Abstract
We aimed to evaluate the expression of sarcosine metabolism-related proteins according to site of metastatic breast cancer, and the clinical implications. Immunohistochemical staining for glycine N-methyltransferase (GNMT), sarcosine dehydrogenase (SARDH), and l-pipecolic acid oxidase (PIPOX) was performed on tissue microarrays from 162 metastatic breast cancer (bone metastases = 47, brain metastases = 39, liver metastases = 24, and lung metastases = 52). Sarcosine metabolism-related proteins showed variable expression with regard to metastatic sites. GNMT was expressed in brain and lung metastases, but not in bone and liver metastases (P = 0.016). In view of the sarcosine metabolic phenotype, high sarcosine and intermediate type were only found in the brain and lung metastases, and low sarcosine type was observed more frequently in bone and lung metastases (P = 0.047). By univariate analysis, PIPOX positivity was correlated with shorter overall survival (OS) (P = 0.031). In lung metastases, PIPOX positivity (P = 0.019) and stromal PIPOX positivity (P = 0.001) were associated with shorter OS. In conclusion, in metastatic breast cancer, sarcosine metabolism-related proteins are differently expressed according to the metastatic site. Expression of GNMT and high sarcosine type are predominantly observed in brain and lung metastases.
Keywords: Breast cancer, metabolism, molecular subtype, sarcosine
Introduction
Breast cancer is associated with high morbidity and mortality, largely due to distant metastasis. Common metastatic sites of breast cancer include the lung, brain, liver, and bone [1,2], and among these, brain and bone metastases have been thoroughly studied [3-8]. In general, reciprocal interaction between tumor cells and host tissue is the main mechanism of metastasis, together with adhesion, proteolysis, invasion, and angiogenesis [2,9]. Because every single tumor shows a different metastatic pattern, the ‘seed and soil’ hypothesis has been proposed as an explanation for how a specific tumor (seed) would survive in a specific visceral organ (soil) [10]. Similarly, metastatic breast cancer has different properties according to metastatic sites. In previous studies, young age, ER (estrogen receptor) negativity, prior lung metastasis, HER-2 (human epidermal growth factor-2) overexpression, EGFR (epidermal growth factor) overexpression, and basal subtype have been shown in brain metastases [5-7]. Lower histologic grade, ER positivity, ER positivity/PR negativity, strand growth pattern, and presence of fibrotic foci in invasive ductal carcinoma were found in bone metastases [4,11,12]. Thus, different characteristics are associated with different metastatic sites.
Sarcosine (N-methylglycine) is a non-proteinogenic amino acid, synthesized from glycine metabolism. Glycine N-methyltransferase (GNMT), sarcosine dehydrogenase (SARDH), and l-pipecolic acid oxidase (PIPOX) are the major enzymes in the sarcosine metabolism pathway. GNMT involves the transfer of a methyl group from S-adenosylmethionine (SAM) to glycine. Sarcosine-metabolizing enzymes, SARDH and PIPOX, convert sarcosine to glycine by oxidative demethylation [13]. Sarcosine is thought to be a potential oncometabolite rather than a non-proteinogenic amino acid. In prostate cancer, sarcosine is a sensitive tumor biomarker and is associated with tumor progression and the metastatic process [14,15]. While increased sarcosine level has been studied in metastatic prostatic cancer [16], sarcosine metabolism in metastatic breast cancer has yet to be investigated. As the tumor has been known to show different properties according to metastatic sites, evaluation of sarcosine metabolism-related proteins in different metastatic sites is required. Hence, we aimed to evaluate the expression of sarcosine metabolism-related proteins according to site of metastatic breast cancer, and its clinical implication.
Materials and methods
Patient selection
Invasive primary breast cancer and metastatic breast cancer to distant organs (liver, lung, brain, and bone) were retrieved from data files of the Department of Pathology of Severance Hospital, Seoul, South Korea. Only patients with a diagnosis of invasive ductal carcinoma were included. A total of 169 cases were included, and 49 cases consisted of paired primary and metastasis carcinomas. All slides were reviewed again and pathologic diagnoses were approved by two pathologists (JSK and WJ). The histological grade was assessed using the Nottingham grading system [17]. This study was approved by the Institutional Review Board of Severance Hospital.
Tissue microarray
On H&E-stained slides of tumors, a representative area was selected and a corresponding spot was marked on the surface of the paraffin block. Using a biopsy needle, the selected area was punched out and a 3-mm tissue core was placed into a 6 × 5 recipient block. Two tissue cores were extracted to minimize extraction bias. Each tissue core was assigned to a unique tissue microarray location number that was linked to a database containing other clinicopathologic data.
