Abstract
Processing of Holliday junctions is essential in recombination. We have identified the gene for the junction-resolving enzyme GEN1 from the thermophilic fungus Chaetomium thermophilum and expressed the N-terminal 487-amino-acid section. The protein is a nuclease that is highly selective for four-way DNA junctions, cleaving 1 nt 3′ to the point of strand exchange on two strands symmetrically disposed about a diagonal axis. CtGEN1 binds to DNA junctions as a discrete homodimer with nanomolar affinity. Analysis of the kinetics of cruciform cleavage shows that cleavage of the second strand occurs an order of magnitude faster than the first cleavage so as to generate a productive resolution event. All these properties are closely similar to those described for bacterial, phage and mitochondrial junction-resolving enzymes. CtGEN1 is also similar in properties to the human enzyme but lacks the problems with aggregation that currently prevent detailed analysis of the latter protein. CtGEN1 is thus an excellent enzyme with which to engage in biophysical and structural analysis of eukaryotic GEN1.
Abbreviations: GFP, green fluorescent protein; TEV, tobacco etch virus; BSA, bovine serum albumin; BSA, bovine serum albumin; EDTA, ethylenediaminetetraacetic acid
Keywords: DNA recombination and repair, Holliday junction resolution, Chaetomium thermophilum, FEN1, thermophilic proteins
Graphical abstract

Highlights
-
•
We have identified the gene for the junction-resolving enzyme GEN1 in the thermophilic fungus C. thermophilum.
-
•
CtGEN1 is a nuclease that is highly selective for the structure of four-way DNA junctions.
-
•
CtGEN1 binds to DNA junctions as a discrete homodimer with nanomolar affinity.
-
•
Second-strand cleavage by CtGEN1 is accelerated to ensure productive resolution.
-
•
In contrast to the human enzyme, CtGEN1 is highly suitable for structural and mechanistic investigation.
Introduction
Homologous genetic recombination performs indispensable functions. In mitotic cells, it provides a mechanism for high-fidelity double-strand break repair and contributes to the resolution of interstrand crosslinks and lesions arising during replication. In meiosis, it enhances genetic diversity and ensures accurate chromosomal segregation by generating a temporary physical linkage between homologous chromosomes. Deficiency in homologous recombination leads to markedly elevated susceptibility to a variety of cancers.
The central intermediate in homologous recombination is a four-way DNA, or Holliday, junction [1], where four DNA helices are connected by the covalent continuity of the strands. The key event in the pathway is the processing of the junction. Holliday junctions formed in cellular chromosomal DNA are processed by two types of process, distinguished by whether or not they involve nucleolytic cleavage. The first process, called “dissolution”, requires the BLM helicase to translocate two adjacent junctions toward each other before they are unlinked by topoisomerase IIIα [2], [3], [4]. This pathway, which does not lead to strand exchange, is probably the primary response to the presence of DNA junctions in mitotically dividing cells, and defects lead to Bloom syndrome [5] that is characterized by genomic instability [6]. Any junctions that persist are processed by mechanisms that are based on the action of nucleases that are selective for the structure of a four-way DNA junction; this second process is called “resolution”. Junction-resolving enzymes have been well characterized in bacteria, phage, archaea and yeast mitochondria (reviewed in Ref. [7]).
In eukaryotic cells, there are a number of nucleases that can act upon branched DNA structures, but in the last 3 years, it has become clear that there are two main nucleolytic activities that are responsible for recognizing and processing of four-way DNA junctions. The first to be identified is GEN1 (Yen1 in yeast) [8], [9], [10], a member of the XPG superfamily of 5′ nucleases that includes EXO1, FEN1 and XPG [11], [12], [13]. In the second pathway, the nuclease making the initial cleavage of the junction is SLX1 in complex with SLX4 that binds a number of nucleases involved in DNA repair [14], [15], [16], [17], [18]. SLX1 is a member of the UvrC family of endonucleases and contains a GIY-YIG element that forms a metal ion-binding active center in a number of nucleases [19], [20]. However, unlike GEN1, SLX1 introduces a single cleavage into the junction; the unilaterally cleaved junction is then the substrate for the MUS81-EME1 nuclease that is also tethered to the SLX4 complex [21], [22]. Acting in isolation, the properties of MUS81-EME1 are akin to those of a flap endonuclease [23], [24], [25], but acting in concert with SLX1 generates a productive resolution. One or other of these two systems is required to be functional for cell viability; GEN1 and SLX4 are synthetically lethal in human cells as a result of unprocessed junctions leading to dysfunctional mitosis [26]. Ectopic expression of human GEN1 has been found to restore the meiotic phenotype of mus81Δ fission yeast [27], and Yen1 resolves persistent DNA junctions in meiotic yeast [28].
The N-terminal section of human GEN1 can act as a single protein in dimeric form to generate the productive resolution of a four-way junction [10]. It thus appears to have properties that are analogous to those of well-characterized junction-resolving enzymes from bacteria, phage, archaea and mitochondria [7]. However, more detailed structural and biophysical analysis of this protein has been hampered by some of its properties. In our hands, all fragments of human GEN1 tested have proved to be polydisperse and fail to form discrete complexes with junctions, and that behavior is evident in other published studies [10]. We therefore sought an ortholog with closely similar sequence and properties that was more suited to quantitative study. To that end, we investigated thermophilic fungi, as thermostable proteins often are well behaved in solution. We identified the GEN1 orthologs from a number of such species and expressed and purified GEN1 enzyme from Chaetomium thermophilum. We find that this protein is very well behaved in solution, binding to DNA junctions in dimeric form and generating bilateral cleavage by accelerating second-strand cleavage. Further investigation reveals that its properties are closely similar to those of non-eukaryotic resolving enzymes, and the enzyme is highly suitable for more detailed analysis.
Results
Identification of a gene encoding GEN1 in C. thermophilum
To identify the GEN1 gene in thermophilic fungi, we extended our previous phylogenetic analysis of XPG superfamily members that covered a set of FEN1, EXO1, XPG and GEN1-like sequences from phylogenetically diverse eukaryotes [9], and we included sequences from the thermophilic fungi C. thermophilum, Myceliophthora thermophile, Talaromyces marneffei, Talaromyces stipitatus and Thielavia terrestris. The full alignment is shown in Supplementary Fig. 1, and an unrooted phylogenetic tree is shown in Supplementary Fig. 2. The analysis confirms that XPG superfamily members from thermophilic fungi fall into four distinct classes represented by the XPG, FEN1, EXO1 and GEN1 nuclease families, allowing us to identify the putative GEN1 of C. thermophilum. An alignment of the N-terminal half of human FEN1 and GEN1 and the proposed C. thermophilum GEN1 is shown in Fig. 1a. Seven strongly conserved acidic amino acids are boxed red; these bind two active-site metal ions in the structure of human FEN1 [13] and are conserved in all the XPG superfamily of nucleases (Supplementary Fig. 1). In addition, lysine K87 (boxed blue) is conserved in all three proteins; this is close to the scissile phosphate in human FEN1 and probably stabilizes the anionic transition state of the hydrolysis reaction. The N-terminal 230 amino acids of the human and deduced C. thermophilum GEN1 sequences that include these conserved acidic residues are 26% identical.
Fig. 1.
Identification and expression of a putative GEN1 sequence from C. thermophilum.
(a ) Alignment of the protein sequences of human (Hs) FEN1 and GEN1 and the putative C. thermophilum (Ct) GEN1, shaded by identity. Seven conserved acidic amino acids involved in metal ion binding in the active site of FEN1 are boxed red, and the conserved lysine is boxed blue.
(b) Expression of CtGEN11–487. The protein fragment was expressed in E. coli as a fusion with GFP that was subsequently cleaved using TEV protease. Purified protein was analyzed by electrophoresis in 10% polyacrylamide containing SDS (Fisher). Tracks: 1, a mixture of proteins as a size marker, with molecular mass (kDa) written on the left; 2, the CtGEN11–487-GFP fusion protein; 3, purified CtGEN11–487; 4, GFP released from the fusion.
