 3 for full listings). Furthermore, although bidirectonal and antisense transcription is a common feature of many genomes35, comparison with binding sites for other transcription factors (Fig. 7a) or non-PcG bound promoters (Fig. 7b) showed a highly significant enrichment of bidirectional transcription at PcG bound sites in most cases, indicating that this feature is favored at PcG regulated genes.
Figure 7. Elements with GEAR box potential are widespread.
(a) Fly: ChIP peaks from21 with CAGE tags from31. Mouse: ChIP peaks from 33,34 compared with CAGE tags from the FANTOM3 CAGE dataset32. Suz12 comparison gave similar results (Supplementary Table 3). % of total peaks overlapping with CAGE tags in the categories shown are indicated. Convergent CAGE tags: tags on opposite strands are transcribed towards each other. Divergent CAGE tags: tags on opposite strands are transcribed away from each other. (b) CAGE tag status of PcG ChIP peaks at promoters was compared to that of non-PcG bound promoters. Fly: data as for (a). Mouse: combined Jarid2 and SUZ12 data from 34 were used for PcG peaks. In most cases, bidirectional transcription occurs significantly more frequently at PcG bound sites than at other sites. The frequency of occurrence of the category of interest (convergent or divergent) compared to all other categories including no transcription, was compared for pairs of data sets (for each comparison, PcG bound was compared to non-PcG bound). p-values (Yates Chi-squared test, one sided). In all cases except caudal, convergent, and Jarid1, divergent, the category of interest was more abundant in PcG than non-PcG bound sites. For fly promoters, convergent, p=0.19; for all other categories, p< 4×106. See also Supplementary Table 2 and Supplementary Table 3, and Methods. (c) Mouse Vgll2 locus: grey, SUZ12 binding in34, black lines, CAGE tag occurrence32. Black bars below plot indicate potential GEAR box elements.
We note that the occurrence of bidirectional transcription is in general lower in the fly than the mouse. This may be due to the fact that fly transcript data were available only from embryos or cell lines, whereas the mouse CAGE tag data were generated from multiple different tissues. For the mouse data set we were able to evaluate tissue- and strand-specificity, showing that over 99% of those PcG sites that transcribe opposite strands do so in different tissues. A notable example is the mouse Vgll2 gene, a functional homolog of the fly vg gene36 (Fig. 7c). The PcG binding sites at this locus transcribe the forward strand in muscle tissue, where the gene is active, and the reverse strand in embryonic tissues, in which the gene is repressed. In summary, this analysis identifies many further sites of PcG regulation in fly and vertebrate genomes, that may switch their function between activation and silencing by switching the strand of their nc transcription.
DISCUSSION
We demonstrate both by analysis of endogenous transcripts, and using ectopic over-expression strategies in vivo, that transcripts from opposite strands of the vg PRE/TRE have opposite effects on PRE/TRE status. However, in vitro, both ncRNAs have equivalent inhibitory effects on the HMTase activity of PRC2. Taking the in vitro and in vivo data together, we propose that specificity of ncRNA interaction with PcG proteins in vivo is not a result of inherently different affinities of PRC2 for different ncRNAs, but of the availability of a given ncRNA (regulated by interactions of that RNA with other molecules) to interact with PRC2 and inhibit its enzymatic activity. We propose that the forward strand ncRNA promotes silencing by facilitating pairing between PRE/TREs. PRE/TRE pairing has been shown to be essential for maximum silencing by the vg PRE/TRE26, and this silencing is genetically dependent on the PcG23,26. Thus we propose that forward strand-induced pairing may facilitate or stabilize PcG mediated pairing-dependent silencing (Fig. 8, left). E(Z) is detected at the vg PRE/TRE in ChIP analyses25 (Supplementary Fig. 2b, Supplementary Fig. 10), but does not interact with the forward strand (Fig. 6e,f). Thus, we propose that E(Z) binds at the silenced PRE/TRE independently of RNA. The forward strand ncRNA may facilitate or stabilize pairing by binding to additional bridging proteins (yellow in Fig. 8). These or other proteins may also prevent binding and inhibition of E(Z) by the RNA.
Figure 8. Proposed model for switching PRE/TRE properties upon strand switching.
See main text for details.
Upon switching to the active state (Fig. 8, right), transcription of the reverse PRE/TRE strand would be incompatible with forward strand transcription, because the reverse transcript runs through the forward strand promoter (Fig. 1b, 2a). Reverse strand transcription may thus destabilize pairing, enabling activation of the PRE/TRE (Fig. 4c,f). In addition, the reverse strand binds E(Z) (Fig. 6e,f), and upon binding, would inhibit E(Z) HMTase activity (Fig. 5a, 6g), and may remove E(Z) from the locus (Fig. 6g). In this way, multiple self-reinforcing events could contribute to stable switching of the PRE/TRE into an active state. The vg PRE/TRE responds to TrxG mutations by loss of activation23. Whether TrxG-dependent activation acts via the ncRNAs will be a key question for future studies.