Immunohistochemistry
The antibodies used for immunohistochemistry in this study are shown in Table 1. Three-micrometer paraffin sections were deparaffinized and rehydrated by xylene and alcohol solution. Immunohistochemistry was performed using the Ventana Discovery XT automated stainer (Ventana Medical System, Tucson, AZ, USA). Antigen retrieval was performed using Cell Conditioning 1 (CC1; citrate buffer pH 6.0, Ventana Medical System). Appropriate positive and negative controls for immunohistochemistry were included.
Table 1.
Source, clone, and dilution of antibodies used in this study
| Antibody | Company | Clone | Dilution |
|---|---|---|---|
| Sarcosine metabolism-related | |||
| GNMT | Abcam, Cambridge, UK | Polyclonal | 1:100 |
| SARDH | Abcam, Cambridge, UK | Polyclonal | 1:100 |
| PIPOX | Abcam, Cambridge, UK | Polyclonal | 1:100 |
| Molecular subtype-related | |||
| ER | Thermo Scientific, San Diego, CA, USA | SP1 | 1:100 |
| PR | DAKO, Glostrup, Denmark | PgR | 1:50 |
| HER-2 | DAKO, Glostrup, Denmark | Polyclonal | 1:1500 |
| Ki-67 | Abcam, Cambridge, UK | MIB | 1:1000 |
GNMT, glycine N-methyltransferase; SARDH, sarcosine dehydrogenase; PIPOX, l-pipecolic acid oxidase; ER, estrogen receptor; PR, progesterone receptor; and HER-2, human epidermal growth factor-2; ER, estrogen receptor; PR, progesterone receptor.
Interpretation of immunohistochemical staining
All immunohistochemical markers were accessed by light microscopy. Pathologic parameters such as ER, PR, and HER-2 status were obtained from the patients’ pathologic reports. A cut-off value of 1% or more positively stained nuclei was used to define ER and PR positivity [18]. HER-2 staining was analyzed according to the American Society of Clinical Oncology (ASCO)/College of American Pathologists (CAP) guidelines using the following categories: 0 = no immunostaining; 1+ = weak incomplete membranous staining, less than 10% of tumor cells; 2+ = complete membranous staining, either uniform or weak in at least 10% of tumor cells; and 3+ = uniform intense membranous staining in at least 30% of tumor cells [19]. HER-2 immunostaining was considered positive when strong (3+) membranous staining was observed whereas cases with 0 to 1+ were regarded as negative. The cases showing 2+ HER-2 expression were evaluated for HER-2 amplification by fluorescent in situ hybridization (FISH).
Immunohistochemical markers for GNMT, SARDH and PIPOX were accessed by light microscopy. Interpretation of immunohistochemical staining was determined by multiplying the proportion of stained cells (0% = 0, 1-29% = 1, 30-100% = 2) with the intensity (negative = 0, weak = 1, moderate = 2, strong = 3). The final scores of 0-1 and 2-6 were interpreted as negative (-) and positive (+), respectively [20]. Ki-67 labeling indices (LI) were scored by counting the number of positively stained nuclei and expressed as a percentage of total tumor cells.
Tumor phenotype classification
In this study, we classified breast cancer phenotypes according to the immunohistochemistry results for ER, PR, HER-2 and Ki-67 and FISH results for HER-2 as follows [21]; Luminal A type: ER and/or PR positive and HER-2 negative and Ki-67 LI < 14%; Luminal B type: (HER-2 negative) ER and/or PR positive and HER-2 negative and Ki-67 LI ≥ 14%, (HER-2 positive) ER and/or PR positive and HER-2 overexpressed and/or amplified; HER-2 overexpression type: ER and PR negative and HER-2 overexpressed and/or amplified; and TNBC type: ER, PR, and HER-2 negative.
Sarcosine metabolism phenotype
Based on the result of GNMT, SARDH, and PIPOX immunohistochemistry, the sarcosine metabolism phenotype was determined as follows; high sarcosine type: GNMT (+)/SARDH and PIPOX (-), low sarcosine type: GNMT (-)/SARDH or PIPOX (+), intermediate sarcosine type: GNMT (+)/SARDH or PIPOX (+), and null type: GNMT (-)/SARDH and PIPOX (-).
