Abstract
REGgamma (REGγ) has been recently found in several types of human cancer, however, its clinical significance in metastasis and prognosis of breast cancer remains unknown. In this study, immunohistochemical staining and western blot analysis were performed to evaluate REGγ expression in both mouse and human breast cancer specimens. We found that in MMTV-PyMT mice, 14 out of 20 (70%) mouse mammary carcinomas were REGγ positive, which was significantly higher than control (0/20, 0%, P < 0.001) and lower than metastatic lung tumour (20/20, 100%, P = 0.027). Further investigation for REGγ expression in 136 human breast cancer tissues with the paired peritumoural normal breast tissues and 140 breast benign disease tissue samples showed that REGγ was undetectable in normal breast tissues and nonmetastatic axillary lymph nodes (ALNs), whereas 111 out of 136 (81.6%) breast cancer tissue samples were REGγ positive, which was significantly higher than breast benign disease tissues (9/140, 6.4%, P < 0.001) and lower than metastatic ALNs (116/116, 100%, P < 0.001). The 5-year disease-free and overall survivals of patients with negative/low level of REGγ were significantly higher than those of patients with high level of REGγ (P < 0.05). Cox regression analyses further indicated that REGγ could serve as a novel independent prognostic factor for breast cancer (OR = 4.369, P = 0.008). Our results suggest that the high expression of REGγ might predict metastasis and poor prognosis in breast cancer.
Keywords: REGgamma, breast cancer, prognosis, survival analysis
Introduction
Breast cancer is the most frequently diagnosed cancer among women worldwide [1,2]. While many breast cancer biomarkers have been identified for clinical practice, it is still difficult to match a particular patient with appropriate treatment because of the heterogeneity of cancer cells. Hence, there is an urgent need to identify new indicators for breast cancer diagnosis and prognosis.
Proteasome activator subunit 3, also known as Ki, PA28gamma or REGgamma (REGγ), was first identified as Ki antigen, which is a nuclear protein targeted by autoantibodies and is found in sera of systemic lupus erythematosus patients [3]. REGγ is a member of 11S proteasome regulator which includes three subunits: alpha, beta and gamma [4-6]. Although the biological roles of REGγ have not been completely understood, it has been reported that REGγ stimulated the proteolytic activity of 20S proteasome core, whereby protein substrates could diffuse into the environment of proteasomes [7,8]. The function of REGγ was elucidated by gene-targeting and while REGγ-deficient mice were born without appreciable abnormalities, but growth retardation and cell specific mitotic defects were observed [9]. The REGγ-deficient embryonic fibroblasts showed impeded entry from G to S phase in cell cycle [10]. Recently, it had been reported that REGγ was highly expressed in multiple human cancers, such as breast cancer [11,12], thyroid cancer [13,14], hepatocellular carcinoma [15] and colorectal cancer [16-18]. In breast cancer, REGγ enhanced the oncogenicity of breast cancer cells by promoting cell growth, inhibiting cell apoptosis, degrading p21 and suppressing activation of NK, suggesting that REGγ is involved in multiple processes of cancer progression [12]. However, the clinical significance of REGγ expression in breast cancer has not been systemically studied. In the present study, we examined the expression of REGγ in mouse mammary tumour virus-polyomavirus middle T antigen transgenic mouse model (MMTV-PyMT), as well as in surgical specimens of human breast cancer, axillary lymph node (ALN), breast benign disease and normal breast tissues, and analyzed the relationship between REGγ expression and prognosis of breast cancer patients as well as clinical pathological parameters. REGγ was highly expressed in breast cancer tissues and metastatic ALNs, but not in normal breast tissues and rarely in breast benign disease tissues. REGγ expression was positively correlated with poor clinical parameters and prognosis of patients with breast cancer. These results suggested that REGγ could be a novel prognostic indicator for breast cancer.
Materials and methods
Mouse samples
MMTV-PyMT mice (FVB/N-Tg MMTV-PyVT 634 Mul/J mice, #002374) and control FVB ones (FVB/NJ, nontransgenic, #001800) were obtained from Jackson Laboratory (Bar Harbor, Maine, USA) and bred in the specific pathogen free level animal facilities. Experimental mice were generated from the same cohort of breeding pairs. All pups were weaned and tail-clipped at age 4 weeks, genotyped for MMTV-PyMT transgene (5’-GGAAAGTCACTAGGAGCAGGG-3’ and 5’-GGAAGCAAGTACTTCACAAGGG-3’). Animal care and use followed National Research Council guide for the laboratory animals.
Patient specimens
A total of 136 breast cancer with paired peritumoural (> 3 cm) normal breast tissue and 140 breast benign disease tissue paraffin specimens were obtained from patients who experienced open surgical excision from December 2008 to April 2009 in breast disease center of Southwest Hospital, Third Military Medical University, Chongqing, China. The paired metastatic ALNs were included as well. As a validation of REGγ expression pattern as detected by immunohistochemical (IHC) staining, another set of fresh breast tissue samples (30 breast cancers and paired peritumoural normal breast tissues, ALNs and 30 breast benign disease tissues) were collected for the western blot analysis. All pathological diagnoses were performed by pathology department of Southwest Hospital. All patients enrolled in this study were completely monitored for 5 years. This study was in agreement with the Helsinki Declaration and approved by the ethics committee of Southwest Hospital, the informed consent forms were signed by all the subjects.
