Abstract
The acidic (leucine-rich) nuclear phosphoprotein 32 kDa (ANP32) family is composed of small, evolutionarily conserved proteins characterized by an N-terminal leucine-rich repeat domain and a C-terminal low-complexity acidic region. The mammalian family members (ANP32A, ANP32B, and ANP32E) are ascribed physiologically diverse functions including chromatin modification and remodelling, apoptotic caspase modulation, protein phosphatase inhibition, as well as regulation of intracellular transport. In addition to reviewing the widespread literature on the topic, we present a concept of the ANP32s as having a whip-like structure. We also present hypotheses that ANP32C and other intronless sequences should not currently be considered bona fide family members, that their disparate necessity in development may be due to compensatory mechanisms, that their contrasting roles in cancer are likely context-dependent, along with an underlying hypothesis that ANP32s represent an important node of physiological regulation by virtue of their diverse biochemical activities.
Keywords: caspase regulation, chromatin regulation, intracellular transport, leucine-rich repeats, low-complexity acidic region, phosphatase inhibition
Introduction
Cell physiology is normally viewed as a number of discrete functional assignments with limited overlap. Cell proliferation, transcription, intracellular transport, and execution of apoptosis are generally studied separately. With the advent of systems biology, scientists are finding interplay between seemingly disparate pathways that are far more significant than originally expected. Therefore, recognition of regulatory nodes that can influence a range of cellular activities will be increasingly important for a holistic understanding of the cell.
As detailed below, there are reasons to believe that the acidic (leucine-rich) nuclear phosphoprotein 32 kDa (ANP32) family of proteins represent one of these critical regulatory nodes, having impacts on transcriptional regulation, protein phosphorylation, intracellular transport, and cell-death pathways. In this review, we aim to bring context to the expanding base of literature on the ANP32 family, a topic that can quickly become complicated given the diverse activities ascribed to the family, the varied gene nomenclature, the existence of annotated intronless sequences of questionable relevance, and indeterminate functional overlap between the family members.
A highly conserved structure of ANP32 proteins through evolution
Although the first cloning of an ANP32 family member was in 1994 1, the extent of identifiable ANP32 family of proteins was first examined in 2005 2. These proteins mostly range in size from 220 to 290 amino acid residues in length and are characterized by an N-terminal leucine-rich repeat (LRR) domain 3 composed of four LRR motifs and a C-terminal low-complexity acidic region (LCAR) of approximately 100 amino acids.
Protein sequences resembling the ANP32 family, i.e. proteins containing both LRR and LCAR sequences, have been annotated in the animal and plant kingdom as well as in protists 2, but not in yeast or other fungi. This observation suggests an early eukaryotic origin of the gene(s) for ANP32 and their loss specifically in fungi. The existence of the LRR and LCAR region on all known ANP32 family members, from the protist Plasmodium to Drosophila to the three vertebrate family members, suggests that both regions play critical roles for survival. Unfortunately, functional conservation in eukaryotes is unclear because, thus far, only animal ANP32 proteins have been studied: specifically, only the Drosophila member, termed Mapmodulin, and the mammalian family members.
The ANP32 structure gets whipped into shape
The structure of the LRR domain has been solved for family members ANP32A and ANP32B in three separate studies 4–6. The resulting models wherein the LRR motifs generate canonical parallel beta-sheet structures are very similar in the different studies 6 and consistent with those proposed earlier 2, suggesting high confidence in the consensus model. A number of protein-protein interactions have been mapped to the LRR of the ANP32 members, including CRM1 7, PP2Ac 8, Ataxin-1 9, Histone H3-H4 6, and Clip170 4.
Although other LCAR-containing proteins are recognizable and the class was described as early as 1986 10,11, ANP32s possess the longest LCARs that we have identified so far. The vertebrate ANP32 LCARs are approximately 100 amino acids long and are composed of 60–75% either glutamatic or aspartic acid. Distantly related species have less acidic content in their ANP32 LCAR (36% for Caenorhabditis elegans to 51% in Plasmodium) but are still recognizable as acidic. Unfortunately, these regions are difficult to categorize precisely. Sequence homology searches do not function effectively, because of the low complexity of the proteins and the fact that such methods mask low-complexity regions 12. Furthermore, the paucity of hydrophobic residues likely prevents tertiary structure in this region, allowing it to remain flexible in solution with a capacity for physical interaction with any positively charged surface. Conceptually, the ANP32 protein structure likely resembles a whip, where the LRR domain constitutes the handle and the LCAR is the thong. Due to the very high anionic content of the ANP32 LCARs, we expect that some ascribed functions will ultimately be viewed as artefacts resulting from non-specific cation binding.
As the name “ANP32” suggests, phosphorylation events on these proteins further increase their acidity. Upon the cloning of rat ANP32A, the researchers recognized a number of potential sites of phosphorylation by Casein kinase II (CKII), Calcium-Calmodulin dependent kinase II, and protein kinase C 13. Phosphorylation by CKII has been verified 14. These and/or other phosphorylation events are predicted to affect protein function 15,16.
Based on the absence of oligomerization domains and the results of overexpression experiments on the founding member ANP32A, these proteins likely exist as monomers 17, although ANP32A isolated from Chinese hamster ovary cells is reported to form a homotrimer through its LRR domain 16. No model currently exists to reconcile these potentially discrepant observations.
SET and other LCAR-containing proteins are related to ANP32s
Intriguingly, several other prominent LCAR-containing factors are multifunctional, including the cellular proteins Nucleophosmin, Nucleoplasmin, high mobility group box (HMGB)1, HMGB2, and SET, as well as the human herpesvirus 8 protein LANA. It is notable that each of these factors has been implicated in regulating chromatin structure in some way, suggesting a primary activity of the LCAR. Among the other multifunctional LCAR-containing proteins, the SET oncoprotein (a.k.a. TAF1β, I2PP2A, PHAPII) is of particular interest for ANP32 biology. Firstly, the ANP32 proteins and SET have been found in different biochemical isolations with shared characteristics 1,18,19 in spite of structural differences in their N-termini. In contrast to the LRR motifs in the N-termini of ANP32s, SET contains a nucleosome-assembly-protein domain. Secondly, SET and ANP32 proteins have been isolated together in higher order complexes 7,20,21. This physical interaction and functional overlap suggest that SET and ANP32 proteins coevolved in particular regulatory pathway(s). Finally, a cell-permeable inhibitor of SET, likely targeting its LCAR 22–24, is being investigated as a cytotoxic anti-cancer agent 25,26. Simultaneous inhibition of ANP32 proteins by this candidate drug has not been excluded.
The intronless ANP32 loci: Expressed and functional?
Controversy exists about the extent of the ANP32 family in mammals. A previous article has suggested eight different ANP32 family members in humans 2, of which six different loci are currently annotated in Genbank. Three conserved family members exist in vertebrates, namely ANP32A (a.k.a. PHAPI, pp32, I1PP2A, LANP, HPPCn, Mapmodulin), ANP32B (a.k.a. SSP29, APRIL, PAL31, PHAPI2), and ANP32E (a.k.a. CPD1, LANP-L, PHAPIII). These have all been isolated in multiple biochemical fractionations 7,19,27–29 and are well represented in transcriptomic and proteomic surveys. The controversy surrounds the nature of intronless loci, ANP32C, and ANP32D. Researchers alternately characterize these sequences as retrogenes – products of retrotransposition that support protein coding – as functional transcribed pseudogenes, or as inert pseudogenes.
Here we examine the evidence for ANP32C, also known as pp32r1, which is the most frequently described of these intronless sequences in the literature 30–35. This open-reading frame has physiological effects when ectopically expressed 17,30–33 but doubts surround its endogenous expression. On a genomic level, ANP32C is not conserved from rodent to human. Mouse and human sequences of ANP32C, present in introns of Ranbp17 and MARCH1, respectively, are more homologous with ANP32A sequences of their own species than with each other 2 suggesting separate and relatively recent origins. This would not preclude functional significance, but it indicates that these should be treated as non-orthologous sequences.
