Abstract
The hepatitis C virus (HCV) core protein is an important causative agent in HCV related hepatocellular carcinoma (HCC). Tumor suppressor gene PTEN appears to act in the liver at the crossroad of processes controlling cell proliferation. In this study we investigated the effect of the HCV core protein on the PTEN pathway in hepatocarcinogenesis. The HCV core was transfected stably into HepG2 cell. The effect of HCV core on cell proliferation and viability were detected by 3-(4, 5)-dimethylthiahiazo-(-z-y1)-3, 5-di-phenytetrazoliumromide (MTT) assay, clonogenic survival assay and Fluorescence Activating Cell Sorter (FACS) analysis. The expressions of PTEN were detected by real time RT-PCR and/or Western blot analysis, also the mechanism of down-regulation of PTEN was explored by western blot, luciferase assay and RNA interference. We found the HCV core promoted cell proliferation, survival and G2/M phase accumulation. It downregulated PTEN at mRNA and protein level and activated PTEN downstream gene Akt accompanied with NF-κB activation. Furthermore, the inhibition of HCV core by its specific shRNAs decreased the effect of growth promotion and G2/M phase arrest, inhibited the expression of nuclear p65 and increased PTEN expression. The activity of PTEN was restored when treated with NF-κB inhibitor PDTC. By luciferase assay we found that NF-κB inhibited PTEN promoter transcription activity directly in HCV core cells, while PDTC was contrary. Our study suggests that HCV proteins could modulate PTEN by activating NF-κB. Furthermore strategies designed to restore the expression of PTEN may be promising therapies for preventing HCV dependent hepatocarcinogenesis.
Keywords: Core protein, hepatocellular carcinoma, hepatitis C virus, PTEN
Introduction
Hepatocellular carcinoma (HCC) is one of the most common malignant tumors in the world especially in China which ranks third among all malignancies both in incidence and in mortality. Hepatitis C virus (HCV) is an important causative agent which often leads to liver cirrhosis and hepatocellular carcinoma. However, the precise role of HCV in hepatocarcinogenesis remains unknown. Among HCV viral proteins, the hepatitis C virus core protein is likely to be important determinant in mediating pathological effects of HCV through several of its multifunctional activities. The HCV core protein has been shown to possess a number of regulatory functions for viral and cellular gene expression as well as oncogenic potential [1,2]. It was reported that transgenic mice expressing the HCV core gene developed hepatocellular carcinoma [3].
The phosphoinositide 3-kinase (PI3K)/Akt signaling pathway plays an important role in carcinogenesis through regulating of cell growth and survival in several cell systems. Among the pathway PTEN is the famous strong negative controller by inhibiting PIP3 and inactivation of Akt phosphorylation [4]. Although frequent loss of expression of the PTEN gene have been found in HCC [5,6], the role of the HCV core protein in the regulation of PTEN in HCC has not been previously studied. Furthermore to understand the role of the HCV core protein in hepatocarcinogenesis, it is necessary to investigate whether the HCV core protein can modulate the PI3/Akt pathway through inhibiting PTEN function during the transformation processes.
In this study, we investigated the effect of the HCV core protein on the expression of PTEN. We reported that the HCV core protein decreased the expression of PTEN through activating nuclear factor-κB (NF-κB) by inhibiting the transcriptional activity of PTEN in its promoter region, which resulting in rapid cell proliferation.
Material and methods
Cell line, plasmids and reagents
The human liver cancer cell line HepG2 was purchased from the Cell Bank of Type Culture Collection of Chinese Academy of Sciences (Shanghai, China). The cells were cultured at 37°C with 5% CO2 and 95% air in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% (v/v) fetal bovine serum (FBS).
The expression vector pEGFP-C3 (Clontech, Palo Alto, CA, USA), which contains both the green fluorescence protein (GFP) coding sequences and a neomycin resistance cassette (Neor), was used as a negative control. pEGFP-Core which expresses the full-length coding sequences of core region (amino acids 1-191) of the HCV 1b genotype were constructed by our group previously. The eukaryotic expression vector pGenesil1 for construction of HCV core shRNA was purchased from Wuhan GeneSil Biotechnology Co. Led (Wuhan, China). The NF-κB inhibitor PDTC was obtained from Sigma (Saint Louis, MO, USA).
In vitro DNA stable transfection
On the day before transfection, 3.0 × 105 cells were seeded into each well of 6-well culture plates. The cells were transfected with pEGFP-Core using LipofectamineTM 2000 (Gibco) in accordance with the instructions of the manufacturer. At 24 h after transfection, the media was replaced with DMEM supplemented with 10% (v/v) FBS and G-418 sulfate (G418) at 800 μg /ml for selection and formation of single colonies of cells. After about 14 days, the single colonies were isolated and maintained in media containing 200 μg/ml of G418. The empty vector pEGFP-C3 transfectants we used as mock group.
