Abstract
Pharmaceutical researchers must evaluate vast numbers of protein sequences and formulate innovative strategies for identifying valid targets and discovering leads against them as a way of accelerating drug discovery. The ever increasing number and diversity of novel protein sequences identified by genomic sequencing projects and the success of worldwide structural genomics initiatives have spurred great interest and impetus in the development of methods for accurate, computationally empowered protein function prediction and active site identification. Previously, in the absence of direct experimental evidence, homology-based protein function annotation remained the gold-standard for in silico analysis and prediction of protein function. However, with the continued exponential expansion of sequence databases, this approach is not always applicable, as fewer query protein sequences demonstrate significant homology to protein gene products of known function. As a result, several non-homology based methods for protein function prediction that are based on sequence features, structure, evolution, biochemical and genetic knowledge have emerged. Herein, we review current bioinformatic programs and approaches for protein function prediction/annotation and discuss their integration into drug discovery initiatives. The development of such methods to annotate protein functional sites and their application to large protein functional families is crucial to successfully utilizing the vast amounts of genomic sequence information available to drug discovery and development processes.
Keywords: function prediction, protein annotation, structural comparison, drug discovery, structural genomics, bioinformatics
INTRODUCTION
The molecular details of protein function are of fundamental importance in designing specific and selective inhibitors or ligands to modulate protein activity as part of the process of developing small-molecule drug candidates. By identifying and characterizing protein structure, function, and active site information early in the discovery process, pharmaceutical researchers can study multiple targets using integrated biological, structural, and chemical methods simultaneously to more rapidly, cost effectively discover selective lead compounds.
Since the emergence of initiatives in the early 1990’s, genome sequencing projects have been exceedingly successful in producing an impressive array of fully sequenced genomes and vast amounts of protein sequence information. The availability of these genome sequences and their associated curated annotations has generated a wealth of information for new avenues of investigation, including drug discovery efforts. However, one of the fundamental challenges for the post-genomic era is to develop methods to incorporate the exponentially growing protein sequence information for thousands of functionally-uncharacterized proteins into large-scale drug discovery strategies. In the human genome and in the genomes of pathogenic agents there will be thousands of potential, unexplored drug targets, without the development of robust methods for the computational prediction of protein function or functional site identification.
Similarly, global protein structure initiatives underway today are providing new high-resolution protein structures [Burley and Bonanno 2003; Weigelt et al. 2008] that are representative of both current and novel pharmaceutical targets and endowing a strong foundation for drug discovery. Representative structures provide templates for comparative modeling and knowledge-based potentials for ab initio structure folding methods so that new structural models can be generated for protein sequences with high sequence similarity (>30%) to expand the known structural space relative to experimentally derived templates [Grant 2009]. The accuracy of these computational models can be detailed enough to provide valuable information in lead development for the structure and chemistry of binding sites identified in the protein structure. However, while structural genomics initiatives continue to produce new protein structures, the current focus is on characterizing the largest number of different folds to have the best possible sampling of structure space. As a consequence, a large number of structures and potential comparative models belong to proteins of unknown function, annotated merely as ‘hypothetical proteins’. This fact has greatly increased the interest in computational methods for functional inference. Herein, we review the research approaches and recently developed tools in the field of computational protein function prediction and discuss the ways these can be integrated into the process of drug discovery.
FUNCTIONAL ANNOTATION OF PROTEINS
Biological function can be highly contextual with different degrees of functional specificity and can be described at many levels ranging from biochemical, process, pathway, organ, or organism levels. To make protein function annotation available universally and for throughput computational processing, there is an essential need to describe the function of any gene product in any organism with a controlled and well-defined vocabulary. Several schemes for classifying protein function have been developed, most notably the Enzyme Commission (EC) Classification, which described enzymatic reactions using four-levels of indentified hierarchy. More recently, to address the need to describe complex protein functions beyond biochemical ones, the open-source Gene Ontology (GO) schema [2009a; Ashburner et al. 2000] has become the standard approach for a controlled vocabulary and a machine-readable ontology for functional annotation. GO comprises a framework of controlled vocabularies describing three aspects of gene product function: molecular function, biological process, and cellular location. This scheme represents the expanded view of protein function, whereby a protein is defined as an element in a network of its interactions. The GO Annotation (GOA) project aims to annotate all of the complete and incomplete proteomes that exist in the SWISS-PROT Protein Knowledgebase sequence database and its supplement, TrEMBL, using defined GO terms [Camon et al. 2003; Camon et al. 2004], as well as evidence codes reflecting how the annotation was obtained or determined. Through such standardization, protein function annotations may be computationally processed and a means for programs to output protein function predictions exists.
PROTEIN FUNCTION PREDICTION METHODS
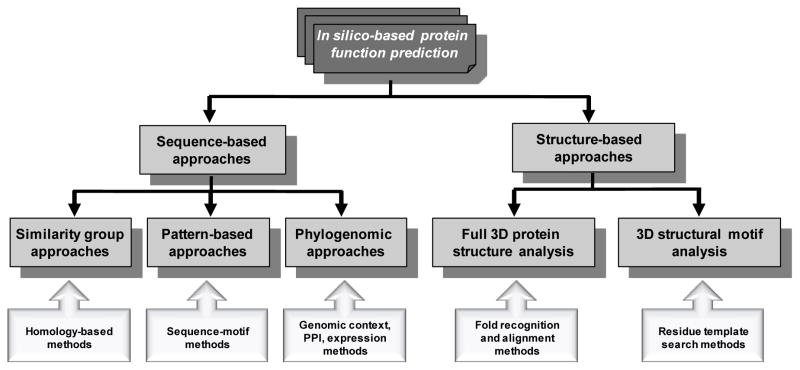
Protein function prediction methods mainly fall into sequence- and structure-based approaches. Herein, we outline the best described bioinformatic technologies for sequence- and structure-based protein function prediction. A schematic overview of these protein function prediction methods is shown in Figure 1. Table 1 lists a number of important databases and collections (sequence, structural, and ontology) that are extremely useful for approaches to protein function annotation.
Figure 1.
A schematic overview of protein function prediction methods. Various approaches to protein function prediction (grey boxes) are described in the text along with various methodologies employed in these approaches (white boxes). Both protein sequences and structures can provide information for family classification and functional inference.
Table 1.
Useful databases for protein function annotation.
Similarity-based approaches
Sequence homology-based methods
The most widely used approach for function prediction is homology transfer. For a given unannotated protein, this approach is based on searches for an annotated homolog (highly sequence-similar) and uses the experimentally verified function of the latter to infer the function of the former. The rationale for this approach is the assumption that two sequences with a high degree of similarity most likely evolved from a common ancestor and thus must have similar function. An important distinction in this context is between orthologous and parologous sequences, however. In general, function tends to be more conserved in orthologs than in paralogs [Theissen 2002], however, there are examples of more functionally divergent orthologs than corresponding paralogs [Punta and Ofran 2008]. Several data bases have been created to identify orthologous genes, for example COGs [Tatusov et al. 1997] and InParanoid [Remm et al. 2001], and also to catalog groups of orthologous genes in a hierarchical manner, such as in OrthoDB [Kriventseva et al. 2008].