At the transcript level, ANP32C is reportedly expressed in a variety of cancers and cell lines 33–35. Unfortunately, the intronless nature makes targeted reverse transcription-PCR very susceptible to DNA contamination and these reports of ANP32C expression do not overtly show the reverse transcriptase controls to assess this potential contamination. A significant tool for examining the human transcriptome is expressed sequence tag (EST) analysis, which has extensive coverage of neoplasias in which ANP32C expression is reported. ANP32C is currently represented by a total of four ESTs compared to 752 for ANP32A, 720 for ANP32B, and 490 for ANP32E. The EST count for ANP32C is also low compared to the functional transcribed pseudogene PTENP1, which is represented by 99 ESTs. With its presence within the MARCH1 gene, the ANP32C locus is almost certainly transcribed, at least as part of the intron, but the stability and functionality of the resulting RNA remains an outstanding question.
The potential translation of ANP32C is also unclear. While its open-reading frame contains a Kozak translational initiation sequence, intact endogenous ANP32C protein has not yet been reported in biochemical isolations. Furthermore each of the peptides annotated for ANP32C in the Human Proteomic Project database portal, “PeptideAtlas” 36, has 100% identity with sequences in ANP32A including one peptide inappropriately described as unique to ANP32C. In contrast, a commercial antiserum was recently used to show ANP32C protein expression 32, suggesting that it is a retrogene. RNA inhibition (RNAi) validation of this antiserum will be essential to determine endogenous expression going forward.
Given what we feel is the lack of solid unbiased evidence for transcription and translation of ANP32C as well as other purported family members, we believe that only ANP32A, ANP32B, and ANP32E should be considered bona fide mammalian ANP32 family members at this time. While there may be means of parental gene regulation associated with these loci, we limit ourselves to the three unquestioned protein-coding ANP32 family members for the purpose of the functional review.
ANP32s: Here, there, and everywhere
The literature describes a startlingly diverse array of biochemical activities for the ANP32 family. Since cellular localization would preclude certain activities, there has been significant attention placed on determining where the ANP32 proteins are located. Unfortunately, not even the localization of these factors is generally accepted, because different reports conclude, variously, that ANP32 proteins are predominantly nuclear 17,37–39, shuttling nuclear-cytoplasmic 7,15,40,41, predominantly cytoplasmic 42–44, on the cell surface 45, or even secreted 46. In the case of ANP32B, a phosphorylation event on Thr244 appears to regulate its nuclear export, because a phospho-site-deficient protein localized exclusively to the nucleus 15. For ANP32A, induction of reactive oxygen species causes a cytoplasmic-to-nuclear translocalization 42. In contrast, a nuclear to cytoplasmic translocalization of ANP32A has been seen in the process of in vitro neuronal differentiation 47; however, this effect was not seen in brain tissue 48 or cultured primary neurons 38. A separate study suggests that apoptotic stimuli can induce translocation to cytoplasm 17, although this may be due to nuclear envelope breakdown.
Whereas some studies are more compelling than others, it is very plausible that these disparate findings with respect to localization reflect different model systems and reagents used. It is clear from these studies that no particular activity of the ANP32 proteins may be excluded based on protein localization.
ANP32 proteins regulate chromatin by various means
From the earliest classification of LCAR-containing proteins, it was evident that they are involved in regulating transcription and chromatin architecture 11. The reported activities of ANP32 proteins in chromatin regulation are diagrammed in Fig. 1. The ANP32 proteins were first noted to function in transcriptional repression upon purification of ANP32A, a member of the inhibitor of histone acetyltransferase (INHAT) complex 21. Further studies revealed that ANP32A blocks histone modification by binding to histone tails and sterically inhibiting acetylation. More specifically, ANP32A preferentially binds to unmodified histone H3 tails 49,50. After this discovery, a number of groups suggested that ANP32A likely modifies activated transcription due to its recruitment to promoters by DNA-binding transcription factors (TFs) 51–54. In one study, ANP32A also facilitated estrogen receptor loading onto DNA 54.
Figure 1.
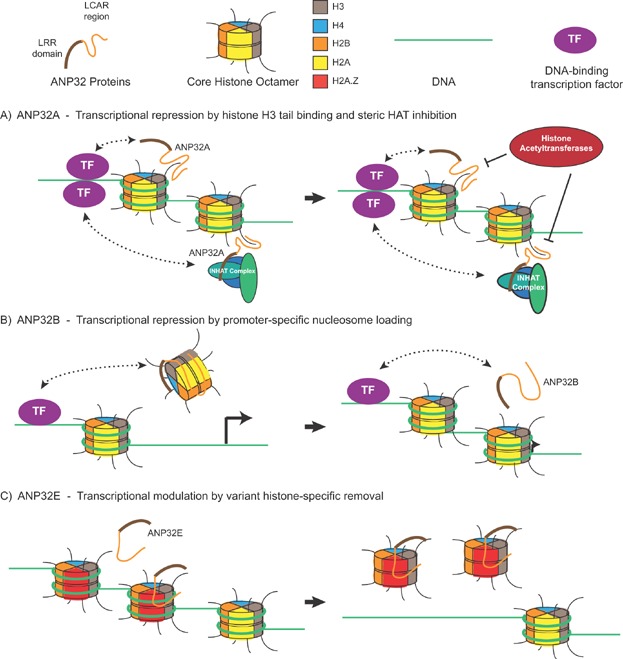
Reported chromatin modification activities of the ANP32 family. Each mammalian ANP32 protein is suggested to modify transcription by regulating chromatin, but by different means. A: ANP32A, either alone or as part of the INHAT complex, is recruited to specific regions of DNA by transcription factors. Here it binds histone-H3 tails and inhibit their modification 49,50. B: ANP32B is recruited by KLF5 to maintain promoter-specific histone occupancy. Abolishing the KLF5-ANP32B interaction correlates with promoter accessibility and transcriptional activation 55. C: ANP32E specifically removes nucleosomes containing variant histone H2A.Z, likely as a means of targeted regulation of transcription 29. DNA-binding transcription factors that may target ANP32E to particular promoters are yet to be identified.
ANP32B has also been found to bind TFs and modulate their activity 55. In this study, Krüppel-like factor 5 (KLF5) binds to ANP32B and is recruited to specific regions of DNA to repress transcription in a promoter-specific manner. In addition to binding histones, recombinant ANP32B demonstrated plasmid-supercoiling activity indicative of histone chaperone activity 55. Further studies mapped a physical interaction between ANP32B LRR region and core histones H3-H4 6.
ANP32E has recently also been shown to have histone chaperone activity. In this case, ANP32E is associated with the p400/Tip60 complex and specifically removes histone H2A.Z from DNA 29,56. Since H2A.Z is associated with transcriptional regulation based on its preferential localization to transcriptional start sites 57,58 and its role in determining transcription responsiveness 59, recruitment of ANP32E can likely alter activated gene transcription. Although this is yet to be shown, deletion of ANP32E in mouse cells changes the profile of H2A.Z occupancy 29. H2A.Z chaperone activity seems to be specific for ANP32E since the H2A.Z interaction maps to a discrete conserved sequence within the LCAR of ANP32E that is not found in ANP32A or ANP32B 29,56. Intriguingly, however, the Drosophila Protein Interaction Map identified variant H2A (Dmel\His2Av), the precursor of mammalian H2A.Z, as a Mapmodulin-interacting factor 60. This suggests a conserved role for ANP32 proteins in regulating H2A.Z placement.
The revelation that ANP32 proteins have histone binding and chaperone activity is another example of shared functions with its binding partner SET. SET, also present in the INHAT complex, independently shows H3 tail binding 49 as well as histone chaperone activity 55,61. These findings suggest that ANP32A recruitment to promoters may likewise provide histone chaperone activity as a means of transcriptional repression.
ANP32 proteins as regulators of cell death pathways
Thus far, only a single biochemical study has isolated all three of the mammalian ANP32 family members 27. In this work, the focus was their capacity to aid activation of the apoptosome, the initiator caspase complex containing procaspase 9, APAF-1, and cytochrome c 27. This and a subsequent study 62 found that ANP32 proteins allowed apoptosome activation at physiological levels of dATP. This finding is now supported by several different studies 17,63–66, while one study showed that the ANP32A LCAR also promotes caspase-3 activation directly 63. Two different groups have posited that ANP32A is part of a regulated positive feedback mechanism in caspase activation, whereby complexed ANP32A is sequestered from caspases until the appropriate cell death stimulus is applied 67,68. In one model, Granzyme A stimulates release of ANP32A concurrent with release of the Granzyme A-activated DNase, NM23H1 69, hence suggesting that it may have a role in caspase-independent cell death. Strangely, however, the different groups provide evidence for ANP32A sequestration in different complexes (Fig. 2). We are sceptical of this model since pro-survival sequestration would imply that no free pool of ANP32A would exist to perform transcriptional control or other noted activities.
