Abstract
This study was designed to learn the expression status of miR-24 and its clinical relevance in patients with acute myeloid leukemia (AML). We detected the miR-24 expression levels using real-time quantitative PCR in 84 AML patients and investigated the clinical significance of miR-24 expression in AML. There was no difference in clinical parameters between cases with miR-24 high expression and with miR-24 low expression. The frequency of miR-24 high expression was higher in patients with t(8;21) than in others (82% (9/11) versus 44% (32/72), P=0.026). The levels of miR-24 expression had no correlation with the mutations of nine genes (FLT3-ITD, NPM1, C-KIT, IDH1/IDH2, DNMT3A, N/K-RAS and C/EBPA). Meanwhile, among the group who obtained CR, the cases with miR-24 high expression had no difference in overall survival (OS) and relapse-free survival (RFS) than those with miR-24 low expression (P=0.612 and 0.665, respectively). These findings implicated that miR-24 high regulation is a common event in AML with t(8;21), and it might serve as a novel and selective therapeutic target for the treatment of AML with t(8;21).
Keywords: miR-24, acute myeloid leukemia, microRNA
Introduction
Acute myeloid leukemia (AML) is a heterogeneous malignant disease, which is characterized by arrested differentiation and abnormal proliferation, leading to bleeding, fatal infection, or organ infiltration [1]. Recurring chromosomal aberrations and gene mutations contribute to the pathogenesis of AML [2]. Recently, many researches have indicated that miRNAs play important roles in myeloid leukemogenesis [3-5].
MicroRNAs (miRNAs) are known as small non-coding single-stranded RNAs of 20-22 nucleotides that regulate the expression of over 60% of all human genes and are involved in pivotal biological processes, including development, differentiation, proliferation, as well as apoptosis [6-10]. A large body of evidences implicate that dysregulation of miRNA expression may take part in oncogenesis of human malignant cancers, such as non-small cell lung cancer (NSCLC), gastrointestinal carcinoma, hepatocellular carcinoma, pancreatic cancer, breast cancer, cervical cancer and the like [11-17]. Particular microRNA signatures have been discovered in myeloid and lymphoid leukemia, and have relationships with the pathogenesis, diagnosis and prognosis of myeloid and lymphoid leukemia, such as miR-124-1, let-7a-3, miR-181, miR-29B [4,18-23].
Many researches have shown miR-24 takes control of cell cycle distribution and apoptosis [24-26]. Overexpression of miR-24 has been found in oral carcinoma and non-small cell lung cancer (NSCLC) [11,27]. MiR-24 was reported to enhance invasion and metastasis in cancer cell [28]. However, the role of miR-24 in AML should be explored more. Here we addressed the question whether miR-24 expression is related to AML.
Materials and methods
Patients and samples
This study included 84 patients who had a diagnosis of de novo AML at the Affiliated People’ Hospital of Jiangsu University. The diagnosis and classification of AML patients were based on French-America-British (FAB) and World Health Organization (WHO) criteria [23,29]. Treatment protocol was described as reported previously [21]. Written informed consent was obtained from all patients. The study was approved by the Institutional Review Board of the Affiliated People’ Hospital of Jiangsu University. The main clinical and laboratory features of the patient cohort were collected in Table 1. 16 healthy donors were collected as controls.
Table 1.