Homology-based approaches require clustering of proteins into evolutionary families using sequence similarity-detection and/or alignment-search tools such as BLAST [Altschul et al. 1990] or tools based on multiple sequence alignments such as PSI-BLAST [Altschul et al. 1997], MAFFT [Katoh et al. 2002], and ProbCons [Do et al. 2005]. Several available resources provide pre-compiled sequence-based family assignments for proteins on a genomic scale, for example, PIRSF [Nikolskaya et al. 2006] in which a set of rules is applied to define primary and curated clusters (protein superfamilies) divided into common domain architectures. Table 2 lists a number of important resources, implementations, and tools for sequence searching and sequence alignment. Full-length similarity methods have been largely superseded by more sophisticated sequence pattern-based methods.
Table 2.
Useful sequence search and alignment programs and servers.
| Program/Server | URL |
|---|---|
| Align | http://bioinfo.mbb.yale.edu/Align/ |
| CLUSTALW2 | http://www.ebi.ac.uk/Tools/clustalw2/index.html |
| COMPASS | http://prodata.swmed.edu/compass/compass.php |
| DALIGN-TX | http://dialign-tx.gobics.de/ |
| FFAS03 | http://ffas.ljcrf.edu |
| HMMER3 | http://hmmer.janelia.org/ |
| MAFFT | http://align.bmr.kyushu-u.ac.jp/mafft/software/ |
| MultAlign | http://mendel.ethz.ch:8080/Server/MultAlign.html |
| MUSCLE | http://www.drive5.com/muscle/ |
| PFP | http://kiharalab.org/web/pfp.php |
| PSI-BLAST | http://blast.ncbi.nlm.nih.gov/Blast.cgi |
| PROBCONS | http://probcons.stanford.edu/ |
| RE-MuSiC | http://140.113.239.131/RE-MUSIC/ |
| SAM-T08 | http://compbio.soe.ucsc.edu/HMM-apps/HMM-applications.html |
| T-coffee | http://www.tcoffee.org/ |
| Espresso/3D-coffee | http://www.tcoffee.org/Projects_home_page/expresso_home_page.html |
Sequence motif and pattern-based methods
Two protein sequences that would not match in sequence searches may still have common sequence signatures that could reveal their functional relatedness. Finding one of these well-characterized motifs in a newly discovered sequence could offer some insights into its function. Several resources classify proteins on the basis of locally conserved sequence patterns (active site motifs), which often reflect the function(s) of the whole protein. The advantage of these profile methods are that they provide greater sensitivity compared to simple sequence-sequence comparisons because the profiles inherently contain both well-conserved residue and variable residue information for the motif/pattern across protein families. The most common type of profile is the hidden Markov model (HMM) and several methods exist for creating them from sequences of whole protein families. Profiling tools based on multiple sequence alignments are PSI-BLAST, HMMER [Bateman et al. 1999], and SAM [2009b], for example. The recent PFP [Hawkins et al. 2009] tool uses three rounds of PSI-BLAST and tolerant sequence similarity thresholds to include the annotations of remote homologues or homologous domains.
The PROSITE [Hulo et al. 2008] resource comprises manually selected biologically important motifs and has three types of signatures: patterns, rules, and profiles. While the two local signatures, patterns and rules, extend over just a few residues, profiles extend to the level of entire domains. PROSITE scans a query sequence against short, position-specific residue profiles that are characteristic of distinct protein families. Another widely used database is Pfam [Bateman et al. 2004; Sammut et al. 2008] comprising motifs that span over entire domains and focuses on the functional aspect of the domain definition. Pfam currently contains more than 10,000 family profiles and covers roughly 75% of UniProt [Wu et al. 2006] sequences, reflecting about half their amino acids. SMART [Letunic et al. 2009] utilizes the same approach and consists of a considerably smaller but completely manually curated set of families, and other well-known functional motif databases include BLOCKS [Henikoff et al. 2000], PRINTS [Attwood et al. 2003] and ELM [Puntervoll et al. 2003]. Other resources, for example CDD [Marchler-Bauer et al. 2009], use externally defined profiles to provide rapid assignments to sequence queries using a BLAST-like engine. The PANTHER [Mi et al. 2005] database distinguishes functional divergence within homologous protein families by defining groups of protein sequences into functional subfamilies. TIGRFAMs [Haft et al. 2003] uses models of full-length proteins and shorter regions at the levels of superfamilies, subfamilies and functional conservation in families of ‘equivalogs’ – sets of homologous proteins conserved with respect to function since their last common ancestor. Table 3 lists these and a number of additional important sequence similarity searching and sequence-based function assignment programs or servers.
Table 3.
Sequence similarity search and sequence-based function assignment methods.
| Database | URL |
|---|---|
| BLOCKS | http://blocks.fhcrc.org/ |
| CDD/CD-Search | http://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi |
| ELM | http://elm.eu.org/ |
| EVEREST | http://www.everest.cs.huji.ac.il/ |
| HHsearch/FHMMmer/HHpred | http://toolkit.lmb.uni-muenchen.de/sections/search |
| InterPro | http://www.ebi.ac.uk/interpro/ |
| PANTHER | http://www.pantherdb.org/ |
| Pfam | http://pfam.sanger.ac.uk/ |
| PIRSF | http://pir.georgetown.edu/pirwww/dbinfo/pirsf.shtml |
| PRINTS | http://www.bioinf.manchester.ac.uk/dbbrowser/PRINTS/index.php |
| ProDom | http://prodom.prabi.fr/prodom/current/html/home.php |
| PROSITE | http://www.expasy.org/prosite/ |
| SMART | http://smart.embl-heidelberg.de/ |
| SUPERFAMILY | http://supfam.cs.bris.ac.uk/SUPERFAMILY/ |
| TIGR | http://www.tigr.org/tdb/mdb/mdbcomplete.html |
| TIGRFAMS | http://www.tigr.org/TIGRFAMs/index.shtml |
Genomic context and phylogenomic-based methods
Genomic context-based prediction, also termed phylogenomic profiling, comprises protocols for predicting protein function based on the observation that proteins with similar inter-genomic profiles are thought to have evolved in tandem and share a common function [Eisen and Fraser 2003; Gomase and Tagore 2009; Sjolander 2004]. The active sites of proteins can be predicted using phylogenetic analysis and assessment of tree-determinant residues [del Sol et al. 2003]. The evolutionary trace (ET) method is well described and uses trees to rank residues by evolutionary importance and map these onto the structure to identify clusters and functional sites [Yao et al. 2003]. Phylomat [Graham et al. 2004] is a motif analysis tool for phylogenomics that scans predicted proteome sets for proteins containing highly conserved amino acid motifs or domains for analysis of the evolutionary history of these motifs/domains. RIO [Zmasek and Eddy 2002], SIFTER [Engelhardt et al. 2005; Zmasek and Eddy 2002], and OrthoStrapper [Storm and Sonnhammer 2002] algorithms search for protein orthologs by inferring gene duplications on a gene tree by comparing it to a species tree, thereby distinguishing orthologous from paralogous events.