Figure 2.
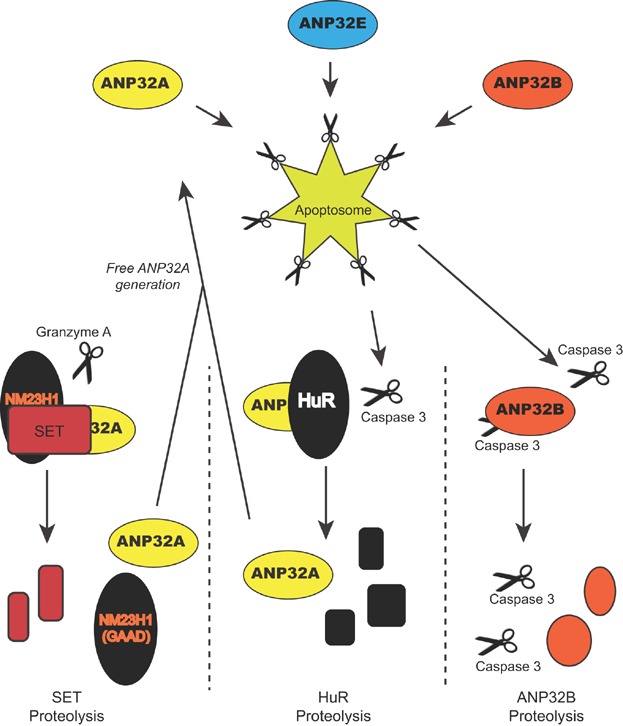
Mammalian ANP32 proteins function in cell-death pathways. Depicted is a summary of four suggested points of involvement for the ANP32 proteins in cell-death pathways. All three ANP32 proteins were isolated as activators of the apoptosome 27. Proteolytic cleavage of ANP32A-sequestering proteins SET and HuR may stimulate a positive feedback loop allowing more apoptosome activation 67,68. ANP32B, as both an inhibitor and substrate of Caspase 3 may represent a separate positive feedback mechanism on caspase activation 71.
In contrast, although ANP32B could activate the apoptosome 27, its depletion by RNAi in cells induces high rates of apoptosis 70–72. This pro-survival activity correlates with ANP32B's reported inhibition of caspase 3 72, the converse of the activity reported for ANP32A. Since we also found that ANP32B is a caspase substrate 71, we propose that ANP32B may be part of a separate positive feedback network for effector-caspase activation (Fig. 2).
ANP32s control phosphatase activity
ANP32A was isolated together with SET as an inhibitor of protein phosphatase 2A (PP2A) 19,73, and each has been reported to bind to protein phosphatase 1 (PP1) independently to affect activity 74. One study has found that ANP32A binding to the signalling lipid molecule, sphingosine, abrogates its PP2A interaction, suggesting a physiological regulation of this activity 75. Although a shared activity with SET would imply involvement of the LCAR, the interaction between ANP32A and PP2A requires the LRR region 8, which is not present on SET. Functionally, the interaction with PP2A is suggested to impact cell proliferation 76, inflammation 75, and neurodegenerative disorders 8,77,78, although these potential consequences invite further in vivo study. Intriguingly, in separate studies ANP32E was able to inhibit PP2A 79, whereas ANP32B was not 72, suggesting either a functional divergence or, more likely, differences in in vitro assay conditions.
ANP32 proteins mediate intracellular transport
Regulating transport within the cell is another activity that ANP32 proteins are reported to possess. As adaptors between the nuclear-export factor CRM1 and the mRNA-binding protein HuR, ANP32A, and ANP32B have been implicated in expediting transport of mRNA strands containing adenosine-rich elements 7. This ANP32 function has been suggested as a means of control of both cellular 7,15 and viral mRNAs 80–82.
In addition to mRNA shuttling, the ANP32 proteins are reported to regulate transport of factors within the cytoplasm. This conclusion comes from the initial characterization of the ANP32A in Chinese hamster ovary cells as a microtubule-associated protein (MAP) interacting factor 16,44,83. Following on this finding, an ANP32A-MAP1B interaction was demonstrated in mammals and shown to impact neuritogenesis 47.
ANP32A: Moonlighting outside the cell
In addition to the role of the ANP32 proteins inside the cell, two groups have suggested a role for ANP32A in the extracellular space. A study in 1998 suggested that ANP32A could be present on the surface of intact peripheral blood mononucleocytes, potentially acting as an HIV receptor by virtue of its ability to bind an HIV gp120 mimic peptide 45.
In a series of papers since 2008, ANP32A has also been reported to act as a growth factor for hepatocytes 41,46,84–86 both in vivo and in vitro. These studies demonstrate that ANP32A is increased in hepatocytes in order to reduce cellular oxidative stress in response to both carbon tetrachloride and ethanol. Since provision of recombinant ANP32A in media in vitro and intravenously in vivo is sufficient to provide this protective activity 84, the activity is presumed to be extracellular. Although a pathway involving sphingosine kinase has been implicated 46,84,86, the hepatocyte receptor for ANP32A has yet to be identified.
Taken together, this unusual array of activities suggests that the ANP32 proteins may constitute a critical regulatory node in cell physiology that should affect viability and normal development. Differential affinities, expression patterns, or controlled complex formation may allow dynamic functions of the ANP32s across time, allowing them to change roles between regulating gene expression, cell signalling, and cell death, depending on changes in cell physiology.
Unequal requirement for ANP32 proteins in development
Gene expression analyses of ANP32 family members suggest that they are upregulated in proliferating tissues and downregulated in the process of terminal differentiation. This appears to be true for each of the mammalian family members to differing degrees. Indeed, both ANP32A and ANP32B were initially cloned based on their abundance in proliferating tissues 28,39, while ANP32E was cloned from the most postnatal cerebellum 79, when tissue is most proliferative. Furthermore, ANP32A and ANP32B expression are associated with proliferative neoangiogenesis 87 and macrophage activation 70, respectively. In addition to licensing proliferation, ANP32 expression may enforce self-renewal capacity as has been hypothesized for a number of systems 88–92. In vitro models also suggest that ANP32A can modulate neuritogenesis either positively 47,78 or negatively 90,93.
To determine their role in development, ANP32 genes have been examined through animal loss-of-function models. Whereas their conservation and reported functions would suggest catastrophic consequences, the resulting phenotypes have so far been quite varied. For two Mapmodulin loss-of-function models, one RNAi and one P-element insertion, the flies are viable in spite of Mapmodulin being the only known Drosophila ANP32 family member 94,95. On the other hand, in C. elegans ANP32 loss of function, i.e. RNAi of T19H12.2 is embryonic lethal 96. No loss-of-function analysis is reported for the other C. elegans ANP32 family member, F33H2.3.
Five mutant-mouse strains carrying Anp32 loss-of-function alleles have so far been generated. Surprisingly, mice deficient in the most prominent family member, Anp32a, demonstrated no apparent phenotype in two separate studies 97,98. Likewise, two alleles of Anp32e are published 98,99 and, apart from a disputed, subtle, neurological phenotype in the gene-trapped mutant 99,100, the mice were apparently normal. On the other hand, the single targeted allele of Anp32b had a severe, albeit complex, phenotype. Anp32b-deficient mice demonstrated a strain-dependent penetrance of perinatal lethality with surviving mice in a genetically mixed background showing growth defects, premature aging, and a wide array of pathologies 101. These mice also demonstrate a role for Anp32a in mouse development that only becomes apparent in the context of Anp32b deficiency 101, strongly suggesting a functional overlap. The same genetic interaction could not be established between Anp32b and Anp32e 101. Intriguingly, the requirement of a particular Anp32 mouse gene seems inversely related to the size of its 3′ untranslated region (UTR). We speculate that 3′UTR regulation may affect the efficiency of compensation by alternate ANP32s. In this model, Anp32b is the most important by virtue of regulatory or context-dependent impediments to Anp32a and Anp32e translation. The physiological contexts in which Anp32a and/or Anp32e play an important role still await discovery. Certainly their conservation among vertebrates implies such importance.