Immunohistochemistry and scoring
Sections were deparaffinized and rehydrated, antigen retrieval was performed in a pressure cooker with sodium citrate buffer (10 mM/L, pH 6.0). The endogenous peroxidase activity was blocked by 3% hydrogen peroxide in methanol. Sections were blocked with serum-free blocker (DAKO, USA, #X0909), and incubated with anti-REGγ monoclonal antibody (Invitrogen, USA, #710800, at 1:600 dilution) for 12 hours at 4°C. Sections were incubated with EnVision + System-HRP antibody (DAKO, USA, #K4010) for 15 minutes at room temperature before DAB (DAKO, USA, #K3467) staining. Finally, sections were counterstained with Mayer’s haematoxylin (DAKO, USA, #S3309). A REGγ-positive section was used as positive control, and the same concentration of non-immune rabbit IgG was applied as negative control.
Immunoreactivity was scored for the extent and intensity of the nuclear staining. Extent of positivity was scored as follows: 0, no positive cells; 1, < 25% positive cells; 2, 25-50% positive cells; 3, 50-75% positive cells and 4, > 75% positive cells. Intensity was scored as follows: 0, no positive staining; 1, weak staining; 2, moderate staining and 3, strong staining. Multiplying extent by intensity gave the REGγ staining scores from negative expression (0), positive low expression (1-6) to positive high expression (> 6) accordingly. The REGγ scoring was valuated independently by two investigators in a blinded fashion, and the scores were thereafter averaged.
Western blot analysis
For tissue protein extraction, samples were snap-frozen in liquid nitrogen and homogenized in RIPA buffer with proteinase inhibitors. Protein was quantified by ELISA plate reader (BIO-TEK Synergy HT, USA). Total protein lysate (30 μg) was separated in 10% SDS-PAGE and transferred to nitrocellulose membranes (BIO-RAD, USA, #162-0115). Membranes were blocked with Blotting-Grade Blocker (BIO-RAD, USA, #170-6404) and incubated with anti-REGγ monoclonal antibody (Invitrogen, USA, #710800, at 1:1000 dilution). Blots were developed by using the enhanced chemiluminescence method (Thermo Scientific, USA, #1856135/6). To verify the equal loading, blots were stripped and re-probed for β-actin.
Statistical analysis
Correlations between REGγ expression and clinicopathological parameters were studied with the chi-square test. Survival rates were estimated by Kaplan-Meier method, survival curves were compared with the Log-rank (Mantel-Cox) test. The Cox proportional hazards regression analysis was applied for patient prognosis. A value of P < 0.05 was considered as a significant difference. GraphPad Prism 5.01 (GraphPad Software, Inc., La Jolla, CA, USA) and SPSS statistics 20 (IBM Corp., Armonk, NY, USA) were used for analyses.
Results
REGγ is highly expressed in MMTV-PyMT mouse mammary carcinomas and metastatic lung tumours
REGγ expression was investigated in MMTV-PyMT mouse mammary carcinoma samples. DNA genotyping was performed to confirm mice genetic backgrounds (Figure 1A). As shown in Figure 1B, all MMTV-PyMT virgin female mice (20/20, 100%) developed mammary gland carcinoma (a) and metastatic lung tumour (b) at age 18 weeks, whereas no tumour was found in the control FVB normal mice (Figure 1C). Figure 1D showed MMTV-PyMT mice sample IHC staining results, low level of REGγ expression in mammary carcinoma (a) was associated with weak REGγ expression in metastatic lung tumour (c) whereas the high level of REGγ expression in mammary carcinoma (b) was correlated with strong REGγ expression in metastatic lung tumour (d). Statistics showed that all MMTV-PyMT mice metastatic lung tumours were REGγ positive, and most of the metastatic tumours (17/20, 85%) had high level of REGγ expression (Figure 1E). The IHC staining showed that mammary gland tissues in all FVB control mice were REGγ negative (0/20), while REGγ positive expression was detected in 14 out of 20 (70%) MMTV-PyMT mouse mammary carcinomas and all the metastatic lung tumours (20/20, 100%). REGγ expression in MMTV-PyMT mouse mammary carcinoma was significantly higher than FVB normal mouse mammary gland tissue (P < 0.001) and lower than metastatic lung tumour (P = 0.027) (Table 1). REGγ protein expression level was next validated by western blot. As shown in Figure 1F and 1G, there was no obvious REGγ protein expression in FVB mouse normal breast tissue and normal lung tissue, but a significant high level of REGγ expression was detected in MMTV-PyMT mouse breast cancer tissue (P = 0.0051) and metastatic lung tumour (P = 0.0190). In a set of MMTV-PyMT mice overall survival analysis (10 mice for each group), mice with the high expression level of REGγ showed lower percent of survival than that of mice with the low expression of REGγ (Figure 1H). These data suggested that MMTV-PyMT was an ideal mouse model to further investigate the biological functions of REGγ in breast cancer research. Moreover, these results further indicated a prognostic role of REGγ in MMTV-PyMT mouse mammary gland carcinoma progression.
Figure 1.