Clinical characteristics at diagnosis of AML patients divided according to miR-24 expression status
| miR-24 high expression | |||
|---|---|---|---|
|
|
|||
| + | - | P | |
| Sex (male/female) | 21/21 | 27/15 | 0.270 |
| Median age at diagnosis, years (range) | 53 (10-86) | 57 (15-87) | 0.164 |
| Median WBC at diagnosis, ×109 L-1 (range) | 8.4 (0.3-185.4) | 11.7 (1.1-528.0) | 0.697 |
| Median hemoglobin at diagnosis, g/L (range) | 75.0 (34.0-131.0) | 77.0 (40.0-138.0) | 0.333 |
| Median platelets at diagnosis, ×109 L-1 (range) | 33.0 (3.0-140.0) | 38.0 (4.0-264.0) | 0.707 |
| FAB | 0.053 | ||
| M1 | 2 | 4 | |
| M2 | 24 | 14 | |
| M3 | 6 | 11 | |
| M4 | 4 | 11 | |
| M5 | 5 | 2 | |
| WHO | 0.080 | ||
| AML with t(8;21) | 9 | 2 | |
| AML with t(15;17) | 6 | 11 | |
| AML without maturation | 2 | 4 | |
| AML with maturation | 15 | 12 | |
| Acute myelomonocytic leukemia | 5 | 11 | |
| Acute monoblastic and monocytic leukemia | 4 | 2 | |
| Karyotype classification | 0.919 | ||
| Favorable | 15 | 13 | |
| Intermediate | 23 | 23 | |
| Poor | 3 | 4 | |
| No date | 1 | 2 | |
| Karyotyping | 0.250 | ||
| Normal | 20 | 20 | |
| t(8;21) | 9 | 2 | |
| t(15;17) | 6 | 11 | |
| Complicated | 3 | 3 | |
| Others | 3 | 4 | |
| No date | 1 | 2 | |
| Gene mutation | |||
| NPM1 (+/-) | 5/37 | 4/37 | 1.000 |
| FLT3 ITD (+/-) | 6/36 | 6/35 | 1.000 |
| C-KIT (+/-) | 1/41 | 0/41 | 1.000 |
| IDH1/IDH2 (+/-) | 3/35 | 1/38 | 0.358 |
| DNMT3A (+/-) | 3/35 | 3/36 | 1.000 |
| NRAS/KRAS (+/-) | 4/34 | 3/36 | 0.711 |
| C/EBPA (+/-) | 6/36 | 3/38 | 0.483 |
| CR (+/-) | 23/18 | 20/19 | 0.823 |
| Median miR-24 expression (range) | 0.33 (0.13-3.31) | 0.01 (0.00-0.12) | < 0.001 |
RNA extraction and reverse transcription
Using the mirVana miRNA isolation kit (Ambion, Austin, TX, USA), we extracted the total RNA. According to the manufacturer’s protocol using miScript Reverse Transcription Kit (Qiagen, Duesseldorf, Germany), total RNA was reverse transcribed to cDNA.
Real-time quantitative PCR
Real-time quantitative PCR (RQ-PCR) was carried out according to the Manufacturer’s instructions using miScript SYBR green PCR kit (Qiagen, catalog no. 218073) with the manufacturer-provided miScript Universal primer and miRNA-specific forward primer: TGGCTCAGTTCAGCAGGAACA (miR-24).
RQ-PCR was performed on a 7300 Thermo cycler (Applied Biosystems, Foster City, CA, USA), using 50 ng of cDNA in a 20 μl reaction volume with 1× QuantiTect SYBR Green PCR Master Mix, 1× miScript Universal Primer, and 1.0 μM of the specific forward primer. PCR program conditions were 95°C for 15 min, followed by 40 cycles at 94°C for 15 s, 55°C for 30 s and 72°C for 34 s. At the end of the PCR cycles, melting program (95°C for 15 s, 60°C for 60 s, 95°C for 15 s, and 60°C for 15 s) was performed to validate the specificity of the expected PCR product. PCR amplicons were also confirmed by direct DNA sequencing in three randomly selected patients. The relative expression level of miR-24 was calculated by the comparative 2-ΔΔCt method using U6 small nuclear RNA level for normalization.
Gene mutation detection
NPM1 and C-KIT mutations were detected by high-resolution melting analysis (HRMA) as reported previously [3]. Briefly, genomic DNA samples were amplified using gene-specific primers. Mutation scanning was performed for PCR products using HRMA with the LightScannerTM platform (Idaho Technology Inc., Salt Lake City, Utah). All positive samples were directly DNA sequenced to confirm the results of HRMA. FLT3 internal tandem duplication (ITD) and C/EBPA mutations were detected using direct DNA sequencing [30,31].