ANP32 proteins in human pathogenesis
Whereas the functions ascribed to this protein family would lead one to believe that ANP32s are critically important, no human pathogenic mutations have yet been identified. ANP32A's interaction with 9 and regulation by 78 mutated, pathogenic Ataxin-1 protein as well as its ability to regulate phosphorylation of the Alzheimer's disease-related protein tau 8 suggest a possible activity in neurodegenerative disorders. In contrast, the functional significance of the physical interaction with ATXN-1 has not been addressed 78, the ANP32A-deficient mice have not demonstrated aberrant behaviour (97, Reilly unpublished), and a polymorphism analysis suggests that ANP32A is not genetically associated with Alzheimer's disease 102.
Studies in mice suggest that most mutations would be tolerated during developmental processes, but the evolutionary sequence conservation suggests that this is not likely the case. It is a curious paradox that may be related to the controlled conditions of laboratory animals. Indeed, there is reason to believe that ANP32 proteins function in the life cycles of a wide variety of viruses, including DNA viruses adenovirus 82,103 and adeno-associated virus 104 as well as retroviruses, Foamy virus 81, Nipah virus 80, and HIV 105. Challenge with such pathogens is typically excluded from laboratory animal colonies.
Contrasting data on the role of ANP32s in cancer
The most prominent association between ANP32s and human pathology is in the case of cancer. Firstly, the activities described above can each be related to cancer. Cell death control, regulated protein phosphatase activity, and epigenetic gene regulation each have a clear and demonstrated role in cancer progression. Indeed, many of the factors with which ANP32s physically interact are established players in human cancer including SET, KLF5, pRB 38,106, NM23H1 20, and Axin-1 107. Potential regulation of any of these factors would imply that ANP32s are important regulators of tumorigenesis.
Secondly, and consistent with this hypothesis, significant direct genetic and epigenetic data exist to suggest that ANP32s are dysregulated in an array of cancers. The results, however, do not identify ANP32s as consistently tumor promoting or tumor suppressive, thus suggesting context dependency. ANP32A is regularly referenced as a tumor suppressor based on early studies that showed it could inhibit cell transformation 17,108–110 as well as its apparent reduced expression in prostate 33 and breast cancer 34. Additionally, ANP32A was also shown to be a positive prognostic marker in non-small-cell lung cancer 65, and reducing its expression increased ras-induced tumorigenicity of NIH3T3 cells 108. These data contrast with other studies that show expression of ANP32A is increased in cancers including prostate 37, colorectal 111, ovarian 112, and liver 113. Furthermore, it is a negative prognostic marker in hepatocellular carcinoma 41, where reducing its expression also reduces xenograft growth 41. ANP32E similarly shows enhanced expression in gastric cancer 114, and is a negative prognostic marker in myeloma 115. However, it is also reported as a positive predictor of follicular lymphoma treatment response 116. For ANP32B, results in breast cancer prognosis suggest that it is a tumor-promoting gene 101, whereas it is also ranked among the highest hits in a tumor-suppressor-rich genome-wide search for recessive cancer genes 117. Certainly the different functions of the ANP32 proteins provide plenty of opportunity to rationalize the genetic and epigenetic evidence. Currently, however, there is no clear theme for ANP32 expression in cancer. Taking their proliferation-related expression together with their roles in activating caspase-mediated cell death, we hypothesize that these proteins are a “double-edged sword” in cancer progression. Within a genetic context of defective apoptotic pathways they may provide proliferative advantage by selective gene expression, whereas in cancer cells with intact apoptotic cell-death pathways their overexpression would drive tumor reduction. This paradigm is now increasingly evident in cancer genetics regarding other factors.
Conclusions and outlook
Figure 3 provides an overview of the homology, functions, and loss-of-function phenotypes of mammalian ANP32s. With the rationale presented, we contend that there are only three real ANP32 genes in vertebrates, namely ANP32A, ANP32B, and ANP32E. We believe that any intronless sequences should be considered pseudogenes until compelling evidence of expression is presented. We also contend that, with some exceptions, the listed functions are shared among the family members. The field still awaits a systematic examination to clarify which, if any, of these activities are exclusive for particular members. With the compelling evidence for the described functions, however, we propose that the ANP32 proteins likely act as important regulators in the cell, providing molecular crosstalk between gene-expression, cell-survival, and cell-signalling pathways. The severity of their loss, however, appears to be gene- and context-dependent. For example, although RNAi against ANP32B induces apoptosis in tissue culture, mice lacking this gene can grow to adulthood. Is the cell able to compensate for individual loss, particularly slow loss, with alternative ANP32 protein usage, as suggested by a theory 118 that redundant genes prevent developmental error? Regardless of the interchangeability of the ANP32 proteins, these factors are clearly providing a regulatory role in the cell, which we believe can be exploited for medical benefit in the future. LCAR regions are already proving potential targets for peptide inhibition. With clearer understanding of the genuine human ANP32 proteins, we can focus on the best strategies and circumstances to fine-tune these multifunctional factors for desired physiological outcome.
Figure 3.
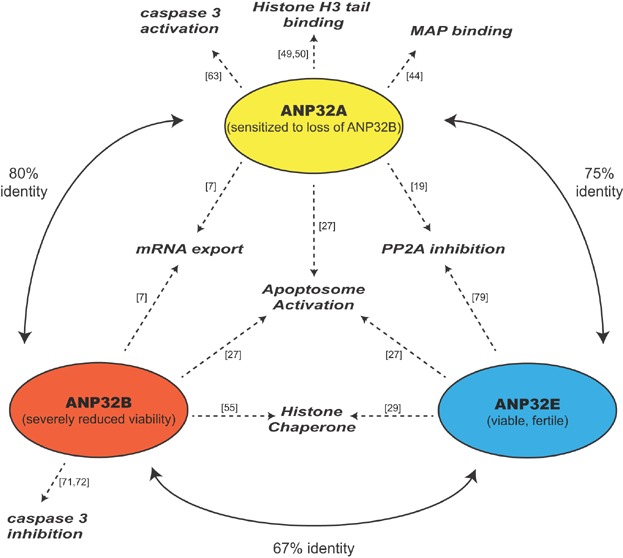
Homology, functions, and phenotypes of the mammalian ANP32 family. Each circle reflects a mammalian ANP32 family member with its respective mouse loss-of-function phenotype in parentheses. Curved solid lines indicate the amino-acid identities of the structured LRR domains between the indicated human ANP32 family members. Dashed arrows represent evidence for observed function, with references provided.
Box 1 Abbreviations and glossary.
LRR: leucine-rich repeat motif normally present in multiples repeats
LCAR: low-complexity acidic region
Retrogene: a DNA sequence produced by retrotransposition supporting protein coding
Pseudogene: defunct genomic sequence with similarity to functional genes but lacking protein-coding capacity
EST: expressed sequence tag
INHAT: inhibitor of histone acetyltransferase complex
Histone chaperone: a protein or complex involved in placement or removal of histones from chromatin
Apoptosome: a complex containing Caspase 9 that initiates caspase-dependent apoptosis in response to cellular stress
Acknowledgments
We would like to thank Dr. Winship Herr and Ms. Vonny Leo for critical editing of the manuscript. PR is supported by National Cancer Centre Research Foundation.