Regular and real-time reverse transcription-polymerase chain reaction (RT-PCR)
Total RNA was isolated using TRIzol reagent (Invitrogen). One microgram of total RNA from each sample was used to synthesize first-strand cDNA using the Revert Aid TM H Minus first strand cDNA synthesis kit (Fermentas). Expression of HCV core mRNA was detected by regular RT-PCR using the following primers: forward: 5’-CGGGATCCATGAGCACGAATCCTAAACCTCAA-3’, reverse: 5’-GGAATTCTTAGGCTGAAGCGGGCACAGT-3’. Each 25 μl PCR reaction mixture contained 1.25 U rTaq, 10 μM sense primer, 10 μM antisense primer, 1 μg of cDNA. The PCR conditions were 95°C for 5 min, followed by 40 cycles of 95°C for 30 s, 52.0°C for 30 s, and 72°C for 50 s, with final 7-min incubation at 72°C. Expression of PTEN mRNA was detected by real-time RT-PCR using the following primers: forward: 5’-GACGAACTGGTGTAATGATATGTGC-3’, reverse: 5’-AAAGTACATGAACTTGTCTTCCCGT-3’. β-actin was used as the internal reference; The primers were: forward: 5’-ATCATGTTTGAGACCTTCAACA-3’, reverse: 5’-CATCTCTTGCTCGAAGTCCA-3’. The real-time RT-PCR reaction mixture contained 7.5 μl of 2 × iQTM SYBR® Green Supermix (Bio-Rad Laboratories), 0.33 M sense primer, 0.33 M antisense primer, 1 μg of cDNA, and water to a final volume of 15 μl. The PCR conditions were 95°C for 3 min, followed by 40 cycles of 94°C for 30 s, 68.0°C for 30 s, and 72°C for 30 s. Relative mRNA expression was calculated using the 2-ΔΔCt method.
MTT analysis
Cells were seeded into 96-well plates at 1000 cells/well and incubated at 37°C. At different time intervals (1, 2, 3, 4 and 5 days), the culture medium was removed and 3-(4, 5)-dimethylthiahiazo-(-z-y1)-3, 5-di-phenytetrazoliumromide (MTT) dye (5 mg/ml) was added into each well, followed by incubaton for a further 4 h. The reaction was stopped by adding dimethyl sulfoxide (DMSO), and the absorbance was measured at a wavelength of 490 nm in a microplate reader (BioRad Model 680, Hercules, California, USA). Experiments were repeated at six independent experiments done at different times.
Clonogenic survival assay
For clonogenic survival assay, cells were seeded in six-well tissue culture dishes with 100 cells each dish. After 14 days, the cells were washed and stained with crystal violet, and the colonies containing > 50 cells were counted. Plating efficiency was calculated by dividing the average number of cell colonies per well by the number of cells plated.
Cell-cycle analysis
Cells were harvested and fixed in cold 70% ethanol and incubated overnight at 4°C. Fixed cells were then resuspended in 100 μl and stained with propidium iodide (PI) (50 μg/ml) after treatment with 100 μl of ribonuclease A (RNase A) (10 mg/ml). The stained cells were analyzed for DNA content using a FACScan instrument (Becton Dickinson, Franklin Lakes, New Jersey, USA).
Western blot analysis
Whole cell lysates or nuclear proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), transferred to polyvinylidene difluoride (PVDF) membranes, probed with primary and secondary antibodies, and visualized by Enhanced chemiluminescence (ECL) (Pierce, Rockford, IL, USA). Antibodies were obtained from Meridian Life Science (Memphis, TN, USA; HCV core protein), NeoMarker (Fremont, Ca, USA; PTEN), Cell Signaling Technologies (Danvers, MA, USA; phospho-Akt (Ser-473), p65 (NF-κB) and IκB-α) and Santa Cruz (Santa Cruz, CA, USA; β-actin).
Luciferase assay
A PCR fragment encompassing the human PTEN promoter was amplified with the primers 5’-ggtaccGAGGAGTGGCACCAGTTTG-3’ (forward) and 5’-ctcgagGCTGCTCAGTGTAGAGGGAA-3’ (reverse). The PCR fragment was cut with the restriction enzymes KpnI and XhoI and inserted in the corresponding sites of the pGL3 promoter reporter vector (Promega, Madison, WI, USA). Cell transfections were conducted. Briefly, 1 × 105 HepG2 cells transfected with HCV core or mock were seeded into each well of 24-well culture plates the day before transfection. The cells were transfected using LipofectamineTM 2000 (Gibco, Grand Island, NY, USA) and luciferase reporter plasmid PTEN promoter. After 5 h of incubation, the transfection mix was replaced with fresh medium with DMEM supplemented with 20% (v/v) FBS and 16 h after transfection the medium was replaced with 60 μmol/L PDTC. Luciferase activity was measured at 12 h after PDTC treatment by using the Dual-Luciferase Reporter Assay System (Promega). Cells were also transfected with pGL3 Control and pGL3 Promoter (Promega) which served as positive and negative controls, respectively.