Bacterial expression of C. thermophilum GEN1
A synthetic gene encoding the N-terminal section of CtGEN1 comprising amino acids 1–487 optimized for Escherichia coli codon usage was inserted into the pET derivative pWaldo [29] and expressed in E. coli BL21(DE3) RIL. The translated protein that resulted from this was a fusion of CtGEN11–487 with green fluorescent protein (GFP) at the C-terminus (the fluorescence of which could be monitored during purification) and an octahistidine tag at the C-terminus (Supplementary Fig. 3). The GFP could be cleaved from the fusion by virtue of an intervening tobacco etch virus (TEV) protease site. The protein was purified by sequential application to Ni2 +-charged metal affinity, heparin and gel-filtration columns. The purified protein migrated as a single band on electrophoresis in polyacrylamide in the presence of SDS (Fig. 1b) even when heavily overloaded (Supplementary Fig. 4). CtGEN11–487 eluted from the gel-filtration column in between bovine serum albumin (BSA) (66 kDa) and carbonic anhydrase (29 kDa) (Supplementary Fig. 5), consistent with the protein existing in monomeric form (calculated molecular mass of 55.1 kDa). No elution corresponding to a dimer of CtGEN11–487 was detected. Rass et al. similarly concluded that human GEN11–527 is monomeric in free solution [10].
C. thermophilum GEN1 is a nuclease that is selective for the structure of four-way DNA junctions
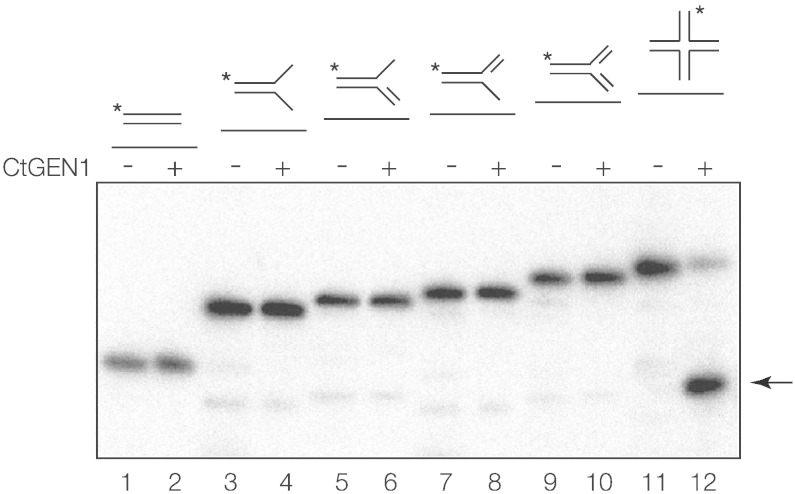
Purified CtGEN11–487 was assessed for nucleolytic activity on a variety of branched DNA substrates including various flap species, a nicked three-way junction and a four-way (4H) junction (Fig. 2 and Supplementary Fig. 6). The four-way junction used was Jbm5 [30] with a 12-bp core of homology that can consequently undergo 12 steps of branch migration. Each construct was incubated at 5 nM with 50 nM protein (single-turnover conditions) for 2 min at 25 °C and the resulting products were examined by gel electrophoresis. Under the conditions of the experiment, it is clear that there is strong cleavage of the four-way junction and very much weaker cleavage of the other species. Thus, CtGEN11–487 is highly selective for the structure of the four-way DNA junction, consistent with a role as a junction-resolving enzyme.
Fig. 2.
Substrate selectivity for CtGEN11–487. A number of branched DNA species were prepared, with one strand radioactively 5′-32P-labeled (indicated by an asterisk). Each was incubated with (even tracks) or without (odd tracks) 50 nM CtGEN11–487 in 10 mM Hepes (pH 7.5) in the presence of 10 mM MgCl2. Potential products were separated by electrophoresis in 6% polyacrylamide and visualized by phosphorimaging. The substrates were duplex (tracks 1 and 2), splayed duplex (tracks 3 and 4), 3′ flap (tracks 5 and 6), 5′ flap (tracks 7 and 8), nicked three-way junction (tracks 9 and 10) and four-way junction Jbm5 (tracks 11 and 12). Only the four-way junction was significantly cleaved, with the product arrowed.
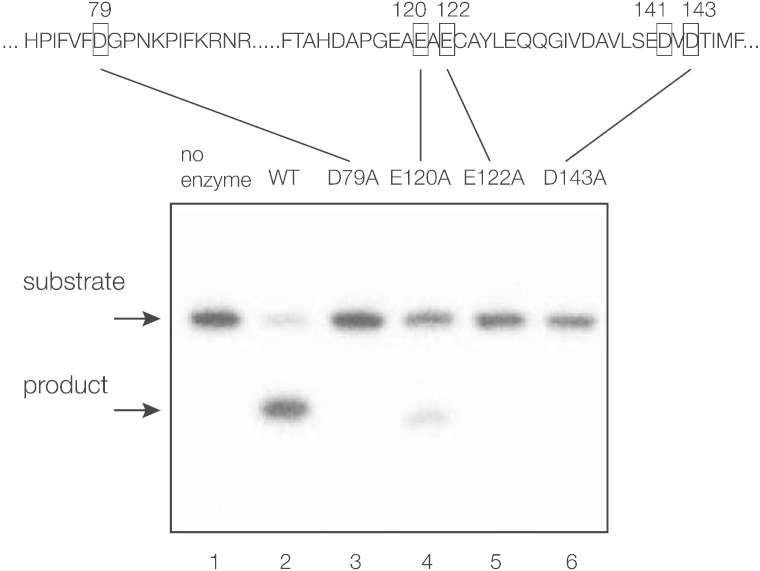
Mutation of conserved acidic amino acids leads to loss of cleavage of four-way DNA junctions
The sequence alignment between CtGEN1 and human FEN1 (Fig. 1a) indicates a number of probable acidic amino acids likely to be involved in catalysis if these proteins are related, viz. D38, D79, E120, E122, E141, D143 and D199 (numbered for CtGEN11–487). We have individually altered four of these by site-directed mutagenesis and tested the junction-cleaving activity in preparations of the proteins (as GFP fusions for preparative convenience). While the wild-type enzyme cleaves the DNA junction, those with D79A, E122A or D143A mutation lead to no cleavage of the junction (Fig. 3), even after prolonged incubation (data not shown). The equivalent carboxylate groups in human FEN1 are ~ 2.3 Å from the catalytic metal ions [13]. E120A has some activity though significantly lower than for the wild-type protein; in FEN1, this carboxylate is 4.8 Å from the metal ion, thus not directly bound. These data provide further evidence that the protein is indeed GEN1 of C. thermophilum.
Fig. 3.
Cleavage activity on a DNA junction by putative active-site mutants of CtGEN11–487. Conserved acidic residues were individually mutated to alanine and the cleavage activity against junction 3 was tested using purified CtGEN11–487-GFP fusions. Junction 3 radioactively 5′-32P-labeled on the x strand was incubated without enzyme (track 1) or with wild type (track 2), D79A (track 3), E120A (track 4), E122A (track 5) or D143A (track 6) GEN1 for 4 min at 37 °C, and any products of cleavage were separated by electrophoresis in a 15% polyacrylamide and visualized by phosphorimaging.
