Abstract
Oncogenic Ras proteins rely on a series of key effector pathways to drive the physiological changes that lead to tumorigenic growth. Of these effector pathways, the RalGEF pathway, which activates the two Ras-related GTPases RalA and RalB, remains the most poorly understood. This review will focus on key developments in our understanding of Ral biology, and will speculate on how aberrant activation of the multiple diverse Ral effector proteins might collectively contribute to oncogenic transformation and other aspects of tumor progression.
Keywords: RalA, RalB, Ras, RalGEF, RalBP1, Exocyst
Introduction
The discovery, in the mid 1980s, that tumors often contained transforming alleles of one of the three Ras genes (HRAS, KRAS, NRAS) led a number of groups to seek to identify novel small GTPases related to Ras. Using an oligonucleotide probe corresponding to a highly conserved region of the Ras proteins to screen a simian cDNA library, Pierre Chardin and Armand Tavitian discovered an open reading frame that shared a high degree of homology with the three Ras genes and named it Ral (Ras-like) [1]. Using the simian cDNA sequence to probe a human pheochromocytoma cDNA library, they then isolated the human Ral cDNA and an additional cDNA with which it shared a high degree of homology (approximately 85% identity at the amino acid level) [2]. These two human Ral proteins were named RalA and RalB. Rals are activated by a family of at least six guanine nucleotide exchange factors (RalGEFs) that include RalGDS, RGL, RGL2/Rlf, RGL3, RalGPS1 and RalGPS2 and inactivated by two GTPase activating proteins, RalGAP1 and RalGAP2 [3,4]. Interest in Ral signaling spiked in the mid 1990s when several groups discovered through yeast 2-hybrid screens that four of the RalGEFs (RalGDS, RGL, RGL2 and RGL3) interact directly with the effector binding region of GTP-bound Ras, and thus potentially mediate some of the pro-tumorigenic signaling in cancers harboring mutations in one of the three Ras genes [5–9]. This interest was tempered in subsequent years however as experiments, performed mostly in murine fibroblasts, showed that Ral played a distinct, but mostly complementary, role in oncogenic Ras-driven cellular transformation, and that most of the heavy lifting was done by the Raf-driven MAP kinase pathway [10,11]. The discovery of tumor-associated mutations in the MAP kinase pathway itself, notably of BRaf, as well as mutations affecting the phosphatidylinositol 3-kinase (PI3K) pathway, another identified Ras effector pathway, lent further support for the primacy of the MAP kinase and PI3K pathways in mediating the oncogenic effects of mutant Ras [12,13].
The attitude towards Ral started to change with a series of studies showing that unlike rodent cells, RalGEFs and Ral play a dominant role in Ras-mediated transformation of several different immortalized human cell lines [14–16]. Using effector domain mutants of Ras that differentially engage downstream effector pathways, it was shown that expression of the E37G mutant of activated RasG12V, which binds to RalGEFs, but not Raf or PI3K, was able to promote the anchorage-independent growth of immortalized human fibroblasts, epithelial cells and astrocytes. The T35S and Y40C mutants, on the other hand, which exclusively engage the MAP Kinase and PI3K pathways, respectively, were unable to promote anchorage-independent growth [14]. These data were in contrast to similar experiments performed in immortalized murine cells, in which the RasG12V,T35S mutant was consistently able to induce more colonies than either of the other two effector domain mutants. Additionally, a membrane targeted RalGEF (Rlf fused to the C-terminal CAAX region of Ras) was able to transform human cells, and inhibition of Ral, using a dominant negative RalAS28N mutant, blocked both RasG12V and RasG12V,E37G-mediated transformation [14]. Subsequent studies showed that RalA, but not RalB, played a dominant role in this RalGEF-mediated transformation, as knockdown of RalA, but not RalB, inhibited RasG12V or RalGEF-induced anchorage independent growth [15,16], and expression of a constitutively active RalA, but not RalB, could weakly transform immortalized human epithelial cells [15]. RalB, on the other hand, was shown to be more important for cell survival and motility, as transformed cell lines, but not non-tumorigenic lines, underwent apoptotic cell death upon knockdown of RalB [16], and knockdown of RalB inhibited transwell migration of cultured human bladder cancer cells [17]. These results were consistent with the finding that while knockdown of RalA, but not RalB, blocked tumor initiation in a panel of pancreatic cancer cell lines, knockdown of RalB inhibited invasion and metastasis of these lines in a tail-vein injection assay [18].