References
- 1.Vaesen M, Barnikol-Watanabe S, Gotz H, Awni LA, et al. Purification and characterization of two putative HLA class II associated proteins: PHAPI and PHAPII. Biol Chem Hoppe Seyler. 1994;375:113–26. doi: 10.1515/bchm3.1994.375.2.113. [DOI] [PubMed] [Google Scholar]
- 2.Matilla A, Radrizzani M. The Anp32 family of proteins containing leucine-rich repeats. Cerebellum. 2005;4:7–18. doi: 10.1080/14734220410019020. [DOI] [PubMed] [Google Scholar]
- 3.Kobe B, Kajava AV. The leucine-rich repeat as a protein recognition motif. Curr Opin Struct Biol. 2001;11:725–32. doi: 10.1016/s0959-440x(01)00266-4. [DOI] [PubMed] [Google Scholar]
- 4.de Chiara C, Kelly G, Frenkiel TA, Pastore A. NMR assignment of the leucine-rich repeat domain of LANP/Anp32a. J Biomol NMR. 2007;38:177. doi: 10.1007/s10858-006-9101-2. [DOI] [PubMed] [Google Scholar]
- 5.Huyton T, Wolberger C. The crystal structure of the tumor suppressor protein pp32 (Anp32a): structural insights into Anp32 family of proteins. Protein Sci. 2007;16:1308–15. doi: 10.1110/ps.072803507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tochio N, Umehara T, Munemasa Y, Suzuki T, et al. Solution structure of histone chaperone ANP32B: interaction with core histones H3-H4 through its acidic concave domain. J Mol Biol. 2010;401:97–114. doi: 10.1016/j.jmb.2010.06.005. [DOI] [PubMed] [Google Scholar]
- 7.Brennan CM, Gallouzi IE, Steitz JA. Protein ligands to HuR modulate its interaction with target mRNAs in vivo. J Cell Biol. 2000;151:1–14. doi: 10.1083/jcb.151.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen S, Li B, Grundke-Iqbal I, Iqbal K. I1PP2A affects tau phosphorylation via association with the catalytic subunit of protein phosphatase 2A. J Biol Chem. 2008;283:10513–21. doi: 10.1074/jbc.M709852200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Matilla A, Koshy BT, Cummings CJ, Isobe T, et al. The cerebellar leucine-rich acidic nuclear protein interacts with ataxin-1. Nature. 1997;389:974–8. doi: 10.1038/40159. [DOI] [PubMed] [Google Scholar]
- 10.Kuehl L, Childers TJ, McCauley RM. The occurrence of extended acidic sequences in nonhistone chromosomal proteins. Arch Biochem Biophys. 1986;248:272–81. doi: 10.1016/0003-9861(86)90424-8. [DOI] [PubMed] [Google Scholar]
- 11.Earnshaw WC. Anionic regions in nuclear proteins. J Cell Biol. 1987;105:1479–82. doi: 10.1083/jcb.105.4.1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ye J, McGinnis S, Madden TL. BLAST: improvements for better sequence analysis. Nucleic Acids Res. 2006;34:W6–9. doi: 10.1093/nar/gkl164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Matsuoka K, Taoka M, Satozawa N, Nakayama H, et al. A nuclear factor containing the leucine-rich repeats expressed in murine cerebellar neurons. Proc Natl Acad Sci USA. 1994;91:9670–4. doi: 10.1073/pnas.91.21.9670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hong R, Macfarlan T, Kutney SN, Seo SB, et al. The identification of phosphorylation sites of pp32 and biochemical purification of a cellular pp32-kinase. Biochemistry. 2004;43:10157–65. doi: 10.1021/bi0493968. [DOI] [PubMed] [Google Scholar]
- 15.Fries B, Heukeshoven J, Hauber I, Gruttner C, et al. Analysis of nucleocytoplasmic trafficking of the HuR ligand APRIL and its influence on CD83 expression. J Biol Chem. 2007;282:4504–15. doi: 10.1074/jbc.M608849200. [DOI] [PubMed] [Google Scholar]
- 16.Ulitzur N, Rancano C, Pfeffer SR. Biochemical characterization of mapmodulin, a protein that binds microtubule-associated proteins. J Biol Chem. 1997;272:30577–82. doi: 10.1074/jbc.272.48.30577. [DOI] [PubMed] [Google Scholar]
- 17.Pan W, da Graca LS, Shao Y, Yin Q, et al. PHAPI/pp32 suppresses tumorigenesis by stimulating apoptosis. J Biol Chem. 2009;284:6946–54. doi: 10.1074/jbc.M805801200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kendall RT, Strungs EG, Rachidi SM, Lee MH, et al. The beta-arrestin pathway-selective type 1A angiotensin receptor (AT1A) agonist [Sar1,Ile4,Ile8]angiotensin II regulates a robust G protein-independent signaling network. J Biol Chem. 2011;286:19880–91. doi: 10.1074/jbc.M111.233080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li M, Guo H, Damuni Z. Purification and characterization of two potent heat-stable protein inhibitors of protein phosphatase 2A from bovine kidney. Biochemistry. 1995;34:1988–96. doi: 10.1021/bi00006a020. [DOI] [PubMed] [Google Scholar]
- 20.Fan Z, Beresford PJ, Oh DY, Zhang D, et al. Tumor suppressor NM23-H1 is a granzyme A-activated DNase during CTL-mediated apoptosis, and the nucleosome assembly protein SET is its inhibitor. Cell. 2003;112:659–72. doi: 10.1016/s0092-8674(03)00150-8. [DOI] [PubMed] [Google Scholar]
- 21.Seo SB, McNamara P, Heo S, Turner A, et al. Regulation of histone acetylation and transcription by INHAT, a human cellular complex containing the set oncoprotein. Cell. 2001;104:119–30. doi: 10.1016/s0092-8674(01)00196-9. [DOI] [PubMed] [Google Scholar]
- 22.Switzer CH, Cheng RY, Vitek TM, Christensen DJ, et al. Targeting SET/I(2)PP2A oncoprotein functions as a multi-pathway strategy for cancer therapy. Oncogene. 2011;30:2504–13. doi: 10.1038/onc.2010.622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Christensen DJ, Ohkubo N, Oddo J, Van Kanegan MJ, et al. Apolipoprotein E and peptide mimetics modulate inflammation by binding the SET protein and activating protein phosphatase 2A. J Immunol. 2011;186:2535–42. doi: 10.4049/jimmunol.1002847. [DOI] [PubMed] [Google Scholar]
- 24.Christensen DJ, Chen Y, Oddo J, Matta KM, et al. SET oncoprotein overexpression in B-cell chronic lymphocytic leukemia and non-Hodgkin lymphoma: a predictor of aggressive disease and a new treatment target. Blood. 2011;118:4150–8. doi: 10.1182/blood-2011-04-351072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Agarwal A, MacKenzie RJ, Pippa R, Eide CA, et al. Antagonism of SET using OP449 enhances the efficacy of tyrosine kinase inhibitors and overcomes drug resistance in myeloid leukemia. Clin Cancer Res. 2014;20:2092–103. doi: 10.1158/1078-0432.CCR-13-2575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fujiwara N, Kawasaki H, Yabe R, Christensen DJ, et al. A potential therapeutic application of SET/I2PP2A inhibitor OP449 for canine T-cell lymphoma. J Vet Med Sci. 2013;75:349–54. doi: 10.1292/jvms.12-0366. [DOI] [PubMed] [Google Scholar]
- 27.Jiang X, Kim HE, Shu H, Zhao Y, et al. Distinctive roles of PHAP proteins and prothymosin-alpha in a death regulatory pathway. Science. 2003;299:223–6. doi: 10.1126/science.1076807. [DOI] [PubMed] [Google Scholar]
- 28.Malek SN, Katumuluwa AI, Pasternack GR. Identification and preliminary characterization of two related proliferation-associated nuclear phosphoproteins. J Biol Chem. 1990;265:13400–9. [PubMed] [Google Scholar]
- 29.Obri A, Ouararhni K, Papin C, Diebold ML, et al. ANP32E is a histone chaperone that removes H2A.Z. from chromatin. Nature. 2014;505:648–53. doi: 10.1038/nature12922. [DOI] [PubMed] [Google Scholar]
- 30.Buddaseth S, Gottmann W, Blasczyk R, Huyton T. Overexpression of the pp32r1 (ANP32C) oncogene or its functional mutant pp32r1Y140H confers enhanced resistance to FTY720 (Finguimod) Cancer Biol Ther. 2013;15:289–96. doi: 10.4161/cbt.27307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Buddaseth S, Gottmann W, Blasczyk R, Huyton T. Dysregulation of cell cycle control caused by overexpression of the oncogene pp32r1 (ANP32C) and the Tyr>His mutant pp32r1Y140H. Biochim Biophys Acta. 2013;1833:1212–21. doi: 10.1016/j.bbamcr.2013.02.001. [DOI] [PubMed] [Google Scholar]