Statistical analyses
All statistical analyses were implemented using spss17.0. Chi-square analysis or Fisher exact test was executed to compare the distinction of categorical variables. The comparison of miRNA expression status between patients and controls was executed using Mann-Whitney test. Survival was analyzed according to the Kaplan-Meier method. All values showed two-sided with a P-value < 0.05 considering statistically significant.
Results
MiR-24 expression in AML
In our 84 samples, miR-24 expression levels represented a continuum ranging from 0.000011 to 130.192 (median 0.123342). To evaluate the impact of miR-24 expression levels on clinical outcome without seeking an optimal cutpoint, patients were divided into low and high expressers according to the median exp-ression level of miR-24.
Association of miR-24 expression with clinical and laboratory characteristics in AML
There was no significant difference in sex, age, WBC, hemoglobin and platelets between patients with and without miR-24 high expression (Table 1). MiR-24 high expression could be observed in each AML subtype analyzed (Table 1). The level of miR-24 was higher in AML with t(8;21) than in t(15;17) (P=0.0127), in normal (P=0.0214), in others (P=0.1237), and in complex (P=0.8802) (Figure 1). Meanwhile, the frequency of miR-24 high expression was higher in patients with t(8;21) than in others (82% (9/11) versus 44% (32/72), P=0.026). There was no significant difference in the mutations of nine genes between low expressers and high expressers (Table 1).
Figure 1.
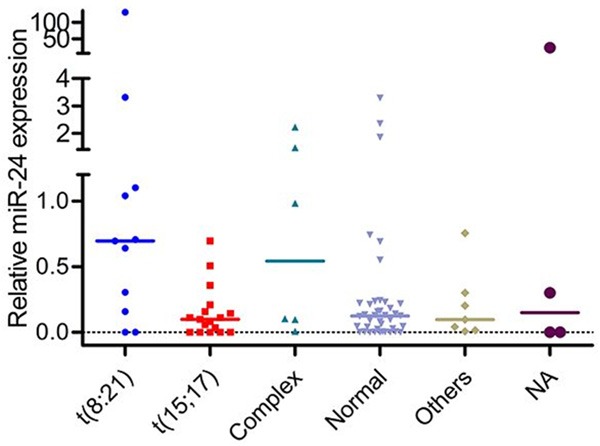
Relative miR-24 expression in AML with different kayotypes. NA: not available. The level of miR-24 was higher in AML with t(8;21) than in t(15;17) (P=0.0127), in normal (P=0.0214), in others (P=0.1237), and in complex (P=0.8802).
Impact of miR-24 expression on outcome of AML patients
To investigate the prognostic impact of miR-24 high expression in AML, survival analysis was performed in 80 cases with follow-up data. There was no definite difference in the rates of CR between patients with and without miR-24 high expression (56% versus 51%, P=0.823). The patients with and without miR-24 high expression were similar in the overall survival (OS) (P=0.929). Among the group who obtained CR, although the OS of AML patients with miR-24 high expression (median 20.5 months, 95% confidence interval 17-32 months) was shorter than those with miR-24 low expression (median 30 months, 95% confidence interval 22-38 months), the difference was not statistically significant (P=0.612) (Figure 2A). In addition, the cases with miR-24 high expression had no difference in relapse-free survival (RFS) than those with miR-24 low expression (P=0.665) (Figure 2B). The size of cases with t(8;21) was small and most cases were still alive, so the OS and RFS could not be analyzed. In cases with cytogenetically normal AML, there was no significant difference in the OS and RFS between patients with and without miR-24 high expression (P=0.532 and 0.772, respectively).
Figure 2.