Given the popularity and relative success of targeting the MAP kinase and PI3K pathway to treat human tumors, these studies sparked a flurry of interest in Ral signaling in a variety of different tumor types in an effort to determine whether this pathway might also present a viable therapeutic opportunity for cancers, especially those in which the prevalence of Ras mutations was high [3]. As a result of these efforts, Ral has been shown to play an important role in a number of different tumor types, including mutant Ras-associated tumors such as pancreatic [15,18,19], colorectal [20], lung [21], melanoma [22,23] and squamous cell carcinoma [24] as well as a number of tumors not typically associated with Ras mutations, such as bladder [25,26] and prostate [27]. While Ral mutations are not commonly found in these tumors, higher than normal Ral activity has been shown to result from a number of different mechanisms, including the loss of microRNA mediated inhibition [28], increased GEF expression [29], and decreased GAP activity [30]. The level of Ral activity, as measured by comparison to a transcriptional signature of Ral-dependent genes, correlates positively with a number of poor prognostic factors, including disease stage, progression to muscle invasion and survival in bladder cancer, and seminal vesicle invasion and androgen-independent progression in prostate cancer [31]. Collectively, these studies highlight the need to better understand how RalA and RalB signal to their downstream effectors to promote tumorigenic processes and to continue to lift the RalGEF-Ral pathway out of the shadow of the more intensely-studied and better-understood MAP kinase and PI3K pathways.
RalA and RalB exhibit both distinct and redundant roles in tumorigenesis
Despite their overall amino acid identity of 80% [2], including 100% identity in the effector-binding region, human RalA and RalB exhibit distinct functions in both normal and pathological biology [17,18,32,33]. This somewhat contrasts the recently reported RalA and RalB knockout mice [34]. The RalA and RalB knockouts did show phenotypic differences, notably in their effects on neuronal development, but while knockout of both RalA and RalB inhibited Ras-driven tumorigenesis, expression of either RalA or RalB was sufficient to support tumorigenesis in an inducible model of non-small cell lung carcinoma [34]. Studies performed on human cells on the other hand have consistently shown that inhibition of RalA alone, but not RalB, is sufficient to inhibit tumor initiation [15,16,18,35], while inhibition of RalB inhibits invasion and metastasis [16–18,36]. While the reasons for this apparent discrepancy in tumorigenic function remain unclear, two models seem most likely. First, that knockout of both proteins [34], or of an upstream GEF [37], is required to inhibit tumorigenesis in mouse models may indicate a functional overlap between rodent RalA and RalB that has been lost in the human proteins. Another possibility is that the same level of functional divergence exists between rodent and human RalA and RalB, but that MAP kinase signaling or some other tumorigenic pathway is sufficient to overcome the loss of RalA in the transformation of rodent, but not human cells. Further experiments will be required to distinguish between these or other possibilities, but until then, we should take caution when interpreting the function of Ral proteins in any one system and be aware of the limitations of both human cell culture and mouse models of tumorigenesis.
The differences in biological function between RalA and RalB have been mapped to the C-terminal 30 amino acid hypervariable region of the two proteins, as engineering a RalB–RalA fusion protein with the C-terminal 30 amino acids of RalA fused to the N-terminal portion of RalB imparts RalA tumorigenic potential [15] and polarized delivery of membrane proteins [32] on RalB. The importance of this C-terminal region highlights the importance of subcellular localization in determining Ral effector usage and function, as this region determines the membranes in the cell with which Ral associates. Both RalA and RalB are geranylgeranylated and associate with membranes through insertion of this 20-carbon lipid into the membrane in addition to interactions between the negatively charged phospholipids and the Lysine and Arginine-rich C-terminal polybasic region [38]. The hypervariable domain is the region of greatest divergence between RalA and RalB, and includes a number of sites of post-translational modification that have been shown to influence Ral subcellular localization and function. RalA is phosphorylated on Serine 194 by the kinase Aurora A [39], and this phosphorylation has been shown to relocalize RalA to internal membranes [40], including the mitochondria, where RalA promotes mitochondrial fission during mitosis [41]. RalA is also phosphorylated on Serine 183, potentially by the kinase PKA [25], and both Serines can be dephosphorylated by the Aβ subunit of the phosphatase PP2A [35]. Mutational analysis of these Serines has indicated that they play a critical role not only in RalA localization but also in tumorigenic function [35,40]. RalB is phosphorylated on Serine 198 by the kinase PKC, and like phosphorylation of RalA, this modification promotes internalization of the protein and is important for its tumorigenic function [25]. How phosphorylation of RalA and RalB changes their subcellular localization has not been formally tested, but it has been proposed for other small GTPases that the addition of a negatively charged phosphate group sufficiently neutralizes the charge of the polybasic region, causing its disassociation from the plasma membrane and allowing it to relocalize to other membranes [42]. In addition to phosphorylation, RalA, but not RalB, has recently been shown to be modified by nondegradative ubiquitination and this modification, like phosphorylation, affects RalA subcellular localization, promoting its association with lipid rafts [43]. Ultimately, these differences in post-translational modification promote distinct subcellular distributions for RalA and RalB, allowing these two proteins to use the same set of effectors to promote different biological outcomes.