CtGEN1 introduces 2-fold symmetrical cleavages into a four-way DNA junction
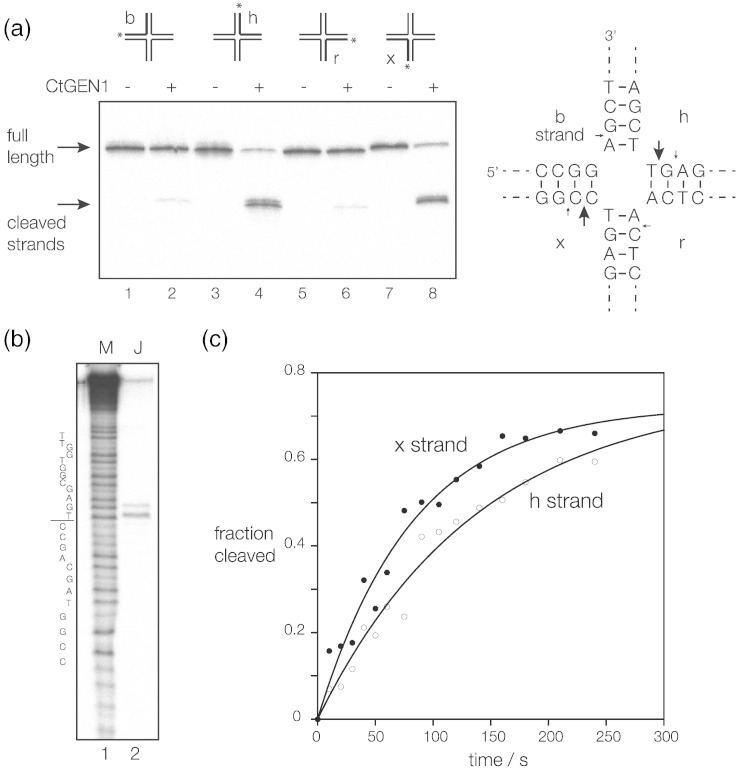
Productive resolution of a junction requires symmetrical cleavage of two strands; thus, we mapped the cleavages introduced into each strand of the well-characterized junction 3 [31] that is not capable of branch migration. The junction was prepared from four strands of 50 bp, generating a version of junction 3 having four arms each of 25 bp in length. Four radioactive forms were made, each individually 5′-32P-labeled on one strand. Each was incubated at a concentration of 1 nM with 50 nM CtGEN11–487 for 10 min. The products were examined by gel electrophoresis under denaturing conditions and phosphorimaging (Fig. 4a). The h and x strands were strongly cleaved and the b and r strands were more weakly cleaved, such that the h and x strands were cleaved ≥ 20-fold greater than the b and r strands.
Fig. 4.
Location and rates of cleavage of the four strands of a four-way junction. Four versions of a junction with arms of 25 bp and a core sequence corresponding to junction 3 were each radioactively 5′-32P-labeled on a single strand. Each was incubated with 100 nM CtGEN11–487 in 10 mM Hepes (pH 7.5), 10 mM MgCl2, 50 mM NaCl, 0.1% BSA and 1 mM DTT at 37 °C for 10 min, and the products were separated by electrophoresis in a 15% polyacrylamide and visualized by phosphorimaging.
(a) Phosphorimage of the gel. Samples were applied in the order b, h, r and x strands labeled, with even- and odd-numbered tracks containing samples with and without enzyme added. All strands were cleaved to some degree, but the h and x strands were cleaved more strongly than the b or r strands.
(b) The product of h-strand cleavage by CtGEN11–487 was electrophoresed in a 15% polyacrylamide sequencing gel in TBE containing 8 M urea alongside a sequence ladder derived from partial chemical degradation of the h strand in order to map the positions of cleavage at nucleotide resolution. The sequence of the h strand is shown on the left, with the position of strand exchange indicated by the line. The positions of cleavage in all four strands are shown on the schematic of the central sequence of the junction, with the larger arrows indicating strong cleavage.
(c) Reaction progress plotted as a function of time for the h (open circles) and x (filled circles) strands. The data have been fitted to single exponential functions (lines).
We compared the products of cleavage of junction 3 with those from an active N-terminal fragment of the human GEN1, HsGEN11–527 (as the C-terminal fusion with GFP, i.e., HsGEN11–527-GFP). In separate reactions, we incubated the same radioactive junctions with HsGEN11–527-GFP and compared the products with those of CtGEN11–487 (Supplementary Fig. 7). Both enzymes cleave at the same sites, with similar patterns of cleavage, in agreement with the sites mapped for human GEN1 by Rass et al. [10]. However, the difference between h, x and b, r cleavages for the human GEN11–527 was less pronounced compared to the CtGEN11–487 products.
Comparison of the position of migration of the CtGEN11–487 product bands with those of a ladder generated from the same strand by chemical depurination shows that the major cleavage occurs 1 nt 3′ to the point of strand exchange (Fig. 4b). This is identical with the position of cleavage of a 5′ flap substrate by FEN1 and other members of that superfamily [32], [33], highlighting the similarity of the two enzymes.
We went on to measure the rates of cleavage of the four strands by CtGEN11–487. The reaction was initiated by addition of 10 mM MgCl2 (final concentration), aliquots were removed at various times and the reaction terminated by denaturation in formamide and the products and substrate were separated by gel electrophoresis under denaturing conditions. The extent of cleavage at each time was quantified by phosphorimaging from which progress curves of fraction of cleavage as a function of time were plotted (Fig. 4c). The data were well fitted by single exponential functions, from which rates of cleavage of 0.008 and 0.01 s− 1 were measured for the h and x strands, respectively.
CtGEN1 binds DNA junctions with high affinity
We have examined binding of CtGEN11–487 to a four-way DNA junction using gel electrophoresis in polyacrylamide under non-denaturing conditions, as previously performed for junction-resolving enzymes of bacteriophage [34], [35], [36], mitochondrial [37], [38], [39], [40] and archaeal [41] origins. Radioactively 5′-32P-labeled junction 3 at 100 pM was incubated with a range of concentrations between 43 pM and 44 nM of CtGEN11–487 and applied to a 6% polyacrylamide gel (Fig. 5a and b) and electrophoresed under non-denaturing conditions at room temperature. The experiment was performed under two different conditions, in the presence of either 1 mM ethylenediaminetetraacetic acid (EDTA) (to chelate trace metal ions) or 1 mM Ca2 +. CtGEN11–487 is catalytically inactive in Ca2 +; thus, in general, we employ this cation in binding experiments. Slower-migrating species were observed with the addition of increasing concentrations of protein in both cases, indicative of complex formation. However, in EDTA, a species of intermediate mobility was observed, whereas in Ca2+, the complex migrated largely as a single retarded species. It is likely that the intermediate species observed in the presence of EDTA is a complex with a single monomer of CtGEN11–487 bound to the junction, while the slower species is a complex with a dimer of CtGEN11–487 (see the following section). The fraction of bound junction was estimated by quantification of bound and unbound species and plotted as a function of protein concentration (Fig. 5c). The isotherms were fitted to a simple binding model, yielding an estimate of the affinity of the GEN1 fragment for a DNA four-way junction of Kd = 10 nM. However, it is clear that the binding is more cooperative than a simple two-state binding process, as previously observed for RuvC [42], Ydc2 [39] and Hjc [41] junction-resolving enzymes. Fitting the data to the standard Hill equation gives a significantly better fit, consistent with a strongly cooperative binding of CtGEN11–487 as a dimeric complex the presence of Ca2 +.
Fig. 5.
Binding of CtGEN11–487 to junction 3. Junction 3 was prepared with 25 bp arms, radioactively 5′-32P-labeled on the x strand. We incubated 82 pM junction with increasing concentrations of CtGEN11–487 in 10 mM Hepes (pH 7.5), 50 mM NaCl, 0.1% BSA and 1 mM DTT with either 1 mM EDTA or 1 mM Ca2 + for 60 min and electrophoresed in 6% polyacrylamide under non-denaturing conditions.
(a and b) Phosphorimages of the gels electrophoresed in (a) 1 mM EDTA and (b) 1 mM Ca2 +. Tracks contain DNA junction incubated with increasing concentrations of CtGEN11–487 left to right. The concentration of CtGEN11–487 is shown over each track. DNA–protein complexes are visible migrating more slowly than the free junction (arrowed right). A complex of intermediate mobility is visible in EDTA, but in Ca2 +, essentially only a single complex is evident.
(c) The fractional intensity of the retarded complex in Ca2 + is plotted as a function of CtGEN11–487 concentration (filled circles). The data have been fitted to two models. A standard binding isotherm (line) gives an affinity Kd = 10 nM, but the data clearly exhibit cooperativity. They have therefore additionally been fitted to the Hill equation (broken line).