- 32.Imamachi K, Higashino F, Kitamura T, Kakuguchi W, et al. pp32r1 controls the decay of the RNA-binding protein HuR. Oncol Rep. 2014;31:1103–8. doi: 10.3892/or.2013.2956. [DOI] [PubMed] [Google Scholar]
- 33.Kadkol SS, Brody JR, Pevsner J, Bai J, et al. Modulation of oncogenic potential by alternative gene use in human prostate cancer. Nat Med. 1999;5:275–9. doi: 10.1038/6488. [DOI] [PubMed] [Google Scholar]
- 34.Kadkol SS, El Naga GA, Brody JR, Bai J, et al. Expression of pp32 gene family members in breast cancer. Breast Cancer Res Treat. 2001;68:65–73. doi: 10.1023/a:1017919507109. [DOI] [PubMed] [Google Scholar]
- 35.Kochevar GJ, Brody JR, Kadkol SS, Murphy KM, et al. Identification of a functional mutation in pp32r1 (ANP32C) Hum Mutat. 2004;23:546–51. doi: 10.1002/humu.20030. [DOI] [PubMed] [Google Scholar]
- 36.Desiere F, Deutsch EW, King NL, Nesvizhskii AI, et al. The PeptideAtlas project. Nucleic Acids Res. 2006;34:D655–8. doi: 10.1093/nar/gkj040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kadkol SS, Brody JR, Epstein JI, Kuhajda FP, et al. Novel nuclear phosphoprotein pp32 is highly expressed in intermediate- and high-grade prostate cancer. Prostate. 1998;34:231–7. doi: 10.1002/(sici)1097-0045(19980215)34:3<231::aid-pros11>3.0.co;2-f. [DOI] [PubMed] [Google Scholar]
- 38.Khan MZ, Vaidya A, Meucci O. CXCL12-mediated regulation of ANP32A/Lanp, a component of the inhibitor of histone acetyl transferase (INHAT) complex, in cortical neurons. J Neuroimmune Pharmacol. 2011;6:163–70. doi: 10.1007/s11481-010-9228-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mutai H, Toyoshima Y, Sun W, Hattori N, et al. PAL31, a novel nuclear protein, expressed in the developing brain. Biochem Biophys Res Commun. 2000;274:427–33. doi: 10.1006/bbrc.2000.3133. [DOI] [PubMed] [Google Scholar]
- 40.Hostetter C, Licata LA, Witkiewicz A, Costantino CL, et al. Cytoplasmic accumulation of the RNA binding protein HuR is central to tamoxifen resistance in estrogen receptor positive breast cancer cells. Cancer Biol Ther. 2008;7:1496–506. doi: 10.4161/cbt.7.9.6490. [DOI] [PubMed] [Google Scholar]
- 41.Zhu BD, Li XL, Liu Y, Chang J, et al. Involvement of hepatopoietin Cn in the development of human hepatocellular carcinoma. Clin Exp Metastasis. 2010;27:571–80. doi: 10.1007/s10585-010-9346-8. [DOI] [PubMed] [Google Scholar]
- 42.Martinvalet D, Zhu P, Lieberman J. Granzyme A induces caspase-independent mitochondrial damage, a required first step for apoptosis. Immunity. 2005;22:355–70. doi: 10.1016/j.immuni.2005.02.004. [DOI] [PubMed] [Google Scholar]
- 43.Tsujio I, Zaidi T, Xu J, Kotula L, et al. Inhibitors of protein phosphatase-2A from human brain structures, immunocytological localization and activities towards dephosphorylation of the Alzheimer type hyperphosphorylated tau. FEBS Lett. 2005;579:363–72. doi: 10.1016/j.febslet.2004.11.097. [DOI] [PubMed] [Google Scholar]
- 44.Ulitzur N, Humbert M, Pfeffer SR. Mapmodulin: a possible modulator of the interaction of microtubule-associated proteins with microtubules. Proc Natl Acad Sci USA. 1997;94:5084–9. doi: 10.1073/pnas.94.10.5084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Callebaut C, Blanco J, Benkirane N, Krust B, et al. Identification of V3 loop-binding proteins as potential receptors implicated in the binding of HIV particles to CD4(+) cells. J Biol Chem. 1998;273:21988–97. doi: 10.1074/jbc.273.34.21988. [DOI] [PubMed] [Google Scholar]
- 46.Chang J, Liu Y, Zhang DD, Zhang DJ, et al. Hepatopoietin Cn suppresses apoptosis of human hepatocellular carcinoma cells by up-regulating myeloid cell leukemia-1. World J Gastroenterol. 2010;16:193–200. doi: 10.3748/wjg.v16.i2.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Opal P, Garcia JJ, Propst F, Matilla A, et al. Mapmodulin/leucine-rich acidic nuclear protein binds the light chain of microtubule-associated protein 1B and modulates neuritogenesis. J Biol Chem. 2003;278:34691–9. doi: 10.1074/jbc.M302785200. [DOI] [PubMed] [Google Scholar]
- 48.Kovacech B, Kontsekova E, Zilka N, Novak P, et al. A novel monoclonal antibody DC63 reveals that inhibitor 1 of protein phosphatase 2A is preferentially nuclearly localised in human brain. FEBS Lett. 2007;581:617–22. doi: 10.1016/j.febslet.2007.01.015. [DOI] [PubMed] [Google Scholar]
- 49.Schneider R, Bannister AJ, Weise C, Kouzarides T. Direct binding of INHAT to H3 tails disrupted by modifications. J Biol Chem. 2004;279:23859–62. doi: 10.1074/jbc.C400151200. [DOI] [PubMed] [Google Scholar]
- 50.Seo SB, Macfarlan T, McNamara P, Hong R, et al. Regulation of histone acetylation and transcription by nuclear protein pp32, a subunit of the INHAT complex. J Biol Chem. 2002;277:14005–10. doi: 10.1074/jbc.M112455200. [DOI] [PubMed] [Google Scholar]
- 51.Cvetanovic M, Rooney RJ, Garcia JJ, Toporovskaya N, et al. The role of LANP and ataxin 1 in E4F-mediated transcriptional repression. EMBO Rep. 2007;8:671–7. doi: 10.1038/sj.embor.7400983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hunter CS, Malik RE, Witzmann FA, Rhodes SJ. LHX3 interacts with inhibitor of histone acetyltransferase complex subunits LANP and TAF-1beta to modulate pituitary gene regulation. PLoS One. 2013;8:e68898. doi: 10.1371/journal.pone.0068898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kadota S, Nagata K. pp32, an INHAT component, is a transcription machinery recruiter for maximal induction of IFN-stimulated genes. J Cell Sci. 2011;124:892–9. doi: 10.1242/jcs.078253. [DOI] [PubMed] [Google Scholar]
- 54.Loven MA, Davis RE, Curtis CD, Muster N, et al. A novel estrogen receptor alpha-associated protein alters receptor-deoxyribonucleic acid interactions and represses receptor-mediated transcription. Mol Endocrinol. 2004;18:2649–59. doi: 10.1210/me.2003-0195. [DOI] [PubMed] [Google Scholar]
- 55.Munemasa Y, Suzuki T, Aizawa K, Miyamoto S, et al. Promoter region-specific histone incorporation by the novel histone chaperone ANP32B and DNA-binding factor KLF5. Mol Cell Biol. 2008;28:1171–81. doi: 10.1128/MCB.01396-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mao Z, Pan L, Wang W, Sun J, et al. Anp32e, a higher eukaryotic histone chaperone directs preferential recognition for H2A.Z. Cell Res. 2014;24:389–99. doi: 10.1038/cr.2014.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bargaje R, Alam MP, Patowary A, Sarkar M, et al. Proximity of H2A.Z. containing nucleosome to the transcription start site influences gene expression levels in the mammalian liver and brain. Nucleic Acids Res. 2012;40:8965–78. doi: 10.1093/nar/gks665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nekrasov M, Amrichova J, Parker BJ, Soboleva TA, et al. Histone H2A.Z. inheritance during the cell cycle and its impact on promoter organization and dynamics. Nat Struct Mol Biol. 2012;19:1076–83. doi: 10.1038/nsmb.2424. [DOI] [PubMed] [Google Scholar]