Ral effectors
The increasingly important role that RalA and RalB play in tumorigenesis underscores the importance of understanding how and when Ral signals to its effector proteins, how this signaling impacts basic cellular processes and which of these effector interactions are the most important for tumorigenesis. The first effector to be identified for Ral was RalBP1 (also known as RLIP76 or RIP1) [44–46]. A clear understanding of RalBP1 function has been hampered by the sheer number of activities and interacting partners associated with it since its discovery. One notable feature initially identified for RalBP1 was GAP activity towards the Rho family GTPases Rac1 and Cdc42, thus giving RalBP1 the potential to impact actin dynamics and the formation of filopodia and membrane ruffling. In addition to this enzymatic activity, RalBP1 also regulates a number of different endocytic pathways, including receptor-mediated endocytosis of proteins such as the epidermal growth factor receptor, insulin receptor and transferrin receptor, through its interactions with POB1, Reps1 and AP2 [47–50]. RalBP1 has been identified as a non-ABC transporter of either unconjugated or glutathione-conjugated electrophilic compounds and thus may be involved in the resistance to small molecule chemotherapeutic drugs such as doxorubicin [51]. This activity requires two ATP binding sites present in the RalBP1 N-terminus. RalBP1 has been shown to promote invadopodia formation downstream of RalB in a GAP-independent fashion [52]. Finally, RalBP1 has been shown to induce mitochondrial fission by promoting the Cyclin B/CDK1-dependent phosphorylation of the GTPase Drp1 and facilitating the recruitment of Drp1 to mitochondrial membranes [41].
With all of these disparate roles, teasing apart the contribution of the Ral–RalBP1 interaction to tumorigenesis has been difficult. Xenograft studies have shown RalBP1 to play an important role in a number of tumor types, including pancreatic [53], prostate [54], colorectal [55], bladder [54] and glioblastoma [56], among others. Hopefully, by identifying new mutants that separate the different functions of RalBP1, in the future we will be able to determine the extent to which each of these functions contributes to tumorigenesis.
Aside from RalBP1, Sec5 and Exo84 are the most well-understood Ral effector proteins. These two proteins are subunits of the octomeric exocyst complex, which is involved in the targeted delivery of secretory vesicles to specific membrane compartments, such as the basolateral surface of polarized epithelial cells [57,58]. Ral regulates exocyst function at the level of localization by directly targeting Sec5 and Exo84 to specific membrane domains [59]. This regulation ultimately contributes to several complex cellular processes including polarized membrane trafficking [32], cytokinesis [60], tight junction formation [61], and tumor cell invasion [59]. Interestingly, exclusive interactions between RalB and Sec5 and between RalB and Exo84 have been shown to play additional roles, independent of the full octameric exocyst complex. RalB–Sec5 recruits and activates the atypical IκB kinase TBK1 to promote cell survival [36]. Normally activated as a part of the innate immune response to certain viruses, TBK1 activation can be coopted by tumor cells to avoid oncogene-induced apoptosis and its activation by RalB may explain, at least in part, the critical role RalB, but not RalA, plays in protection against apoptosis [33,36]. Interestingly, a separate RalB–Exo84 complex promotes autophagosome assembly under starvation conditions through the recruitment of ULK1 and Beclin1-VPS34 [62]. As autophagy is known to be important for Ras-driven tumorigenesis [63,64], this function of RalB may also contribute to its tumorigenic function.
Certain functions of RalA and RalB, such as their roles in radioresistance [65], are mediated through their interactions with neither RalBP1 nor the Exocyst components, suggesting that additional effectors of Ral contribute to their tumorigenic properties. Several such effectors have been identified, as well as proteins that bind to Ral in a GTP-independent fashion, but their roles in mediating Ral signaling have not been extensively explored. These proteins include the actin-crosslinking protein filamin [66], the transcription factor ZONAB [67] and the important signaling molecules phospholipase D [68] and phospholipase C delta 1 [69], among others. Further characterization of these interactions will be necessary to determine whether and how they contribute to the pro-tumorigenic functions of RalA and RalB. This characterization of Ral-effector interactions will be greatly aided by additional structural studies of RalA and RalB with and without bound effector proteins [70,71]. These studies should identify key residues and ultimately allow for the generation of effector domain mutants such as the ones that have been so useful for the dissection of the Ras effector pathways [72,73].
Conclusions
While the complexities of how RalA and RalB coordinate their interactions with multiple diverse effector proteins and how the spatio-temporal regulation of these interactions contributes to the process of tumorigenesis remain unclear, a number of unifying themes have begun to emerge that may guide the research moving forward. One commonality is that both RalBP1 and the exocyst act as meeting sites on which a diverse set of protein complexes assemble. A large protein with a number of protein-protein interaction domains, RalBP1 has been shown to act as a bridge between CyclinB/Cdk1 and at least two of its substrates, Drp1 [41] and Epsin [74], and also promotes the assembly of the endocytic machinery [48,50]. Similarly, Exo84 brings together the core components for autophagosome assembly [62] while Exo84 and Sec5 promote assembly of the multi-subunit exocyst [57,58]. RalA and RalB, through their interactions with these effectors, are able to couple the assembly of these complexes with their delivery to specific subcellular locations using the very specific and regulatable targeting information encoded in their hypervariable C-termini, such as the delivery of phosphorylated Drp1 to mitochondria, or the delivery of secretory vesicles to the basolateral surface of epithelial cells. This dual functionality of Ral proteins and their effectors, the ability to assemble large protein machines and then deliver them to specific locations, is ideal for the type of complex, membrane-altering events that require a relatively large level of spatially-targeted biophysical engineering and with which Ral proteins are often involved. These events include targeted exocytosis, receptor-mediated endocytosis, the formation of filopodia and membrane ruffles, the formation of autophagosomes, the fission of mitochondria and the division of the cell through cytokinesis.