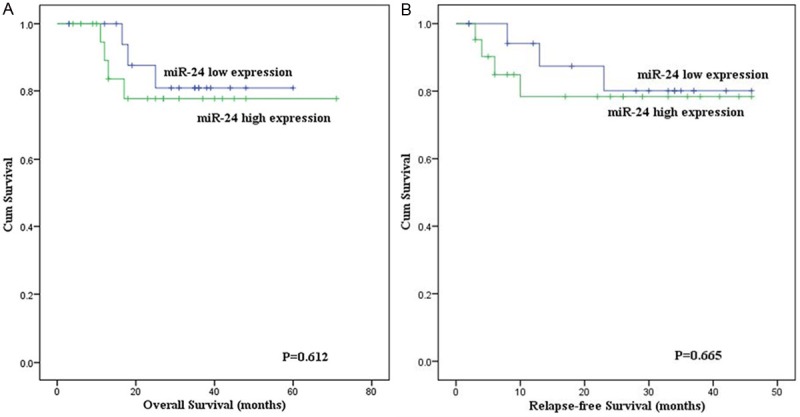
Overall and relapse-free survival of AML patients obtained complete remission. A. Overall survival; B. Relapse-free survival.
Discussion
High endogenous expression levels of miR-24 were more abundant in myeloid cells compared with lymphoid cells [32]. Overexpression of miR-24 has been found in oral carcinoma, non-small cell lung cancer (NSCLC) and breast cancer [11,27,33]. The function of miRNAs was highly tissue-dependent, which means that in different types of tissues one specific miRNA might get involved in different functions [26]. In our study, we didn’t see obvious difference between AML patients and controls. Meanwhile, we didn’t find miR-24 expression had any influences on outcome of AML patients. Though the OS of AML patients with miR-24 high expression (median 20.5 months, 95% confidence interval 17-32 months) was shorter than those with miR-24 low expression (median 30 months, 95% confidence interval 22-38 months), the difference was not statistically significant (` =0.612) due to the small size of samples. Furthermore, more cases should be analyzed to further confirm its clinical significance in AML.
In our previous studies, we found some microRNAs had close relationships with AML involving t(8;21) as well [3,21]. However, lacking of definite mechanisms, regarding microRNA and AML with t(8;21), it seemed that there were no clinically meaningful findings. Recently, few studies have reported the expression status of miR-24 in AML. Zaidi et al. have reported that miR-24 may act as a novel and selective therapeutic target for the treatment of AML. The t(8;21)-encoded AML1-ETO hold the miR-24-23-27 locus and control miR-24 transcription. Disruption of Runx1/AML1 subnuclear localization, by a chromosomal translocation t(8;21), is connected to the etiology of acute myeloid leukemia. Modified Runx1 subnuclear targeting by leukemia-related translocation t(8;21) transcriptionally deregulates the miR-24. By activating a miR-24/MKP-7/MAP kinase network, modified Runx1 subnuclear targeting may enhance proliferation and block granulocytic differentiation [34]. In the present study, we investigated the expression status of miR-24 in patients with AML. Interestingly, high expression of miR-24 showed more frequently in AML patients with translocation t(8;21) than in others (82% (9/11) versus 44% (32/72), P=0.026). Zaidi et al. provided exact evidences for us to prove that miR-24 played an important role in AML involving t(8;21) translocation. It was a coincidence that the experiment we conducted corroborated association between miR-24 and AML with t(8;21) in clinic. Nowadays, a growing body of evidences showed microRNAs really had close relationships with this chromosomal translocation [5,35-37]. According to those researches, they focused on the AML1/ETO, which is a fusion protein having functions of inhibiting differentiation and apoptosis, and triggering signals for cell proliferation [38,39]. Next, we should take attentions to this fusion protein then we study the networks between microRNA and AML with t(8;21) translocation. Taken together with existing evidence from microRNA and AML with t(8;21) studies, the current results support a relationship between microRNA and AML with t(8;21) translocation, which merits further study.
In summary, we suggest that high expression of miR-24 in AML patients might imply miR-24 can serve as a valuable source for biomarker discovery and validation in AML patients with t(8;21), meanwhile, miR-24 might serve as a novel and selective therapeutic target for the treatment of AML patients with t(8;21).