- 59.Coleman-Derr D, Zilberman D. Deposition of histone variant H2A.Z. within gene bodies regulates responsive genes. PLoS Genet. 2012;8:e1002988. doi: 10.1371/journal.pgen.1002988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Guruharsha KG, Obar RA, Mintseris J, Aishwarya K, et al. Drosophila protein interaction map (DPiM): a paradigm for metazoan protein complex interactions. Fly. 2012;6:246–53. doi: 10.4161/fly.22108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gamble MJ, Erdjument-Bromage H, Tempst P, Freedman LP, et al. The histone chaperone TAF-I/SET/INHAT is required for transcription in vitro of chromatin templates. Mol Cell Biol. 2005;25:797–807. doi: 10.1128/MCB.25.2.797-807.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kim HE, Jiang X, Du F, Wang X. PHAPI, CAS, and Hsp70 promote apoptosome formation by preventing Apaf-1 aggregation and enhancing nucleotide exchange on Apaf-1. Mol Cell. 2008;30:239–47. doi: 10.1016/j.molcel.2008.03.014. [DOI] [PubMed] [Google Scholar]
- 63.Hill MM, Adrain C, Duriez PJ, Creagh EM, et al. Analysis of the composition, assembly kinetics and activity of native Apaf-1 apoptosomes. EMBO J. 2004;23:2134–45. doi: 10.1038/sj.emboj.7600210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Fan Z, Zhang H, Zhang Q. Tumor suppressor pp32 represses cell growth through inhibition of transcription by blocking acetylation and phosphorylation of histone H3 and initiating its proapoptotic activity. Cell Death Differ. 2006;13:1485–94. doi: 10.1038/sj.cdd.4401825. [DOI] [PubMed] [Google Scholar]
- 65.Hoffarth S, Zitzer A, Wiewrodt R, Hahnel PS, et al. pp32/PHAPI determines the apoptosis response of non-small-cell lung cancer. Cell Death Differ. 2008;15:161–70. doi: 10.1038/sj.cdd.4402256. [DOI] [PubMed] [Google Scholar]
- 66.Schafer ZT, Parrish AB, Wright KM, Margolis SS, et al. Enhanced sensitivity to cytochrome c-induced apoptosis mediated by PHAPI in breast cancer cells. Cancer Res. 2006;66:2210–8. doi: 10.1158/0008-5472.CAN-05-3923. [DOI] [PubMed] [Google Scholar]
- 67.Beresford PJ, Zhang D, Oh DY, Fan Z, et al. Granzyme A activates an endoplasmic reticulum-associated caspase-independent nuclease to induce single-stranded DNA nicks. J Biol Chem. 2001;276:43285–93. doi: 10.1074/jbc.M108137200. [DOI] [PubMed] [Google Scholar]
- 68.Mazroui R, Di Marco S, Clair E, von Roretz C, et al. Caspase-mediated cleavage of HuR in the cytoplasm contributes to pp32/PHAP-I regulation of apoptosis. J Cell Biol. 2008;180:113–27. doi: 10.1083/jcb.200709030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Chakravarti D, Hong R. SET-ting the stage for life and death. Cell. 2003;112:589–91. doi: 10.1016/s0092-8674(03)00151-x. [DOI] [PubMed] [Google Scholar]
- 70.Shen LF, Cheng H, Tsai MC, Kuo HS, et al. PAL31 may play an important role as inflammatory modulator in the repair process of the spinal cord injury rat. J Neurochem. 2009;108:1187–97. doi: 10.1111/j.1471-4159.2008.05865.x. [DOI] [PubMed] [Google Scholar]
- 71.Shen SM, Yu Y, Wu YL, Cheng JK, et al. Downregulation of ANP32B, a novel substrate of caspase-3, enhances caspase-3 activation and apoptosis induction in myeloid leukemic cells. Carcinogenesis. 2010;31:419–26. doi: 10.1093/carcin/bgp320. [DOI] [PubMed] [Google Scholar]
- 72.Sun W, Kimura H, Hattori N, Tanaka S, et al. Proliferation related acidic leucine-rich protein PAL31 functions as a caspase-3 inhibitor. Biochem Biophys Res Commun. 2006;342:817–23. doi: 10.1016/j.bbrc.2006.02.026. [DOI] [PubMed] [Google Scholar]
- 73.Li M, Makkinje A, Damuni Z. Molecular identification of I1PP2A, a novel potent heat-stable inhibitor protein of protein phosphatase 2A. Biochemistry. 1996;35:6998–7002. doi: 10.1021/bi960581y. [DOI] [PubMed] [Google Scholar]
- 74.Katayose Y, Li M, Al-Murrani SW, Shenolikar S, et al. Protein phosphatase 2A inhibitors, I(1)(PP2A) and I(2)(PP2A), associate with and modify the substrate specificity of protein phosphatase 1. J Biol Chem. 2000;275:9209–14. doi: 10.1074/jbc.275.13.9209. [DOI] [PubMed] [Google Scholar]
- 75.Habrukowich C, Han DK, Le A, Rezaul K, et al. Sphingosine interaction with acidic leucine-rich nuclear phosphoprotein-32A (ANP32A) regulates PP2A activity and cyclooxygenase (COX)-2 expression in human endothelial cells. J Biol Chem. 2010;285:26825–31. doi: 10.1074/jbc.M110.147058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Yu LG, Packman LC, Weldon M, Hamlett J, et al. Protein phosphatase 2A, a negative regulator of the ERK signaling pathway, is activated by tyrosine phosphorylation of putative HLA class II-associated protein I (PHAPI)/pp32 in response to the antiproliferative lectin, jacalin. J Biol Chem. 2004;279:41377–83. doi: 10.1074/jbc.M400017200. [DOI] [PubMed] [Google Scholar]
- 77.Costanzo RV, Vila-Ortiz GJ, Perandones C, Carminatti H, et al. Anp32e/Cpd1 regulates protein phosphatase 2A activity at synapses during synaptogenesis. Eur J Neurosci. 2006;23:309–24. doi: 10.1111/j.1460-9568.2005.04555.x. [DOI] [PubMed] [Google Scholar]
- 78.Sanchez I, Pinol P, Corral-Juan M, Pandolfo M, et al. A novel function of Ataxin-1 in the modulation of PP2A activity is dysregulated in the spinocerebellar ataxia type 1. Hum Mol Genet. 2013;22:3425–37. doi: 10.1093/hmg/ddt197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Radrizzani M, Vila-Ortiz G, Cafferata EG, Di Tella MC, et al. Differential expression of CPD1 during postnatal development in the mouse cerebellum. Brain Res. 2001;907:162–74. doi: 10.1016/s0006-8993(01)02351-4. [DOI] [PubMed] [Google Scholar]
- 80.Bauer A, Neumann S, Karger A, Henning AK, et al. ANP32B is a nuclear target of henipavirus M proteins. PLoS One. 2014;9:e97233. doi: 10.1371/journal.pone.0097233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Bodem J, Schied T, Gabriel R, Rammling M, et al. Foamy virus nuclear RNA export is distinct from that of other retroviruses. J Virol. 2011;85:2333–41. doi: 10.1128/JVI.01518-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Higashino F, Aoyagi M, Takahashi A, Ishino M, et al. Adenovirus E4orf6 targets pp32/LANP to control the fate of ARE-containing mRNAs by perturbing the CRM1-dependent mechanism. J Cell Biol. 2005;170:15–20. doi: 10.1083/jcb.200405112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Itin C, Ulitzur N, Muhlbauer B, Pfeffer SR. Mapmodulin, cytoplasmic dynein, and microtubules enhance the transport of mannose 6-phosphate receptors from endosomes to the trans-golgi network. Mol Biol Cell. 1999;10:2191–7. doi: 10.1091/mbc.10.7.2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Cui CP, Zhang DJ, Shi BX, Du SJ, et al. Isolation and functional identification of a novel human hepatic growth factor: hepatopoietin Cn. Hepatology. 2008;47:986–95. doi: 10.1002/hep.22126. [DOI] [PubMed] [Google Scholar]
- 85.Cui CP, Wei P, Liu Y, Zhang DJ, et al. The protective role of Hepatopoietin Cn on liver injury induced by carbon tetrachloride in rats*. Hepatol Res. 2009;39:200–6. doi: 10.1111/j.1872-034X.2008.00447.x. [DOI] [PubMed] [Google Scholar]
- 86.Liu Y, Saiyan S, Men TY, Gao HY, et al. Hepatopoietin Cn reduces ethanol-induced hepatoxicity via sphingosine kinase 1 and sphingosine 1-phosphate receptors. J Pathol. 2013;230:365–76. doi: 10.1002/path.4194. [DOI] [PubMed] [Google Scholar]
- 87.Zippo A, De Robertis A, Bardelli M, Galvagni F, et al. Identification of Flk-1 target genes in vasculogenesis: Pim-1 is required for endothelial and mural cell differentiation in vitro. Blood. 2004;103:4536–44. doi: 10.1182/blood-2003-11-3827. [DOI] [PubMed] [Google Scholar]