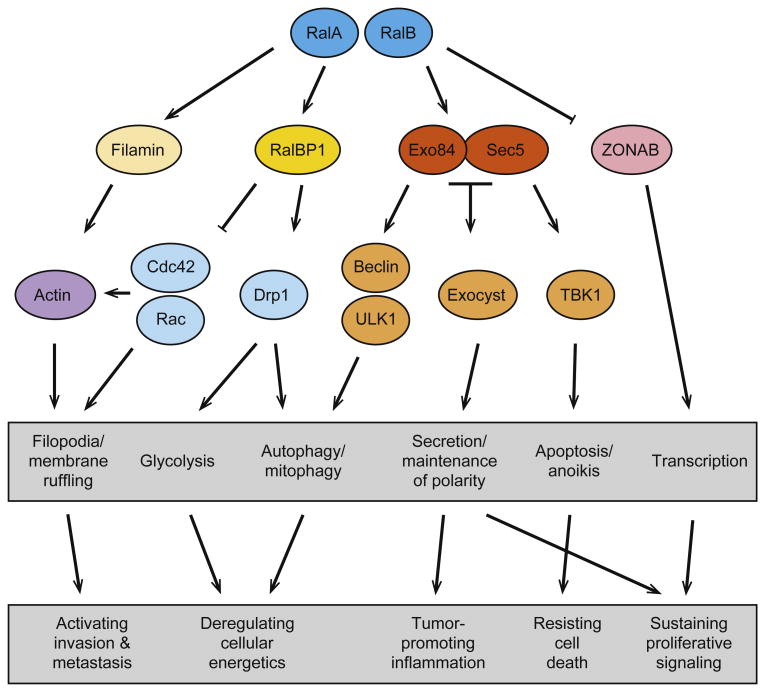
As these and other themes come into better focus, we will hopefully begin to form a more complete picture of how activation of RalA and RalB collectively contribute to the tumorigenic phenotype. Hanahan and Weinberg have famously identified 10 hallmarks of human cancer [75], dysregulated processes required for tumors to progress. To understand Ral function in human tumors it will be useful to put the aberrant activation of Ral in tumor cells, and the aberrant engagement of these effector pathways, in the context of these processes (Fig. 1). Ultimately, with better tools, including new tumor models and better separation-of-function mutants, we will able to parse out the contributions each of the diverse Ral pathways to tumorigenesis and hopefully use this information to develop novel treatments for Ras-driven cancers.
Fig. 1.
Potential role of RalA, RalB and their effectors on tumorigenesis. When activated, RalA and RalB engage a diverse set of effector proteins that are involved in a variety of biological processes. Disruption, or inappropriate activation of these processes has the potential to impact a number of the hallmarks of cancer proposed by Hanahan and Weinberg [62].
References
- 1.Chardin P, Tavitian A, et al. The ral gene: a new ras related gene isolated by the use of a synthetic probe. EMBO J. 1986;5:2203–2208. doi: 10.1002/j.1460-2075.1986.tb04485.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chardin P, Tavitian A, et al. Coding sequences of human ralA and ralB cDNAs. Nucl Acids Res. 1989;17:4380. doi: 10.1093/nar/17.11.4380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Neel NF, Martin TD, Stratford JK, Zand TP, Reiner DJ, Der CJ, et al. The RalGEF-Ral Effector signaling network: the road less traveled for anti-Ras drug discovery. Genet Cancer. 2011;2:275–287. doi: 10.1177/1947601911407329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shirakawa R, Fukai S, Kawato M, Higashi T, Kondo H, Ikeda T, et al. Tuberous sclerosis tumor suppressor complex-like complexes act as GTPase-activating proteins for Ral GTPases. J Biol Chem. 2009;284:21580–21588. doi: 10.1074/jbc.M109.012112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shao H, Andres DA, et al. A novel RalGEF-like protein, RGL3, as a candidate effector for rit and Ras. J Biol Chem. 2000;275:26914–26924. doi: 10.1074/jbc.M002241200. [DOI] [PubMed] [Google Scholar]
- 6.Peterson SN, Trabalzini L, Brtva TR, Fischer T, Altschuler DL, Martelli P, et al. Identification of a novel RalGDS-related protein as a candidate effector for Ras and Rap1. J Biol Chem. 1996;271:29903–29908. doi: 10.1074/jbc.271.47.29903. [DOI] [PubMed] [Google Scholar]
- 7.Spaargaren M, Bischoff JR, et al. Identification of the guanine nucleotide dissociation stimulator for Ral as a putative effector molecule of R-ras, H-ras, K-ras, and Rap. Proc Nat Acad Sci U S A. 1994;91:12609–12613. doi: 10.1073/pnas.91.26.12609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kikuchi A, Demo SD, Ye ZH, Chen YW, Williams LT, et al. ralGDS family members interact with the effector loop of ras p21. Mol Cell Biol. 1994;14:7483–7491. doi: 10.1128/mcb.14.11.7483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hofer F, Fields S, Schneider C, Martin GS, et al. Activated Ras interacts with the Ral guanine nucleotide dissociation stimulator. Proc Nat Acad Sci U S A. 1994;91:11089–11093. doi: 10.1073/pnas.91.23.11089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Urano T, Emkey R, Feig LA, et al. Ral-GTPases mediate a distinct downstream signaling pathway from Ras that facilitates cellular transformation. EMBO J. 1996;15:810–816. [PMC free article] [PubMed] [Google Scholar]
- 11.White MA, Vale T, Camonis JH, Schaefer E, Wigler MH, et al. A role for the Ral guanine nucleotide dissociation stimulator in mediating Ras-induced transformation. J Biol Chem. 1996;271:16439–16442. doi: 10.1074/jbc.271.28.16439. [DOI] [PubMed] [Google Scholar]