Acknowledgements
This study was supported by National Natural Science Foundation of China (81172592, 81270630).
Disclosure of conflict of interest
None.
References
- 1.Estey E, Döhner H. Acute myeloid leukaemia. Lancet. 2006;368:1894–1907. doi: 10.1016/S0140-6736(06)69780-8. [DOI] [PubMed] [Google Scholar]
- 2.Chen J, Odenike O, Rowley JD. Leukaemogenesis: more than mutant genes. Nat Rev Cancer. 2010;10:23–36. doi: 10.1038/nrc2765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Qian J, Lin J, Qian W, Ma JC, Qian SX, Li Y, Yang J, Li JY, Wang CZ, Chai HY, Chen XX, Deng ZQ. Overexpression of miR-378 is frequent and may affect treatment outcomes in patients with acute myeloid leukemia. Leuk Res. 2013;37:765–768. doi: 10.1016/j.leukres.2013.03.014. [DOI] [PubMed] [Google Scholar]
- 4.Emmrich S, Katsman-Kuipers JE, Henke K, Khatib ME, Jammal R, Engeland F, Dasci F, Zwaan CM, den Boer ML, Verboon L, Stary J, Baruchel A, de Haas V, Danen-van Oorschot AA, Fornerod M, Pieters R, Reinhardt D, Klusmann JH, van den Heuvel-Eibrink MM. miR-9 is a tumor suppressor in pediatric AML with t(8;21) Leukemia. 2014;28:1022–1032. doi: 10.1038/leu.2013.357. [DOI] [PubMed] [Google Scholar]
- 5.Hager M, Pedersen CC, Larsen MT, Andersen MK, Hother C, Grønbæk K, Jarmer H, Borregaard N, Cowland JB. MicroRNA-130a-mediated down-regulation of Smad4 contributes to reduced sensitivity to TGF-beta1 stimulation in granulocytic precursors. Blood. 2011;118:6649–6659. doi: 10.1182/blood-2011-03-339978. [DOI] [PubMed] [Google Scholar]
- 6.Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6:857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 7.Visone R, Croce CM. MiRNAs and cancer. Am J Pathol. 2009;174:1131–1138. doi: 10.2353/ajpath.2009.080794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schotte D, Pieters R, Den Boer ML. MicroRNAs in acute leukemia: from biological players to clinical contributors. Leukemia. 2012;26:1–12. doi: 10.1038/leu.2011.151. [DOI] [PubMed] [Google Scholar]
- 9.Deng Z, Yang X, Fang L, Rutnam ZJ, Yang BB. Misprocessing and functional arrest of microRNAs by miR-Pirate: roles of miR-378 and miR-17. Biochem J. 2013;450:375–386. doi: 10.1042/BJ20120722. [DOI] [PubMed] [Google Scholar]
- 10.Deng Z, Du WW, Fang L, Shan SW, Qian J, Lin J, Qian W, Ma J, Rutnam ZJ, Yang BB. The intermediate filament vimentin mediates microRNA miR-378 function in cellular self-renewal by regulating the expression of the Sox2 transcription factor. J Biol Chem. 2013;288:319–331. doi: 10.1074/jbc.M112.418830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Franchina T, Amodeo V, Bronte G, Savio G, Ricciardi GR, Picciotto M, Russo A, Giordano A, Adamo V. Circulating miR-22, miR-24 and miR-34a as novel predictive biomarkers to pemetrexed-based chemotherapy in advanced non-small cell lung cancer. J Cell Physiol. 2014;229:97–99. doi: 10.1002/jcp.24422. [DOI] [PubMed] [Google Scholar]