Phylogenomics refers to the application of phylogenetic information to genomic studies and considers the evolutionary history of homologs in the prediction of function to increase the accuracy of annotation transfer. Annotation transfer is performed from the closest ortholog, rather than from the most similar sequence. Additionally, in the prokaryote genomes, the loci of functionally related proteins tend to be chromosomally co-localized. Several phylogenomic-based protein function prediction methods thus combine co-evolution and chromosomal proximity observations into function prediction algorithms, such as in Phydbac2 [Enault et al. 2004].
Expression-based prediction methods
Following the rationale of co-location, genes involved in similar cellular functions tend to be co-transcribed. Thus, from the analysis of gene expression arrays, unknown genes co-expressed with genes of known function may be functionally annotated through co-transcriptional associations. Unlike sequence motif-based approaches which center on molecular function, expression-based predictions can be useful for the annotation of the cellular aspect of protein function [Sleator and Walsh]. An extension of this concept is that most cellular processes are carried out by groups of physically interacting proteins and therefore, interacting proteins may have similar cellular functions. If so, protein-protein interaction (PPI) data may represent great potential for facilitating protein function annotation. Several PPI databases are available including the STRING [Jensen et al. 2009], DIP [Lehne and Schlitt 2009], GRID [Breitkreutz et al. 2003], MINT [Zanzoni et al. 2002], OPHID [Brown and Jurisica 2005], HAPPI [Chen et al. 2009b], HPRD [Keshava Prasad et al. 2009] and PIPs [McDowall et al. 2009] databases, as well as servers for mining and predicting protein-protein interactions, such as PPISearch [Chen et al. 2009a], PRISM [Keskin et al. 2008], and PPI Finder [He et al. 2009].
As sequence databases continue to expand, the homology-based transfer approach losses utility. As a direct consequence of exponential sequence expansion of unannotated novel sequence data from large-scale genomic sequencing projects, the number of clustered similar proteins for which no single annotated reference sequence exists is rapidly growing. Indeed, it has been estimated that <35% of all proteins can be annotated automatically with homology-based transfer, while >30% of all proteins cannot [Rost et al. 2003].
Structure-based approaches
As homologous proteins evolve, their three-dimensional structures often remain more conserved than their sequences. Proteins sharing similar function often have similar overall protein folds as a result of descent from a common ancestral protein. Additionally, similarities in protein structure can be more reliable than sequence similarities for grouping together distant homologs [Brenner et al. 1996; Rost 1997]. Many protein sequences that exhibit little or no sequence similarity [Gherardini and Helmer-Citterich 2008; Watson et al. 2005] still retain significant structural similarity due to evolutionary constraints, and thus structure is a powerful potential indicator of function [Bartlett et al. 2003; Todd et al. 2001]. Additionally, as function is critically related to structure, the structure of a protein directly suggests the mechanistic determinants of its function [Watson et al. 2005].
Methods for predicting protein function from three-dimensional structure can be classified according to the level of protein structure and specificity at which they perform their analysis, ranging from analysis of the overall protein fold to the identification of highly specific three-dimensional clusters (motifs and patterns) of functional residues. In the cases of the latter, existing methods can be classified generally into two groups: those that use comparative approaches to look for the presence of structural motifs associated with known biochemical function, and those methods consisting of analyzing the physicochemical characteristics of a protein surface to identify patches that have features (e.g. shape, electrostatic properties, etc.) characteristic of functional sites [Gherardini and Helmer-Citterich 2008]. Overall, structural comparison methods can identify even distant evolutionary relationships between proteins and make the identification of independently evolved sites possible. In general, it is suggested to use more than one method since different methods may find different valid matches [Kolodny et al. 2005].
Comparative structure-based approaches
Similar to sequence comparison methods, structural comparison methods can be classified as either global or local searches. Global comparison algorithms are mainly used in protein structure classification and to identify evolutionary links between distant homologues. They can also be used for function prediction, however one should caution that the relationship between fold and function is extremely complex and numerous examples are known of folds supporting a great variety of functions [42]. Proteins sharing similar functions often have similar global structure folds. Finding a fold match serves as the first approach in structure-based functional prediction. Computational tools that can scan the Protein Data Bank (PDB) [Berman et al. 2002; Berman et al. 2000] for global structural similarity or structure classification databases SCOP [Andreeva et al. 2004; Hubbard et al. 1999] and CATH [Greene et al. 2007; Orengo et al. 1997] given a query sequence using structural alignment methods include, CE [Guda et al. 2004; Shindyalov and Bourne 2001], DALI/FSSP [Holm et al. 2006; Holm and Sander 1993], FATCAT [Ye and Godzik 2004], PAST [Taubig et al. 2006], and FAST [Zhu and Weng 2005], Matras [Kawabata 2003], GRATH [Harrison et al. 2003], and FragBag [Budowski-Tal et al.], along with others. Table 4 lists the various programs, servers, and databases, described here and elsewhere for use in structural comparisons and structure-based protein function prediction. The recent Annolite [Marti-Renom et al. 2007] program was developed specifically as a structure-alignment based tool for protein function prediction given a query structure using annotation transfer from similar structures. An important assessment of several structure alignment servers has been performed and concluded that multiplicity in efforts for structure alignments, using multiple methods and algorithms, generates more accurate results than any single approach [Novotny et al. 2004].
Table 4.
Useful structural comparison and structure-based function assignment methods.
Arguably, the function of a protein depends more on the identity and location of a few residues comprising the active site than on the overall fold. In order to directly analyze and compare the residues effectively involved in protein function, local structural comparison methods have been developed. Local structural comparison refers to detecting similar three-dimensional arrangements or motifs (patterns) of a small set of residues, in the context of different global protein folds. As such, in local structure comparisons, one can either compare two entire protein structures looking for local similarities, or one can use a pre-defined structural template, which represents the spatial arrangement of a local functional residue motif, to screen a structure.
Residue template local search methods
Local structural comparison refers to identifying a similar three-dimensional arrangement of a small set of residues or spatial sub-regions within the protein structure, possibly in the context of completely different protein structures (folds). In applying such algorithms, one can either compare two entire protein structures in search for local similarities or use a pre-defined structural template to screen a structure. A template represents the spatial arrangement of the residues involved in some biochemical function and can be regarded as a three-dimensional extension of the linear sequence motif idea. Often the specific arrangement/conformation of the residues is crucial to the performance of the function and remains strongly conserved. The various methods available for local structure comparison differ essentially in two aspects: the way the protein structure is represented and the computational strategy that is used to search for similarities. Structure arrangements or patterns range from three-dimensional shapes dissociated from the amino acids to a string of characters representing amino acids and their physical environment. The level of detail in the representation ranges from very approximate, to elaborate schemes that take into account the presence of different chemical groups along the amino acid side chains.