Statistical analysis
Data were statistically processed using SPSS for Windows, version 12.0 (SPSS Inc., Chicago, IL, USA). Correlation analysis of immunostaining results between primary breast cancer and metastatic breast cancer were calculated by McNemar test. Student’s t and Fisher’s exact tests were used to examine any differences in continuous and categorical variables, respectively. A corrected p-value and the Bonferroni method were used for multiple comparisons. Statistical significance was assumed when P < 0.05. Kaplan-Meier survival curves and log-rank statistics were employed to evaluate time to tumor metastasis and time to survival. Multivariate regression analysis was performed using a Cox proportional hazards model.
Results
Patients’ clinicopathologic characteristics
Of 162 patients, 47 (29.0%) had bone metastases, 30 (18.5%) had brain metastases, 24 (14.8%) had liver metastases, and 52 (32.1%) had lung metastases (Table 2). There was a higher frequency of ER-positive/PR-positive in bone and liver metastases (P < 0.001), and HER-2-positive in brain metastases (P = 0.017). Luminal A type was predominant in bone and liver metastases, and TNBC was predominant in brain and lung metastases (P < 0.001).
Table 2.
Basal clinicopathologic characteristics of breast cancer metastasis according to metastatic site
| Parameters | Total n = 162 (%) | Bone metastasis n = 47 (%) | Brain metastasis n = 39 (%) | Liver metastasis n = 24 (%) | Lung metastasis n = 52 (%) | P-value |
|---|---|---|---|---|---|---|
| Age (years) | 0.022 | |||||
| ≤ 50 | 81 (50.0) | 27 (57.4) | 17 (43.6) | 6 (25.0) | 31 (59.6) | |
| > 50 | 81 (50.0) | 20 (42.6) | 22 (56.4) | 18 (75.0) | 21 (40.4) | |
| ER | < 0.001 | |||||
| Negative | 69 (42.6) | 8 (17.0) | 26 (66.7) | 6 (25.0) | 29 (55.8) | |
| Positive | 93 (57.4) | 39 (83.0) | 13 (33.3) | 18 (75.0) | 23 (44.2) | |
| PR | < 0.001 | |||||
| Negative | 109 (67.3) | 23 (48.9) | 38 (97.4) | 12 (50.0) | 36 (69.2) | |
| Positive | 53 (32.7) | 24 (51.1) | 1 (2.6) | 12 (50.0) | 16 (30.8) | |
| HER-2 | 0.017 | |||||
| Negative | 114 (70.4) | 38 (80.9) | 20 (51.3) | 19 (79.2) | 37 (71.2) | |
| Positive | 48 (29.6) | 9 (19.1) | 19 (48.7) | 5 (20.8) | 15 (28.8) | |
| Molecular subtype | < 0.001 | |||||
| Luminal A | 67 (41.4) | 33 (70.2) | 4 (10.3) | 15 (62.5) | 15 (28.8) | |
| Luminal B | 27 (16.7) | 7 (14.9) | 9 (23.1) | 3 (12.5) | 8 (15.4) | |
| HER-2 | 30 (18.5) | 5 (10.6) | 12 (30.8) | 3 (12.5) | 10 (19.2) | |
| TNBC | 38 (23.5) | 2 (4.3) | 14 (35.9) | 3 (12.5) | 19 (36.5) | |
| Patient death | 53 (32.7) | 23 (48.9) | 11 (28.2) | 7 (29.2) | 12 (23.1) | 0.040 |
ER, estrogen receptor; PR, progesterone receptor; HER-2, human epidermal growth factor-2; TNBC, triple negative breast cancer.
Expression of sarcosine metabolism-related proteins in metastatic breast cancer according to metastatic sites
When expression of sarcosine metabolism-related protein was correlated with metastatic sites, GNMT expression was only observed in brain and lung metastases, not in bone and liver metastases (P = 0.016, Table 3 and Figure 1). Based on sarcosine metabolism-related proteins expression, high sarcosine and intermediate sarcosine type were found only in brain and lung metastases, and low sarcosine type was largely found in bone and lung metastases (P = 0.047, Table 4). In a set of 49-paired primary and metastatic tumors, the expression of sarcosine metabolism-related proteins was not statistically different (Table 5).
Table 3.