- 88.Anisimov SV, Tarasov KV, Tweedie D, Stern MD, et al. SAGE identification of gene transcripts with profiles unique to pluripotent mouse R1 embryonic stem cells. Genomics. 2002;79:169–76. doi: 10.1006/geno.2002.6687. [DOI] [PubMed] [Google Scholar]
- 89.Brody JR, Kadkol SS, Hauer MC, Rajaii F, et al. pp32 reduction induces differentiation of TSU-Pr1 cells. Am J Pathol. 2004;164:273–83. doi: 10.1016/S0002-9440(10)63117-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kular RK, Cvetanovic M, Siferd S, Kini AR, et al. Neuronal differentiation is regulated by leucine-rich acidic nuclear protein (LANP), a member of the inhibitor of histone acetyltransferase complex. J Biol Chem. 2009;284:7783–92. doi: 10.1074/jbc.M806150200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Walensky LD, Coffey DS, Chen TH, Wu TC, et al. A novel M(r) 32,000 nuclear phosphoprotein is selectively expressed in cells competent for self-renewal. Cancer Res. 1993;53:4720–6. [PubMed] [Google Scholar]
- 92.Yu Y, Shen SM, Zhang FF, Wu ZX, et al. Acidic leucine-rich nuclear phosphoprotein 32 family member B (ANP32B) contributes to retinoic acid-induced differentiation of leukemic cells. Biochem Biophys Res Commun. 2012;423:721–5. doi: 10.1016/j.bbrc.2012.06.025. [DOI] [PubMed] [Google Scholar]
- 93.Mutz D, Weise C, Mechai N, Hofmann W, et al. Integrin alpha3beta1 interacts with I1PP2A/lanp and phosphatase PP1. J Neurosci Res. 2006;84:1759–70. doi: 10.1002/jnr.21078. [DOI] [PubMed] [Google Scholar]
- 94.Buszczak M, Paterno S, Lighthouse D, Bachman J, et al. The carnegie protein trap library: a versatile tool for Drosophila developmental studies. Genetics. 2007;175:1505–31. doi: 10.1534/genetics.106.065961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Mummery-Widmer JL, Yamazaki M, Stoeger T, Novatchkova M, et al. Genome-wide analysis of Notch signalling in Drosophila by transgenic RNAi. Nature. 2009;458:987–92. doi: 10.1038/nature07936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Rual JF, Ceron J, Koreth J, Hao T, et al. Toward improving Caenorhabditis elegans phenome mapping with an ORFeome-based RNAi library. Genome Res. 2004;14:2162–8. doi: 10.1101/gr.2505604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Opal P, Garcia JJ, McCall AE, Xu B, et al. Generation and characterization of LANP/pp32 null mice. Mol Cell Biol. 2004;24:3140–9. doi: 10.1128/MCB.24.8.3140-3149.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Reilly PT, Afzal S, Wakeham A, Haight J, et al. Generation and characterization of the Anp32e-deficient mouse. PLoS One. 2010;5:e13597. doi: 10.1371/journal.pone.0013597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Kular RK, Gogliotti RG, Opal P. Cpd-1 null mice display a subtle neurological phenotype. PLoS One. 2010;5:e12649. doi: 10.1371/journal.pone.0012649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Wong P, Leo VI, Low M, Mak TW, et al. Targeted ANP32E mutant mice do not demonstrate obvious movement defects. PLoS One. 2013;8:e63815. doi: 10.1371/journal.pone.0063815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Reilly PT, Afzal S, Gorrini C, Lui K, et al. Acidic nuclear phosphoprotein 32kDa (ANP32)B-deficient mouse reveals a hierarchy of ANP32 importance in mammalian development. Proc Natl Acad Sci USA. 2011;108:10243–8. doi: 10.1073/pnas.1106211108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Vazquez-Higuera JL, Mateo I, Sanchez-Juan P, Rodriguez-Rodriguez E, et al. Genetic variation in the tau protein phosphatase-2A pathway is not associated with Alzheimer's disease risk. BMC Res Notes. 2011;4:327. doi: 10.1186/1756-0500-4-327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Harada JN, Shevchenko A, Pallas DC, Berk AJ. Analysis of the adenovirus E1B-55K-anchored proteome reveals its link to ubiquitination machinery. J Virol. 2002;76:9194–206. doi: 10.1128/JVI.76.18.9194-9206.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Pegoraro G, Marcello A, Myers MP, Giacca M. Regulation of adeno-associated virus DNA replication by the cellular TAF-I/set complex. J Virol. 2006;80:6855–64. doi: 10.1128/JVI.00383-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Yan N, Cherepanov P, Daigle JE, Engelman A, et al. The SET complex acts as a barrier to autointegration of HIV-1. PLoS Pathog. 2009;5:e1000327. doi: 10.1371/journal.ppat.1000327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Adegbola O, Pasternack GR. Phosphorylated retinoblastoma protein complexes with pp32 and inhibits pp32-mediated apoptosis. J Biol Chem. 2005;280:15497–502. doi: 10.1074/jbc.M411382200. [DOI] [PubMed] [Google Scholar]
- 107.Stelzl U, Worm U, Lalowski M, Haenig C, et al. A human protein-protein interaction network: a resource for annotating the proteome. Cell. 2005;122:957–68. doi: 10.1016/j.cell.2005.08.029. [DOI] [PubMed] [Google Scholar]
- 108.Bai J, Brody JR, Kadkol SS, Pasternack GR. Tumor suppression and potentiation by manipulation of pp32 expression. Oncogene. 2001;20:2153–60. doi: 10.1038/sj.onc.1204294. [DOI] [PubMed] [Google Scholar]
- 109.Brody JR, Kadkol SS, Mahmoud MA, Rebel JM, et al. Identification of sequences required for inhibition of oncogene-mediated transformation by pp32. J Biol Chem. 1999;274:20053–5. doi: 10.1074/jbc.274.29.20053. [DOI] [PubMed] [Google Scholar]
- 110.Chen TH, Brody JR, Romantsev FE, Yu JG, et al. Structure of pp32, an acidic nuclear protein which inhibits oncogene-induced formation of transformed foci. Mol Biol Cell. 1996;7:2045–56. doi: 10.1091/mbc.7.12.2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Shi H, Hood KA, Hayes MT, Stubbs RS. Proteomic analysis of advanced colorectal cancer by laser capture microdissection and two-dimensional difference gel electrophoresis. J Proteomics. 2011;75:339–51. doi: 10.1016/j.jprot.2011.07.025. [DOI] [PubMed] [Google Scholar]
- 112.Ouellet V, Le Page C, Guyot MC, Lussier C, et al. SET complex in serous epithelial ovarian cancer. Int J Cancer. 2006;119:2119–26. doi: 10.1002/ijc.22054. [DOI] [PubMed] [Google Scholar]
- 113.Li C, Ruan HQ, Liu YS, Xu MJ, et al. Quantitative proteomics reveal up-regulated protein expression of the SET complex associated with hepatocellular carcinoma. J Proteome Res. 2012;11:871–85. doi: 10.1021/pr2006999. [DOI] [PubMed] [Google Scholar]
- 114.Tsukamoto Y, Uchida T, Karnan S, Noguchi T, et al. Genome-wide analysis of DNA copy number alterations and gene expression in gastric cancer. J Pathol. 2008;216:471–82. doi: 10.1002/path.2424. [DOI] [PubMed] [Google Scholar]
- 115.Walker BA, Leone PE, Chiecchio L, Dickens NJ, et al. A compendium of myeloma-associated chromosomal copy number abnormalities and their prognostic value. Blood. 2010;116:e56–65. doi: 10.1182/blood-2010-04-279596. [DOI] [PubMed] [Google Scholar]
- 116.Bjorck E, Ek S, Landgren O, Jerkeman M, et al. High expression of cyclin B1 predicts a favorable outcome in patients with follicular lymphoma. Blood. 2005;105:2908–15. doi: 10.1182/blood-2004-07-2721. [DOI] [PubMed] [Google Scholar]
- 117.Volinia S, Mascellani N, Marchesini J, Veronese A, et al. Genome wide identification of recessive cancer genes by combinatorial mutation analysis. PLoS One. 2008;3:e3380. doi: 10.1371/journal.pone.0003380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Nowak MA, Boerlijst MC, Cooke J, Smith JM. Evolution of genetic redundancy. Nature. 1997;388:167–71. doi: 10.1038/40618. [DOI] [PubMed] [Google Scholar]