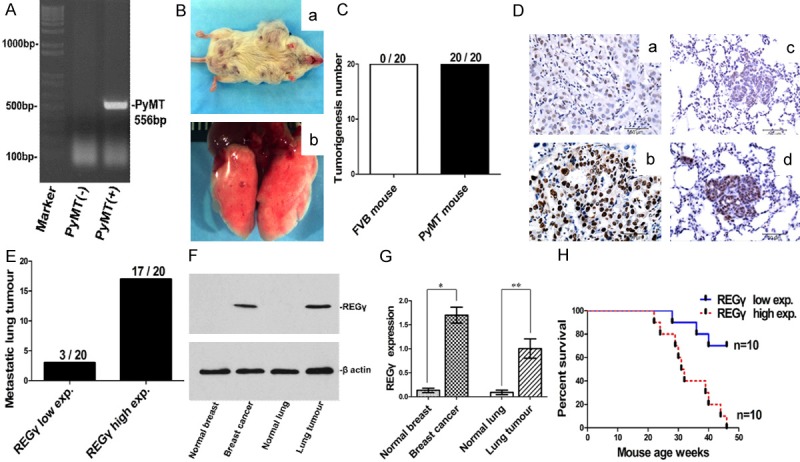
REGγ expression in mouse mammary carcinoma and metastatic lung tumour. A. DNA genotyping showing MMTV-PyMT transgene band at 556 bp. B. Mammary carcinoma (a) and metastatic lung tumour (b) in MMTV-PyMT mice. C. Tumorigenesis was found in all MMTV-PyMT mice (20/20, 100%) whereas no tumour was found in FVB control mice (0/20, 0%). D. Representative REGγ IHC staining images for mouse mammary cacinoma and metastatic lung tumour (bar = 50 μm). REGγ low expression in mammary carcinoma (a) was associated with low level of REGγ expression in metastatic lung tumour (c), and REGγ high expression in mammary carcinoma (b) was correlated with high level of REGγ expression in the metastatic lung tumour (d). E. Statistical analysis showed that all metastatic lung tumours were REGγ positive (20/20, 100%): 17 metastatic tumours had high level of REGγ expression and 3 tumours had low level of REGγ expression. F. Representative Western blot result of REGγ expression in mouse fresh tissue. G. Densitometry analysis showing relative expression of REGγ as detected by Western blot in fresh samples (20 mice for each group). Statistical analysis showed the significant difference between normal breast tissue and breast cancer tissue (P = 0.0051), as well as between normal lung tissue and metastatic lung tumour (P = 0.0190). H. Survival analysis in a set of MMTV-PyMT mouse (10 mice for each group) showed that mice with high expression of REGγ had significantly poor survival than those with low expression of REGγ (Log-rank Mantel-Cox Test, P = 0.0013, Hazard Ratio = 0.1448, 95% CI of ratio is 0.04474 to 0.4686).
Table 1.
REGγ expression in mouse mammary gland, mammary carcinoma and lung tumour
| REGγ expression | ||||||
|---|---|---|---|---|---|---|
|
|
||||||
| Case | Neg. | low | high | Positive (%) | P-value | |
| Normal mammary gland | 20 | 20 | 0 | 0 | 0/20 (0%) | |
| < 0.001 | ||||||
| Mammary carcinoma | 20 | 6 | 5 | 9 | 14/20 (70%) | |
| 0.027 | ||||||
| Metastatic lung tumour | 20 | 0 | 3 | 17 | 20/20 (100%) | |
The expression of REGγ in mouse mammary carcinoma was significantly higher than normal mammary gland tissue (P < 0.001) and lower than metastatic lung tumour (P = 0.027).
REGγ is highly expressed in human breast cancer tissue
Out of the 136 breast cancer samples evaluated by IHC, 25 specimens (18.4%) were REGγ negative while 111 specimens (81.6%) were REGγ positive. REGγ was undetectable in normal breast tissues and normal ALN (Figure 2Aa and 2Ab). Breast cancer tissue with low expression level of REGγ (c) was associated with low level of REGγ expression in ALN (d), whereas breast cancer tissue with high expression level of REGγ (e) was correlated with high level of REGγ expression in ALN (f). The positive rate of REGγ expression in breast cancer tissue (111/136, 81.6%) was significantly higher than normal breast tissue (0/136, 0%) as well as breast benign disease tissue (9/140, 6.4%) (P < 0.001), and lower than metastatic ALN (116/116, 100%) (P < 0.001) (Table 2). Consistent with the IHC data, western blot analysis in human fresh tissue samples did not detect REGγ protein expression in normal breast tissue and normal ALN. In contrast to control normal tissues, a significant higher level of REGγ expression was detected in breast cancer tissue (P = 0.0024) as well as in metastatic ALN (P = 0.0007) (Figure 2B, 2C). The expression of REGγ in human breast tissue was in accordance with its expression pattern in metastatic ALN, which suggested a potential prognostic role of REGγ in human breast cancer.
Figure 2.