Expression of metabolism-related proteins in tumor cell compartments of breast cancer metastasis according to metastatic site
| Parameters | Total n = 162 (%) | Bone metastasis n = 47 (%) | Brain metastasis n = 39 (%) | Liver metastasis n = 24 (%) | Lung metastasis n = 52 (%) | P-value |
|---|---|---|---|---|---|---|
| GNMT (T) | 0.016 | |||||
| Negative | 153 (94.4) | 47 (100.0) | 37 (94.9) | 24 (100.0) | 45 (86.5) | |
| Positive | 9 (5.6) | 0 (0.0) | 2 (5.1) | 0 (0.0) | 7 (13.5) | |
| SARDH (T) | 0.730 | |||||
| Negative | 158 (97.5) | 45 (95.7) | 38 (97.4) | 24 (100.0) | 51 (98.1) | |
| Positive | 4 (2.5) | 2 (4.3) | 1 (2.8) | 0 (0.0) | 1 (1.9) | |
| PIPOX (T) | 0.069 | |||||
| Negative | 125 (77.2) | 35 (74.5) | 35 (89.5) | 20 (83.3) | 35 (67.3) | |
| Positive | 37 (22.8) | 12 (25.5) | 4 (10.3) | 4 (16.7) | 17 (32.7) | |
| PIPOX (S) | 0.533 | |||||
| Negative | 152 (93.8) | 43 (91.5) | 36 (92.3) | 24 (100.0) | 49 (94.2) | |
| Positive | 10 (6.2) | 4 (8.5) | 3 (7.7) | 0 (0.0) | 3 (5.8) |
GNMT, glycine N-methyltransferase; SARDH, sarcosine dehydrogenase; PIPOX, l-pipecolic acid oxidase; T, tumor; S, stroma.
Figure 1.
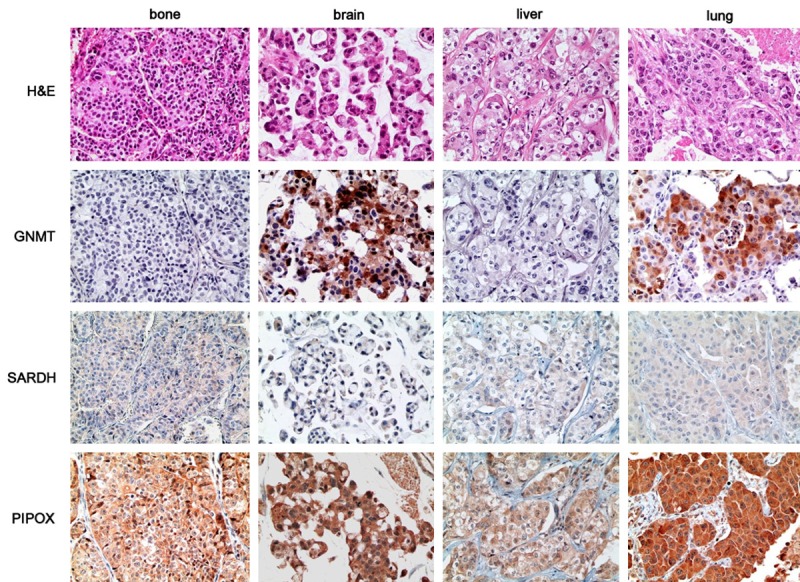
Expression of sarcosine metabolism-related proteins in metastatic breast cancer according to metastatic sites. GNMT expression is only found in brain and lung metastases, and is absent in bone and liver metastases. PIPOX is highly expressed in in brain and lung metastases.
Table 4.
Sarcosine-related metabolic phenotypes of breast cancer metastasis according to metastatic site
| Parameters | Total n = 162 (%) | Bone metastasis n = 47 (%) | Brain metastasis n = 39 (%) | Liver metastasis n = 24 (%) | Lung metastasis n = 52 (%) | p-value |
|---|---|---|---|---|---|---|
| Metabolic type | 0.047 | |||||
| High sarcosine type | 5 (3.1) | 0 (0.0) | 1 (2.6) | 0 (0.0) | 4 (7.7) | |
| Low sarcosine type | 36 (22.2) | 13 (27.7) | 4 (10.3) | 4 (16.7) | 15 (28.8) | |
| Intermediate sarcosine type | 4 (2.5) | 0 (0.0) | 1 (2.6) | 0 (0.0) | 3 (5.8) | |
| Null type | 117 (72.2) | 34 (72.3) | 33 (84.6) | 20 (83.3) | 30 (57.7) |
Table 5.