- 12.Osborne JK, Zaganjor E, Cobb MH, et al. Signal control through Raf: in sickness and in health. Cell Res. 2012;22:14–22. doi: 10.1038/cr.2011.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chalhoub N, Baker SJ, et al. PTEN and the PI3-kinase pathway in cancer. Annu Rev Pathol. 2009;4:127–150. doi: 10.1146/annurev.pathol.4.110807.092311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hamad NM, Elconin JH, Karnoub AE, Bai W, Rich JN, Abraham RT, et al. Distinct requirements for Ras oncogenesis in human versus mouse cells. Genes Dev. 2002;16:2045–2057. doi: 10.1101/gad.993902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lim KH, Baines AT, Fiordalisi JJ, Shipitsin M, Feig LA, Cox AD, et al. Activation of RalA is critical for Ras-induced tumorigenesis of human cells. Cancer Cell. 2005;7:533–545. doi: 10.1016/j.ccr.2005.04.030. [DOI] [PubMed] [Google Scholar]
- 16.Chien Y, White MA, et al. RAL GTPases are linchpin modulators of human tumour-cell proliferation and survival. EMBO Rep. 2003;4:800–806. doi: 10.1038/sj.embor.embor899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Oxford G, Owens CR, Titus BJ, Foreman TL, Herlevsen MC, Smith SC, et al. RalA and RalB: antagonistic relatives in cancer cell migration. Cancer Res. 2005;65:7111–7120. doi: 10.1158/0008-5472.CAN-04-1957. [DOI] [PubMed] [Google Scholar]
- 18.Lim KH, O'Hayer K, Adam SJ, Kendall SD, Campbell PM, Der CJ, et al. Divergent roles for RalA and RalB in malignant growth of human pancreatic carcinoma cells. Curr Biol. 2006;16:2385–2394. doi: 10.1016/j.cub.2006.10.023. [DOI] [PubMed] [Google Scholar]
- 19.Vigil D, Martin TD, Williams F, Yeh JJ, Campbell SL, Der CJ, et al. Aberrant overexpression of the Rgl2 Ral small GTPase-specific guanine nucleotide exchange factor promotes pancreatic cancer growth through Ral-dependent and Ral-independent mechanisms. J Biol Chem. 2010;285:34729–34740. doi: 10.1074/jbc.M110.116756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Martin TD, Samuel JC, Routh ED, Der CJ, Yeh JJ, et al. Activation and involvement of Ral GTPases in colorectal cancer. Cancer Res. 2011;71:206–215. doi: 10.1158/0008-5472.CAN-10-1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Male H, Patel V, Jacob MA, Borrego-Diaz E, Wang K, Young DA, et al. Inhibition of RalA signaling pathway in treatment of non-small cell lung cancer. Lung Cancer. 2012;77:252–259. doi: 10.1016/j.lungcan.2012.03.007. [DOI] [PubMed] [Google Scholar]
- 22.Mishra PJ, Ha L, Rieker J, Sviderskaya EV, Bennett DC, Oberst MD, et al. Dissection of RAS downstream pathways in melanomagenesis: a role for Ral in transformation. Oncogene. 2010;29:2449–2456. doi: 10.1038/onc.2009.521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zipfel PA, Brady DC, Kashatus DF, Ancrile BD, Tyler DS, Counter CM, et al. Ral activation promotes melanomagenesis. Oncogene. 2010;29:4859–4864. doi: 10.1038/onc.2010.224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sowalsky AG, Alt-Holland A, Shamis Y, Garlick JA, Feig LA, et al. RalA suppresses early stages of Ras-induced squamous cell carcinoma progression. Oncogene. 2010;29:45–55. doi: 10.1038/onc.2009.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang H, Owens C, Chandra N, Conaway MR, Brautigan DL, Theodorescu D, et al. Phosphorylation of RalB is important for bladder cancer cell growth and metastasis. Cancer Res. 2010;70:8760–8769. doi: 10.1158/0008-5472.CAN-10-0952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Smith SC, Oxford G, Baras AS, Owens C, Havaleshko D, Brautigan DL, et al. Expression of ral GTPases, their effectors, and activators in human bladder cancer. Clin Cancer Res. 2007;13:3803–3813. doi: 10.1158/1078-0432.CCR-06-2419. [DOI] [PubMed] [Google Scholar]
- 27.Yin J, Pollock C, Tracy K, Chock M, Martin P, Oberst M, et al. Activation of the RalGEF/Ral pathway promotes prostate cancer metastasis to bone. Mol Cell Biol. 2007;27:7538–7550. doi: 10.1128/MCB.00955-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fei J, Li Y, Zhu X, Luo X, et al. miR-181a post-transcriptionally downregulates oncogenic RalA and contributes to growth inhibition and apoptosis in chronic myelogenous leukemia (CML) PLoS One. 2012;7:e32834. doi: 10.1371/journal.pone.0032834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Osei-Sarfo K, Martello L, Ibrahim S, Pellicer A, et al. The human Rgr oncogene is overexpressed in T-cell malignancies and induces transformation by acting as a GEF for Ras and Ral. Oncogene. 2011;30:3661–3671. doi: 10.1038/onc.2011.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Saito R, Shirakawa R, Nishiyama H, Kobayashi T, Kawato M, Kanno T, et al. Downregulation of Ral GTPase-activating protein promotes tumor invasion and metastasis of bladder cancer. Oncogene. 2013;32:894–902. doi: 10.1038/onc.2012.101. [DOI] [PubMed] [Google Scholar]