CtGEN1 binds to DNA junctions as a dimer
The results in the previous section suggest that CtGEN11–487 binds as a dimer, but we sought a more conclusive test of this. We have shown previously for other junction-resolving enzymes that, provided subunit exchange is possible, electrophoretic examination of complexes using mixtures of the enzyme and longer versions generated by fusion with another protein domain together can provide information on the oligomeric state of the protein in the complex [35], [36], [37], [39], [43]. For these experiments, we used the C-terminal fusion of GFP with CtGEN11–487. The experiment was performed in two ways. First, we incubated radioactively labeled junction 3 with 40 nM CtGEN11–487 (i.e., the non-fusion) and made five further equivalent incubations with addition of the CtGEN11–487-GFP fusion at individual concentrations between 25 and 125 nM (Fig. 6a). Incubation with CtGEN11–487 resulted in a single retarded band of junction–protein complex. However, addition of the CtGEN11–487-GFP led to three complex bands, with the relative intensity of the slowest increasing with the molar fraction of the fusion protein. This indicates that the protein binds as a dimer to the junction, thereby generating complexes with three combinations of the two proteins, that is, non-fusion + non-fusion, non-fusion + fusion and fusion + fusion, with fastest, intermediate and slowest mobilities, respectively.
Fig. 6.
CtGEN11–487 binds to a DNA junction as dimer. Complexes have been formed with CtGEN11–487 alone and fused to GFP. These have been analyzed by gel electrophoresis in 6% polyacrylamide. In each case, free junction was electrophoresed in track 1. The experiment was performed in two ways.
(a) We incubated 160 nM junction with 40 nM CtGEN11–487 in 10 mM Hepes (pH 7.5), 50 mM NaCl, 0.1% BSA, 1 mM DTT and 1 mM CaCl2 (track 2), with increasing concentrations of CtGEN1-GFP fusion (tracks 3–7; concentrations indicated above each track).
(b) We incubated 80 nM junction with 40 nM CtGEN1-GFP fusion in 10 mM Hepes (pH 7.5), 50 mM NaCl, 0.1% BSA, 1 mM DTT and 1 mM CaCl2 (track 2), with increasing concentrations of CtGEN11–487 fusion (tracks 3–7; concentrations indicated above each track).
Note that, in each case, three retarded complexes are observed, consistent with CtGEN11–487 binding as a dimer to the DNA junction.
The experiment was also carried out with a fixed 30 nM concentration of CtGEN11–487-GFP fusion and addition of increasing concentrations (30–180 nM) of CtGEN11–487 (Fig. 6b). Once again, three species of different mobility were observed with equimolar CtGEN11–487 and CtGEN11–487-GFP, but as the mole fraction of CtGEN11–487 increased, only the fastest species was observed. This confirms the dimeric nature of the protein in the complex. It is furthermore apparent from these data that the C-terminal fusion of GFP lowers the affinity of CtGEN11–487 for the four-way junction.
CtGEN1 exhibits acceleration of second-strand cleavage
A resolving enzyme must introduce bilateral cleavage within the lifetime of the protein–DNA complex in order to generate complete resolution of a four-way junction. We have previously used the cleavage of a supercoil-stabilized cruciform structure to demonstrate bilateral cleavage by phage, bacterial and yeast mitochondrial resolving enzymes [36], [44], [45], [46], [47]. The cruciform structure is intrinsically unstable except in a negatively supercoiled circular DNA [48] and is rapidly reabsorbed if the covalent continuity of the circle is broken at any point. If the enzyme makes a single cleavage in the DNA, the cruciform can only remain extruded if its integrity is preserved by the DNA–protein interactions in the complex. If the protein dissociates before second-strand cleavage occurs, the cruciform is reabsorbed and no substrate remains for a second cleavage reaction. We have applied this approach to the analysis of junction cleavage by CtGEN11–487.
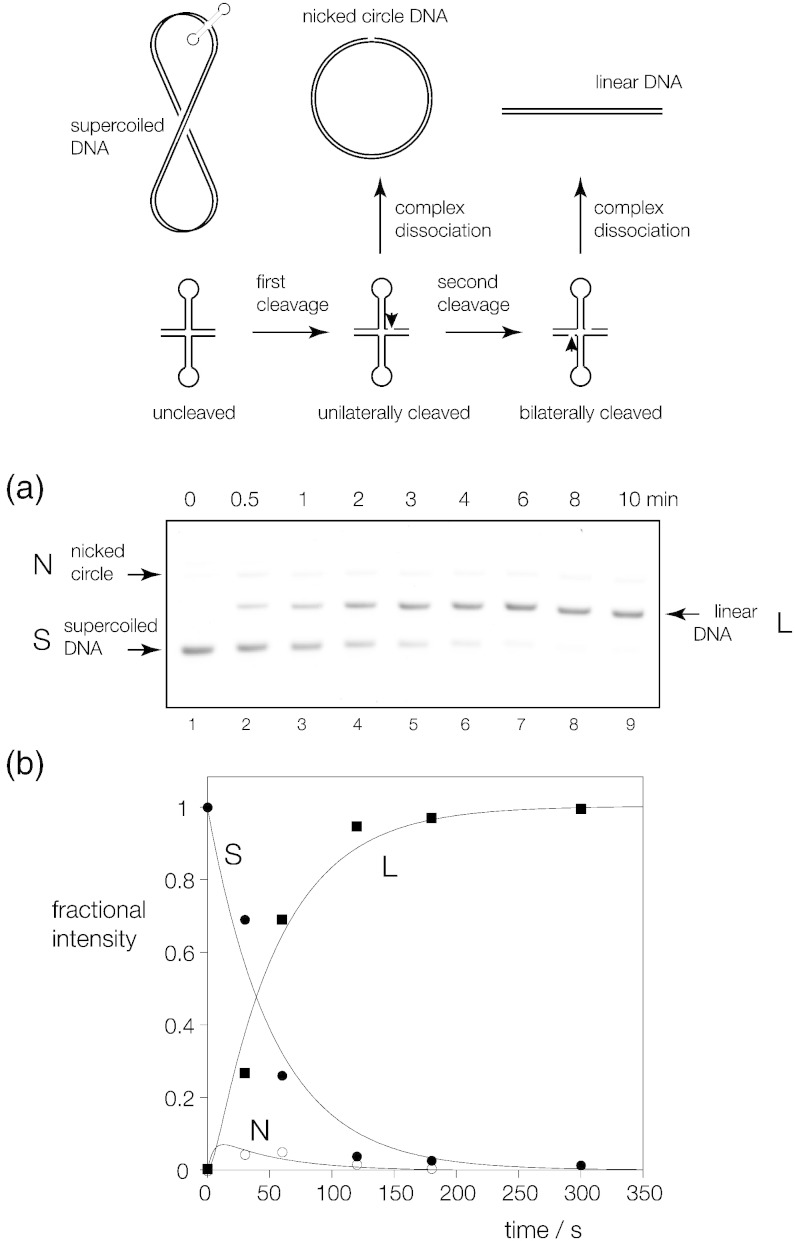
For this purpose, we used a 34-bp cruciform that is stably extruded in the plasmid pBHR3 (Supplementary Fig. 8) extracted from E. coli in exponential growth (Fig. 7). Supercoiled pBHR3 was incubated CtGEN11–487 and aliquots were removed at different times. The topological state of the plasmid was then examined by gel electrophoresis in 1% agarose (Fig. 7a). Uncleaved supercoiled plasmid migrates as the fastest species. Any unilateral cleavage by the enzyme converts the plasmid into a nicked circle, which migrates slowly. By contrast, bilateral cleavage generates a linear DNA product of intermediate mobility.
Fig. 7.
Analysis of bilateral cleavage in a DNA junction using a supercoil-stabilized cruciform substrate. The principle of the experiment is shown in the scheme (top). A cruciform structure contains a four-way junction that is a substrate for resolving enzymes. However, the cruciform requires negative supercoiling for stabilization. Unilateral cleavage of the junction followed by dissociation of the protein leads to the formation of a nicked circle in which the cruciform substrate is no longer present. By contrast, subsequent second cleavage within the lifetime of the complex generates a linear product as the cruciform is now bilaterally cleaved.