Several databases and search algorithms have been developed including Catalytic Site Atlas (CSA) [Porter et al. 2004], pdbFun [Ausiello et al. 2005], PDBSiteScan [Ivanisenko et al. 2004; Ivanisenko et al. 2005], SuMo [Jambon et al. 2005], pvSOAR [Binkowski et al. 2004], SARIG [Amitai et al. 2004], FEATURE [Wei and Altman 1998; Wei and Altman 2003], JESS [Barker and Thornton 2003], RIGOR [Kleywegt 1999] and PatchFinder [Nimrod et al. 2005]. The well-known PINTS [Stark and Russell 2003] allows comparison of a protein structure against a database of patterns or a PDB format pattern against a pattern database. As other examples, Phunctioner [Pazos and Sternberg 2004] extracts conserved residues and uses GO annotation as a core element of its assignment and validation, while SuMo [Jambon et al. 2005] uses a stereo-chemical group representation for residues arranged in triangles and graph theory to superimpose them for data base searching. FEATURE [Wei and Altman 1998] defines subdomains in the protein structure as a series of concentric spheres or as a three-dimensional cubic lattice, while seqFEATURE [Wei and Altman 2003] enables the creation of structure patterns from known sequence patterns. To overcome the difficulty that structural templates must be derived manually, methods for the automatic discovery of structural motifs characterizing a protein family have been developed [Bandyopadhyay et al. 2006; Polacco and Babbitt 2006; Wangikar et al. 2003].
Approaches based on physiochemical characteristics and structure calculations
In general, these methods are based on the observation that the functional patches of a protein have unique physicochemical features which set them apart from the protein surface and exhibit function. The aim of these methods is usually to predict either the location of a ligand-binding or active site. Numerous algorithms employ the notion that functional sites are usually located in clefts on the protein surface [Laskowski et al. 1996]. This basic idea is used either directly to predict the location of functional sites, or as a first step to identify candidate residues before further scoring procedures are applied. Methods for identifying cavities, clefts, pockets and surfaces in a protein include PASS [Brady and Stouten 2000], CASTp [Dundas et al. 2006], LIGSITEcsc [Huang and Schroeder 2006], VICE [Tripathi and Kellogg], SURFNET/SURFNET-ConSurf [Glaser et al. 2006; Laskowski 1995], BSAlign [Aung and Tong 2008], CAVER [Petrek et al. 2006], pvSPAR [Binkowski et al. 2004] and PocketPicker [Weisel et al. 2007], among others. Besides being located in clefts, active site residues are reported as being close to the centroid of the structure, having a destabilizing effect on the structure, interacting with a high number of residues of the same protein, having perturbed pKa values and inducing clusters in the electrostatic potential around the protein. All of these observations have been used to develop methods aimed at the inference of active site location from structure. Electrostatic calculations have also been used to predict DNA binding sites, combined with the analysis of the curvature of the molecular surface and the detection of specific structural motifs.
Combining multiple methods and multiple data sources
Since protein function is a multifaceted concept, its comprehensive prediction and characterization requires data from many sources. Recognizing the power and thoroughness in predictive multiplicity, several recent methods or applications, also referred to as ‘metasevers’ in some cases, have been developed to integrate information pertaining to function such as structure, sequence information, physiochemical features, and protein interaction data and also to provide a consensus view that can better identify the most likely functional predictions. This approach has been used by ProtFun [Jensen et al. 2003] which combines numerous different sequence-based methods to generate GO term predictions. InterPro [Hunter et al. 2009] integrates together predictive models or ‘signatures’ representing protein domains, families and functional sites from multiple, diverse databases and predicts the occurrence of functional domains, repeats and important sites. Another resource, ProFunc [Laskowski et al. 2005] uses varied sequence and structure-based methods, combined with the identification of active and binding sites and integrates them with interaction data and knowledge of genomic sequences to yield a comprehensive prediction summary of the most likely GO term-represented functions. ProKnow [Pal and Eisenberg 2005] considers structural features that are associated with specific functions in addition to sequence motifs, fold similarity, templates, and interaction data. Joined Assembly of Function Annotations [Friedberg et al. 2006], or JAFA, is a metaserver that surveys several function-prediction servers with a query sequence, returning a summary of predicted GO terms.
APPLICATION OF PROTEIN FUNCTION IN DRUG DISCOVERY INITIATIVES
One of the most important challenges for computational biology and drug discovery efforts has been to predict the function of previously uncharacterized proteins for which there is no known experimental three-dimensional structure [Betz et al. 2002; Chanda and Caldwell 2003; Ofran et al. 2005]. New protein sequences with the potential to be involved in important genetic and parasitic disease are being discovered at rapid pace and spurring the need to predict their structures in order to understand their function and investigate their potential as therapeutic targets. Despite significant, emerging advances in our understanding of the relationship between structure and function in this era of structural genomics, the identification of new drug targets and the successful development of potent and specific therapeutic drugs is still a slow and resource expensive process. However, with the remarkable research efforts by many into the development of accurate methods for protein function prediction, homology molecular modeling, virtual screening of chemical libraries by docking experiments, and new avenues for drug design and optimization, certain future success in drug discovery initiatives that implement these methods will lead to promising drug candidates more rapidly.
Having a comprehensive, functionally annotated sequence database cataloging of all the members of a given gene family in the human genome allows one to integrate a very different perspective into the drug-discovery process. The availability of the complete sets of related genes for a given target and a representative subset of protein structures allows one to build three-dimensional models for the entire protein family and to map the interactions of a given substrate/inhibitor and specific residues in the target even when detailed structural data is not available. Specificity predictions at the target level can also be applied to target selection. Knowing the sequences and structures of the target and those proteins that are physiochemically and/or structurally related to the target in the inhibitor-binding site can be essential in the evaluation of the target. Having this information in hand for all targets within a biological pathway would allow one to use the specificity prediction to guide target selection to the most appropriate and specific target in the pathway.
Optimization of a lead compound to a clinical development candidate involves iterative cycles to improve targeting and specificity. There is a significant need to understand fully how small molecules interact with other targets, outside of the target of therapeutic value. Knowing the detailed receptor-ligand interactions within a binding pocket and having a catalog of all the motifs, patterns, and functional residue subsets found in all sequences allows the prediction of the relatedness of various targets for broad arrays of inhibitors. With such model sets, a comprehensive evaluation of inhibitor modifications at certain positions can reveal interactions with increased specificity for the target over others. Furthermore, having the complete set of residues that provide key inhibitor interactions across a catalog of functionally related sequences enables the prediction of inhibitor specificity significantly earlier in the discovery process. Genomic data can then be used to focus inhibitor design towards areas of the novel target molecule that might facilitate engineering of inhibitor/ligand specificity.
We have witnessed significant changes to the pharmaceutical drug discovery process over the past few years. Protein function prediction can have an impact on target selection and on various stages of lead compound design [Betz et al. 2002] [Chanda and Caldwell 2003]. One change that is impacting strongly on the development process in its early stages is the shift from focusing on optimizing the chemistry of a small number of targets to focusing on the validation process of a few thousand druggable targets [Ofran et al. 2005]. With only a small fraction of proteins used as drug targets, the goal is to explore the space of druggable proteins and reveal the relationship between them and the chemistry space, and to use computational tools to address this challenge.
It is becoming apparent that an important goal in pharmacological targeting will be to identify and select disease modifying nodes or functional hubs that control or link several desired targets within a biochemical network. Functional knowledge about each potential target will be crucial to fully evaluating complex biochemical networks, their integrated protein functions, and their respective role in disease. With the poor coverage of all potential targets by direct experimental structures and experimental functional annotations, functional information from computational or in silico function prediction methods might be the most readily available means to provide critical functional information to inform such network-based targeting approaches. Protein function prediction will have an impact on adding novel gene products to the considered target space and on narrowing down the number of potential targets, as well, if applied in the early stages of discovery.