- 12.Jiao F, Jin Z, Wang L, Wang L. Research and clinical applications of molecular biomarkers in gastrointestinal carcinoma (Review) Biomed Rep. 2013;1:819–827. doi: 10.3892/br.2013.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Salvi A, Abeni E, Portolani N, Barlati S, De Petro G. Human hepatocellular carcinoma cell-specific miRNAs reveal the differential expression of miR-24 and miR-27a in cirrhotic/non-cirrhotic HCC. Int J Oncol. 2013;42:391–402. doi: 10.3892/ijo.2012.1716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Song S, Zhou J, He S, Zhu D, Zhang Z, Zhao H, Wang Y, Li D. Expression levels of microRNA- 375 in pancreatic cancer. Biomed Rep. 2013;1:393–398. doi: 10.3892/br.2013.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Deng ZQ, Qian J, Liu FQ, Lin J, Shao R, Yin JY, Tang Q, Zhang M, He L. Expression level of miR-93 in formalin-fixed paraffin-embedded tissues of breast cancer patients. Genet Test Mol Biomarkers. 2014;18:366–70. doi: 10.1089/gtmb.2013.0440. [DOI] [PubMed] [Google Scholar]
- 16.Deng ZQ, Yin JY, Tang Q, Liu FQ, Qian J, Lin J, Shao R, Zhang M, He L. Over-expression of miR-98 in FFPE tissues might serve as a valuable source for biomarker discovery in breast cancer patients. Int J Clin Exp Pathol. 2014;7:1166–1171. [PMC free article] [PubMed] [Google Scholar]
- 17.Wang X, Tang S, Le SY, Lu R, Rader JS, Meyers C, Zheng ZM. Aberrant expression of oncogenic and tumor-suppressive microRNAs in cervical cancer is required for cancer cell growth. PLoS One. 2008;3:e2557. doi: 10.1371/journal.pone.0002557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Landgraf P, Rusu M, Sheridan R, Sewer A, Iovino N, Aravin A, Pfeffer S, Rice A, Kamphorst AO, Landthaler M, Lin C, Socci ND, Hermida L, Fulci V, Chiaretti S, Foà R, Schliwka J, Fuchs U, Novosel A, Müller RU, Schermer B, Bissels U, Inman J, Phan Q, Chien M, Weir DB, Choksi R, De Vita G, Frezzetti D, Trompeter HI, Hornung V, Teng G, Hartmann G, Palkovits M, Di Lauro R, Wernet P, Macino G, Rogler CE, Nagle JW, Ju J, Papavasiliou FN, Benzing T, Lichter P, Tam W, Brownstein MJ, Bosio A, Borkhardt A, Russo JJ, Sander C, Zavolan M, Tuschl T. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell. 2007;129:1401–1414. doi: 10.1016/j.cell.2007.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Volinia S, Calin GA, Liu CG, Ambs S, Cimmino A, Petrocca F, Visone R, Iorio M, Roldo C, Ferracin M, Prueitt RL, Yanaihara N, Lanza G, Scarpa A, Vecchione A, Negrini M, Harris CC, Croce CM. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci U S A. 2006;103:2257–2261. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen XX, Lin J, Qian J, Qian W, Yang J, Ma JC, Deng ZQ, Xie D, An C, Tang CY, Qian Z. Dysregulation of miR-124-1 predicts favorable prognosis in acute myeloid leukemia. Clin Biochem. 2014;47:63–66. doi: 10.1016/j.clinbiochem.2013.09.020. [DOI] [PubMed] [Google Scholar]
- 21.Li Y, Lin J, Yang J, Qian J, Qian W, Yao DM, Deng ZQ, Liu Q, Chen XX, Xie D, An C, Tang CY. Overexpressed let-7a-3 is associated with poor outcome in acute myeloid leukemia. Leuk Res. 2013;37:1642–1647. doi: 10.1016/j.leukres.2013.09.022. [DOI] [PubMed] [Google Scholar]