(a) The supercoiled DNA substrate and the nicked and linear products are readily separated by electrophoresis in 1% agarose. Supercoiled plasmid pBHR3 was incubated with 200 nM CtGEN11–487 in 10 mM Hepes (pH 7.5), 50 mM NaCl, 1 mM DTT and 0.1% BSA at 37 °C. After the cleavage reaction was initiated by addition of MgCl2 to 10 mM, samples were removed at different times, protein was removed by treatment with proteinase K and electrophoresed in a 1% agarose gel and DNA was fluorescently stained. A fluoroimage is shown. With time, the supercoiled DNA is converted to linear product, with a low intensity of nicked circular DNA appearing as a transient intermediate.
(b) The intensities of the three species were quantified and plotted as a function of time (lower). These data are fitted to the integrated rate Eqs. (3), (4), (5) shown in the main text.
Incubation of pBHR3 with CtGEN11–487 results in loss of supercoiled DNA and formation of linear product, indicative of bilateral cleavage occurring within the lifetime of the enzyme–junction complex. However, a smaller fraction of nicked circular DNA is also visible at early times. The intensities of the different species have been quantified and plotted as a function of time (Fig. 7b). This shows the conversion of supercoiled to linear DNA product over the time course. Nicked circular DNA appears as a transient intermediate, being formed at early times and subsequently being converted to linear product. The cruciform substrate would be unstable in an unconstrained nicked circle; thus, it must be preserved within the bound complex with the resolving enzyme.
We have analyzed these data in terms of a kinetic model, whereby the first cleavage occurs at a rate k1, followed by cleavage of the other strand with rate k2. In this scheme, we make no attempt to distinguish the identity of the strand that is cleaved first. Thus, the rates of change of supercoiled DNA (concentration [S]t at time t) and nicked circular DNA ([N]t at time t) will be given by:
| (1) |
| (2) |
Integration of these equations by standard analytical methods leads to the kinetic rate equations expressing the instantaneous fractional concentrations of the three species as:
| (3) |
| (4) |
and by conservation, the fractional concentration of linear species [L]t at time t will be:
| (5) |
The measured intensities of the three species have been fitted to these equations (Fig. 7b), giving the rates k1 = 0.019 and k2 = 0.20 min− 1. From these rates, we see that the rate of cleavage of the nicked species is 11-fold greater than that of the supercoiled DNA; that is, there is an acceleration of second-strand cleavage relative to the first. The effect of this is to ensure that bilateral cleavage occurs within the lifetime of the enzyme–junction complex.
Discussion
We have identified the ortholog of GEN1 from the thermophilic fungus C. thermophilum. The sequence aligns with human GEN1 and with the putative genes for the other XPG superfamily of 5′ nucleases EXO1, FEN1 and XPG. The alignments identify the probable active-site residues, some of which we have confirmed by mutagenesis. Expression of a fragment of the CtGEN1 protein reveals that it possesses all the properties expected of a junction-resolving enzyme.
CtGEN11–487 exhibits a strong selectivity for the structure of the four-way DNA junction and cleaves with a rate of ~ 0.5 min− 1. The strands are cleaved predominantly 1 nt 3′ to the point of strand exchange, analogous to the positions of cleavage of 5′ flap structures by FEN1 [32], [33], further strengthening assignment of the CtGEN1 gene. CtGEN11–487 binds to DNA junctions with nanomolar affinity to form a discrete complex. Use of C-terminal protein fusions shows that the bound form is a dimer (although monomeric in free solution), consistent with the observed cleavage pattern. In the absence of added metal ions, some binding in monomeric form is observed at intermediate protein concentrations, but in the presence of Ca2 +, the complex contains a dimer of the protein, consistent with the requirement for two spatially separated cleavages to generate a productive resolution of the junction. Under the latter, more physiologically relevant conditions, binding occurs with strong cooperativity. Evidently, the C-terminal section of CtGEN1 is not required for the binding in dimeric form, although we note that C-terminal fusion with GFP substantially weakens the affinity, and thus, in principle, the deleted section of the protein might modulate the dimerization.
CtGEN11–487 introduces bilateral cleavage into the junction within the lifetime of the enzyme–junction complex. Kinetic analysis of supercoiled cruciform cleavage shows that the rate of second-strand cleavage is greater than that of the first by a factor of 11. Acceleration of the second-strand cleavage increases the probability that both cleavages occur before dissociation of the bound complex and thus provides a kinetic mechanism that will tend to ensure a productive resolution of the junction. The yeast mitochondrial enzyme Cce1 exhibits closely similar behavior, with a 10-fold acceleration of second-strand cleavage [46].
The two sets of cleavages observed with junction 3 are consistent with the formation of alternative 2-fold symmetrical complexes in which the h and x or the b and r strands are cleaved, with the former predominating over the latter by a ratio of ~ 25:1. This pattern of cleavage (i.e., cleavage intensity b = r ≠ h = x) is commonly observed in other well-studied junction-resolving enzymes, where a dimeric protein binds to a potentially 4-fold symmetrical junction lowering the symmetry to a single dyad axis.
All these properties are closely similar to those of the well-characterized junction-resolving enzymes of bacteriophage [34], [35], [36], [45], bacterial [47], [49], mitochondrial [37], [38], [39], [40], [46] and archaeal [30], [41], [50] origins. Thus, eukaryotic GEN1 seems to be a canonical junction-resolving enzyme in all its properties analyzed to date. This is in marked contrast to the alternative pathway, involving the complex SLX1-SLX4-MUS81-EME1, requiring at least four different proteins and two nuclease activities to generate the two cleavages essential for resolution [21], [22].
As frequently observed with proteins derived from thermophilic organisms, CtGEN11–487 is very well behaved in solution. Of particular note, this protein is free of the aggregation properties that have hindered the analysis of the human protein. Whereas the human protein typically binds to DNA junctions to generate multiple different complexes (found both by Rass et al. [10] and in our own unpublished observations), CtGEN11–487 binding results in the formation of a discrete complex with a DNA junction. This makes this an excellent subject for quantitative biophysical, structural and mechanistic studies. We are presently performing crystallization trials with the enzyme and have obtained crystals of a complex of CtGEN11–487 bound to a DNA junction that are in the process of optimization.
Materials and Methods
Bioinformatic analysis of XPG family proteins
XPG superfamily sequences were aligned using JALVIEW employing the MAFT algorithm [51]. An unrooted phylogenetic tree was generated using the Splitstree program [52] and employing the neighbor joining method.
Cloning and expression of a gene encoding CtGEN1
A synthetic gene expressing CtGEN11–487 was obtained from GeneArt (Life Technologies) and cloned into pWaldo, a variant of the pET28 [29]. This generates CtGEN11–487 with GFP at its C-terminus linked via a site for TEV protease and with an octahistidine C-terminal peptide to facilitate purification (Supplementary Fig. 3). Plasmids were transformed into BL21(DE3) RIL immediately before cell preparation and were grown in Luria Broth medium supplemented with kanamycin and chloramphenicol. Cells were initially grown at 37 °C until the absorbance at 600 nm (A600) of the cell culture reached 0.4 whereupon the cells were transferred to 20 °C. Once the culture reached A600 = 0.6, expression of CtGEN11–487 was induced by addition of 0.1 mM IPTG and incubated for a further ~ 20 h. Cells were harvested by centrifugation at 5000g, resuspended (at 3 ml/g of cell paste) on ice and lysed in 50 mM NaH2PO4 (pH 8.0), 1 M NaCl, 1 mg/ml lysozyme and 0.2% Triton X100. The cellular extract was sonicated and clarified by centrifugation at 20,000g for 30 min at 4 °C. The extract was then applied to Ni2 +-IDA metal chelate resin (Generon) and eluted in 50 mM NaH2PO4 (pH 8.0) and 1 M NaCl with a gradient of 0–0.5 M imidazole. Elution was monitored by the green color of GFP, and CtGEN11–487 eluted around 150 mM imidazole. The protein was dialyzed against 25 mM Hepes (pH 7.5), 0.1 M NaCl and 1 mM DTT, and 1/10 (mol/mol) of TEV protease was added to the protein extract. The protein extract was applied to a Hitrap Heparin HP column (GE healthcare) and eluted in a gradient of 0.1–1 M NaCl in a background of 25 mM Hepes (pH 7.5). GFP does not bind to the heparin column under these conditions and CtGEN11–487 eluted at ~ 0.4 M NaCl. The protein was then subjected to gel filtration by application to a Superose 12 column in 25 mM Hepes (pH 7.5) and 1 M NaCl. The peak fraction of CtGEN11–487 was dialyzed against 25 mM Hepes (pH 7.5), 0.1 M NaCl and 50% glycerol and was stored at − 80 °C. The concentration of CtGEN11–487 was estimated by absorbance at 280 nm with an estimated A280 = 50,600 M− 1 cm− 1 (per monomer). Protein was analyzed by electrophoresis in 10% EZ-RUN™ SDS polyacrylamide gels (Fisher Scientific) following the manufacturer's instructions at 200 V for 50 min. Gels were stained using Quick Coomassie stain (Generon) and destained in water.