SUMMARY
To understand the function of whole proteomes in nature is one of the grand goals of molecular biology. The implications of this knowledge will have a tremendous impact on understanding the biochemical details of molecular processes, the molecular basis of diseases and the non-trivial relationships between structure and function. As the available protein structural and experimental data grow and computational methods to exploit this information improve, as is the strong case for emerging methods for protein function prediction, the derived knowledge will have positive consequences for future successful drug discovery initiatives. Here we have reviewed current approaches and programs for protein function prediction and discussed their integration into drug discovery initiatives. The development of such methods to annotate protein functional sites and their application to large protein functional families will be crucial to successfully utilizing the vast amounts of genomic sequence information available to drug discovery and development processes.
References
- The Universal Protein Resource (UniProt) Nucleic Acids Res. 2010;38(Database issue):D142–8. doi: 10.1093/nar/gkp846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Gene Ontology’s Reference Genome Project: a unified framework for functional annotation across species. PLoS Comput Biol. 2009a;5(7):e1000431. doi: 10.1371/journal.pcbi.1000431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Universal Protein Resource (UniProt) 2009. Nucleic Acids Res. 2009b;37(Database issue):D169–74. doi: 10.1093/nar/gkn664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215(3):403–10. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25(17):3389–402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amitai G, Shemesh A, Sitbon E, Shklar M, Netanely D, Venger I, Pietrokovski S. Network analysis of protein structures identifies functional residues. J Mol Biol. 2004;344(4):1135–46. doi: 10.1016/j.jmb.2004.10.055. [DOI] [PubMed] [Google Scholar]
- Andreeva A, Howorth D, Brenner SE, Hubbard TJ, Chothia C, Murzin AG. SCOP database in 2004: refinements integrate structure and sequence family data. Nucleic Acids Res. 2004;32(Database issue):D226–9. doi: 10.1093/nar/gkh039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25(1):25–9. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Attwood TK, Bradley P, Flower DR, Gaulton A, Maudling N, Mitchell AL, Moulton G, Nordle A, Paine K, Taylor P, et al. PRINTS and its automatic supplement, prePRINTS. Nucleic Acids Res. 2003;31(1):400–2. doi: 10.1093/nar/gkg030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aung Z, Tong JC. BSAlign: a rapid graph-based algorithm for detecting ligand-binding sites in protein structures. Genome Inform. 2008;21:65–76. [PubMed] [Google Scholar]
- Ausiello G, Zanzoni A, Peluso D, Via A, Helmer-Citterich M. pdbFun: mass selection and fast comparison of annotated PDB residues. Nucleic Acids Res. 2005;33(Web Server issue):W133–7. doi: 10.1093/nar/gki499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandyopadhyay D, Huan J, Liu J, Prins J, Snoeyink J, Wang W, Tropsha A. Structure-based function inference using protein family-specific fingerprints. Protein Sci. 2006;15(6):1537–43. doi: 10.1110/ps.062189906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker JA, Thornton JM. An algorithm for constraint-based structural template matching: application to 3D templates with statistical analysis. Bioinformatics. 2003;19(13):1644–9. doi: 10.1093/bioinformatics/btg226. [DOI] [PubMed] [Google Scholar]
- Bartlett GJ, Todd AE, Thornton JM. Inferring protein function from structure. Methods Biochem Anal. 2003;44:387–407. [PubMed] [Google Scholar]
- Bateman A, Birney E, Durbin R, Eddy SR, Finn RD, Sonnhammer EL. Pfam 3.1: 1313 multiple alignments and profile HMMs match the majority of proteins. Nucleic Acids Res. 1999;27(1):260–2. doi: 10.1093/nar/27.1.260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bateman A, Coin L, Durbin R, Finn RD, Hollich V, Griffiths-Jones S, Khanna A, Marshall M, Moxon S, Sonnhammer EL, et al. The Pfam protein families database. Nucleic Acids Res. 2004;32(Database issue):D138–41. doi: 10.1093/nar/gkh121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berman HM, Battistuz T, Bhat TN, Bluhm WF, Bourne PE, Burkhardt K, Feng Z, Gilliland GL, Iype L, Jain S, et al. The Protein Data Bank. Acta Crystallogr D Biol Crystallogr. 2002;58(Pt 61):899–907. doi: 10.1107/s0907444902003451. [DOI] [PubMed] [Google Scholar]
- Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. The Protein Data Bank. Nucleic Acids Res. 2000;28(1):235–42. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betz SF, Baxter SM, Fetrow JS. Function first: a powerful approach to post-genomic drug discovery. Drug Discov Today. 2002;7(16):865–71. doi: 10.1016/s1359-6446(02)02398-x. [DOI] [PubMed] [Google Scholar]
- Binkowski TA, Freeman P, Liang J. pvSOAR: detecting similar surface patterns of pocket and void surfaces of amino acid residues on proteins. Nucleic Acids Res. 2004;32(Web Server issue):W555–8. doi: 10.1093/nar/gkh390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brady GP, Jr, Stouten PF. Fast prediction and visualization of protein binding pockets with PASS. J Comput Aided Mol Des. 2000;14(4):383–401. doi: 10.1023/a:1008124202956. [DOI] [PubMed] [Google Scholar]
- Breitkreutz BJ, Stark C, Tyers M. The GRID: the General Repository for Interaction Datasets. Genome Biol. 2003;4(3):R23. doi: 10.1186/gb-2003-4-3-r23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner SE, Chothia C, Hubbard TJ, Murzin AG. Understanding protein structure: using scop for fold interpretation. Methods Enzymol. 1996;266:635–43. doi: 10.1016/s0076-6879(96)66039-x. [DOI] [PubMed] [Google Scholar]
- Brown KR, Jurisica I. Online predicted human interaction database. Bioinformatics. 2005;21(9):2076–82. doi: 10.1093/bioinformatics/bti273. [DOI] [PubMed] [Google Scholar]
- Budowski-Tal I, Nov Y, Kolodny R. FragBag, an accurate representation of protein structure, retrieves structural neighbors from the entire PDB quickly and accurately. Proc Natl Acad Sci U S A. 107(8):3481–6. doi: 10.1073/pnas.0914097107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burley SK, Bonanno JB. Structural genomics. Methods Biochem Anal. 2003;44:591–612. [PubMed] [Google Scholar]
- Camon E, Barrell D, Brooksbank C, Magrane M, Apweiler R. The Gene Ontology Annotation (GOA) Project--Application of GO in SWISS-PROT, TrEMBL and InterPro. Comp Funct Genomics. 2003;4(1):71–4. doi: 10.1002/cfg.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camon E, Barrell D, Lee V, Dimmer E, Apweiler R. The Gene Ontology Annotation (GOA) Database--an integrated resource of GO annotations to the UniProt Knowledgebase. In Silico Biol. 2004;4(1):5–6. [PubMed] [Google Scholar]