Genome-wide analysis identifies many sites that may share functional features with the vg PRE/TRE. We propose that these elements can exist in a “neutral” state, in which neither they nor their associated genes are transcribed (this is consistent with our observation that the vg PRE/TRE is not transcribed in tissues or at embryonic stages that do not express the vg mRNA, Supplementary Fig. 1). However, in tissues that have the potential to transcribe the gene, the element may be switched either to forward or reverse mode, thereby boosting either silencing or activation. This may serve to sharpen spatial expression boundaries, to stabilize gene expression states, or to accelerate kinetics of activation or repression37.
In conclusion, this work provides a novel paradigm linking forward and reverse nc transcription to dynamic and developmentally regulated switching of PRE/TRE properties, and thus to the maintenance of cell identities during development. Furthermore, the demonstration that any RNA is a potent inhibitor of PRC2 enzymatic activity in vitro, but that only specific RNAs are able to bind and inhibit PRC2 in vivo, strongly implies that specific RNAs are masked in vivo from interacting with PRC2. This provides an enormous potential for regulated and reversible RNA-mediated inhibition of local PRC2 activity.
METHODS
PRE/TRE transcript mapping
Mapping of 5' ends of PRE/TRE transcripts (Fig. 1b, Supplementary Fig. 2) was performed using 5' RACE kit (version 2.0, Invitrogen) according to manufacturers instructions. Each transcript was mapped from at least two independent cDNA clones generated from at least two different gene specific primers (GSP1). Primers used for 5' RACE analysis are given in Supplementary Table 1. For 3' end mapping, the A-rich nature of the PRE/TRE transcripts precluded the use of 3' RACE with oligo dT primers. Instead, 3' ends were mapped by primer walking. Strand-specific cDNA synthesis was performed using a single 5' primer in combination with different 3' primers, designed to anneal consecutively towards the 3' end of each transcript at intervals of 50 to 100 bp (Supplementary Fig. 3, Supplementary Table 1). The 3' end was designated as the position of the last primer that gave a product.
Double RNA in situ hybridization
PCR products from embryonic cDNA for the vg gene or the vg PRE/TRE ncRNA were cloned in both orientations into PCR XL Topo (Invitrogen). For in situ hybridization, RNA probes were in vitro transcribed from each cDNA strand. One probe was Dig labeled (Boehringer), detected with sheep Anti-Digoxigenin-AP (Roche) and visualized with FastRed (Sigma). The other probe was labeled with fluorescein and detected with primary antibody: Mouse anti-fluorescein (Roche), secondary antibody: goat anti-mouse-HRP (Invitrogen) and visualized with Alexa Fluor 488 Tyramide (Invitrogen) using Tyramide Signal Amplification kit (TSA™, Invitrogen) according to manufacturers instructions. Label swaps were performed to ensure specificity. In the images shown, vg mRNA was detected with Fast Red, and PRE/TRE transcripts were detected with TSA. Images were taken using confocal microscopy, with LSM 700 Axioimager (larval tissues) or LSM 510 Axiovert 200M (embryos) (Zeiss).
3D quantification of RNA in situ hybridization signals
3D quantification (Fig. 3 o,p) was performed using IMARIS® x64 7.4.0 (Dec 14 2011), Bitplane Scientific Software. Z-stack images were uploaded into the software. Surfaces were generated by defining contours around areas expressing the enGAL4 driver or areas not expressing the driver. The contours were drawn manually in each slice of the Z-stack. Based on these the software calculated the volume of the surface in the Z-stack, and quantified the intensity sum of the vg mRNA signal within this volume. For the animation shown in Supplementary Movie 1, again the Z-stack image was uploaded into the software and key frames were added manually (e.g. rotation, removing of surfaces and channels).
Transgenic flies
The transgenic vg PRE/TRE lines and control lines are described in26 or are derived from those constructs as described below. All constructs contain a UAS, hsp70 minimal promoter, PRE/TRE (or no PRE/TRE in controls), and a mw reporter. The landing site is marked with yellow. Landing site 1 in this study corresponds to landing site 2 in26 (Cytological location chr. 2R, 46E1 genomic position 5,965,083). Landing site 2 in this study corresponds to landing site 3 in26 (Cytological location chr. 2R, 58F4, genomic position 18,549,410). pKC27vg.fwd (Fig. 2; 6a-f; Supplementary Figure 3), carries 1.6 kb of the vg PRE/TRE cloned into pKC27mw downstream of the UAS cassette and hsp70 promoter (described in detail in26). pKC27vg.rev (Fig. 6a-f) was generated by introducing XbaI sites upstream and downstream of the vgPRE/TRE in the pKC27vg.fwd plasmid by site-directed mutagenesis (QuikChange® II Site-Directed Mutagenesis Kit, Stratagene) and subsequent excision and re-ligation of the vgPRE, such that the PRE/TRE was in the opposite orientation to that in pKC27vg.fwd (direction was validated by sequencing the XbaI junction sites). In pKC27vg.fwd' (Fig. 3b, 3k-n, 6g, Supplementary Fig. 7, Supplementary Fig. 10) and pKC27vg.rev' (Fig. 3b, 3g-j, 6g, Supplementary Fig. 10), the UAS-PRE/TRE cassette was inverted with respect to the mw reporter so that induced transcription drives away from mw. These constructs were assembled by single-step in vitro recombination40 and validated by Sanger sequencing. Primer sequences and constructs are available on request.