- 31.Smith SC, Baras AS, Owens CR, Dancik G, Theodorescu D, et al. Transcriptional signatures of Ral GTPase are associated with aggressive clinicopathologic characteristics in human cancer. Cancer Res. 2012;72:3480–3491. doi: 10.1158/0008-5472.CAN-11-3966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shipitsin M, Feig LA, et al. RalA but not RalB enhances polarized delivery of membrane proteins to the basolateral surface of epithelial cells. Mol Cell Biol. 2004;24:5746–5756. doi: 10.1128/MCB.24.13.5746-5756.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Falsetti SC, Wang DA, Peng H, Carrico D, Cox AD, Der CJ, et al. Geranylgeranyltransferase I inhibitors target RalB to inhibit anchorage-dependent growth and induce apoptosis and RalA to inhibit anchorage-independent growth. Mol Cell Biol. 2007;27:8003–8014. doi: 10.1128/MCB.00057-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Peschard P, McCarthy A, Leblanc-Dominguez V, Yeo M, Guichard S, Stamp G, et al. Genetic deletion of RALA and RALB small GTPases reveals redundant functions in development and tumorigenesis. Curr Biol. 2012;22:2063–2068. doi: 10.1016/j.cub.2012.09.013. [DOI] [PubMed] [Google Scholar]
- 35.Sablina AA, Chen W, Arroyo JD, Corral L, Hector M, Bulmer SE, et al. The tumor suppressor PP2A Abeta regulates the RalA GTPase. Cell. 2007;129:969–982. doi: 10.1016/j.cell.2007.03.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chien Y, Kim S, Bumeister R, Loo YM, Kwon SW, Johnson CL, et al. RalB GTPase-mediated activation of the IkappaB family Kinase TBK1 couples innate immune signaling to tumor cell survival. Cell. 2006;127:157–170. doi: 10.1016/j.cell.2006.08.034. [DOI] [PubMed] [Google Scholar]
- 37.González-García A, Pritchard CA, Paterson HF, Mavria G, Stamp G, Marshall CJ, et al. RalGDS is required for tumor formation in a model of skin carcinogenesis. Cancer Cell. 2005;7:219–226. doi: 10.1016/j.ccr.2005.01.029. [DOI] [PubMed] [Google Scholar]
- 38.Kinsella BT, Erdman RA, Maltese WA, et al. Carboxyl-terminal isoprenylation of ras-related GTP-binding proteins encoded by rac1, rac2, and ralA. J Biol Chem. 1991;266:9786–9794. [PubMed] [Google Scholar]
- 39.Wu JC, Chen TY, Yu CTR, Tsai SJ, Hsu JM, Tang MJ, et al. Identification of V23RalA-Ser194 as a critical mediator for Aurora-A-induced cellular motility and transformation by small pool expression screening. J Biol Chem. 2005;280:9013–9022. doi: 10.1074/jbc.M411068200. [DOI] [PubMed] [Google Scholar]
- 40.Lim KH, Brady DC, Kashatus DF, Ancrile BB, Der CJ, Cox AD, et al. Aurora-A phosphorylates, activates, and relocalizes the small GTPase RalA. Mol Cell Biol. 2010;30:508–523. doi: 10.1128/MCB.00916-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kashatus DF, Lim KH, Brady DC, Pershing NLK, Cox AD, Counter CM, et al. RALA and RALBP1 regulate mitochondrial fission at mitosis. Nat Cell Biol. 2011;13:1108–1115. doi: 10.1038/ncb2310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bivona TG, Quatela SE, Bodemann BO, Ahearn IM, Soskis MJ, Mor A, et al. PKC regulates a farnesyl-electrostatic switch on K-Ras that promotes its association with Bcl-XL on mitochondria and induces apoptosis. Mol Cell. 2006;21:481–493. doi: 10.1016/j.molcel.2006.01.012. [DOI] [PubMed] [Google Scholar]
- 43.Neyraud V, Aushev VN, Hatzoglou A, Meunier B, Cascone I, Camonis J, et al. RalA and RalB proteins are ubiquitinated GTPases, and ubiquitinated RalA increases lipid raft exposure at the plasma membrane. J Biol Chem. 2012;287:29397–29405. doi: 10.1074/jbc.M112.357764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jullien-Flores V, Dorseuil O, Romero F, Letourneur F, Saragosti S, Berger R, et al. Bridging Ral GTPase to Rho pathways. RLIP76, a Ral effector with CDC42/Rac GTPase-activating protein activity. J Biol Chem. 1995;270:22473–22477. doi: 10.1074/jbc.270.38.22473. [DOI] [PubMed] [Google Scholar]