Correlation of expression of metabolism-related proteins between primary and metastatic breast cancer according to metastatic site
| Parameters | Total | Bone metastasis | Brain metastasis | Liver metastasis | Lung metastasis | |||||
|---|---|---|---|---|---|---|---|---|---|---|
|
| ||||||||||
| n = 49 (%) | P-value | n = 13 (%) | P-value | n = 9 (%) | P-value | n = 4 (%) | P-value | n = 23 (%) | P-value | |
| GNMT | 1.000 | 1.000 | N/A | N/A | 0.500 | |||||
| (+) → (+) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | |||||
| (+) → (-) | 1 (2.0) | 1 (7.7) | 0 (0.0) | 0 (0.0) | 0 (0.0) | |||||
| (-) → (+) | 2 (4.1) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 2 (8.7) | |||||
| (-) → (-) | 46 (93.9) | 12 (92.3) | 9 (100.0) | 4 (100.0) | 21 (91.3) | |||||
| SARDH | 1.000 | 1.000 | 1.000 | N/A | 1.000 | |||||
| (+) → (+) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | |||||
| (+) → (-) | 1 (2.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 1 (4.3) | |||||
| (-) → (+) | 2 (4.1) | 1 (7.7) | 1 (11.1) | 0 (0.0) | 0 (0.0) | |||||
| (-) → (-) | 46 (93.9) | 12 (92.3) | 8 (88.9) | 4 (100.0) | 22 (95.7) | |||||
| PIPOX | 1.000 | 0.375 | 0.500 | N/A | 1.000 | |||||
| (+) → (+) | 4 (8.2) | 1 (7.7) | 0 (0.0) | 0 (0.0) | 3 (13.0) | |||||
| (+) → (-) | 8 (16.3) | 4 (30.8) | 0 (0.0) | 0 (0.0) | 4 (17.4) | |||||
| (-) → (+) | 7 (14.3) | 1 (7.7) | 2 (22.2) | 0 (0.0) | 4 (17.4) | |||||
| (-) → (-) | 30 (61.2) | 7 (53.8) | 7 (77.8) | 4 (100.0) | 12 (52.2) | |||||
(+) and (-) indicate protein expression levels that are positive and negative, respectively. Arrow indicates the change of protein expression from the primary tumor to the metastasis. GNMT, glycine N-methyltransferase; SARDH, sarcosine dehydrogenase; PIPOX, l-pipecolic acid oxidase; N/A, not available.
Correlation between expression of sarcosine metabolism-related proteins and pathologic factors
Among sarcosine metabolism phenotypes, the high sarcosine type demonstrated higher Ki-67 LI (P = 0.031). With regard to metastatic site, higher Ki-67 LI was associated with stromal PIPOX expression in bone metastases (P = 0.031). In lung metastases, ER negativity was associated with PIPOX positivity (P = 0.009) and non-null type (P = 0.002, Figure 2).
Figure 2.
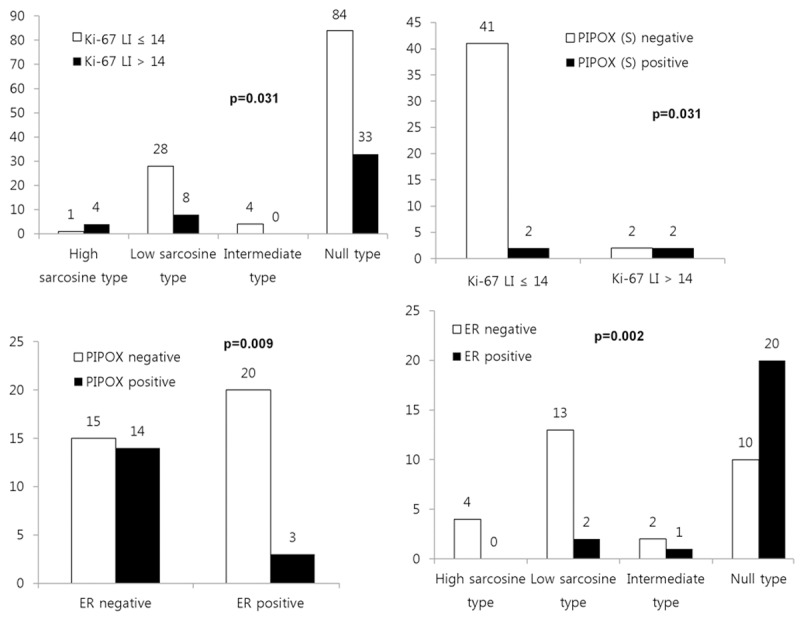
Correlation between pathologic factors and expression of sarcosine metabolism-related proteins.
Impact of sarcosine metabolism-related protein expression on patient prognosis
PIPOX positivity (P = 0.031) and TNBC type (P = 0.002) were associated with shorter OS by univariate analysis (Table 6 and Figure 3). When risk factors for shorter OS in each metastatic site were assessed by univariate analysis, TNBC type (P < 0.001) in bone metastases, and PIPOX positivity (P = 0.019) and stromal PIPOX positivity (P = 0.001) in lung metastases, were significant risk factors (Figure 3).