RNA extraction, cDNA synthesis and qPCR analysis
Total RNA was extracted from dissected larval tissues and dechorionated embryos using the High Pure RNA isolation kit (Roche). Embryonic RNA was additionally treated with DNAse (Turbo DNAse, Ambion). cDNA was synthesized using the Superscript II kit (Invitrogen) according to manufacturers instructions. For random priming (Fig. 6c,e, Supplementary Fig. 1) priming was performed with equal quantities of oligo dT and random decamer primers (Ambion). For strand-specific cDNA synthesis (all other Figures), cDNA was primed with a cocktail containing 0.2 pmol each of appropriate primers, for detection of TBP forward strand (for normalization), and other transcripts of interest (see Supplementary Table 2 for primer sequences). For the transactivation experiment (Fig. 4), primers for the Timp transcript were included as control for embryo age. For RNA IPs (Fig. 6e,f), primers to GFP were included to control for transfection efficiency. For all strand-specific cDNAs, appropriate controls for DNA contamination and mispriming were included. Annealing and cDNA synthesis were performed at 50°C, and after heat inactivation of the RT enzyme, RNA was removed by RNAse H (NEB) prior to PCR analysis. qPCR analysis was performed using SYBR Green JumpStart Taq ReadyMix (Sigma) in a Realplex mastercycler (Eppendorf).
3D DNA FISH
Probe labeling was carried out using the Invitrogen Fish-Tag DNA green and orange Kit following the protocol without modifications. Whole fly genomic DNA was used as PCR template. See Supplementary Table 2 for primer sequences. Homozygous transgenic fly lines were crossed to homozygous enGAL4 flies as described above, and 3rd instar larval wing discs of the progeny (heterozygous for both the transgene and the GAL4 driver) were dissected in PBS and transferred to an eppendorf tube for immediate fixation in 4% PFA in PBT for 5 mins on a rotating wheel. Pre-hybridization was carried out as in41, except that the rehydration steps were omitted, proceeding directly to RNaseA treatment. Hybridization and post-hybridization washes were performed as in41. The samples were subsequently counterstained with DAPI (1:1000 in PBT) for 10 min on a rotating wheel. The samples were washed once with PBT, rinsed once in PBS and mounted in a drop of ProLong Antifade Kit (Invitrogen). 3D stacks were imaged by confocal microscopy. Deconvolved images were analyzed by automated image analysis implemented in Definiens XD software.
For each disc, at least three non-overlapping images were acquired within the posterior enGAL4 expression domain, and at least three images within the opposite anterior domain not expressing enGAL4. The positions of the domains were defined from morphological features with reference to in situ images such as those shown in Figure 3g and l. For each disc, data from the three anterior images and the three posterior images were combined, giving a single data set for each half of each disc (these data are shown in Supplementary Fig. 7). Within single discs, tissue and cell growth occurs uniformly42,43, thus we reason that the comparison of distances in the anterior and posterior compartments is informative. However we observed that different discs had different sizes, and thus they may have different cell sizes, which in turn may affect nuclear size and thus the average distances between loci for different discs. To account for such differences, data for a given genotype were normalised using the mean of distances observed in the anterior compartment. Thus for disc 1 in each panel of Supplementary Figure 7 the distances shown are the actual observed distances, whilst those of discs 2 and 3 were multiplied by a factor representing the difference in mean distance between the anterior compartment of each disc to that of disc 1. The correction factors for discs 2 and 3 respectively are as follows: Supplementary Figure 7a (control): 1.04 and 1.17; Supplementary Figure 7b (reverse strand over-expression): 0.93 and 0.76; Supplementary Figure 7c (forward strand over-expression) 0.94 and 0.92. These normalised data are given in the data Table attached to the legend of Supplementary Figure 7.
For the plots in Figure 3b, the data were further normalised between genotypes (these data are given in the table attached to Fig. 3 legend). Thus for the pKC27mw control line in Figure 3b, the distances shown are the actual observed distances, whilst those of the other two transgenic lines were multiplied by a factor representing the difference in mean distance between the anterior compartment of line to that of the control. The data from pKC27vg.rev' were multiplied by 1.38; those for pKC27vg.fwd', were multiplied by 1.29.
Fly crosses
The w;P{en GAL4M616} (chrII) driver line was provided by Barry Dickson. Homozygous enGAL4 flies were crossed to homozygous transgenic lines carrying UAS, vg PRE/TRE, or control lines with UAS and no PRE/TRE. 3rd instar larval wing discs of progeny were dissected and subjected to double RNA in situ hybridization or 3D DNA FISH as described above. For the RNAi control shown in Supplementary Figure 8, the VDRC line 16896, expressing an RNAi hairpin against the vg mRNA, was used instead of the UAS vg PRE/TRE lines described above44. For ChIP (Fig. 6g), a homozygous daGAL4 (chrIII) driver line provided by Jean Maurice Dura, was crossed to appropriate homozygous transgenic lines.