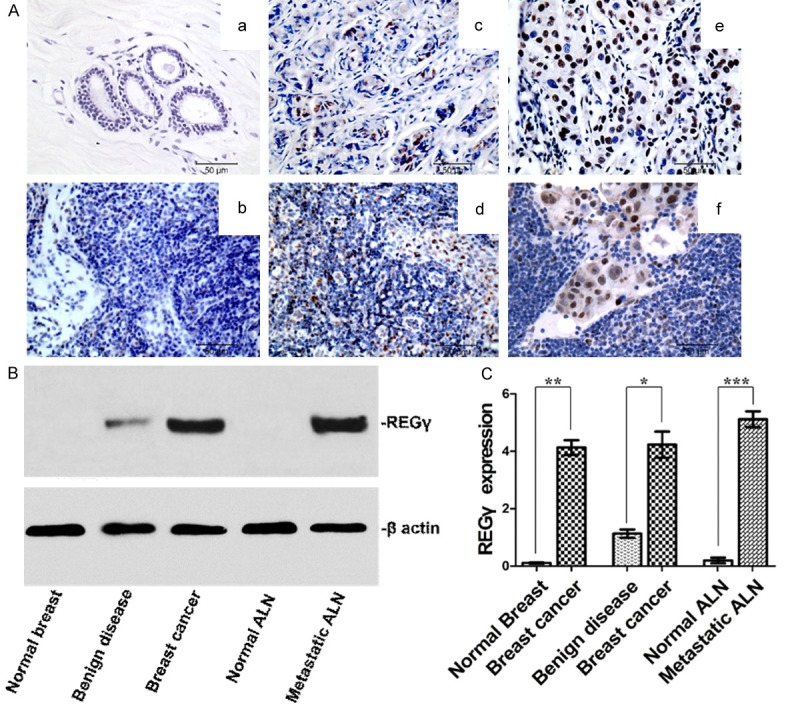
REGγ expression in human normal breast tissue, normal axillary lymph node (ALN), breast cancer tissue and metastatic ALN. A. Representative REGγ IHC staining images (bar = 50 μm). No REGγ expression was found in normal breast tissue (a) and normal ALN (b). Breast cancer tissue with low expression level of REGγ (c) was associated with low level of REGγ expression in ALN (d). Breast cancer tissue with high expression level of REGγ (e) was correlated with high level of REGγ expression in ALN (f). B. Representative western blot result of REGγ expression in a different set of human fresh tissue. C. Densitometry analysis showing relative expression of REGγ as detected by western blot (30 for each). Statistical analysis showed significant difference between normal breast and breast cancer (P = 0.0012), breast benign disease and breast cancer (P = 0.0024), as well as between normal ALN and metastatic ALN (P = 0.0007).
Table 2.
REGγ expression in human breast tissue and metastatic axillary lymph node
| REGγ expression | ||||||
|---|---|---|---|---|---|---|
|
|
||||||
| Case | Neg. | Low | High | Positive % | P-value | |
| Benign disease | 140 | 131 | 9 | 0 | 6.4% | |
| 0.008 | ||||||
| Normal breast | 136 | 136 | 0 | 0 | 0% | |
| < 0.001 | ||||||
| Breast cancer | 136 | 25 | 52 | 59 | 81.6% | |
| < 0.001 | ||||||
| Metastatic ALN | 116 | 0 | 29 | 87 | 100% | |
ALN, axillary lymph node. REGγ expression in breast benign disease tissue was significantly higher than normal breast (P = 0.008). REGγ expression in breast cancer was significantly higher than normal breast tissues (P < 0.001) and lower than metastatic ALN (P < 0.001).
REGγ expression is positively correlated with poor clinicopathological features
Using chi-square test, REGγ expression level did not correlate with breast cancer patient age (mean age = 50) (P = 0.702), patient menopausal state (P = 0.171), progesterone receptor (PR) status (P = 0.211) or human epidermal growth factor receptor-2 (HER-2) status (P = 0.557). However, REGγ expression was positively correlated with breast tumour size (P < 0.001), ALN metastasis state (P < 0.001), ALN metastasis number (P < 0.001), tumour TNM stage (P = 0.038), histological differentiation (P = 0.001) and estrogen receptor-α (ERα) status (P < 0.001). Our data indicated a significant positive correlation between REGγ expression and the poor clinicopathological features in patients with breast cancer (Table 3).
Table 3.
Relationship between REGγ expression and cancer patient clinicopathological features
| Characteristics | REGγ expression | |||
|---|---|---|---|---|
|
|
||||
| Case (%) | Negative (%) | Positive (%) | P-value | |
| Age | 0.702 | |||
| ≤ 50 | 76 (55.9%) | 23 (30.3%) | 53 (69.7%) | |
| > 50 | 60 (44.1%) | 20 (33.3%) | 40 (66.7%) | |
| Menopausal state | 0.171 | |||
| No | 80 (58.8%) | 42 (52.5%) | 38 (47.5%) | |
| Yes | 56 (41.2%) | 36 (64.3%) | 20 (35.7%) | |
| Tumour size | < 0.001 | |||
| T1 + T2 | 84 (61.8%) | 58 (69.0%) | 26 (31.0%) | |
| T3 + T4 | 52 (38.2%) | 17 (32.7%) | 35 (67.3%) | |
| ALN metastasis state | < 0.001 | |||
| No | 20 (14.7%) | 20 (100%) | 0 (0%) | |
| Yes | 116 (85.3%) | 0 (0%) | 116 (100%) | |
| ALN metastasis number | < 0.001 | |||
| < 4 | 66 (48.5%) | 46 (69.7%) | 20 (30.3%) | |
| ≥ 4 | 70 (51.5%) | 16 (22.9%) | 54 (77.1%) | |
| TNM Stage | 0.038 | |||
| 0 + I | 79 (58.1%) | 39 (49.4%) | 40 (50.6%) | |
| II + III | 57 (41.9%) | 18 (31.6%) | 39 (68.4%) | |
| Histological differentiation | 0.001 | |||
| well | 67 (49.3%) | 36 (53.7%) | 31 (46.3%) | |
| Poor + moderate | 69 (50.7%) | 18 (26.1%) | 51 (73.9%) | |
| Estrogen receptor-α | < 0.001 | |||
| Negative | 59 (43.4%) | 33 (55.9%) | 26 (44.1%) | |
| Positive | 77 (56.6%) | 12 (15.6%) | 65 (84.4%) | |
| Progesterone receptor | 0.211 | |||
| Negative | 58 (42.6%) | 27 (46.6%) | 31 (53.4%) | |
| Positive | 78 (57.4%) | 28 (35.9%) | 50 (64.1%) | |
| HER-2 | 0.557 | |||
| Negative | 92 (67.6%) | 34 (37.0%) | 58 (63.0%) | |
| Positive | 44 (32.4%) | 14 (31.9%) | 30 (68.1%) | |
ALN, axillary lymph node. HER-2, Human epidermal growth factor receptor-2. P = Statistical significance by chi-square test. A two-sided P < 0.05 was considered to be statistically significant.