Table 6.
Univariate analysis of the impact of expression of metabolism-related proteins in metastatic breast cancers on overall survival by the log-rank test
| Parameters | Total n = 162 (%) | Bone metastasis n = 47 (%) | Brain metastasis n = 39 (%) | Liver metastasis n = 24 (%) | Lung metastasis n = 52 (%) | |||||
|---|---|---|---|---|---|---|---|---|---|---|
|
| ||||||||||
| Mean survival (95% CI) months | P-value | Mean survival (95% CI) months | P-value | Mean survival (95% CI) months | P-value | Mean survival (95% CI) months | P-value | Mean survival (95% CI) months | P-value | |
| GNMT (T) | 0.310 | N/A | 0.473 | N/A | 0.407 | |||||
| Negative | 108 (93-122) | N/A | 109 (86-131) | N/A | 123 (95-151) | |||||
| Positive | 132 (91-173) | N/A | 31 (7-55) | N/A | 144 (106-183) | |||||
| SARDH (T) | 0.795 | 0.769 | N/A | N/A | N/A | |||||
| Negative | 110 (96-124) | 83 (62-104) | N/A | N/A | N/A | |||||
| Positive | 80 (51-110) | 44 (43-45) | N/A | N/A | N/A | |||||
| PIPOX (T) | 0.031 | 0.614 | 0.321 | 0.156 | 0.019 | |||||
| Negative | 118 (102-133) | 87 (63-110) | 110 (87-133) | 90 (70-109) | 145 (117-172) | |||||
| Positive | 80 (55-106) | 58 (41-75) | 58 (16-100) | 51 (26-76) | 92 (53-132) | |||||
| PIPOX (S) | 0.194 | N/A | 0.690 | N/A | 0.001 | |||||
| Negative | 113 (98-127) | N/A | 108 (86-131) | N/A | 139 (115-162) | |||||
| Positive | 51 (31-72) | N/A | 40 (27-52) | N/A | 37 (2-71) | |||||
| Metabolic type | 0.131 | N/A | N/A | N/A | N/A | |||||
| High sarcosine type | 72 (48-96) | N/A | N/A | N/A | N/A | |||||
| Low sarcosine type | 59 (46-72) | N/A | N/A | N/A | N/A | |||||
| Intermediate sarcosine type | 128 (63-192) | N/A | N/A | N/A | N/A | |||||
| Null type | 116 (100-132) | N/A | N/A | N/A | N/A | |||||
| Molecular subtypes | 0.002 | < 0.001 | 0.081 | N/A | N/A | |||||
| Luminal A | 105 (86-124) | 84 (62-107) | 55 (10-100) | N/A | N/A | |||||
| Luminal B | 140 (111-170) | 60 (26-93) | 138 (112-164) | N/A | N/A | |||||
| HER-2 | 134 (109-158) | 62 (47-77) | 79 (60-97) | N/A | N/A | |||||
| TNBC | 51 (38-64) | 3 (2-4) | 31 (22-39) | N/A | N/A | |||||
GNMT, glycine N-methyltransferase; SARDH, sarcosine dehydrogenase; PIPOX, l-pipecolic acid oxidase; T, tumor; S, stroma; ER, estrogen receptor; PR, progesterone receptor; HER-2, human epidermal growth factor-2; TNBC, triple negative breast cancer; N/A, not available; CI, confidence interval.
Figure 3.
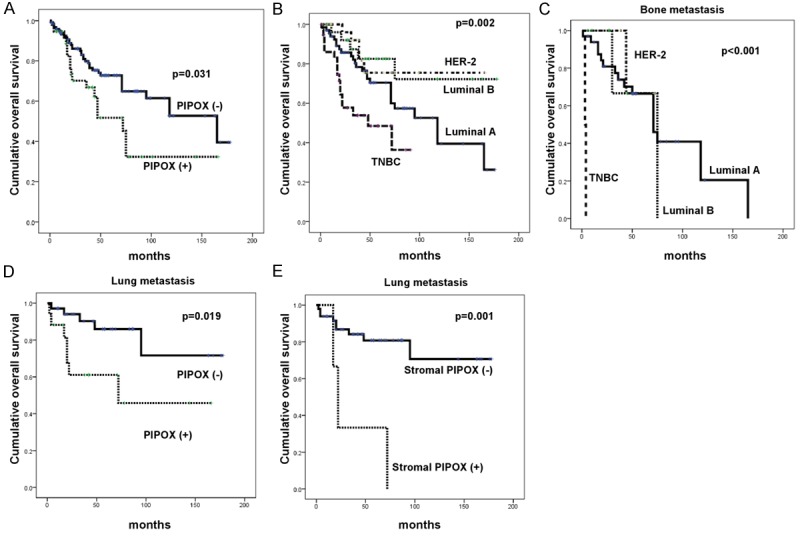
Impact of the expression of sarcosine metabolism-related proteins on patient prognosis in metastatic breast cancer (A, B), bone metastasis (C), and lung metastasis (D, E).