Synthesis of DNA oligonucleotides and construction of junction species
Oligonucleotides were synthesized using β-cyanoethyl phosphoramidite chemistry. Fully deprotected oligonucleotides were purified by gel electrophoresis in polyacrylamide gels [10–20% (w/v) depending upon oligonucleotide length] in 90 mM Tris–borate (pH 8.5) and 2 mM EDTA (TBE buffer) containing 8 M urea and were recovered by electroelution and ethanol precipitation. Helical junctions were assembled by mixing stoichiometric quantities of strands (all sequences shown in Supplementary Fig. 6) and were annealed by incubation in 20 mM Tris–HCl (pH 8) and 50 mM NaCl for 5 min at 85 °C, followed by slow cooling. These were purified by electrophoresis under non-denaturing conditions in 8% polyacrylamide in TBE buffer for 8 h at 20 °C and were recovered by electroelution and ethanol precipitation.
Analysis of cleavage of branched DNA species by CtGEN1
A variety of flap-containing and branched DNA species were tested for cleavage by CtGEN11–487. We incubated 50 nM CtGEN11–487 with 5–10 nM DNA radioactively 5′-32P-labeled on one strand in 10 mM Hepes (pH 7.5), 10 mM MgCl2, 50 mM NaCl, 0.1 mg/ml BSA and 1 mM DTT for 1 h. Any reaction was terminated by addition of 50 mM EDTA and 1 μg/ml proteinase K (final concentrations). Substrates and products separated by electrophoresis in 6% (29:1) polyacrylamide in TBE buffer at 120 V for 4 h. Gels were dried and exposed to storage phosphor screens, imaged using a Fuji BAS 1500 phosphorimager and quantified using MacBAS software.
Mapping the cleavage site on junction 3 by CtGEN1
The product of cleavage of junction 3 radioactively 5′-32P-labeled on one strand was analyzed by electrophoresis alongside the same strand after depurination. This was performed by incubation in 10 μl of 50 mM sodium citrate (pH 4) and 0.1 mM EDTA for 15 min at 80 °C as described by Wang et al. [53]. The strand was then cleaved at depurinated positions by addition of 115 μl of water and 15 μl of 10 M piperidine followed by incubation at 90 °C for a further 30 min. DNA was precipitated with isopropanol followed by NaCl and ethanol. Electrophoresis was performed using 15% sequencing gels in TBE buffer containing 8 M urea.
Analysis of cleavage a four-way DNA junction using point mutants of CtGEN1
D79, E120, E122 and D143 were individually converted into alanine into CtGEN11–487 by PCR of the gene using the Q5 site-directed mutagenesis kit (BioLab) following the manufacturer's instructions. Mutations were verified by DNA sequencing. Mutant CtGEN11–487 was expressed and purified as a fusion with GFP as described above, except that the incubation with TEV protease and gel-filtration steps were omitted. Wild type and mutant CtGEN11–487-GFP eluted in a similar manner from the heparin column. Protein concentration was estimated by absorbance at 280 nm using A280 = 72,550 M− 1 cm− 1 (per monomer). Analysis of junction cleavage was performed as described below except that junction was incubated with 200 nM CtGEN11–487-GFP in the presence of 10 μg/ml of calf thymus DNA for 4 min at 37 °C.
Analysis of cleavage kinetics of a four-way DNA junction by CtGEN1
Cleavage kinetics were analyzed under single-turnover conditions. We incubated 50–100 nM CtGEN11–487 with 5–10 nM junction 3 radioactively 5′-32P-labeled on one strand in 10 mM Hepes (pH 7.5), 50 mM NaCl, 0.1% BSA and 1 mM DTT for 1 h. Cleavage was initiated by addition of 10 mM MgCl2 and aliquots were removed at chosen times and EDTA was added to 50 mM final concentration to terminate the reaction. One volume of 90% formamide was added and the samples were then denatured at 90 °C for 15 min. before separation of substrates and products by electrophoresis in a 15% (19:1) polyacrylamide gel in TBE containing 8 M urea at 80 W. Gels were dried and exposed to storage phosphor screens and quantified using a Fuji BAS 1500 phosphorimager using MacBAS software. The fraction of DNA cleaved at time t (Ft) was fitted by nonlinear regression analysis to the equation:
| (6) |
where Ff is the fraction of DNA cleaved at the end of the reaction and kc is the rate of cleavage.
Cleavage of cruciform substrates in supercoiled DNA
The cruciform-containing plasmid pBHR3 was generated by ligating the oligonucleotides 5′ AATTCAAGGGGCTGTATATATATATATATATATATATACAGCCTGAGCG and 5′ GATCCGCTCAGGCTGTATATATATATATATATATATATACAGCCCCTTG between the EcoRI and BamHI sites of pAT153 (Supplementary Fig. 8). The central alternating adenine-thymine repeated sequence ensures facile extrusion of the cruciform in negatively supercoiled DNA [54]. This was transformed into E. coli DH5α and grown to A600 = 0.6 at 37 °C. The plasmid was amplified by treatment with 150 μg/ml chloramphenicol overnight. Plasmid DNA was isolated using Qiagen maxiprep purification kit at 4 °C and purified by two rounds of isopycnic CsCl/ethidium bromide ultracentrifugation. Supercoiled DNA was recovered by side puncture, ethidium bromide was removed by extraction with n-butanol and the DNA was subjected to extensive dialysis against 20 mM Tris (pH 8.0) and 1 mM EDTA to remove CsCl. DNA concentration was measured spectrophotometrically.
We preincubated 10 nM pBHR3 plasmid with 200 nM CtGEN1 in 10 mM Hepes (pH 7.5), 50 mM NaCl, 1 mM DTT and 0.1% BSA for 3 min at 37 °C before the cleavage reaction was initiated by addition of MgCl2 to a final concentration 10 mM. Aliquots were removed at specific times and 50 mM EDTA was added to terminate the reaction. We added 1 μl proteinase K and we incubated the samples overnight at room temperature. Supercoiled, linear and nicked DNAs were separated by electrophoresis in 1% agarose gels in TBE buffer. Gels were stained with Safeview nucleic acid stain (NBS Biological Ltd), extensively washed with water and scanned using a FLA-2000 fluorescent image analyzer (Fuji) using excitation at 473 nm with an emission filter at 580 nm.
Analysis of binding of CtGEN1 to DNA four-way junctions
Radioactively-labelled J3 (at concentrations given in the text) was incubated with increasing concentrations of CtGEN11–487 in 10 mM Hepes (pH 7.5), 50 mM NaCl, 0.1% BSA and 1 mM DTT with either 1 mM EDTA or 1 mM Ca2 + for 1 h at 20 °C. After the addition of Ficoll-400 to 2.5%, the samples were immediately loaded onto a 6% (37:1) polyacrylamide gel in TBE and subjected to electrophoresis for 4 h. Gels were dried onto Whatman 3MM paper and analyzed by phosphorimaging as described above. Data were analyzed as fraction DNA bound (fb) versus protein concentration and fitted by nonlinear regression analysis to the equation:
| (7) |
where Kd is the dissociation constant, Pt is the total protein dimer concentration and Dt is the total DNA concentration. The data were also fitted to the Hill equation for cooperative binding:
| (8) |
where n is the Hill coefficient.