Construction of HCV core shRNA
The double-stranded shRNA (short-hairpin RNA, shRNA) fragments containing nine base loop regions, a terminator sequence, and restriction enzyme sites (BamHI and HindIII) specifically targeted HCV core gene nt 370-390 with the published sequence (GenBank accession no. AF054247) as Liu et al previous described [7], and they were cloned into the BamHI and HindIII restriction enzyme site position in the eukaryotic expression vector pGenesil1 carrying green fluorescent protein and the Neor gene. The specific shRNA sequences as follows: sense: 5’-GATCCAAACCAAACGTAACACCAACCTTCAAGACG GGTTGGTGTTACGTTTGGTTTTTTTTTGTCGACA-3’; antisense: 5’-AGCTTGTCGACAAAAAAAAACCAAACGTAACACCAACCCGTCTTGAAGGTTGGTGTTACGTTTGGTTTG-3’. Scrambled-shRNA was used asnegative control for silencing and the sequencesas follows: sense: 5’-GATCCGACTTCATAAGGCGCATGCTTCAAGACGGCATGCGCCTTATGAAGTCTTTTTTGTCGACA-3’; antisense: 5’-AGCTTGTCGACAAAAAAGACTTCATAAGGCGCATGCCGTCTTGAAGCATGCGCCTTATGAAGTCG-3’.
RNA interference
To confirm the effects of HCV core on PTEN expression and cell proliferation further, the transformant with core were transfected with core specific or scrambled shRNAs. Briefly, 3 × 105 HepG2 cells transfected with HCV core were seeded into each well of 6-well culture plates the day before transfection. Cells were transiently transfected with core specific or scrambled shRNAs in serum free media using Lipofectamine™ 2000 (Invitrogen) at a ratio of 1:3 (plasmid: Lipofectamine) according to the manufacturer’s protocol. After 6 hrs incubation at 37°C in 5% CO2, the complete culture medium was added to the cells. Cells were then harvested for gene expression, protein analysis and Cell-cycle analysis after 48 hours.
Statistical analysis
Statistical analysis was performed using SPSS software for Windows. All analyzes used two-sided hypothesis tests. The differences between experimental groups were determined using the t-test and one-way analysis of variance (ANOVA).
Results
HCV core enhanced cell growth and survival in vitro
The pEGFP-core which can express the HCV core protein was stably transfected into human liver cancer cell line HepG2. By RT-PCR and Western blotting analysis we found that there were high HCV core mRNA and protein expression in the stable transfectant while no HCV core expression in pEGFP-C3 cells (mock group) (Figures 1, 2).
Figure 1.
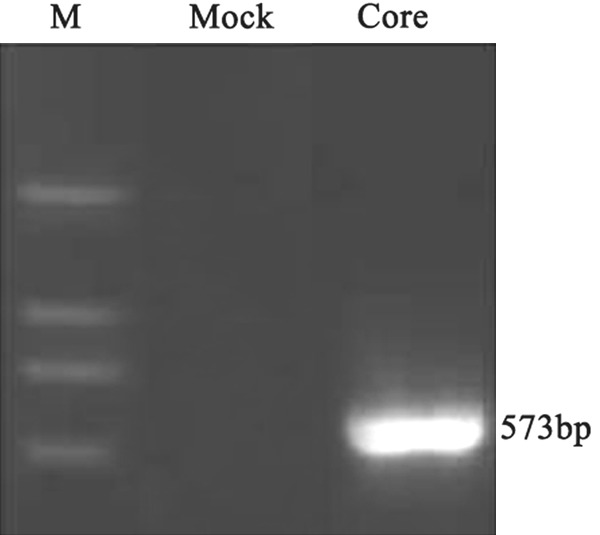
RT-PCR analysis showed a high HCV core mRNA expression in the HCV core stable transfectant. Mock: cells transfected with empty vector pEGFP-C3; Core: cells transfected with HCV core.
Figure 2.
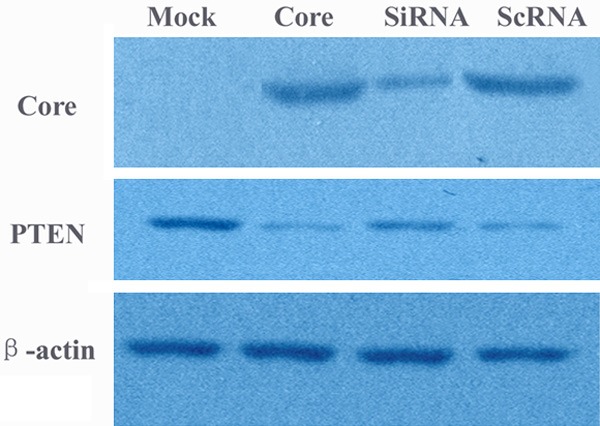
Western blotting analysis for HCV core and PTEN expression. Mock: cells transfected with empty vector pEGFP-C3; Core: cells transfected with HCV core; SiRNA: transient transfection of core specific shRNAs in the HCV core transformant; ScRNA: transient transfection of scrambled shRNAs in the HCV core transformant.
We carried out an MTT assay and found that the absorbance from pEGFP-core transfectants between the second day and fifth day was significantly higher than that from mock transfectants (P < 0.05) indicating that HCV core expression caused an increase in the cell proliferation (Figure 3). Also the viability of cells was assessed by a clonogenic survival assay. As shown in Figure 4, the plating efficiency in the cells expressing core protein (81.0 ± 2.0) was increased compared to the mock group (57.0 ± 3.0). Fluorescence Activating Cell Sorter (FACS) analysis revealed that HepG2 core positive cells displayed an accumulation of cells in G2/M phase (33% ± 1.25 vs. 12% ± 2.46 of control cells) (Figure 5).
Figure 3.