- Chanda SK, Caldwell JS. Fulfilling the promise: drug discovery in the post-genomic era. Drug Discov Today. 2003;8(4):168–74. doi: 10.1016/s1359-6446(02)02595-3. [DOI] [PubMed] [Google Scholar]
- Chen CC, Lin CY, Lo YS, Yang JM. PPISearch: a web server for searching homologous protein-protein interactions across multiple species. Nucleic Acids Res. 2009a;37(Web Server issue):W369–75. doi: 10.1093/nar/gkp309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JY, Mamidipalli S, Huan T. HAPPI: an online database of comprehensive human annotated and predicted protein interactions. BMC Genomics. 2009b;10(Suppl 1):S16. doi: 10.1186/1471-2164-10-S1-S16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Sol A, Pazos F, Valencia A. Automatic methods for predicting functionally important residues. J Mol Biol. 2003;326(4):1289–302. doi: 10.1016/s0022-2836(02)01451-1. [DOI] [PubMed] [Google Scholar]
- Do CB, Mahabhashyam MS, Brudno M, Batzoglou S. ProbCons: Probabilistic consistency-based multiple sequence alignment. Genome Res. 2005;15(2):330–40. doi: 10.1101/gr.2821705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dundas J, Ouyang Z, Tseng J, Binkowski A, Turpaz Y, Liang J. CASTp: computed atlas of surface topography of proteins with structural and topographical mapping of functionally annotated residues. Nucleic Acids Res. 2006;34(Web Server issue):W116–8. doi: 10.1093/nar/gkl282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisen JA, Fraser CM. Phylogenomics: intersection of evolution and genomics. Science. 2003;300(5626):1706–7. doi: 10.1126/science.1086292. [DOI] [PubMed] [Google Scholar]
- Enault F, Suhre K, Poirot O, Abergel C, Claverie JM. Phydbac2: improved inference of gene function using interactive phylogenomic profiling and chromosomal location analysis. Nucleic Acids Res. 2004;32(Web Server issue):W336–9. doi: 10.1093/nar/gkh365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelhardt BE, Jordan MI, Muratore KE, Brenner SE. Protein molecular function prediction by Bayesian phylogenomics. PLoS Comput Biol. 2005;1(5):e45. doi: 10.1371/journal.pcbi.0010045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedberg I, Harder T, Godzik A. JAFA: a protein function annotation meta-server. Nucleic Acids Res. 2006;34(Web Server issue):W379–81. doi: 10.1093/nar/gkl045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gherardini PF, Helmer-Citterich M. Structure-based function prediction: approaches and applications. Brief Funct Genomic Proteomic. 2008;7(4):291–302. doi: 10.1093/bfgp/eln030. [DOI] [PubMed] [Google Scholar]
- Glaser F, Morris RJ, Najmanovich RJ, Laskowski RA, Thornton JM. A method for localizing ligand binding pockets in protein structures. Proteins. 2006;62(2):479–88. doi: 10.1002/prot.20769. [DOI] [PubMed] [Google Scholar]
- Gomase VS, Tagore S. Phylogenomics: evolution and genomics intersection. Int J Bioinform Res Appl. 2009;5(5):548–63. doi: 10.1504/IJBRA.2009.028682. [DOI] [PubMed] [Google Scholar]
- Graham WV, Tcheng DK, Shirk AL, Attene-Ramos MS, Welge ME, Gaskins HR. Phylomat: an automated protein motif analysis tool for phylogenomics. J Proteome Res. 2004;3(6):1289–91. doi: 10.1021/pr0499040. [DOI] [PubMed] [Google Scholar]
- Grant MA. Protein structure prediction in structure-based ligand design and virtual screening. Comb Chem High Throughput Screen. 2009;12(10):940–60. doi: 10.2174/138620709789824718. [DOI] [PubMed] [Google Scholar]
- Greene LH, Lewis TE, Addou S, Cuff A, Dallman T, Dibley M, Redfern O, Pearl F, Nambudiry R, Reid A, et al. The CATH domain structure database: new protocols and classification levels give a more comprehensive resource for exploring evolution. Nucleic Acids Res. 2007;35(Database issue):D291–7. doi: 10.1093/nar/gkl959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guda C, Lu S, Scheeff ED, Bourne PE, Shindyalov IN. CE-MC: a multiple protein structure alignment server. Nucleic Acids Res. 2004;32(Web Server issue):W100–3. doi: 10.1093/nar/gkh464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haft DH, Selengut JD, White O. The TIGRFAMs database of protein families. Nucleic Acids Res. 2003;31(1):371–3. doi: 10.1093/nar/gkg128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison A, Pearl F, Sillitoe I, Slidel T, Mott R, Thornton J, Orengo C. Recognizing the fold of a protein structure. Bioinformatics. 2003;19(14):1748–59. doi: 10.1093/bioinformatics/btg240. [DOI] [PubMed] [Google Scholar]
- Hawkins T, Chitale M, Luban S, Kihara D. PFP: Automated prediction of gene ontology functional annotations with confidence scores using protein sequence data. Proteins. 2009;74(3):566–82. doi: 10.1002/prot.22172. [DOI] [PubMed] [Google Scholar]
- He M, Wang Y, Li W. PPI finder: a mining tool for human protein-protein interactions. PLoS One. 2009;4(2):e4554. doi: 10.1371/journal.pone.0004554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff JG, Pietrokovski S, McCallum CM, Henikoff S. Blocks-based methods for detecting protein homology. Electrophoresis. 2000;21(9):1700–6. doi: 10.1002/(SICI)1522-2683(20000501)21:9<1700::AID-ELPS1700>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- Holm L, Kaariainen S, Wilton C, Plewczynski D. Using Dali for structural comparison of proteins. Curr Protoc Bioinformatics. 2006;Chapter 5(Unit 5):5. doi: 10.1002/0471250953.bi0505s14. [DOI] [PubMed] [Google Scholar]
- Holm L, Sander C. Protein structure comparison by alignment of distance matrices. J Mol Biol. 1993;233(1):123–38. doi: 10.1006/jmbi.1993.1489. [DOI] [PubMed] [Google Scholar]
- Huang B, Schroeder M. LIGSITEcsc: predicting ligand binding sites using the Connolly surface and degree of conservation. BMC Struct Biol. 2006;6:19. doi: 10.1186/1472-6807-6-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubbard TJ, Ailey B, Brenner SE, Murzin AG, Chothia C. SCOP: a Structural Classification of Proteins database. Nucleic Acids Res. 1999;27(1):254–6. doi: 10.1093/nar/27.1.254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hulo N, Bairoch A, Bulliard V, Cerutti L, Cuche BA, de Castro E, Lachaize C, Langendijk-Genevaux PS, Sigrist CJ. The 20 years of PROSITE. Nucleic Acids Res. 2008;36(Database issue):D245–9. doi: 10.1093/nar/gkm977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter S, Apweiler R, Attwood TK, Bairoch A, Bateman A, Binns D, Bork P, Das U, Daugherty L, Duquenne L, et al. InterPro: the integrative protein signature database. Nucleic Acids Res. 2009;37(Database issue):D211–5. doi: 10.1093/nar/gkn785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanisenko VA, Pintus SS, Grigorovich DA, Kolchanov NA. PDBSiteScan: a program for searching for active, binding and posttranslational modification sites in the 3D structures of proteins. Nucleic Acids Res. 2004;32(Web Server issue):W549–54. doi: 10.1093/nar/gkh439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanisenko VA, Pintus SS, Grigorovich DA, Kolchanov NA. PDBSite: a database of the 3D structure of protein functional sites. Nucleic Acids Res. 2005;33(Database issue):D183–7. doi: 10.1093/nar/gki105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jambon M, Andrieu O, Combet C, Deleage G, Delfaud F, Geourjon C. The SuMo server: 3D search for protein functional sites. Bioinformatics. 2005;21(20):3929–30. doi: 10.1093/bioinformatics/bti645. [DOI] [PubMed] [Google Scholar]