Acknowledgement
We thank our colleagues for discussion and Cancer Research UK (C28/A4959) and Wellcome Trust (077012/Z/05/Z) for financial support in the laboratories of D.M.J.L. and A.G., respectively.
Edited by J. Berger
Footnotes
Supplementary data to this article can be found online at http://dx.doi.org/10.1016/j.jmb.2014.10.008.
Appendix A. Supplementary data
Supplementary material.
References
- 1.Holliday R. A mechanism for gene conversion in fungi. Genet Res. 1964;5:282–304. doi: 10.1017/S0016672308009476. [DOI] [PubMed] [Google Scholar]
- 2.Ellis N.A., Groden J., Ye T.Z., Straughen J., Lennon D.J., Ciocci S. The Bloom's syndrome gene product is homologous to RecQ helicases. Cell. 1995;83:655–666. doi: 10.1016/0092-8674(95)90105-1. [DOI] [PubMed] [Google Scholar]
- 3.Wu L., Hickson I.D. The Bloom's syndrome helicase suppresses crossing over during homologous recombination. Nature. 2003;426:870–874. doi: 10.1038/nature02253. [DOI] [PubMed] [Google Scholar]
- 4.Cejka P., Plank J.L., Bachrati C.Z., Hickson I.D., Kowalczykowski S.C. Rmi1 stimulates decatenation of double Holliday junctions during dissolution by Sgs1-Top3. Nat Struct Mol Biol. 2010;17:1377–1382. doi: 10.1038/nsmb.1919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bloom D. Congenital telangiectatic erythema resembling lupus erythematosus in dwarfs; probably a syndrome entity. AMA Am J Dis Child. 1954;88:754–758. [PubMed] [Google Scholar]
- 6.Chaganti R.S., Schonberg S., German J. A manyfold increase in sister chromatid exchanges in Bloom's syndrome lymphocytes. Proc Natl Acad Sci U S A. 1974;71:4508–4512. doi: 10.1073/pnas.71.11.4508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Déclais A.C., Lilley D.M.J. New insight into the recognition of branched DNA structure by junction-resolving enzymes. Curr Opin Struct Biol. 2008;18:86–95. doi: 10.1016/j.sbi.2007.11.001. [DOI] [PubMed] [Google Scholar]
- 8.Ip S.C., Rass U., Blanco M.G., Flynn H.R., Skehel J.M., West S.C. Identification of Holliday junction resolvases from humans and yeast. Nature. 2008;456:357–361. doi: 10.1038/nature07470. [DOI] [PubMed] [Google Scholar]
- 9.Bailly A.P., Freeman A., Hall J., Declais A.C., Alpi A., Lilley D.M. The Caenorhabditis elegans homolog of Gen1/Yen1 resolvases links DNA damage signaling to DNA double-strand break repair. PLoS Genet. 2010;6:e1001025. doi: 10.1371/journal.pgen.1001025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rass U., Compton S.A., Matos J., Singleton M.R., Ip S.C., Blanco M.G. Mechanism of Holliday junction resolution by the human GEN1 protein. Genes Dev. 2010;24:1559–1569. doi: 10.1101/gad.585310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee B.I., Wilson D.M. The RAD2 domain of human exonuclease 1 exhibits 5′ to 3′ exonuclease and flap structure-specific endonuclease activities. J Biol Chem. 1999;274:37763–37769. doi: 10.1074/jbc.274.53.37763. [DOI] [PubMed] [Google Scholar]
- 12.Orans J., McSweeney E.A., Iyer R.R., Hast M.A., Hellinga H.W., Modrich P. Structures of human exonuclease 1 DNA complexes suggest a unified mechanism for nuclease family. Cell. 2011;145:212–223. doi: 10.1016/j.cell.2011.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tsutakawa S.E., Classen S., Chapados B.R., Arvai A.S., Finger L.D., Guenther G. Human flap endonuclease structures, DNA double-base flipping, and a unified understanding of the FEN1 superfamily. Cell. 2011;145:198–211. doi: 10.1016/j.cell.2011.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Munoz I.M., Hain K., Déclais A.-C., Gardiner M., Toh G.W., Sanchez-Pulido L. Coordination of structure–specific nucleases by human SLX4/BTBD12 is essential for DNA repair. Mol Cell. 2009;35:116–127. doi: 10.1016/j.molcel.2009.06.020. [DOI] [PubMed] [Google Scholar]
- 15.Fekairi S., Scaglione S., Chahwan C., Taylor E.R., Tissier A., Coulon S. Human SLX4 is a Holliday junction resolvase subunit that binds multiple DNA repair/recombination endonucleases. Cell. 2009;138:78–89. doi: 10.1016/j.cell.2009.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Svendsen J.M., Smogorzewska A., Sowa M.E., O'Connell B.C., Gygi S.P., Elledge S.J. Mammalian BTBD12/SLX4 assembles a Holliday junction resolvase and is required for DNA repair. Cell. 2009;138:63–77. doi: 10.1016/j.cell.2009.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Andersen S.L., Bergstralh D.T., Kohl K.P., LaRocque J.R., Moore C.B., Sekelsky J. Drosophila MUS312 and the vertebrate ortholog BTBD12 interact with DNA structure-specific endonucleases in DNA repair and recombination. Mol Cell. 2009;35:128–135. doi: 10.1016/j.molcel.2009.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Agostinho A., Meier B., Sonneville R., Jagut M., Woglar A., Blow J. Combinatorial regulation of meiotic holliday junction resolution in C. elegans by HIM-6 (BLM) helicase, SLX-4, and the SLX-1, MUS-81 and XPF-1 nucleases. PLoS Genet. 2013;9:e1003591. doi: 10.1371/journal.pgen.1003591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Aravind L., Walker D.R., Koonin E.V. Conserved domains in DNA repair proteins and evolution of repair systems. Nucleic Acids Res. 1999;27:1223–1242. doi: 10.1093/nar/27.5.1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mak A.N., Lambert A.R., Stoddard B.L. Folding, DNA recognition, and function of GIY-YIG endonucleases: crystal structures of R.Eco29kI. Structure. 2010;18:1321–1331. doi: 10.1016/j.str.2010.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Castor D., Nair N., Déclais A.C., Lachaud C., Toth R., Macartney T.J. Cooperative control of Holliday junction resolution and DNA repair by the SLX1 and MUS81-EME1 nucleases. Mol Cell. 2013;52:221–233. doi: 10.1016/j.molcel.2013.08.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wyatt H.D., Sarbajna S., Matos J., West S.C. Coordinated actions of SLX1-SLX4 and MUS81-EME1 for Holliday junction resolution in human cells. Mol Cell. 2013;52:234–247. doi: 10.1016/j.molcel.2013.08.035. [DOI] [PubMed] [Google Scholar]
- 23.Boddy M.N., Gaillard P.H., McDonald W.H., Shanahan P., Yates J.R., Russell P. Mus81-Eme1 are essential components of a Holliday junction resolvase. Cell. 2001;107:537–548. doi: 10.1016/s0092-8674(01)00536-0. [DOI] [PubMed] [Google Scholar]
- 24.Chen X.B., Melchionna R., Denis C.M., Gaillard P.H., Blasina A., Van de Weyer I. Human Mus81-associated endonuclease cleaves Holliday junctions in vitro. Mol Cell. 2001;8:1117–1127. doi: 10.1016/s1097-2765(01)00375-6. [DOI] [PubMed] [Google Scholar]
- 25.Gaillard P.H., Noguchi E., Shanahan P., Russell P. The endogenous Mus81-Eme1 complex resolves Holliday junctions by a nick and counternick mechanism. Mol Cell. 2003;12:747–759. doi: 10.1016/s1097-2765(03)00342-3. [DOI] [PubMed] [Google Scholar]