- 45.Cantor SB, Urano T, Feig LA, et al. Identification and characterization of Ral-binding protein 1, a potential downstream target of Ral GTPases. Mol Cell Biol. 1995;15:4578–4584. doi: 10.1128/mcb.15.8.4578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Park SH, Weinberg RA, et al. A putative effector of Ral has homology to Rho/Rac GTPase activating proteins. Oncogene. 1995;11:2349–2355. [PubMed] [Google Scholar]
- 47.Yamaguchi A, Urano T, Goi T, Feig LA, et al. An Eps homology (EH) domain protein that binds to the Ral-GTPase target, RalBP1. J Biol Chem. 1997;272:31230–31234. doi: 10.1074/jbc.272.50.31230. [DOI] [PubMed] [Google Scholar]
- 48.Nakashima S, Morinaka K, Koyama S, Ikeda M, Kishida M, Okawa K, et al. Small G protein Ral and its downstream molecules regulate endocytosis of EGF and insulin receptors. EMBO J. 1999;18:3629–3642. doi: 10.1093/emboj/18.13.3629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ikeda M, Ishida O, Hinoi T, Kishida S, Kikuchi A, et al. Identification and characterization of a novel protein interacting with Ral-binding protein 1, a putative effector protein of Ral. J Biol Chem. 1998;273:814–821. doi: 10.1074/jbc.273.2.814. [DOI] [PubMed] [Google Scholar]
- 50.Jullien-Flores V, Mahé Y, Mirey G, Leprince C, Meunier-Bisceuil B, Sorkin A, et al. RLIP76, an effector of the GTPase Ral, interacts with the AP2 complex: involvement of the Ral pathway in receptor endocytosis. J Cell Sci. 2000;113(Pt 16):2837–2844. doi: 10.1242/jcs.113.16.2837. [DOI] [PubMed] [Google Scholar]
- 51.Awasthi S, Cheng J, Singhal SS, Saini MK, Pandya U, Pikula S, et al. Novel function of human RLIP76: ATP-dependent transport of glutathione conjugates and doxorubicin. Biochemistry. 2000;39:9327–9334. doi: 10.1021/bi992964c. [DOI] [PubMed] [Google Scholar]
- 52.Neel NF, Rossman KL, Martin TD, Hayes TK, Yeh JJ, Der CJ, et al. The RalB small GTPase mediates formation of invadopodia through a GTPase-activating protein-independent function of the RalBP1/RLIP76 effector. Mol Cell Biol. 2012;32:1374–1386. doi: 10.1128/MCB.06291-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Leake K, Singhal J, Nagaprashantha LD, Awasthi S, Singhal SS, et al. RLIP76 Regulates PI3K/Akt signaling and chemo-radiotherapy resistance in pancreatic cancer. PLoS One. 2012;7:e34582. doi: 10.1371/journal.pone.0034582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wu Z, Owens C, Chandra N, Popovic K, Conaway M, Theodorescu D, et al. RalBP1 is necessary for metastasis of human cancer cell lines. Neoplasia. 2010;12:1003–1012. doi: 10.1593/neo.101080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mollberg NM, Steinert G, Aigner M, Hamm A, Lin FJ, Elbers H, et al. Overexpression of RalBP1 in colorectal cancer is an independent predictor of poor survival and early tumor relapse. Cancer Biol Ther. 2012;13:694–700. doi: 10.4161/cbt.20087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang Q, Wang JY, Zhang XP, Lv ZW, Fu D, Lu YC, et al. RLIP76 is overexpressed in human glioblastomas and is required for proliferation, tumorigenesis and suppression of apoptosis. Carcinogenesis. 2013 doi: 10.1093/carcin/bgs401. [DOI] [PubMed] [Google Scholar]
- 57.Moskalenko S, Henry DO, Rosse C, Mirey G, Camonis JH, White MA, et al. The exocyst is a Ral effector complex. Nat Cell Biol. 2002;4:66–72. doi: 10.1038/ncb728. [DOI] [PubMed] [Google Scholar]
- 58.Sugihara K, Asano S, Tanaka K, Iwamatsu A, Okawa K, Ohta Y, et al. The exocyst complex binds the small GTPase RalA to mediate filopodia formation. Nat Cell Biol. 2002;4:73–78. doi: 10.1038/ncb720. [DOI] [PubMed] [Google Scholar]