Radioactive histone methyltransferase assay (Drosophila PRC2)
Drosophila PRC2 showed minimal activity on peptide substrates and recombinant mononucleosomes, precluding its use in the DELFIA assay (see below), and was therefore assayed on HeLa polynucleosomes using 3H-S-Adenosylmethionine. Drosophila PRC2 was combined at a final concentration of 20 or 50nM with a final concentration of ~1.4μM 3H-S-Adenosylmethionine and 1 μg (= 0.4 nM) HeLa polynucleosomes (Reaction Biology, Cat # HMT-35-160). Reactions were performed in assay buffer (12 mM Hepes, pH 7.9, 0.24 mM EDTA, 12% Glycerol, 4mM DTT, 2.5 mM MgCl2 and 30 mM KCl) in a final volume of 20 μl. Inhibition experiments were performed with final concentrations of RNA ranging from 1nM to 70nM. PRE/TRE RNAs were made by in vitro transcription of a 1.6 kb RNA from the PRE/TRE (Fig. 2a), in forward or reverse orientation. Control RNAs were made by in vitro transcription of 1.6 kb fragment amplified from the pCR-XL-TOPO plasmid (Invitrogen) using primers ctr1.fwd and ctr1.rev (control 1) or from the pWalium20 plasmid45 using primers ctr2.fwd and ctr2.rev (control 2). Samples were incubated for 60 minutes at 30°C and stopped by addition of SDS loading buffer prior to loading on a 4-12% NuPage Bis-Tris gel (Invitrogen). Western blotting, amido black staining (Sigma) and autoradiography were performed as described46. Quantification of amino black signal and autoradiography films was performed using ImageJ software (NIH).
DELFIA histone methyltransferase assay (human PRC2)
To detect human PRC2 activity in vitro, 2.5μl of human PRC2 (Reaction Biology Cat # HMT-25-114 final concentration = 155 nM) were combined with final concentrations of 16μM S-Adenosylmethionine and 500nM biotinylated H3 peptide, spanning residues 21-40 of histone H3 (H2N-ATKAARKSAPATGGVKKPHR-NTPEG-Biotin; New England Peptide). Reactions were performed in assay buffer (50 mM Tris-HCl pH 9.0, 50 mM NaCl, 1 mM DTT) in a final volume of 25μl. Mono-, di-, or trimethylated H3K27 peptides served as controls (New England Peptide). Inhibition experiments were performed with a total of 5μg RNA (= 380 nM final concentration) or 10uM GSK126 (Chemietek), added prior to the enzyme. Samples were incubated for 60 minutes at 30°C and transferred onto a streptavidin-coated 384-well plate (Perkin Elmer, Cat# CC11-H10), incubated for 60 min at room temperature to allow biotinylated peptides to bind, and washed 3x with wash buffer II (50mM Tris pH 7.5, 150mM NaCl, 0.05% Tween20). Primary mouse-anti-H3K27me3 antibody (#61017, Active Motif, 1:1000) and Europium-labeled secondary antibody (#AD0207, Perkin Elmer, 200ng/ml final concentration, EU-N1-anti-mouse-IgG) were diluted in FI buffer (50mM Tris pH7.5, 150mM NaCl, 0.05% Tween40, 25mM DTPA, 0.2% BSA, 0.05% bovine y-globulins) and added to the wells. Plates were incubated for 60 minutes, and washed 3x with wash buffer II. After addition of Enhancement Solution (Perkin Elmer) plates were incubated for 45 minutes and fluorescence was measured on an EnVision plate reader with excitation at 340 nm and emission at 615 nm (Perkin Elmer).
RNA immunoprecipitation
Plasmids transfected are as follows: pKC27.Tub.GFP; pKC27.Tub.E(Z)::GFP are described in47. Transgenic flies carrying the Tub.E(Z)::GFP construct were generated. The Tub.E(Z)::GFP transgene gives full genetic rescue of lethal E(z) mutations47. pKC27vg.fwd and pKC27vg.rev (Fig. 6a) are described under “Transgenic flies” above.
For strand expression controls, 3 × 105 S2 cells per well were seeded in a 24 well plate. After 24h, 250ng pKC27.Tub.EGFP-nls, 300ng either pKC27vg.fwd or pKC27vg.rev, and 450ng carrier DNA were transfected using Fugene HD transfection reagent (Promega), at a Fugene:DNA ratio of 1:5. After 24h, cells were directly lysed in Trizol (Invitrogen) and RNA extraction was performed according to the manufacturer's instructions (Invitrogen). Isolated RNA was treated with Turbo DNase (Invitrogen) according to the manufacturer’s protocol. Strand-specific cDNA synthesis and qPCR analysis was performed using TBP and PRE/TRE core primers as described above (see Supplementary Table 2 for primers). For the quantification of the input RNA (Fig. 6c), strand-specific cDNA synthesis and RT-qPCR was performed and quantified as %TBP after having controlled to exclude DNA contamination. The forward strand was transcribed typically 3-20 fold higher than the reverse strand (data not shown). Data were normalised to highest expressed strand, typically between 200 and 1000% of TBP. For RNA immunoprecipitations, for each transfection, 3.5 × 107 cells per dish were seeded in three 15cm dishes. After 24h, cells were co-transfected with 25.5 μg of either pKC27.Tub.GFP or pKC27.Tub.E(Z)::GFP and 30.6 μg of either pKC27vg.fwd or pKC27vg.rev, using Fugene HD as described above. Cells were harvested and lysed according to48, and immunoprecipitation of GFP or E(Z)::GFP was performed using GFP Trap agarose beads (Chromotek). Samples were processed according to the GFP Trap manufacturer's protocol with the following modifications: Beads were equilibrated using NT2 buffer48. Washing steps were performed according to48.