REGγ expression is correlated with poor clinical prognosis
At the last follow up point (April 30, 2014), there were 101 breast cancer patients alive while 35 deceased. None of breast benign disease patients experienced recurrence or progression to invasive cancer. The Kaplan-Meier survival curves demonstrated 5-year survival rate of breast cancer patients. The period covered range from 0 to 64 months after the initial surgery. Significant difference in breast cancer patient clinical outcome was found between REGγ negative expression and REGγ low expression groups (OS, Log-rank Mantel-Cox Test, P = 0.0406. Hazard Ratio = 0.2913, 95% CI of ratio is 0.08157 to 1.040. DFS, Log-rank Mantel-Cox Test, P = 0.0405. Hazard Ratio = 2.432, 95% CI of ratio is 0.9813 to 6.027) as well as between REGγ low expression and REGγ high expression groups (OS, Log-rank Mantel-Cox Test, P = 0.0351. Hazard Ratio = 0.4949, 95% CI of ratio is 0.2516 to 0.9736. DFS, Log-rank Mantel-Cox Test, P < 0.0001. The median survival time in REGγ low expression group is 57 mon-ths, and 14 months in REGγ high expression group, Hazard Ratio = 4.071, 95% CI of ratio is 3.505 to 4.638) (Figure 3A, 3B). In breast cancer patient, the Cox proportional hazards regression analysis data indicated that the expression level of REGγ could be an independent prognostic factor for breast cancer (P = 0.008, OR = 4.369, 95% CI of ratio is 1.482 to 12.880). ERα positive was a protective factor (OR = 0.152, 95% CI of ratio is 0.073 to 0.312) and ALN metastasis was a risk factor (OR = 1.648, 95% CI of ratio is 1.141 to 2.382) for breast cancer patients in this study (Figure 3C-E). Collectively, based on the current survival analysis results, a significant positive correlation was established between REGγ positive expression and breast cancer patient poor clinical outcomes, which suggesting an important prognostic significance of REGγ expression in breast cancer.
Figure 3.
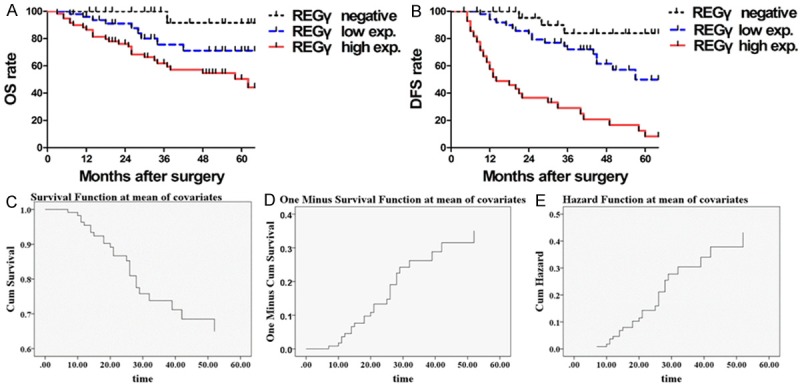
The relationship between REGγ expression and prognoses of patients with breast cancer. A. The Kaplan-Meier graph for overall survival (OS) rate for breast cancer patients: patients with low expression of REGγ had longer OS than those with high expression of REGγ (Log-rank Mantel-Cox Test, P = 0.0351. Hazard Ratio = 0.4949, 95% CI of ratio is 0.2516 to 0.9736) and poor OS rate compare to REGγ negative patients (Log-rank Mantel-Cox Test, P = 0.0406. Hazard Ratio = 0.2913, 95% CI of ratio is 0.08157 to 1.040). B. Kaplan-Meier graph for disease-free survival (DFS) for patients: patients with low expression of REGγ had longer DFS than those with high expression of REGγ (Log-rank Mantel-Cox Test, P < 0.0001. The median survival time in REGγ low expression group was 57 months, and 14 months in REGγ high expression group, Hazard Ratio = 4.071, 95% CI of ratio is 3.505 to 4.638), however, has shorter DFS as compared to those with negative expression of REGγ (Log-rank Mantel-Cox Test, P = 0.0405. Hazard Ratio = 2.432, 95% CI of ratio is 0.9813 to 6.027). As showed by survival function diagram (C), one minus survival function diagram (D) and hazard function diagram (E), REGγ expression played role as an independent prognostic factor for the breast cancer (Cox Regression analyses, P = 0.008, OR = 4.369, 95% CI is 1.482 to 12.880).