- 26.Garner E., Kim Y., Lach F.P., Kottemann M.C., Smogorzewska A. Human GEN1 and the SLX4-associated nucleases MUS81 and SLX1 are essential for the resolution of replication-induced Holliday junctions. Cell Rep. 2013;5:207–215. doi: 10.1016/j.celrep.2013.08.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lorenz A., West S.C., Whitby M.C. The human Holliday junction resolvase GEN1 rescues the meiotic phenotype of a Schizosaccharomyces pombe mus81 mutant. Nucleic Acids Res. 2010;38:1866–1873. doi: 10.1093/nar/gkp1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Matos J., Blanco M.G., Maslen S., Skehel J.M., West S.C. Regulatory control of the resolution of DNA recombination intermediates during meiosis and mitosis. Cell. 2011;147:158–172. doi: 10.1016/j.cell.2011.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Waldo G.S., Standish B.M., Berendzen J., Terwilliger T.C. Rapid protein-folding assay using green fluorescent protein. Nat Biotechnol. 1999;17:691–695. doi: 10.1038/10904. [DOI] [PubMed] [Google Scholar]
- 30.Kvaratskhelia M., White M.F. An archaeal Holliday junction resolving enzyme from Sulfolobus solfataricus exhibits unique properties. J Mol Biol. 2000;295:193–202. doi: 10.1006/jmbi.1999.3363. [DOI] [PubMed] [Google Scholar]
- 31.Duckett D.R., Murchie A.I.H., Diekmann S., von Kitzing E., Kemper B., Lilley D.M.J. The structure of the Holliday junction and its resolution. Cell. 1988;55:79–89. doi: 10.1016/0092-8674(88)90011-6. [DOI] [PubMed] [Google Scholar]
- 32.Hosfield D.J., Mol C.D., Shen B., Tainer J.A. Structure of the DNA repair and replication endonuclease and exonuclease FEN-1: coupling DNA and PCNA binding to FEN-1 activity. Cell. 1998;95:135–146. doi: 10.1016/s0092-8674(00)81789-4. [DOI] [PubMed] [Google Scholar]
- 33.Finger L.D., Blanchard M.S., Theimer C.A., Sengerova B., Singh P., Chavez V. The 3′-flap pocket of human flap endonuclease 1 is critical for substrate binding and catalysis. J Biol Chem. 2009;284:22184–22194. doi: 10.1074/jbc.M109.015065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Duckett D.R., Giraud-Panis M.-E., Lilley D.M.J. Binding of the junction-resolving enzyme bacteriophage T7 endonuclease I to DNA: separation of binding and catalysis by mutation. J Mol Biol. 1995;246:95–107. doi: 10.1006/jmbi.1994.0069. [DOI] [PubMed] [Google Scholar]
- 35.Pöhler J.R.G., Giraud-Panis M.-J.E., Lilley D.M.J. T4 endonuclease VII selects and alters the structure of the four-way DNA junction; binding of a resolution-defective mutant enzyme. J Mol Biol. 1996;260:678–696. doi: 10.1006/jmbi.1996.0430. [DOI] [PubMed] [Google Scholar]
- 36.Giraud-Panis M.-J.E., Lilley D.M.J. Structural recognition and distortion by the DNA junction-resolving enzyme RusA. J Mol Biol. 1998;278:117–133. doi: 10.1006/jmbi.1998.1681. [DOI] [PubMed] [Google Scholar]
- 37.White M.F., Lilley D.M.J. The structure-selectivity and sequence-preference of the junction-resolving enzyme CCE1 of Saccharomyces cerevisiae. J Mol Biol. 1996;257:330–341. doi: 10.1006/jmbi.1996.0166. [DOI] [PubMed] [Google Scholar]
- 38.White M.F., Lilley D.M.J. The resolving enzyme CCE1 of yeast opens the structure of the four-way DNA junction. J Mol Biol. 1997;266:122–134. doi: 10.1006/jmbi.1996.0795. [DOI] [PubMed] [Google Scholar]
- 39.White M.F., Lilley D.M.J. Characterization of a Holliday junction resolving enzyme from Schizosaccharomyces pombe. Mol Cell Biol. 1997;17:6465–6471. doi: 10.1128/mcb.17.11.6465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.White M.F., Lilley D.M.J. Interaction of the resolving enzyme YDC2 with the four-way DNA junction. Nucleic Acids Res. 1998;26:5609–5616. doi: 10.1093/nar/26.24.5609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fogg J.M., Kvaratskhelia M., White M.F., Lilley D.M.J. Distortion of DNA junctions imposed by the binding of resolving enzymes: a fluorescence study. J Mol Biol. 2001;313:751–764. doi: 10.1006/jmbi.2001.5081. [DOI] [PubMed] [Google Scholar]
- 42.Fogg J.M., Schofield M.J., White M.F., Lilley D.M.J. Sequence and functional-group specificity for cleavage of DNA junctions by RuvC of Escherichia coli. Biochemistry. 1999;38:11349–11358. doi: 10.1021/bi990926n. [DOI] [PubMed] [Google Scholar]
- 43.Parkinson M.J., Pöhler J.R.G., Lilley D.M.J. Catalytic and binding mutants of the junction-resolving enzyme endonuclease I of bacteriophage T7: role of acidic residues. Nucleic Acids Res. 1999;27:682–689. doi: 10.1093/nar/27.2.682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Giraud-Panis M.-J.E., Lilley D.M.J. Near-simultaneous DNA cleavage by the subunits of the junction-resolving enzyme T4 endonuclease VII. EMBO J. 1997;16:2528–2534. doi: 10.1093/emboj/16.9.2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Parkinson M.J., Lilley D.M.J. The junction-resolving enzyme T7 endonuclease I: quaternary structure and interaction with DNA. J Mol Biol. 1997;270:169–178. doi: 10.1006/jmbi.1997.1128. [DOI] [PubMed] [Google Scholar]
- 46.Fogg J.M., Schofield M.J., Déclais A.-C., Lilley D.M.J. The yeast resolving enzyme CCE1 makes sequential cleavages in DNA junctions within the lifetime of the complex. Biochemistry. 2000;39:4082–4089. doi: 10.1021/bi992785v. [DOI] [PubMed] [Google Scholar]
- 47.Fogg J.M., Lilley D.M.J. Ensuring productive resolution by the junction-resolving enzyme RuvC: large enhancement of second-strand cleavage rate. Biochemistry. 2001;39:16125–16134. doi: 10.1021/bi001886m. [DOI] [PubMed] [Google Scholar]
- 48.Lilley D.M.J., Hallam L.R. Thermodynamics of the ColE1 cruciform. Comparisons between probing and topological experiments using single topoisomers. J Mol Biol. 1984;180:179–200. doi: 10.1016/0022-2836(84)90436-4. [DOI] [PubMed] [Google Scholar]
- 49.Bennett R.J., West S.C. Structural analysis of the RuvC-Holliday junction complex reveals an unfolded junction. J Mol Biol. 1995;252:213–226. doi: 10.1006/jmbi.1995.0489. [DOI] [PubMed] [Google Scholar]
- 50.Komori K., Sakae S., Fujikane R., Morikawa K., Shinagawa H., Ishino Y. Biochemical characterization of the Hjc holliday junction resolvase of Pyrococcus furiosus. Nucleic Acids Res. 2000;28:4544–4551. doi: 10.1093/nar/28.22.4544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Clamp M., Cuff J., Searle S.M., Barton G.J. The Jalview Java alignment editor. Bioinformatics. 2004;20:426–427. doi: 10.1093/bioinformatics/btg430. [DOI] [PubMed] [Google Scholar]
- 52.Huson D.H. SplitsTree: analyzing and visualizing evolutionary data. Bioinformatics. 1998;14:68–73. doi: 10.1093/bioinformatics/14.1.68. [DOI] [PubMed] [Google Scholar]
- 53.Wang H., Glansdorff N., Charlier D. The arginine repressor of Escherichia coli K-12 makes direct contacts to minor and major groove determinants of the operators. J Mol Biol. 1998;277:805–824. doi: 10.1006/jmbi.1998.1632. [DOI] [PubMed] [Google Scholar]
- 54.Greaves D.R., Patient R.K., Lilley D.M.J. Facile cruciform formation by an (A-T)34 sequence from a Xenopus globin gene. J Mol Biol. 1985;185:461–478. doi: 10.1016/0022-2836(85)90064-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material.