For western blotting, input, unbound and IP fractions were run on NuPage Mini-Gels (Invitrogen) and blotted on Immobilon-P PVDF membranes (Millipore) according to standard protocols. Primary antibodies: anti-αTubulin (Sigma-Aldrich T5168, 1:20000), anti GFP (Clonetech JL-8, 1:2000). Signal was detected on film with HRP-coupled secondary antibodies and ECL (GE Healthcare). For RNA analysis, RNA extraction from beads was performed using Trizol. Random primed (Fig. 6c,e) or strand-specific (Fig. 6f) cDNA synthesis and qPCR was performed as described above.
RNA IP levels (Fig. 6e) were quantified by random-primed cDNA synthesis and qRT-PCR. Data were normalised to input and GFP mRNA levels, and were controlled to exclude DNA contamination. Data are shown normalized to RNA IP levels in GFP IP. Despite that fact that the forward strand was transcribed typically 3-20 fold higher than the reverse strand after transfection of equivalent amounts of each plasmid (see “quantification of input RNA” above), the GFP::E(Z) RNA IP consistently resulted in pulldown of the RNA from the reverse but not the forward strand plasmid transfection. Quantification of forward and reverse strand after GFP::E(Z) IP upon transfection of the reverse plasmid (Fig. 6f) was performed by strand-specific cDNA synthesis and qRT-PCR. Data was controlled to exclude DNA contamination and normalized to %TBP in input.
ChIP
Chromatin fixation and immunoprecipitation were performed essentially as described in49. 0.2 g of embryos were dechorionated with 1.4 % sodium hypochloride and crosslinked with 1.8 % formaldehyde for 15 min at room temperature. Cross-linked embryos were sonicated to produce chromatin fragments of an average size of 200–500 bp. Soluble chromatin was separated from insoluble material by centrifugation. The supernatant containing chromatin of 50-60 μg DNA was used for immunoprecipitation. Antibodies: PC (2 μg per IP): d-220 sc-25762 from St. Cruz Biotech, E(Z) (10 μl per IP): from Richard Jones50, H3K27me3 (4 μg per IP): CS200603 (Milipore).
Preparation of ChIP-seq libraries
Sequencing libraries were prepared with the KAPA Library Preparation Kit Illumina series (KK8201) according to the manufacturer's instructions. After adapter ligation, library fragments of 250-800 bp were isolated from an agarose gel. The DNA was PCR amplified with Illumina primers for 15 cycles, purified, and loaded on an Illumina flow cell for cluster generation. Libraries were sequenced on the Illumina HiSeq 2000. Reads were aligned to the Drosophila genome (BDGP R5/dm3) using bowtie 0.12.5 51.
Bioinformatic analysis of bidirectional transcription at PcG targets
For Drosophila analysis, ChIP data from the modENCODE project21 and from20 were both compared to CAGE and RACE data from31 and to MACE data from20. ChIP data sets are specified in Supplementary Table 2. The modENCODE data for the PcG proteins E(Z), PHO, PC and PSC, and for the transcription factors Hairy and Caudal from several developmental stages were downloaded from the modENDCODE website. Peaks from the same protein (Caudal or Hairy) or group of proteins (PcG) were merged using MultOvl52. For overlapping peaks the region of was overlap was merged to a single peak. Data files from20 were downloaded from the GEO database (GEO Accession: GSE24521).
For mouse analysis, ChIP data from33 (Sox2) and34 (Jarid1, Jarid2 and Suz12), were each compared to CAGE tag data from the FANTOM3 CAGE database32. Mouse ESC PcG binding data from34 were downloaded from the GEO database (GEO Accession: GSE18776). The peaks for Jarid2 and Suz12 as defined by the authors were used for further analyses. Mouse ESC Sox2 binding data was downloaded from GEO (Accession: GSE11431) and the same peak finding method and parameters were applied to the data as in34, using the QuEST peak finder53. The FANTOM3 CAGE summary dataset was downloaded from the FANTOM3 website. The original MM5 coordinates were transformed to MM9 using the LiftOver tool from UCSC
For promoter analyses in both mouse and fly, promoters were defined as -2kb and +500bp from the annotated TSS. Overlapping promoters were merged and defined as PcG bound if one of the above listed PcG ChIP peaks overlapped (by at least one base) with the promoter.
To compare CAGE, RACE or MACE tags with ChIP peaks or promoters, the following classes of transcript tags were defined: “Mono”: transcription peaks in one direction only (one or more MACE/CAGE peaks allowed). “Divergent”: transcription peaks in both directions, where the first start coordinate of + strand transcription is larger than the first start coordinate of the – strand. “Convergent”: transcription peaks in both directions, where first start coordinate on the + strand is smaller than the first start coordinate of the - strand. Custom R/Bioconductor scripts were used to find the overlap between CAGE and/or MACE peaks with ChIP peaks or promoters and to perform statistical analysis of the contingencies54. For both mouse and fly, the identified regions with both PcG or TF binding and MACE/CAGE tags in both directions (mono, divergent and convergent) were annotated using ‘fuge’ (CSF Vienna) to the dm3 refSeq genes annotation (downloaded October 2011 from UCSC, or the MM9 refSeq genes (downloaded April 2010 from UCSC).