- Jensen LJ, Kuhn M, Stark M, Chaffron S, Creevey C, Muller J, Doerks T, Julien P, Roth A, Simonovic M, et al. STRING 8--a global view on proteins and their functional interactions in 630 organisms. Nucleic Acids Res. 2009;37(Database issue):D412–6. doi: 10.1093/nar/gkn760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen LJ, Ussery DW, Brunak S. Functionality of system components: conservation of protein function in protein feature space. Genome Res. 2003;13(11):2444–9. doi: 10.1101/gr.1190803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Misawa K, Kuma K, Miyata T. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002;30(14):3059–66. doi: 10.1093/nar/gkf436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawabata T. MATRAS: A program for protein 3D structure comparison. Nucleic Acids Res. 2003;31(13):3367–9. doi: 10.1093/nar/gkg581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keshava Prasad TS, Goel R, Kandasamy K, Keerthikumar S, Kumar S, Mathivanan S, Telikicherla D, Raju R, Shafreen B, Venugopal A, et al. Human Protein Reference Database--2009 update. Nucleic Acids Res. 2009;37(Database issue):D767–72. doi: 10.1093/nar/gkn892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keskin O, Nussinov R, Gursoy A. PRISM: protein-protein interaction prediction by structural matching. Methods Mol Biol. 2008;484:505–21. doi: 10.1007/978-1-59745-398-1_30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleywegt GJ. Recognition of spatial motifs in protein structures. J Mol Biol. 1999;285(4):1887–97. doi: 10.1006/jmbi.1998.2393. [DOI] [PubMed] [Google Scholar]
- Kolodny R, Koehl P, Levitt M. Comprehensive evaluation of protein structure alignment methods: scoring by geometric measures. J Mol Biol. 2005;346(4):1173–88. doi: 10.1016/j.jmb.2004.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriventseva EV, Rahman N, Espinosa O, Zdobnov EM. OrthoDB: the hierarchical catalog of eukaryotic orthologs. Nucleic Acids Res. 2008;36(Database issue):D271–5. doi: 10.1093/nar/gkm845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laskowski RA. SURFNET: a program for visualizing molecular surfaces, cavities, and intermolecular interactions. J Mol Graph. 1995;13(5):323–30. 307–8. doi: 10.1016/0263-7855(95)00073-9. [DOI] [PubMed] [Google Scholar]
- Laskowski RA, Luscombe NM, Swindells MB, Thornton JM. Protein clefts in molecular recognition and function. Protein Sci. 1996;5(12):2438–52. doi: 10.1002/pro.5560051206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laskowski RA, Watson JD, Thornton JM. ProFunc: a server for predicting protein function from 3D structure. Nucleic Acids Res. 2005;33(Web Server issue):W89–93. doi: 10.1093/nar/gki414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehne B, Schlitt T. Protein-protein interaction databases: keeping up with growing interactomes. Hum Genomics. 2009;3(3):291–7. doi: 10.1186/1479-7364-3-3-291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letunic I, Doerks T, Bork P. SMART 6: recent updates and new developments. Nucleic Acids Res. 2009;37(Database issue):D229–32. doi: 10.1093/nar/gkn808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchler-Bauer A, Anderson JB, Chitsaz F, Derbyshire MK, DeWeese-Scott C, Fong JH, Geer LY, Geer RC, Gonzales NR, Gwadz M, et al. CDD: specific functional annotation with the Conserved Domain Database. Nucleic Acids Res. 2009;37(Database issue):D205–10. doi: 10.1093/nar/gkn845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marti-Renom MA, Rossi A, Al-Shahrour F, Davis FP, Pieper U, Dopazo J, Sali A. The AnnoLite and AnnoLyze programs for comparative annotation of protein structures. BMC Bioinformatics. 2007;8(Suppl 4):S4. doi: 10.1186/1471-2105-8-S4-S4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDowall MD, Scott MS, Barton GJ. PIPs: human protein-protein interaction prediction database. Nucleic Acids Res. 2009;37(Database issue):D651–6. doi: 10.1093/nar/gkn870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi H, Lazareva-Ulitsky B, Loo R, Kejariwal A, Vandergriff J, Rabkin S, Guo N, Muruganujan A, Doremieux O, Campbell MJ, et al. The PANTHER database of protein families, subfamilies, functions and pathways. Nucleic Acids Res. 2005;33(Database issue):D284–8. doi: 10.1093/nar/gki078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikolskaya AN, Arighi CN, Huang H, Barker WC, Wu CH. PIRSF family classification system for protein functional and evolutionary analysis. Evol Bioinform Online. 2006;2:197–209. [PMC free article] [PubMed] [Google Scholar]
- Nimrod G, Glaser F, Steinberg D, Ben-Tal N, Pupko T. In silico identification of functional regions in proteins. Bioinformatics. 2005;21(Suppl 1):i328–37. doi: 10.1093/bioinformatics/bti1023. [DOI] [PubMed] [Google Scholar]
- Novotny M, Madsen D, Kleywegt GJ. Evaluation of protein fold comparison servers. Proteins. 2004;54(2):260–70. doi: 10.1002/prot.10553. [DOI] [PubMed] [Google Scholar]
- Ofran Y, Punta M, Schneider R, Rost B. Beyond annotation transfer by homology: novel protein-function prediction methods to assist drug discovery. Drug Discov Today. 2005;10(21):1475–82. doi: 10.1016/S1359-6446(05)03621-4. [DOI] [PubMed] [Google Scholar]
- Orengo CA, Michie AD, Jones S, Jones DT, Swindells MB, Thornton JM. CATH--a hierarchic classification of protein domain structures. Structure. 1997;5(8):1093–108. doi: 10.1016/s0969-2126(97)00260-8. [DOI] [PubMed] [Google Scholar]
- Pal D, Eisenberg D. Inference of protein function from protein structure. Structure. 2005;13(1):121–30. doi: 10.1016/j.str.2004.10.015. [DOI] [PubMed] [Google Scholar]
- Pazos F, Sternberg MJ. Automated prediction of protein function and detection of functional sites from structure. Proc Natl Acad Sci U S A. 2004;101(41):14754–9. doi: 10.1073/pnas.0404569101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrek M, Otyepka M, Banas P, Kosinova P, Koca J, Damborsky J. CAVER: a new tool to explore routes from protein clefts, pockets and cavities. BMC Bioinformatics. 2006;7:316. doi: 10.1186/1471-2105-7-316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polacco BJ, Babbitt PC. Automated discovery of 3D motifs for protein function annotation. Bioinformatics. 2006;22(6):723–30. doi: 10.1093/bioinformatics/btk038. [DOI] [PubMed] [Google Scholar]