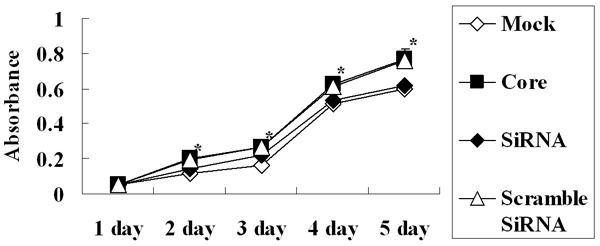
HCV core enhanced cell proliferation. MTT assay demonstrated that the absorbance from HCV core transfectant was higher than that from mock while the inhibition of HCV core by its specific shRNAs decreased the effect of growth promotion. Each data point represents the mean ± SD of six independent experiments. *P < 0.05 between groups.
Figure 4.
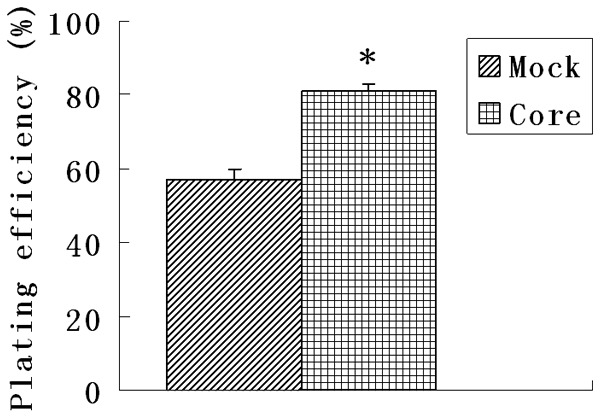
HCV core enhanced cell survival. The viability of cells was assessed by a clonogenic survival assay. As shown in the figure the plating efficiency in the cells expressing core protein increased compared to the mock groups. Each data point represents the mean ± SD of three independent experiments. *P < 0.05.
Figure 5.
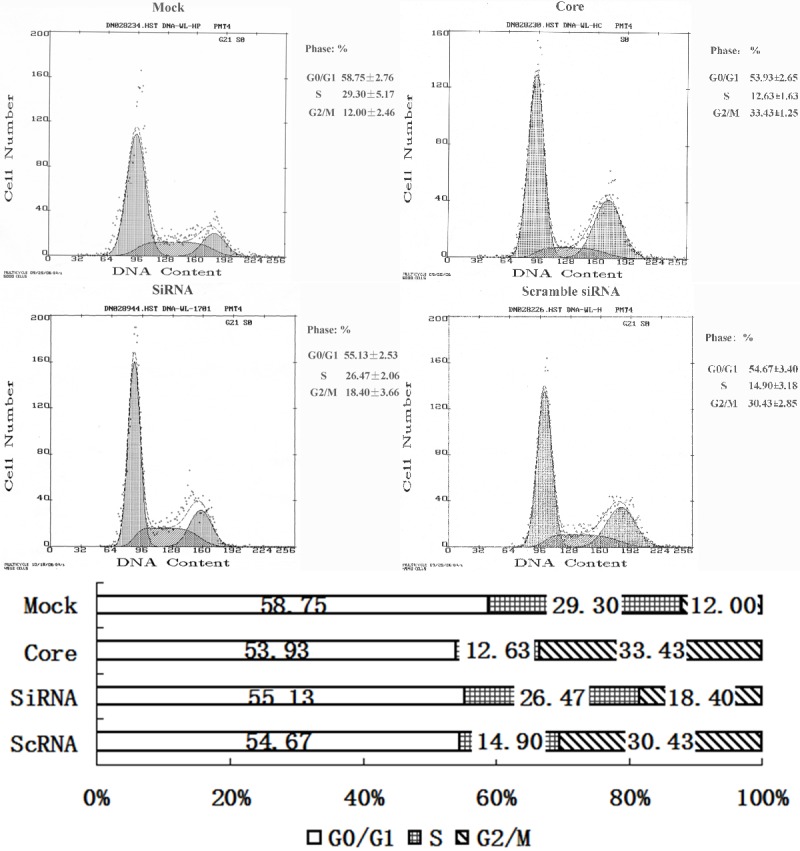
Fluorescence Activating Cell Sorter (FACS) analysis revealed that HepG2 core positive cells displayed an accumulation of cells in G2/M phase while HCV core specific shRNAs suppressed G2/M phase accumulation (P < 0.05).
HCV core decreased PTEN expression
By real time RT-PCR and Western blot, we also found that PTEN gene could be obviously down-regulated both at mRNA and protein level in core-group while there was high PTEN expression in mock cells (Figures 2, 6). The mRNA level in core-group reduced about 11.1 times compared to the mock-group (P < 0.05). In addition, considering the inhibition of PTEN on the activation of AKT the expression of phosphorylated Akt (pAkt) was carried out by Western blot and we found a reduction in PTEN expression by HCV core protein resulted in the activation of AKT protein with an increase in AKT phosphorylated form (pAkt) in HCV core-expressing cells (Figure 7).
Figure 6.