Supplementary Material
ACKNOWLEDGEMENTS
We thank S. Gasser for discussions and for critical reading of the manuscript. We thank M. Rehmsmeier and members of our labs for discussions, P. Pasierbek for advice and training on imaging, P. A. Steffen for the E(Z) and E(Z)::GFP constructs, C. Ehrhardt and E. Dworschak for technical assistance, B. Dickson for the enGAL4 driver line, J.M. Dura for the daGAL4 driver line, I. Tamir for bioinformatic analysis of ChIP-Seq data and sharing the “Fuge” algortithm, F. Bantignies for advice on 3D DNA FISH, R. Jones for providing E(Z) antibody, the Vienna Campus Support Facility (CSF) for library preparation, deep sequencing, and purification of Drosophila PRC2. This work was funded by the Austrian Academy of Sciences, by EC grants EU FP6 Network of Excellence “The Epigenome” (to L.R) and the EU FP7 Network of Excellence “Epigenesys” (to L.R), and by an FWF Austrian Science Fund grant (P21525-B20 to L.R).
Footnotes
AUTHOR CONTRIBUTIONS
A.L. and L.R. initiated the project; L.R, V.A.H and A.L designed the experiments; J.T. performed bioinformatic analysis of genome-wide datasets; G.S. and K.A. performed automated image analysis of 3D DNA FISH; V.A.H., A.L., H.O., C.A., F.R., B.B., A.D., M.R., H.B.S and L.R. conducted the experiments and analyzed the data; S.K. supervised B.B.; A.P. supervised M.R.; M.L.V. and J.A.S. provided purified Drosophila PRC2 and polynucleosome substrates; L.R. prepared the manuscript with inputs from V.A.H., J.T, A.L, B.B. and H.O.
The authors declare no competing financial interests.
URLs
Vienna Campus Support Facility, http://www.csf.ac.at/ Austrian Academy of Sciences, http://www.oeaw.ac.at/ EU FP6 Network of Excellence “The Epigenome”, http://www.epigenome-noe.net/ EU FP7 Network of Excellence “Epigenesys”, http://www.epigenesys.eu/ FWF Austrian Science Fund, http://www.fwf.ac.at/en/index.asp Definiens XD software, http://www.definiens.com/product-services/definiens-xd-product-suite.html CSF Vienna, http://www.csf.ac.at/ Vienna Drosophila Resource Center (VDRC), http://stockcenter.vdrc.at modENCODE, http://www.modencode.org/ FANTOM3 CAGE summary dataset, http://fantom31p.gsc.riken.jp/cage_analysis/export/mm5 LiftOver tool, https://genome.ucsc.edu/cgi-bin/hgLiftOver R/Bioconductor, http://www.R-project.org/ dm3 and MM9 refSeq genes annotation, http://genome.ucsc.edu/
DATA ACCESS
ChIP seq data have been deposited in GEO and are available with the accession code GSE59192.
REFERENCES
- 1.Di Croce L, Helin K. Transcriptional regulation by Polycomb group proteins. Nat Struct Mol Biol. 2013;20:1147–55. doi: 10.1038/nsmb.2669. [DOI] [PubMed] [Google Scholar]
- 2.Steffen PA, Ringrose L. What are memories made of? How Polycomb and Trithorax proteins mediate epigenetic memory. Nat Rev Mol Cell Biol. 2014;15:340–56. doi: 10.1038/nrm3789. [DOI] [PubMed] [Google Scholar]
- 3.Muller J, Kassis JA. Polycomb response elements and targeting of Polycomb group proteins in Drosophila. Curr Opin Genet Dev. 2006;16:476–84. doi: 10.1016/j.gde.2006.08.005. [DOI] [PubMed] [Google Scholar]
- 4.Hekimoglu B, Ringrose L. Non-coding RNAs in polycomb/trithorax regulation. RNA Biol. 2009;6:129–37. doi: 10.4161/rna.6.2.8178. [DOI] [PubMed] [Google Scholar]
- 5.Spitale RC, Tsai MC, Chang HY. RNA templating the epigenome: long noncoding RNAs as molecular scaffolds. Epigenetics. 2011;6:539–43. doi: 10.4161/epi.6.5.15221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brockdorff N. Noncoding RNA and Polycomb recruitment. RNA. 2013;19:429–42. doi: 10.1261/rna.037598.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rinn JL, et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129:1311–23. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhao J, Sun BK, Erwin JA, Song JJ, Lee JT. Polycomb proteins targeted by a short repeat RNA to the mouse X chromosome. Science. 2008;322:750–6. doi: 10.1126/science.1163045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kanhere A, et al. Short RNAs are transcribed from repressed polycomb target genes and interact with polycomb repressive complex-2. Mol Cell. 2010;38:675–88. doi: 10.1016/j.molcel.2010.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhao J, et al. Genome-wide identification of polycomb-associated RNAs by RIP-seq. Mol Cell. 2010;40:939–53. doi: 10.1016/j.molcel.2010.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang KC, et al. A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature. 2011;472:120–4. doi: 10.1038/nature09819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guil S, et al. Intronic RNAs mediate EZH2 regulation of epigenetic targets. Nat Struct Mol Biol. 2012 doi: 10.1038/nsmb.2315. [DOI] [PubMed] [Google Scholar]