- 22.Li Z, Huang H, Li Y, Jiang X, Chen P, Arnovitz S, Radmacher MD, Maharry K, Elkahloun A, Yang X, He C, He M, Zhang Z, Dohner K, Neilly MB, Price C, Lussier YA, Zhang Y, Larson RA, Le Beau MM, Caligiuri MA, Bullinger L, Valk PJ, Delwel R, Lowenberg B, Liu PP, Marcucci G, Bloomfield CD, Rowley JD, Chen J. Up-regulation of a HOXA-PBX3 homeobox-gene signature following down-regulation of miR-181 is associated with adverse prognosis in patients with cytogenetically abnormal AML. Blood. 2012;119:2314–2324. doi: 10.1182/blood-2011-10-386235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xiong Y, Li Z, Ji M, Tan AC, Bemis J, Tse JV, Huang G, Park J, Ji C, Chen J, Bemis LT, Bunting KD, Tse W. MIR29B regulates expression of MLLT11 (AF1Q), an MLL fusion partner, and low MIR29B expression associates with adverse cytogenetics and poor overall survival in AML. Br J Haematol. 2011;153:753–757. doi: 10.1111/j.1365-2141.2011.08662.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lal A, Navarro F, Maher CA, Maliszewski LE, Yan N, O'Day E, Chowdhury D, Dykxhoorn DM, Tsai P, Hofmann O, Becker KG, Gorospe M, Hide W, Lieberman J. miR-24 Inhibits cell proliferation by targeting E2F2, MYC, and other cell-cycle genes via binding to “seedless” 3’UTR microRNA recognition elements. Mol Cell. 2009;35:610–625. doi: 10.1016/j.molcel.2009.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen L, Zhang A, Li Y, Zhang K, Han L, Du W, Yan W, Li R, Wang Y, Wang K, Pu P, Jiang T, Jiang C, Kang C. MiR-24 regulates the proliferation and invasion of glioma by ST7L via beta-catenin/Tcf-4 signaling. Cancer Lett. 2013;329:174–180. doi: 10.1016/j.canlet.2012.10.025. [DOI] [PubMed] [Google Scholar]
- 26.Akbari Moqadam F, Boer JM, Lange-Turenhout EA, Pieters R, den Boer ML. Altered expression of miR-24, miR-126 and miR-365 does not affect viability of childhood TCF3-rearranged leukemia cells. Leukemia. 2014;28:1008–14. doi: 10.1038/leu.2013.308. [DOI] [PubMed] [Google Scholar]
- 27.Lin SC, Liu CJ, Lin JA, Chiang WF, Hung PS, Chang KW. miR-24 up-regulation in oral carcinoma: positive association from clinical and in vitro analysis. Oral Oncol. 2010;46:204–208. doi: 10.1016/j.oraloncology.2009.12.005. [DOI] [PubMed] [Google Scholar]
- 28.Du WW, Fang L, Li M, Yang X, Liang Y, Peng C, Qian W, O’Malley YQ, Askeland RW, Sugg SL, Qian J, Lin J, Jiang Z, Yee AJ, Sefton M, Deng Z, Shan SW, Wang CH, Yang BB. MicroRNA miR-24 enhances tumor invasion and metastasis by targeting PTPN9 and PTPRF to promote EGF signaling. J Cell Sci. 2013;126:1440–1453. doi: 10.1242/jcs.118299. [DOI] [PubMed] [Google Scholar]
- 29.Bennett JM, Catovsky D, Daniel MT, Flandrin G, Galton DA, Gralnick HR, Sultan C. Proposed revised criteria for the classification of acute myeloid leukemia. A report of the French-American-British Cooperative Group. Ann Intern Med. 1985;103:620–625. doi: 10.7326/0003-4819-103-4-620. [DOI] [PubMed] [Google Scholar]
- 30.Kottaridis PD, Gale RE, Frew ME, Harrison G, Langabeer SE, Belton AA, Walker H, Wheatley K, Bowen DT, Burnett AK, Goldstone AH, Linch DC. The presence of a FLT3 internal tandem duplication in patients with acute myeloid leukemia (AML) adds important prognostic information to cytogenetic risk group and response to the first cycle of chemotherapy: analysis of 854 patients from the United Kingdom Medical Research Council AML 10 and 12 trials. Blood. 2001;98:1752–1759. doi: 10.1182/blood.v98.6.1752. [DOI] [PubMed] [Google Scholar]