Discussion
We examined the expression of sarcosine metabolism-related proteins in metastatic breast cancer according to the metastatic sites. GNMT was highly expressed in brain and lung metastases compared to the bone and liver metastases. Previous study has shown higher expression of sarcosine metabolism-related proteins in tumors with HER-2 molecular subtype [22]. Since brain and lung metastases contained a higher fraction of HER-2 type in the current study, higher expression of sarcosine metabolism-related proteins in brain and lung metastases was expected, and the result was concordant. Albeit there has been no previous study of sarcosine metabolism in metastatic breast cancer, a study of prostate cancer showed an association between increased sarcosine level and cancer progression. Benign prostate cells acquired an invasive phenotype with administration of sarcosine [13]. Furthermore, patients with metastatic prostatic cancer revealed an elevated serum sarcosine level [16]. Therefore, it is considered that the sarcosine level is related to tumor progression in prostate cancer. We evaluated the expression level of GNMT, SARDH and PIPOX as surrogate markers of sarcosine level in the tissue from metastatic breast cancer. In a previous study, higher expression of GNMT and lower expression of SARDH and PIPOX was found in the tissue of prostate cancer compared to the normal prostate [14]. Considering that GNMT is a sarcosine-generating enzyme, and SARDH and PIPOX are sarcosine-metabolizing enzymes, the expression levels of GNMT, SARDH, and PIPOX were well-correlated with the sarcosine level in the tissue [14]. In the current study, the high sarcosine type (GNMT (+)/SARDH and PIPOX (-)) was exclusively found in brain and lung metastases, and a higher sarcosine level in brain and lung metastases was suspected, but needs to be validated.
Khan et al. observed repression of tumor growth as a result of sarcosine inhibition [14]. Thus, it is expected that patients with metastatic breast cancer harboring higher sarcosine metabolism-related protein expression would benefit from target therapy that inhibits the sarcosine metabolism pathway. In the present study, GNMT expression was elevated in brain and lung metastases, suggesting that sarcosine inhibition is a candidate therapeutic approach that should be validated in further study.
In conclusion, expression of sarcosine metabolism-related proteins in metastatic breast cancer varied according to the metastatic site. Brain and lung metastases demonstrated higher expression of GNMT and a larger fraction of high-sarcosine type than other sites.
Acknowledgements
This research was supported by the Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education, Science and Technology (2012R1A1A1002886).
Disclosure of conflict of interest
None.
References
- 1.Weil RJ, Palmieri DC, Bronder JL, Stark AM, Steeg PS. Breast cancer metastasis to the central nervous system. Am J Pathol. 2005;167:913–920. doi: 10.1016/S0002-9440(10)61180-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Woodhouse EC, Chuaqui RF, Liotta LA. General mechanisms of metastasis. Cancer. 1997;80:1529–1537. doi: 10.1002/(sici)1097-0142(19971015)80:8+<1529::aid-cncr2>3.3.co;2-#. [DOI] [PubMed] [Google Scholar]
- 3.Abali H, Celik I. High incidence of central nervous system involvement in patients with breast cancer treated with epirubicin and docetaxel. Am J Clin Oncol. 2002;25:632–633. doi: 10.1097/00000421-200212000-00023. [DOI] [PubMed] [Google Scholar]
- 4.Colleoni M, O’Neill A, Goldhirsch A, Gelber RD, Bonetti M, Thurlimann B, Price KN, Castiglione-Gertsch M, Coates AS, Lindtner J, Collins J, Senn HJ, Cavalli F, Forbes J, Gudgeon A, Simoncini E, Cortes-Funes H, Veronesi A, Fey M, Rudenstam CM. Identifying breast cancer patients at high risk for bone metastases. J. Clin. Oncol. 2000;18:3925–3935. doi: 10.1200/JCO.2000.18.23.3925. [DOI] [PubMed] [Google Scholar]