- 13.Davidovich C, Zheng L, Goodrich KJ, Cech TR. Promiscuous RNA binding by Polycomb repressive complex 2. Nat Struct Mol Biol. 2013;20:1250–7. doi: 10.1038/nsmb.2679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cifuentes-Rojas C, Hernandez AJ, Sarma K, Lee JT. Regulatory Interactions between RNA and Polycomb Repressive Complex 2. Mol Cell. 2014 doi: 10.1016/j.molcel.2014.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cohen B, Simcox AA, Cohen SM. Allocation of the thoracic imaginal primordia in the Drosophila embryo. Development. 1993;117:597–608. doi: 10.1242/dev.117.2.597. [DOI] [PubMed] [Google Scholar]
- 16.Williams JA, Bell JB, Carroll SB. Control of Drosophila wing and haltere development by the nuclear vestigial gene product. Genes Dev. 1991;5:2481–95. doi: 10.1101/gad.5.12b.2481. [DOI] [PubMed] [Google Scholar]
- 17.Deng H, Hughes SC, Bell JB, Simmonds AJ. Alternative requirements for Vestigial, Scalloped, and Dmef2 during muscle differentiation in Drosophila melanogaster. Mol Biol Cell. 2009;20:256–69. doi: 10.1091/mbc.E08-03-0288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bernard F, et al. Control of apterous by vestigial drives indirect flight muscle development in Drosophila. Dev Biol. 2003;260:391–403. doi: 10.1016/s0012-1606(03)00255-0. [DOI] [PubMed] [Google Scholar]
- 19.Guss KA, Mistry H, Skeath JB. Vestigial expression in the Drosophila embryonic central nervous system. Dev Dyn. 2008;237:2483–9. doi: 10.1002/dvdy.21664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Enderle D, et al. Polycomb preferentially targets stalled promoters of coding and noncoding transcripts. Genome Res. 2011;21:216–26. doi: 10.1101/gr.114348.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kharchenko PV, et al. Comprehensive analysis of the chromatin landscape in Drosophila melanogaster. Nature. 2011;471:480–5. doi: 10.1038/nature09725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee N, Maurange C, Ringrose L, Paro R. Suppression of Polycomb group proteins by JNK signalling induces transdetermination in Drosophila imaginal discs. Nature. 2005;438:234–7. doi: 10.1038/nature04120. [DOI] [PubMed] [Google Scholar]
- 23.Perez L, et al. Enhancer-PRE communication contributes to the expansion of gene expression domains in proliferating primordia. Development. 2011;138:3125–34. doi: 10.1242/dev.065599. [DOI] [PubMed] [Google Scholar]
- 24.Oktaba K, et al. Dynamic regulation by polycomb group protein complexes controls pattern formation and the cell cycle in Drosophila. Dev Cell. 2008;15:877–89. doi: 10.1016/j.devcel.2008.10.005. [DOI] [PubMed] [Google Scholar]
- 25.Schwartz YB, et al. Genome-wide analysis of Polycomb targets in Drosophila melanogaster. Nat Genet. 2006;38:700–5. doi: 10.1038/ng1817. [DOI] [PubMed] [Google Scholar]
- 26.Okulski H, Druck B, Bhalerao S, Ringrose L. Quantitative analysis of polycomb response elements (PREs) at identical genomic locations distinguishes contributions of PRE sequence and genomic environment. Epigenetics Chromatin. 2011;4:4. doi: 10.1186/1756-8935-4-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bate M. The embryonic development of larval muscles in Drosophila. Development. 1990;110:791–804. doi: 10.1242/dev.110.3.791. [DOI] [PubMed] [Google Scholar]
- 28.DeVido SK, Kwon D, Brown JL, Kassis JA. The role of Polycomb-group response elements in regulation of engrailed transcription in Drosophila. Development. 2008;135:669–76. doi: 10.1242/dev.014779. [DOI] [PubMed] [Google Scholar]
- 29.Bantignies F, et al. Polycomb-dependent regulatory contacts between distant Hox loci in Drosophila. Cell. 2011;144:214–26. doi: 10.1016/j.cell.2010.12.026. [DOI] [PubMed] [Google Scholar]
- 30.Kaneko S, Son J, Shen SS, Reinberg D, Bonasio R. PRC2 binds active promoters and contacts nascent RNAs in embryonic stem cells. Nat Struct Mol Biol. 2013;20:1258–64. doi: 10.1038/nsmb.2700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hoskins RA, et al. Genome-wide analysis of promoter architecture in Drosophila melanogaster. Genome Res. 2011;21:182–92. doi: 10.1101/gr.112466.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Katayama S, Kanamori M, Hayashizaki Y. Integrated analysis of the genome and the transcriptome by FANTOM. Brief Bioinform. 2004;5:249–58. doi: 10.1093/bib/5.3.249. [DOI] [PubMed] [Google Scholar]
- 33.Chen X, et al. Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell. 2008;133:1106–17. doi: 10.1016/j.cell.2008.04.043. [DOI] [PubMed] [Google Scholar]