Discussion
As a proteasomal regulator, REGγ has been reported to be associated with the degradation of both oncogenic and tumour suppressing proteins such as HCV core protein [19,20], SRC-3 [21], p16, p19, p21 [22,23], and p53 [24-26]. Despite the recent progress in REGγ-related studies, the biological function and tissue distribution of this protein remained unclear, especially in breast cancer research. In the present study, we showed direct proof that REGγ is a candidate biomarker to distinguish malignant and normal breast tissues in both human and MMTV-PyMT mouse samples. As shown by western blot analyses, REGγ protein was highly expressed in human breast cancer tissue, metastatic ALN, MMTV-PyMT mouse mammary gland carcinoma as well as in metastatic lung tumour, which is highly consistent with the results from IHC staining. The capability of REGγ expression level to be used as biomarker to distinguish breast cancers from normal breast and breast benign disease tissues suggested that REGγ has potential to be used as a new indicator for breast cancer. Our findings that REGγ was highly expressed in human breast cancer tissue and metastatic tumour were in accordance with the previous REGγ-related studies [11,12]. Compared with the reports in which polyclonal PA28γ antibody was used, we had improved IHC technique to detect REGγ expression by using anti-REGγ monoclonal antibody, which we have shown to yield more accurate and credible results. So far, we did not find the correlation between REGγ expression and HER-2 expression status, which is different with the previous report [11]. Considering that both studies only used a small sample size in research, the further larger sample size investigation is still needed for better understating the relationship between REGγ expression and HER-2 status. Interestingly, our data showed a significant high positive rate of REGγ expression in ERα positive human breast cancer tissue samples (65/77, 84.4%) compared to ERα negative ones (26/59, 44.1%) (Table 3), which may shed light on the therapeutic significance of REGγ in breast cancer endocrine therapy, and we will further investigate the possible mechanisms in our future intensive studies.
We also examined the expression pattern of REGγ in MMTV-PyMT transgenic mouse, which is a long-term used mammary gland carcinoma mouse model. Mammary gland specific expression of PyMT under the control of MMTV promoter/enhancer resulted in widespread transformation of mouse mammary epithelium and the development of multifocal mammary adenocarcinomas as well as the metastatic lesions in lungs [27-29]. MMTV-PyMT mouse has short tumorigenesis latency, high penetrance and high incidence of metastatic lung tumour occurring independently of pregnancy and with reproducible kinetics of progression [30-35]. In our study, the expression level of REGγ in MMTV-PyMT mouse mammary gland carcinoma and metastatic lung tumour were comparable to that in human breast cancer tissue and the metastatic ALN. REGγ expression in MMTV-PyMT mouse was exact mimic of human breast cancer tumorigenesis and progression, as well it implied a potential role of REGγ to be a new indicator for mammary gland carcinomas in this model. These results further implied that MMTV-PyMT mouse will be a relevant model to further investigate the biological functions of REGγ in breast cancer pathogenesis and progression.
REGγ expression level was positively correlated with the poor clinicopathological features, such as bigger breast tumour size, higher TNM stage, poor histological differentiation and more number of metastatic ALNs in breast cancer patients. In addition, patients with REGγ high level expression exhibited lower DFS and OS rate compared with REGγ negative/low level expression breast cancer patients. In comparison to earlier REGγ-related investigations, our study is the first report which demonstrates that high level of REGγ expression is positively correlated with poor clinical outcomes in breast cancer patients with complete 5-year follow up data. These findings not only improved current knowledge of REGγ biological functions, and will also shed light on therapeutic potential of REGγ for the future investigations.
In view of our results, this study is consistent with previous reports that REGγ is highly expressed in human breast cancers. We subsequently addressed the significance of REGγ expression in breast cancer as a new biological marker for the prognostic application.
Acknowledgements
This work was supported by China Scholarship Council (CSC) joint PhD Dissertation Research Program from Chinese Ministry of Education (No. 2010761014).
Disclosure of conflict of interest
None.
References
- 1.Ferlay J, Soerjomataram I, Ervik M, Dikshit R, Eser S, Mathers C, Rebelo M, Parkin DM, Forman D, Bray F. GLOBOCAN 2012 v1.0, Cancer Incidence and Mortality Worldwide: IARC Cancer Base No. 11 [Internet] Lyon, France: International Agency for Research on Cancer. Available at: http: //www.iarc.fr.