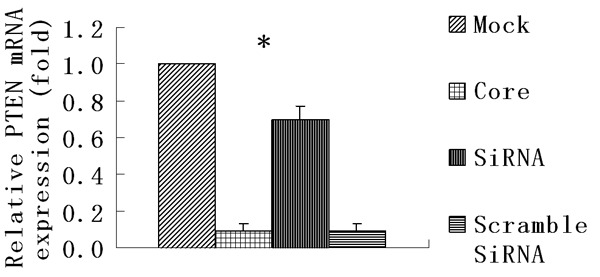
HCV core decreased PTEN expression at mRNA level by real time RT-PCR analysis. Each data point represents the mean ± SD of three independent experiments. *P < 0.05.
Figure 7.
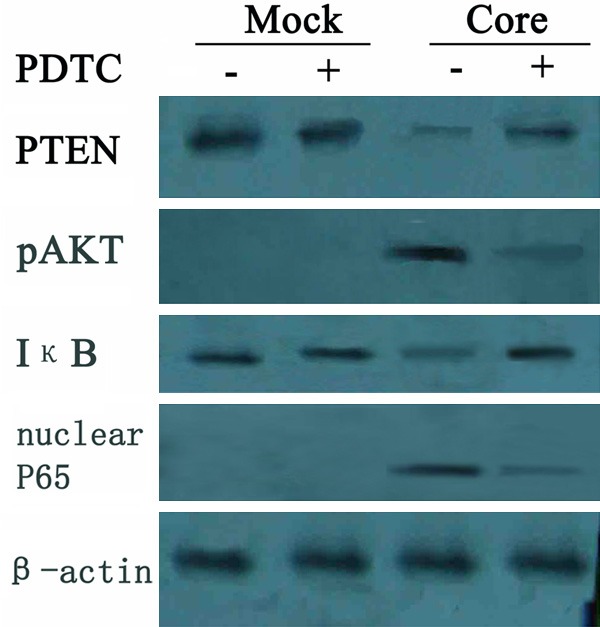
HCV core down-regulated PTEN through activating NF-κB. Western blot detection revealed a decrease in both PTEN and IκB (inhibitor of NF-κB) expression and an increase in pAkt and nuclear p65 (activated NF-κB) in HCV core group. But the changes were suppressed when treated with NF-κB inhibitor PDTC. In the mock group no obvious difference were found with PDTC treatment.
HCV core down-regulated PTEN through activating NF-κB
NF-κB is a key transcription factor controlling a variety of cellular functions. The pivotal roles of it have been described in abnormal cell growth, immune and inflammatory responses through the regulation of pro- and anti-inflammatory genes as well as in oncogenesis. Its activity is regulated by IκB (inhibitor of NF-κB) isoforms by complexing with NF-κB in the cytoplasm and preventing nuclear translocation. As reported HCV core protein can markedly activate nuclear factor-κB (NF-κB) in hepatic cells [8-10], we hypothesized that the down-regulation of PTEN expression by HCV core may be induced by the activation of NF-κB.
By Western blot detection we found that concomitant with PTEN decrease, a similar reduction in IκB with an obvious increase of nuclear p65 was also observed, demonstrating HCV core induced IκB (inhibitor of NF-κB) degradation and then activated NF-κB. But the reduction of PTEN was suppressed when treated with NF-κB inhibitor PDTC with a decrease expression in pAkt in HCV core cells (Figure 7).
In addition, we constructed a reporter plasmid containing 1,100 bp of the human PTEN promoter which containing six possible NF-κB binding sites (NCBI accession no. NM_000314) inserted upstream of the luciferase reporter gene. By luciferase assay we found the relative luciferase activity decreased to 0.41 ± 0.04 in core-group and resumed to 0.86 ± 0.08 when treated with PDTC while the relative luciferase activity in mock-group and mock-group with PDTC treatment were 1.20 ± 0.09 and 1.14 ± 0.19. There was a significant decrease of luciferase activity in the HCV core cells compared with the mock cells and the luciferase activity in the HCV core cells was increased when treated with NF-κB inhibitor PDTC, while no significant response was observed in mock cells with PDTC treatment (Figure 8). These results demonstrated that HCV core protein could down-express PTEN by activating NF-κB which may inhibit the transcriptional activity of PTEN in its promoter region.
Figure 8.
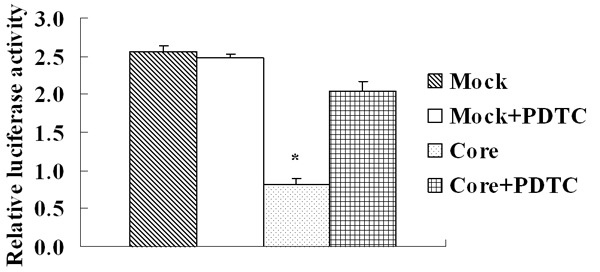
Luciferase assay displayed that there was a significant decrease of luciferase activity in the HCV core cells compared with the mock cells and the luciferase activity in the HCV core cells was increased when treated with NF-κB inhibitor PDTC, while no significant response was observed in mock cells with PDTC treatment. Each data point represents the mean ± SD of three independent experiments. *P < 0.05.