- 31.Lin LI, Chen CY, Lin DT, Tsay W, Tang JL, Yeh YC, Shen HL, Su FH, Yao M, Huang SY, Tien HF. Characterization of CEBPA mutations in acute myeloid leukemia: most patients with CEBPA mutations have biallelic mutations and show a distinct immunophenotype of the leukemic cells. Clin Cancer Res. 2005;11:1372–1379. doi: 10.1158/1078-0432.CCR-04-1816. [DOI] [PubMed] [Google Scholar]
- 32.Kong KY, Owens KS, Rogers JH, Mullenix J, Velu CS, Grimes HL, Dahl R. MIR-23A microRNA cluster inhibits B-cell development. Exp Hematol. 2010;38:629–640. e621. doi: 10.1016/j.exphem.2010.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yin JY, Deng ZQ, Liu FQ, Qian J, Lin J, Tang Q, Wen XM, Zhou JD, Zhang YY, Zhu XW. Association between mir-24 and mir-378 in formalin-fixed paraffin-embedded tissues of breast cancer. Int J Clin Exp Pathol. 2014;7:4261–7. [PMC free article] [PubMed] [Google Scholar]
- 34.Zaidi SK, Dowdy CR, van Wijnen AJ, Lian JB, Raza A, Stein JL, Croce CM, Stein GS. Altered Runx1 subnuclear targeting enhances myeloid cell proliferation and blocks differentiation by activating a miR-24/MKP-7/MAPK network. Cancer Res. 2009;69:8249–8255. doi: 10.1158/0008-5472.CAN-09-1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li Y, Gao L, Luo X, Wang L, Gao X, Wang W, Sun J, Dou L, Li J, Xu C, Wang L, Zhou M, Jiang M, Zhou J, Caligiuri MA, Nervi C, Bloomfield CD, Marcucci G, Yu L. Epigenetic silencing of microRNA- 193a contributes to leukemogenesis in t(8;21) acute myeloid leukemia by activating the PTEN/PI3K signal pathway. Blood. 2013;121:499–509. doi: 10.1182/blood-2012-07-444729. [DOI] [PubMed] [Google Scholar]
- 36.Mrozek K, Marcucci G, Paschka P, Bloomfield CD. Advances in molecular genetics and treatment of core-binding factor acute myeloid leukemia. Curr Opin Oncol. 2008;20:711–718. doi: 10.1097/CCO.0b013e32831369df. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brioschi M, Fischer J, Cairoli R, Rossetti S, Pezzetti L, Nichelatti M, Turrini M, Corlazzoli F, Scarpati B, Morra E, Sacchi N, Beghini A. Down-regulation of microRNAs 222/221 in acute myelogenous leukemia with deranged core-binding factor subunits. Neoplasia. 2010;12:866–876. doi: 10.1593/neo.10482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gardini A, Cesaroni M, Luzi L, Okumura AJ, Biggs JR, Minardi SP, Venturini E, Zhang DE, Pelicci PG, Alcalay M. AML1/ETO oncoprotein is directed to AML1 binding regions and co-localizes with AML1 and HEB on its targets. PLoS Genet. 2008;4:e1000275. doi: 10.1371/journal.pgen.1000275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Steffen B, Knop M, Bergholz U, Vakhrusheva O, Rode M, Köhler G, Henrichs MP, Bulk E, Hehn S, Stehling M, Dugas M, Bäumer N, Tschanter P, Brandts C, Koschmieder S, Berdel WE, Serve H, Stocking C, Müller-Tidow C. AML1/ETO induces self-renewal in hematopoietic progenitor cells via the Groucho-related amino-terminal AES protein. Blood. 2011;117:4328–4337. doi: 10.1182/blood-2009-09-242545. [DOI] [PubMed] [Google Scholar]