- 2.Bray F, Ren JS, Masuyer E, Ferlay J. Global estimates of cancer prevalence for 27 sites in the adult population in 2008. Int J Cancer. 2013;132:1133–1145. doi: 10.1002/ijc.27711. [DOI] [PubMed] [Google Scholar]
- 3.Nikaido T, Shimada K, Shibata M, Hata M, Sakamoto M, Takasaki Y, Sato C, Takahashi T, Nishida Y. Cloning and nucleotide sequence of cDNA for Ki antigen, a highly conserved nuclear protein detected with sera from patients with systemic lupus erythematosus. Clin Exp Immunol. 1990;79:209–214. doi: 10.1111/j.1365-2249.1990.tb05180.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ma CP, Slaughter CA, DeMartino GN. Identification, purification, and characterization of a protein activator (PA28) of the 20 S proteasome (macropain) J Biol Chem. 1992;267:10515–10523. [PubMed] [Google Scholar]
- 5.Dubiel W, Pratt G, Ferrell K, Rechsteiner M. Purification of an 11 S regulator of the multicatalytic protease. J Biol Chem. 1992;267:22369–22377. [PubMed] [Google Scholar]
- 6.Tanahashi N, Yokota K, Ahn JY, Chung CH, Fujiwara T, Takahashi E, DeMartino GN, Slaughter CA, Toyonaga T, Yamamura K, Shimbara N, Tanaka K. Molecular properties of the proteasome activator PA28 family proteins and gamma-interferon regulation. Genes Cells. 1997;2:195–211. doi: 10.1046/j.1365-2443.1997.d01-308.x. [DOI] [PubMed] [Google Scholar]
- 7.Gao X, Li J, Pratt G, Wilk S, Rechsteiner M. Purification procedures determine the proteasome activation properties of REG gamma (PA28 gamma) Arch Biochem Biophys. 2004;425:158–164. doi: 10.1016/j.abb.2004.03.021. [DOI] [PubMed] [Google Scholar]
- 8.Mao I, Liu J, Li X, Luo H. REGgamma, a proteasome activator and beyond. Cell Mol Life Sci. 2008;65:3971–3980. doi: 10.1007/s00018-008-8291-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Murata S, Kawahara H, Tohma S, Yamamoto K, Kasahara M, Nabeshima Y, Tanaka K, Chiba T. Growth retardation in mice lacking the proteasome activator PA28gamma. J Biol Chem. 1999;274:38211–38215. doi: 10.1074/jbc.274.53.38211. [DOI] [PubMed] [Google Scholar]
- 10.Barton LF, Runnels HA, Schell TD, Cho Y, Gibbons R, Tevethia SS, Deepe GS Jr, Monaco JJ. Immune defects in 28-kDa proteasome activator gamma-deficient mice. J Immunol. 2004;172:3948–3954. doi: 10.4049/jimmunol.172.6.3948. [DOI] [PubMed] [Google Scholar]
- 11.Wang X, Tu S, Tan J, Tian T, Ran L, Rodier JF, Ren G. REG gamma: a potential marker in breast cancer and effect on cell cycle and proliferation of breast cancer cell. Med Oncol. 2011;28:31–41. doi: 10.1007/s12032-010-9546-8. [DOI] [PubMed] [Google Scholar]
- 12.Tian M, Xiaoyi W, Xiaotao L, Guosheng R. Proteasomes reactivator REG gamma enchances oncogenicity of MDA-MB-231 cell line via promoting cell proliferation and inhibiting apoptosis. Cell Mol Biol (Noisy-le-grand) 2009;55(Suppl):OL1121–1131. [PubMed] [Google Scholar]
- 13.Okamura T, Taniguchi S, Ohkura T, Yoshida A, Shimizu H, Sakai M, Maeta H, Fukui H, Ueta Y, Hisatome I, Shigemasa C. Abnormally high expression of proteasome activator-gamma in thyroid neoplasm. J Clin Endocrinol Metab. 2003;88:1374–1383. doi: 10.1210/jc.2002-021413. [DOI] [PubMed] [Google Scholar]
- 14.Zhang M, Gan L, Ren GS. REGgamma is a strong candidate for the regulation of cell cycle, proliferation and the invasion by poorly differentiated thyroid carcinoma cells. Braz J Med Biol Res. 2012;45:459–465. doi: 10.1590/S0100-879X2012007500035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jia HL, Ye QH, Qin LX, Budhu A, Forgues M, Chen Y, Liu YK, Sun HC, Wang L, Lu HZ, Shen F, Tang ZY, Wang XW. Gene expression profiling reveals potential biomarkers of human hepatocellular carcinoma. Clin Cancer Res. 2007;13:1133–1139. doi: 10.1158/1078-0432.CCR-06-1025. [DOI] [PubMed] [Google Scholar]
- 16.Roessler M, Rollinger W, Mantovani-Endl L, Hagmann ML, Palme S, Berndt P, Engel AM, Pfeffer M, Karl J, Bodenmuller H, Ruschoff J, Henkel T, Rohr G, Rossol S, Rosch W, Langen H, Zolg W, Tacke M. Identification of PSME3 as a novel serum tumor marker for colorectal cancer by combining two-dimensional polyacrylamide gel electrophoresis with a strictly mass spectrometry-based approach for data analysis. Mol Cell Proteomics. 2006;5:2092–2101. doi: 10.1074/mcp.M600118-MCP200. [DOI] [PubMed] [Google Scholar]
- 17.Hong Y, Ho KS, Eu KW, Cheah PY. A susceptibility gene set for early onset colorectal cancer that integrates diverse signaling pathways: implication for tumorigenesis. Clin Cancer Res. 2007;13:1107–1114. doi: 10.1158/1078-0432.CCR-06-1633. [DOI] [PubMed] [Google Scholar]