Inhibition of core protein expression by shRNAs reduced cell growth and PTEN expression
Next we utilized gene silencing against HCV core to investigate its involvement in the regulation of cell growth and PTEN expression. Transient transfection of core specific shRNAs after 48 hrs showed a dramatic reduction of protein expression of HCV core (Figure 2) in the HCV core transformant. MTT revealed that the inhibition of HCV core by its specific shRNAs decreased the effect of growth promotion by HCV core and also the effect of G2/M phase arrest was suppressed (Figures 3 and 5). The effect of HCV core shRNAs on mRNA expression level of PTEN in HCV core transfected cells was further determined. We found HCV core shRNAs dramatically resumed PTEN mRNA expression for 7.8 fold compared to the HCV core group (P < 0.05) (Figure 6). Complementary to increased mRNA levels, PTEN protein expression was also improved with HCV core shRNAs (Figure 2). Besides, a similar reduction of nuclear p65 was also observed as compared to control scramble shRNA (Figure 9), demonstrating activated NF-κB was inhibited partly.
Figure 9.
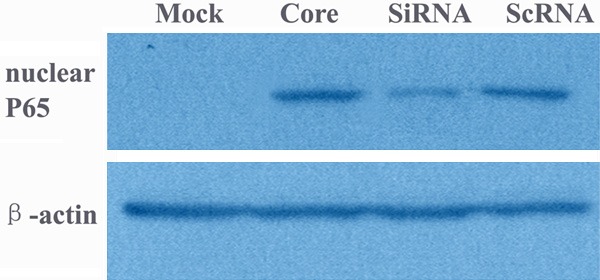
Western blotting analysis for nuclear p65. Mock: cells transfected with empty vector pEGFP-C3; Core: cells transfected with HCV core; SiRNA: transient transfection of core specific shRNAs in the HCV core transformant; ScRNA: transient transfection of scrambled shRNAs in the HCV core transformant.
Discussion
PTEN (phosphatase and tensin homolog deleted in chromosome 10) is a tumor suppressor gene which is the first found tumor suppressor gene functioned with a dual-specificity lipid and protein phosphatase. PTEN negatively controls the phosphoinositide 3-kinase/AKT signaling pathway involved in the regulation of cell growth and survival. A reduced PTEN function determines a marked increase in PIP3 and activation of AKT survival signaling pathways, leading to inhibition of apoptosis and contributing to tumor formation [4,11]. Frequent genetic alterations and loss of expression of the PTEN gene have been found in a variety of human cancers including HCC [5,6]. Yet, despite the important role played by PTEN in cellular processes and the extensive characterization of mutations in human cancers, only a few reports have evaluated the regulation of PTEN expression. A TNF-α-induced down-regulation of PTEN mediated through a TNF-α/NIK/NF-κB pathway has been described [12-14]. Another study has suggested that the Hepatitis B virus-X protein (HBx) could downregulate PTEN mRNA and protein expression leading to uncontrolled Akt activation in Chang liver cells [15,16].
The hepatitis C virus core protein is likely to be important determinant in mediating pathological effects of HCV [1,2]. It was also reported that transgenic mice expressing the HCV core gene developed hepatocellular carcinoma. The core protein of HCV can modulate Rb pathway through pRb down-regulation and induce the expression of FAS ligand and the activity of NF-κB [17,18]. But studies about HCV mediate alterations of PTEN expression and subsequent hepatocytes dysfunctions are scarce.
In this study, we found that the HCV core downregulated PTEN at the transcriptional levels and activated its downstream gene Akt. The HepG2 cells expressing HCV core protein also obtained stronger activity of growth and survival and G2/M phase cells accumulation that were similar to the results of others with core transfected HepG2 cells [19,20], but in contrast to results for Huh-7 cells expressing core [21]. This results were also in accordance with the results in PTEN deficient HCC cells [22] suggesting a possible role of HCV core protein in carcinogenesis by inhibiting the expressing of PTEN and activating Akt. RNA interference with HCV core specific shRNAs further proved that inhibition of HCV core could decrease growth promotion and suppress G2/M phase accumulation. Also the mRNA and protein expression level of PTEN were resumed when HCV core protein was inhibited.
As far as we know, there isn’t a direct effect of the core protein on PTEN and none binding sites of HCV core could been found in PTEN promoter. We deemed the decrease of PTEN by HCV core protein might via regulating other factors. Considering that NF-κB could down-regulate PTEN at the transcriptional levels [13,14,23] and NF-κB played an important role in the process of HCV core carcinogenesis [8,10,24], we presumed if HCV core protein modulated PTEN through activating NF-κB. We found an activation of NF-κB was accompanied with PTEN decrease in HCV core expressing cells. The inhibition of HCV core protein expression by its specific shRNAs also inhibited the expression of nuclear p65 and increased PTEN expression indicated that depression of HCV core could suppress the activation of NF-κB and then resume PTEN activity. Furthermore, we also found that the activity of PTEN was restored when treated with NF-κB inhibitor PDTC with an increase expression of PTEN and a decrease expression in pAkt. In addition, by luciferase assay we demonstrated that NF-κB inhibited PTEN promoter transcription activity directly in HCV core cells, while NF-κB inhibitor PDTC was contrary. These results indicated that HCV core protein down-regulated PTEN mediated through activating NF-κB.
It is worth noting that the downstream inhibited Akt by PTEN can also induce NF-κB activity [4,25,26]. Akt can activate IKK (IκB kinase) by phosphorylation which promotes dissociation of IκB from NF-κB, and then induces NF-κB activity and translocates into the nucleus [27]. The HCV core down-regulates PTEN through activating NF-κB and increases pAkt also suggests an interesting positive feedback loop, where activated Akt further facilitates NF-κB activation. Such a feedback loop may augment HCV core activity in tumor growth and invasion.