- 34.Peng JC, et al. Jarid2/Jumonji coordinates control of PRC2 enzymatic activity and target gene occupancy in pluripotent cells. Cell. 2009;139:1290–302. doi: 10.1016/j.cell.2009.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ietswaart R, Wu Z, Dean C. Flowering time control: another window to the connection between antisense RNA and chromatin. Trends Genet. 2012;28:445–53. doi: 10.1016/j.tig.2012.06.002. [DOI] [PubMed] [Google Scholar]
- 36.Chen HH, Maeda T, Mullett SJ, Stewart AF. Transcription cofactor Vgl-2 is required for skeletal muscle differentiation. Genesis. 2004;39:273–9. doi: 10.1002/gene.20055. [DOI] [PubMed] [Google Scholar]
- 37.Pelechano V, Steinmetz LM. Gene regulation by antisense transcription. Nat Rev Genet. 2013;14:880–93. doi: 10.1038/nrg3594. [DOI] [PubMed] [Google Scholar]
- 38.Williams JA, Paddock SW, Vorwerk K, Carroll SB. Organization of wing formation and induction of a wing-patterning gene at the dorsal/ventral compartment boundary. Nature. 1994;368:299–305. doi: 10.1038/368299a0. [DOI] [PubMed] [Google Scholar]
- 39.Kim J, et al. Integration of positional signals and regulation of wing formation and identity by Drosophila vestigial gene. Nature. 1996;382:133–8. doi: 10.1038/382133a0. [DOI] [PubMed] [Google Scholar]
- 40.Gibson DG, et al. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods. 2009;6:343–5. doi: 10.1038/nmeth.1318. [DOI] [PubMed] [Google Scholar]
- 41.Gemkow MJ, Buchenau P, Arndt-Jovin DJ. FISH in whole-mount Drosophila embryos. RNA: activation of a transcriptional locus, DNA: gene architecture and expression. Bioimaging. 1996;4:107–120. [Google Scholar]
- 42.Milan M, Campuzano S, Garcia-Bellido A. Cell cycling and patterned cell proliferation in the wing primordium of Drosophila. Proc Natl Acad Sci U S A. 1996;93:640–5. doi: 10.1073/pnas.93.2.640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Aegerter-Wilmsen T, Aegerter CM, Hafen E, Basler K. Model for the regulation of size in the wing imaginal disc of Drosophila. Mech Dev. 2007;124:318–26. doi: 10.1016/j.mod.2006.12.005. [DOI] [PubMed] [Google Scholar]
- 44.Dietzl G, et al. A genome-wide transgenic RNAi library for conditional gene inactivation in Drosophila. Nature. 2007;448:151–6. doi: 10.1038/nature05954. [DOI] [PubMed] [Google Scholar]
- 45.Wang JW, Beck ES, McCabe BD. A modular toolset for recombination transgenesis and neurogenetic analysis of Drosophila. PLoS One. 2012;7:e42102. doi: 10.1371/journal.pone.0042102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rai AN, et al. Elements of the polycomb repressor SU(Z)12 needed for histone H3-K27 methylation, the interface with E(Z), and in vivo function. Mol Cell Biol. 2013;33:4844–56. doi: 10.1128/MCB.00307-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Steffen PA, et al. Quantitative in vivo analysis of chromatin binding of Polycomb and Trithorax group proteins reveals retention of ASH1 on mitotic chromatin. Nucleic Acids Res. 2013;41:5235–50. doi: 10.1093/nar/gkt217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Keene JD, Komisarow JM, Friedersdorf MB. RIP-Chip: the isolation and identification of mRNAs, microRNAs and protein components of ribonucleoprotein complexes from cell extracts. Nat Protoc. 2006;1:302–7. doi: 10.1038/nprot.2006.47. [DOI] [PubMed] [Google Scholar]
- 49.Orlando V, Jane EP, Chinwalla V, Harte PJ, Paro R. Binding of trithorax and Polycomb proteins to the bithorax complex: dynamic changes during early Drosophila embryogenesis. Embo J. 1998;17:5141–50. doi: 10.1093/emboj/17.17.5141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Carrington EA, Jones RS. The Drosophila Enhancer of zeste gene encodes a chromosomal protein: examination of wild-type and mutant protein distribution. Development. 1996;122:4073–83. doi: 10.1242/dev.122.12.4073. [DOI] [PubMed] [Google Scholar]
- 51.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Aszodi A. MULTOVL: fast multiple overlaps of genomic regions. Bioinformatics. 2012;28:3318–9. doi: 10.1093/bioinformatics/bts607. [DOI] [PubMed] [Google Scholar]
- 53.Valouev A, et al. Genome-wide analysis of transcription factor binding sites based on ChIP-Seq data. Nat Methods. 2008;5:829–34. doi: 10.1038/nmeth.1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gentleman RC, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fiedler T, Rehmsmeier M. jPREdictor: a versatile tool for the prediction of cis-regulatory elements. Nucleic Acids Res. 2006;34:W546–50. doi: 10.1093/nar/gkl250. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.