- Porter CT, Bartlett GJ, Thornton JM. The Catalytic Site Atlas: a resource of catalytic sites and residues identified in enzymes using structural data. Nucleic Acids Res. 2004;32(Database issue):D129–33. doi: 10.1093/nar/gkh028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Punta M, Ofran Y. The rough guide to in silico function prediction, or how to use sequence and structure information to predict protein function. PLoS Comput Biol. 2008;4(10):e1000160. doi: 10.1371/journal.pcbi.1000160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puntervoll P, Linding R, Gemund C, Chabanis-Davidson S, Mattingsdal M, Cameron S, Martin DM, Ausiello G, Brannetti B, Costantini A, et al. ELM server: A new resource for investigating short functional sites in modular eukaryotic proteins. Nucleic Acids Res. 2003;31(13):3625–30. doi: 10.1093/nar/gkg545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remm M, Storm CE, Sonnhammer EL. Automatic clustering of orthologs and in-paralogs from pairwise species comparisons. J Mol Biol. 2001;314(5):1041–52. doi: 10.1006/jmbi.2000.5197. [DOI] [PubMed] [Google Scholar]
- Rost B. Protein structures sustain evolutionary drift. Fold Des. 1997;2(3):S19–24. doi: 10.1016/s1359-0278(97)00059-x. [DOI] [PubMed] [Google Scholar]
- Rost B, Liu J, Nair R, Wrzeszczynski KO, Ofran Y. Automatic prediction of protein function. Cell Mol Life Sci. 2003;60(12):2637–50. doi: 10.1007/s00018-003-3114-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sammut SJ, Finn RD, Bateman A. Pfam 10 years on: 10,000 families and still growing. Brief Bioinform. 2008;9(3):210–9. doi: 10.1093/bib/bbn010. [DOI] [PubMed] [Google Scholar]
- Shindyalov IN, Bourne PE. A database and tools for 3-D protein structure comparison and alignment using the Combinatorial Extension (CE) algorithm. Nucleic Acids Res. 2001;29(1):228–9. doi: 10.1093/nar/29.1.228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sjolander K. Phylogenomic inference of protein molecular function: advances and challenges. Bioinformatics. 2004;20(2):170–9. doi: 10.1093/bioinformatics/bth021. [DOI] [PubMed] [Google Scholar]
- Sleator RD, Walsh P. An overview of in silico protein function prediction. Arch Microbiol. 192(3):151–5. doi: 10.1007/s00203-010-0549-9. [DOI] [PubMed] [Google Scholar]
- Stark A, Russell RB. Annotation in three dimensions. PINTS: Patterns in Non-homologous Tertiary Structures. Nucleic Acids Res. 2003;31(13):3341–4. doi: 10.1093/nar/gkg506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storm CE, Sonnhammer EL. Automated ortholog inference from phylogenetic trees and calculation of orthology reliability. Bioinformatics. 2002;18(1):92–9. doi: 10.1093/bioinformatics/18.1.92. [DOI] [PubMed] [Google Scholar]
- Tatusov RL, Koonin EV, Lipman DJ. A genomic perspective on protein families. Science. 1997;278(5338):631–7. doi: 10.1126/science.278.5338.631. [DOI] [PubMed] [Google Scholar]
- Taubig H, Buchner A, Griebsch J. PAST: fast structure-based searching in the PDB. Nucleic Acids Res. 2006;34(Web Server issue):W20–3. doi: 10.1093/nar/gkl273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theissen G. Secret life of genes. Nature. 2002;415(6873):741. doi: 10.1038/415741a. [DOI] [PubMed] [Google Scholar]
- Todd AE, Orengo CA, Thornton JM. Evolution of function in protein superfamilies, from a structural perspective. J Mol Biol. 2001;307(4):1113–43. doi: 10.1006/jmbi.2001.4513. [DOI] [PubMed] [Google Scholar]
- Tripathi A, Kellogg GE. A novel and efficient tool for locating and characterizing protein cavities and binding sites. Proteins. 78(4):825–42. doi: 10.1002/prot.22608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wangikar PP, Tendulkar AV, Ramya S, Mali DN, Sarawagi S. Functional sites in protein families uncovered via an objective and automated graph theoretic approach. J Mol Biol. 2003;326(3):955–78. doi: 10.1016/s0022-2836(02)01384-0. [DOI] [PubMed] [Google Scholar]
- Watson JD, Laskowski RA, Thornton JM. Predicting protein function from sequence and structural data. Curr Opin Struct Biol. 2005;15(3):275–84. doi: 10.1016/j.sbi.2005.04.003. [DOI] [PubMed] [Google Scholar]
- Wei L, Altman RB. Recognizing protein binding sites using statistical descriptions of their 3D environments. Pac Symp Biocomput. 1998:497–508. [PubMed] [Google Scholar]
- Wei L, Altman RB. Recognizing complex, asymmetric functional sites in protein structures using a Bayesian scoring function. J Bioinform Comput Biol. 2003;1(1):119–38. doi: 10.1142/s0219720003000150. [DOI] [PubMed] [Google Scholar]
- Weigelt J, McBroom-Cerajewski LD, Schapira M, Zhao Y, Arrowsmith CH. Structural genomics and drug discovery: all in the family. Curr Opin Chem Biol. 2008;12(1):32–9. doi: 10.1016/j.cbpa.2008.01.045. [DOI] [PubMed] [Google Scholar]
- Weisel M, Proschak E, Schneider G. PocketPicker: analysis of ligand binding-sites with shape descriptors. Chem Cent J. 2007;1:7. doi: 10.1186/1752-153X-1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu CH, Apweiler R, Bairoch A, Natale DA, Barker WC, Boeckmann B, Ferro S, Gasteiger E, Huang H, Lopez R, et al. The Universal Protein Resource (UniProt): an expanding universe of protein information. Nucleic Acids Res. 2006;34(Database issue):D187–91. doi: 10.1093/nar/gkj161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao H, Kristensen DM, Mihalek I, Sowa ME, Shaw C, Kimmel M, Kavraki L, Lichtarge O. An accurate, sensitive, and scalable method to identify functional sites in protein structures. J Mol Biol. 2003;326(1):255–61. doi: 10.1016/s0022-2836(02)01336-0. [DOI] [PubMed] [Google Scholar]
- Ye Y, Godzik A. FATCAT: a web server for flexible structure comparison and structure similarity searching. Nucleic Acids Res. 2004;32(Web Server issue):W582–5. doi: 10.1093/nar/gkh430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanzoni A, Montecchi-Palazzi L, Quondam M, Ausiello G, Helmer-Citterich M, Cesareni G. MINT: a Molecular INTeraction database. FEBS Lett. 2002;513(1):135–40. doi: 10.1016/s0014-5793(01)03293-8. [DOI] [PubMed] [Google Scholar]
- Zhu J, Weng Z. FAST: a novel protein structure alignment algorithm. Proteins. 2005;58(3):618–27. doi: 10.1002/prot.20331. [DOI] [PubMed] [Google Scholar]
- Zmasek CM, Eddy SR. RIO: analyzing proteomes by automated phylogenomics using resampled inference of orthologs. BMC Bioinformatics. 2002;3:14. doi: 10.1186/1471-2105-3-14. [DOI] [PMC free article] [PubMed] [Google Scholar]