Taken together these results suggest that HCV proteins could modulate PTEN by activating NF-κB. These data suggest a possible role of HCV core protein in carcinogenesis by inhibiting the expressing of tumor suppressor gene. Furthermore strategies designed to restore the expression of PTEN may be promising therapies for preventing HCV dependent hepatocarcinogenesis.
Acknowledgements
This study was supported by the National Nature Science Foundation of China (Grant No. 81201759) and the Applied & Basic Research Funds of Yunnan Province, China (Grant No. 2012FB210).
Disclosure of conflict of interest
None.
References
- 1.Siavoshian S, Abraham JD, Kieny MP, Schuster C. HCV core, NS3, NS5A and NS5B proteins modulate cell proliferation independently from p53 expression in hepatocarcinoma cell lines. Arch Virol. 2004;149:323–36. doi: 10.1007/s00705-003-0205-7. [DOI] [PubMed] [Google Scholar]
- 2.Watashi K, Shimotohno K. The roles of hepatitis C virus proteins in modulation of cellular functions: a novel action mechanism of the HCV core protein on gene regulation by nuclear hormone receptors. Cancer Sci. 2003;94:937–43. doi: 10.1111/j.1349-7006.2003.tb01381.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tanaka N, Moriya K, Kiyosawa K, Koike K, Aoyama T. Hepatitis C virus core protein induces spontaneous and persistent activation of peroxisome proliferator-activated receptor alpha in transgenic mice: implications for HCV-associated hepatocarcinogenesis. Int J Cancer. 2008;122:124–31. doi: 10.1002/ijc.23056. [DOI] [PubMed] [Google Scholar]
- 4.Asano T, Yao Y, Zhu J, Li D, Abbruzzese JL, Reddy SA. The PI 3-kinase/Akt signaling pathway is activated due to aberrant Pten expression and targets transcription factors NF-kappaB and c-Myc in pancreatic cancer cells. Oncogene. 2004;23:8571–80. doi: 10.1038/sj.onc.1207902. [DOI] [PubMed] [Google Scholar]
- 5.Wang L, Wang WL, Zhang Y, Guo SP, Zhang J, Li QL. Epigenetic and genetic alterations of PTEN in hepatocellular carcinoma. Hepatol Res. 2007;37:389–96. doi: 10.1111/j.1872-034X.2007.00042.x. [DOI] [PubMed] [Google Scholar]
- 6.Peyrou M, Bourgoin L, Foti M. PTEN in liver diseases and cancer. World J Gastroenterol. 2010;16:4627–33. doi: 10.3748/wjg.v16.i37.4627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu M, Ding H, Zhao P, Qin Z, Gao J, Cao M, Luan J, Wu W, Qi Z. RNA interference effectively inhibits mRNA accumulation and protein expression of hepatitisC virus core and E2 genes in human cells. Biosci Biotechnol Biochem. 2006;70:2049–55. doi: 10.1271/bbb.60001. [DOI] [PubMed] [Google Scholar]
- 8.Sato Y, Kato J, Takimoto R, Takada K, Kawano Y, Miyanishi K, Kobune M, Sato Y, Takayama T, Matunaga T, Niitsu Y. Hepatitis C virus core protein promotes proliferation of human hepatoma cells through enhancement of transforming growth factor alpha expression via activation of nuclear factor-kappaB. Gut. 2006;55:1801–8. doi: 10.1136/gut.2005.070417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tai DI, Tsai SL, Chen YM, Chuang YL, Peng CY, Sheen IS, Yeh CT, Chang KS, Huang SN, Kuo GC, Liaw YF. Activation of nuclear factor kappaB in hepatitis C virus infection: implications for pathogenesis and hepatocarcinogenesis. Hepatology. 2000;31:656–64. doi: 10.1002/hep.510310316. [DOI] [PubMed] [Google Scholar]
- 10.Park J, Kang W, Ryu SW, Kim WI, Chang DY, Lee DH, Park do Y, Choi YH, Choi K, Shin EC, Choi C. Hepatitis C virus infection enhances TNFalpha-induced cell death via suppression of NF-kappaB. Hepatology. 2012;56:831–40. doi: 10.1002/hep.25726. [DOI] [PubMed] [Google Scholar]
- 11.Vinciguerra M, Foti M. PTEN at the crossroad of metabolic diseases and cancer in the liver. Ann Hepatol. 2008;7:192–9. [PubMed] [Google Scholar]
- 12.Wang Q, Zhou Y, Wang X, Chung DH, Evers BM. Regulation of PTEN expression in intestinal epithelial cells by c-Jun NH2-terminal kinase activation and nuclear factor-kappaB inhibition. Cancer Res. 2007;67:7773–81. doi: 10.1158/0008-5472.CAN-07-0187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dan HC, Adli M, Baldwin AS. Regulation of mammalian target of rapamycin activity in PTEN-inactive prostate cancer cells by I kappa B kinase alpha. Cancer Res. 2007;67:6263–9. doi: 10.1158/0008-5472.CAN-07-1232. [DOI] [PubMed] [Google Scholar]