- 18.Jiang X, Tan J, Li J, Kivimae S, Yang X, Zhuang L, Lee PL, Chan MT, Stanton LW, Liu ET, Cheyette BN, Yu Q. DACT3 is an epigenetic regulator of Wnt/beta-catenin signaling in colorectal cancer and is a therapeutic target of histone modifications. Cancer Cell. 2008;13:529–541. doi: 10.1016/j.ccr.2008.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Suzuki R, Moriishi K, Fukuda K, Shirakura M, Ishii K, Shoji I, Wakita T, Miyamura T, Matsuura Y, Suzuki T. Proteasomal turnover of hepatitis C virus core protein is regulated by two distinct mechanisms: a ubiquitin-dependent mechanism and a ubiquitin-independent but PA28gamma-dependent mechanism. J Virol. 2009;83:2389–2392. doi: 10.1128/JVI.01690-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Moriishi K, Shoji I, Mori Y, Suzuki R, Suzuki T, Kataoka C, Matsuura Y. Involvement of PA28gamma in the propagation of hepatitis C virus. Hepatology. 2010;52:411–420. doi: 10.1002/hep.23680. [DOI] [PubMed] [Google Scholar]
- 21.Li X, Lonard DM, Jung SY, Malovannaya A, Feng Q, Qin J, Tsai SY, Tsai MJ, O’Malley BW. The SRC-3/AIB1 coactivator is degraded in an ubiquitin- and ATP-independent manner by the REGgamma proteasome. Cell. 2006;124:381–392. doi: 10.1016/j.cell.2005.11.037. [DOI] [PubMed] [Google Scholar]
- 22.Li X, Amazit L, Long W, Lonard DM, Monaco JJ, O’Malley BW. Ubiquitin- and ATP-independent proteolytic turnover of p21 by the REGgamma-proteasome pathway. Mol Cell. 2007;26:831–842. doi: 10.1016/j.molcel.2007.05.028. [DOI] [PubMed] [Google Scholar]
- 23.Chen X, Barton LF, Chi Y, Clurman BE, Roberts JM. Ubiquitin-independent degradation of cell-cycle inhibitors by the REGgamma proteasome. Mol Cell. 2007;26:843–852. doi: 10.1016/j.molcel.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang Z, Zhang R. Proteasome activator PA28 gamma regulates p53 by enhancing its MDM2-mediated degradation. EMBO J. 2008;27:852–864. doi: 10.1038/emboj.2008.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu J, Yu G, Zhao Y, Zhao D, Wang Y, Wang L, Liu J, Li L, Zeng Y, Dang Y, Wang C, Gao G, Long W, Lonard DM, Qiao S, Tsai MJ, Zhang B, Luo H, Li X. REGgamma modulates p53 activity by regulating its cellular localization. J Cell Sci. 2010;123:4076–4084. doi: 10.1242/jcs.067405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ali A, Wang Z, Fu J, Ji L, Liu J, Li L, Wang H, Chen J, Caulin C, Myers JN, Zhang P, Xiao J, Zhang B, Li X. Differential regulation of the REGgamma-proteasome pathway by p53/TGF-beta signalling and mutant p53 in cancer cells. Nat Commun. 2013;4:2667. doi: 10.1038/ncomms3667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Guy CT, Cardiff RD, Muller WJ. Induction of mammary tumors by expression of polyomavirus middle T oncogene: a transgenic mouse model for metastatic disease. Mol Cell Biol. 1992;12:954–961. doi: 10.1128/mcb.12.3.954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lin EY, Jones JG, Li P, Zhu L, Whitney KD, Muller WJ, Pollard JW. Progression to malignancy in the polyoma middle T oncoprotein mouse breast cancer model provides a reliable model for human diseases. Am J Pathol. 2003;163:2113–2126. doi: 10.1016/S0002-9440(10)63568-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fluck MM, Schaffhausen BS. Lessons in signaling and tumorigenesis from polyomavirus middle T antigen. Microbiol Mol Biol Rev. 2009;73:542–563. doi: 10.1128/MMBR.00009-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stewart TA, Pattengale PK, Leder P. Spontaneous mammary adenocarcinomas in transgenic mice that carry and express MTV/myc fusion genes. Cell. 1984;38:627–637. doi: 10.1016/0092-8674(84)90257-5. [DOI] [PubMed] [Google Scholar]
- 31.Sinn E, Muller W, Pattengale P, Tepler I, Wallace R, Leder P. Coexpression of MMTV/v-Ha-ras and MMTV/c-myc genes in transgenic mice: synergistic action of oncogenes in vivo. Cell. 1987;49:465–475. doi: 10.1016/0092-8674(87)90449-1. [DOI] [PubMed] [Google Scholar]
- 32.Schoenenberger CA, Andres AC, Groner B, van der Valk M, LeMeur M, Gerlinger P. Targeted c-myc gene expression in mammary glands of transgenic mice induces mammary tumours with constitutive milk protein gene transcription. EMBO J. 1988;7:169–175. doi: 10.1002/j.1460-2075.1988.tb02797.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sandgren EP, Schroeder JA, Qui TH, Palmiter RD, Brinster RL, Lee DC. Inhibition of mammary gland involution is associated with transforming growth factor alpha but not c-myc-induced tumorigenesis in transgenic mice. Cancer Res. 1995;55:3915–3927. [PubMed] [Google Scholar]
- 34.Cardiff RD, Anver MR, Gusterson BA, Hennighausen L, Jensen RA, Merino MJ, Rehm S, Russo J, Tavassoli FA, Wakefield LM, Ward JM, Green JE. The mammary pathology of genetically engineered mice: the consensus report and recommendations from the Annapolis meeting. Oncogene. 2000;19:968–988. doi: 10.1038/sj.onc.1203277. [DOI] [PubMed] [Google Scholar]
- 35.Maglione JE, Moghanaki D, Young LJ, Manner CK, Ellies LG, Joseph SO, Nicholson B, Cardiff RD, MacLeod CL. Transgenic Polyoma middle-T mice model premalignant mammary disease. Cancer Res. 2001;61:8298–8305. [PubMed] [Google Scholar]