- 59.Spiczka KS, Yeaman C, et al. Ral-regulated interaction between Sec5 and paxillin targets exocyst to focal complexes during cell migration. J Cell Sci. 2008;121:2880–2891. doi: 10.1242/jcs.031641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cascone I, Selimoglu R, Ozdemir C, Del Nery E, Yeaman C, White M, et al. Distinct roles of RalA and RalB in the progression of cytokinesis are supported by distinct RalGEFs. EMBO J. 2008;27:2375–2387. doi: 10.1038/emboj.2008.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hazelett CC, Sheff D, Yeaman C, et al. RalA and RalB differentially regulate development of epithelial tight junctions. Mol Biol Cell. 2011;22:4787–4800. doi: 10.1091/mbc.E11-07-0657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bodemann BO, Orvedahl A, Cheng T, Ram RR, Ou YH, Formstecher E, et al. RalB and the exocyst mediate the cellular starvation response by direct activation of autophagosome assembly. Cell. 2011;144:253–267. doi: 10.1016/j.cell.2010.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kim MJ, Woo SJ, Yoon CH, Lee JS, An S, Choi YH, et al. Involvement of autophagy in oncogenic K-Ras-induced malignant cell transformation. J Biol Chem. 2011;286:12924–12932. doi: 10.1074/jbc.M110.138958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Guo JY, Chen HY, Mathew R, Fan J, Strohecker AM, Karsli-Uzunbas G, et al. Activated Ras requires autophagy to maintain oxidative metabolism and tumorigenesis. Genes Dev. 2011;25:460–470. doi: 10.1101/gad.2016311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kidd AR, Snider JL, Martin TD, Graboski SF, Der CJ, Cox AD, et al. Ras-related small GTPases RalA and RalB regulate cellular survival after ionizing radiation. Int J Radiat Oncol Biol Phys. 2010;78:205–212. doi: 10.1016/j.ijrobp.2010.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ohta Y, Suzuki N, Nakamura S, Hartwig JH, Stossel TP, et al. The small GTPase RalA targets filamin to induce filopodia. Proc Nat Acad Sci U S A. 1999;96:2122–2128. doi: 10.1073/pnas.96.5.2122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Frankel P, Aronheim A, Kavanagh E, Balda MS, Matter K, Bunney TD, et al. RalA interacts with ZONAB in a cell density-dependent manner and regulates its transcriptional activity. EMBO J. 2005;24:54–62. doi: 10.1038/sj.emboj.7600497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Jiang H, Luo JQ, Urano T, Frankel P, Lu Z, Foster DA, et al. Involvement of Ral GTPase in v-Src-induced phospholipase D activation. Nature. 1995;378:409–412. doi: 10.1038/378409a0. [DOI] [PubMed] [Google Scholar]
- 69.Sidhu RS, Clough RR, Bhullar RP, et al. Regulation of phospholipase C-delta1 through direct interactions with the small GTPase Ral and calmodulin. J Biol Chem. 2005;280:21933–21941. doi: 10.1074/jbc.M412966200. [DOI] [PubMed] [Google Scholar]
- 70.Fenwick RB, Prasannan S, Campbell LJ, Nietlispach D, Evetts KA, Camonis J, et al. Solution structure and dynamics of the small GTPase RalB in its active conformation: significance for effector protein binding. Biochemistry. 2009;48:2192–2206. doi: 10.1021/bi802129d. [DOI] [PubMed] [Google Scholar]
- 71.Fenwick RB, Campbell LJ, Rajasekar K, Prasannan S, Nietlispach D, Camonis J, et al. The RalB–RLIP76 complex reveals a novel mode of ral-effector interaction. Structure. 2010;18:985–995. doi: 10.1016/j.str.2010.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Khosravi-Far R, White MA, Westwick JK, Solski PA, Chrzanowska-Wodnicka M, Van Aelst L, et al. Oncogenic Ras activation of Raf/mitogen-activated protein kinase-independent pathways is sufficient to cause tumorigenic transformation. Mol Cell Biol. 1996;16:3923–3933. doi: 10.1128/mcb.16.7.3923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.White MA, Nicolette C, Minden A, Polverino A, Van Aelst L, Karin M, et al. Multiple Ras functions can contribute to mammalian cell transformation. Cell. 1995;80:533–541. doi: 10.1016/0092-8674(95)90507-3. [DOI] [PubMed] [Google Scholar]
- 74.Rossé C, L'Hoste S, Offner N, Picard A, Camonis J, et al. RLIP, an effector of the Ral GTPases, is a platform for Cdk1 to phosphorylate epsin during the switch off of endocytosis in mitosis. J Biol Chem. 2003;278:30597–30604. doi: 10.1074/jbc.M302191200. [DOI] [PubMed] [Google Scholar]
- 75.Hanahan D, Weinberg RA, et al. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]