- 14.Kim S, Domon-Dell C, Kang J, Chung DH, Freund JN, Evers BM. Down-regulation of the tumor suppressor PTEN by the tumor necrosis factor-alpha/nuclear factor-kappaB (NF-kappaB)-inducing kinase/NF-kappaB pathway is linked to a default IkappaB-alpha autoregulatory loop. J Biol Chem. 2004;279:4285–91. doi: 10.1074/jbc.M308383200. [DOI] [PubMed] [Google Scholar]
- 15.Kang-Park S, Im JH, Lee JH, Lee YI. PTEN modulates hepatitis B virus-X protein induced survival signaling in Chang liver cells. Virus Res. 2006;122:53–60. doi: 10.1016/j.virusres.2006.06.010. [DOI] [PubMed] [Google Scholar]
- 16.Chung TW, Lee YC, Ko JH, Kim CH. Hepatitis B Virus X protein modulates the expression of PTEN by inhibiting the function of p53, a transcriptional activator in liver cells. Cancer Res. 2003;63:3453–8. [PubMed] [Google Scholar]
- 17.Ruggieri A, Murdolo M, Rapicetta M. Induction of FAS ligand expression in a human hepatoblastoma cell line by HCV core protein. Virus Res. 2003;97:103–10. doi: 10.1016/j.virusres.2003.08.004. [DOI] [PubMed] [Google Scholar]
- 18.Cho J, Baek W, Yang S, Chang J, Sung YC, Suh M. HCV core protein modulates Rb pathway through pRb down-regulation and E2F-1 up-regulation. Biochim Biophys Acta. 2001;1538:59–66. doi: 10.1016/s0167-4889(00)00137-3. [DOI] [PubMed] [Google Scholar]
- 19.Wen F, Abdalla M, Aloman C, Xiang J, Ahmad I, Walewski J, McCormick M, Brown K, Branch A, Spitz D, Britigan B, Schmidt W. Increased prooxidant production and enhanced susceptibility to glutathione depletion in HepG2cells co-expressing HCV core protein and CYP2E1. J Med Virol. 2004;72:230–40. doi: 10.1002/jmv.10567. [DOI] [PubMed] [Google Scholar]
- 20.Huang S, Xie Y, Yang P, Chen P, Zhang L. HCV core protein-induced down-regulation of microRNA-152 promoted aberrant proliferation by regulating Wnt1 in HepG2 cells. PLoS One. 2014;9:e81730. doi: 10.1371/journal.pone.0081730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Okuda M, Li K, Beard M, Showalter L, Scholle F, Lemon S, Weinman S. Mitochondrial injury, oxidative stress, and antioxidant gene expression are induced by hepatitisC virus core protein. Gastroenterology. 2002;122:366–75. doi: 10.1053/gast.2002.30983. [DOI] [PubMed] [Google Scholar]
- 22.Ruan ZP, Xu R, Lv Y, Tian T, Wang WJ, Guo H, Nan KJ. PTEN enhances the sensitivity of human hepatocellular carcinoma cells to sorafenib. Oncol Res. 2012;20:113–21. doi: 10.3727/096504012x13477145152995. [DOI] [PubMed] [Google Scholar]
- 23.Lee YR, Shim HJ, Yu HN, Song EK, Park J, Kwon KB, Park JW, Rho HW, Park BH, Han MK, Kim JS. Dimethylsulfoxide induces upregulation of tumor suppressor protein PTEN through nuclear factor-kappaB activation in HL-60 cells. Leuk Res. 2005;29:401–5. doi: 10.1016/j.leukres.2004.09.010. [DOI] [PubMed] [Google Scholar]
- 24.Kanda T, Yokosuka O, Nagao K, Saisho H. State of hepatitis C viral replication enhances activation of NF-kB- and AP-1-signaling induced by hepatitis B virus X. Cancer Lett. 2006;234:143–8. doi: 10.1016/j.canlet.2005.03.030. [DOI] [PubMed] [Google Scholar]
- 25.Koul D, Takada Y, Shen R, Aggarwal BB, Yung WK. PTEN enhances TNF-induced apoptosis through modulation of nuclear factor-kappaB signaling pathway in human glioma cells. Biochem Biophys Res Commun. 2006;350:463–71. doi: 10.1016/j.bbrc.2006.09.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mayo MW, Madrid LV, Westerheide SD, Jones DR, Yuan XJ, Baldwin AS Jr, Whang YE. PTEN blocks tumor necrosis factor-induced NF-kappa B-dependent transcription by inhibiting the transactivation potential of the p65 subunit. J Biol Chem. 2002;277:11116–25. doi: 10.1074/jbc.M108670200. [DOI] [PubMed] [Google Scholar]
- 27.Kim JH, Lee G, Cho YL, Kim CK, Han S, Lee H, Choi JS, Choe J, Won MH, Kwon YG, Ha KS, Kim YM. Desmethylanhydroicaritin inhibits NF-kappaB-regulated inflammatory gene expression by modulating the redox-sensitive PI3K/PTEN/Akt pathway. Eur J Pharmacol. 2009;602:422–31. doi: 10.1016/j.ejphar.2008.10.062. [DOI] [PubMed] [Google Scholar]