- 5.Evans AJ, James JJ, Cornford EJ, Chan SY, Burrell HC, Pinder SE, Gutteridge E, Robertson JF, Hornbuckle J, Cheung KL. Brain metastases from breast cancer: identification of a high-risk group. Clin Oncol (R Coll Radiol) 2004;16:345–349. doi: 10.1016/j.clon.2004.03.012. [DOI] [PubMed] [Google Scholar]
- 6.Gaedcke J, Traub F, Milde S, Wilkens L, Stan A, Ostertag H, Christgen M, von Wasielewski R, Kreipe HH. Predominance of the basal type and HER-2/neu type in brain metastasis from breast cancer. Mod Pathol. 2007;20:864–870. doi: 10.1038/modpathol.3800830. [DOI] [PubMed] [Google Scholar]
- 7.Hicks DG, Short SM, Prescott NL, Tarr SM, Coleman KA, Yoder BJ, Crowe JP, Choueiri TK, Dawson AE, Budd GT, Tubbs RR, Casey G, Weil RJ. Breast cancers with brain metastases are more likely to be estrogen receptor negative, express the basal cytokeratin CK5/6, and overexpress HER2 or EGFR. Am J Surg Pathol. 2006;30:1097–1104. doi: 10.1097/01.pas.0000213306.05811.b9. [DOI] [PubMed] [Google Scholar]
- 8.Lorincz T, Toth J, Badalian G, Timar J, Szendroi M. HER-2/neu genotype of breast cancer may change in bone metastasis. Pathol Oncol Res. 2006;12:149–152. doi: 10.1007/BF02893361. [DOI] [PubMed] [Google Scholar]
- 9.Nicolson GL. Organ specificity of tumor metastasis: role of preferential adhesion, invasion and growth of malignant cells at specific secondary sites. Cancer Metastasis Rev. 1988;7:143–188. doi: 10.1007/BF00046483. [DOI] [PubMed] [Google Scholar]
- 10.Paget S. The distribution of secondary growths in cancer of the breast. Lancet. 1889;1:571–572. [PubMed] [Google Scholar]
- 11.Hasebe T, Imoto S, Yokose T, Ishii G, Iwasaki M, Wada N. Histopathologic factors significantly associated with initial organ-specific metastasis by invasive ductal carcinoma of the breast: a prospective study. Hum Pathol. 2008;39:681–693. doi: 10.1016/j.humpath.2007.09.012. [DOI] [PubMed] [Google Scholar]
- 12.Wei B, Wang J, Bourne P, Yang Q, Hicks D, Bu H, Tang P. Bone metastasis is strongly associated with estrogen receptor-positive/progesterone receptor-negative breast carcinomas. Hum Pathol. 2008;39:1809–1815. doi: 10.1016/j.humpath.2008.05.010. [DOI] [PubMed] [Google Scholar]
- 13.Sreekumar A, Poisson LM, Rajendiran TM, Khan AP, Cao Q, Yu J, Laxman B, Mehra R, Lonigro RJ, Li Y, Nyati MK, Ahsan A, Kalyana-Sundaram S, Han B, Cao X, Byun J, Omenn GS, Ghosh D, Pennathur S, Alexander DC, Berger A, Shuster JR, Wei JT, Varambally S, Beecher C, Chinnaiyan AM. Metabolomic profiles delineate potential role for sarcosine in prostate cancer progression. Nature. 2009;457:910–914. doi: 10.1038/nature07762. [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- 14.Khan AP, Rajendiran TM, Ateeq B, Asangani IA, Athanikar JN, Yocum AK, Mehra R, Siddiqui J, Palapattu G, Wei JT, Michailidis G, Sreekumar A, Chinnaiyan AM. The role of sarcosine metabolism in prostate cancer progression. Neoplasia. 2013;15:491–501. doi: 10.1593/neo.13314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Baum CE, Price DK, Figg WD. Sarcosine as a potential prostate cancer biomarker and therapeutic target. Cancer Biol Ther. 2010;9:341–342. doi: 10.4161/cbt.9.5.11310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lucarelli G, Ditonno P, Bettocchi C, Spilotros M, Rutigliano M, Vavallo A, Galleggiante V, Fanelli M, Larocca AM, Germinario CA, Maiorano E, Selvaggi FP, Battaglia M. Serum sarcosine is a risk factor for progression and survival in patients with metastatic castration-resistant prostate cancer. Future Oncol. 2013;9:899–907. doi: 10.2217/fon.13.50. [DOI] [PubMed] [Google Scholar]
- 17.Elston CW, Ellis IO. Pathological prognostic factors in breast cancer. I. The value of histological grade in breast cancer: experience from a large study with long-term follow-up. Histopathology. 1991;19:403–410. doi: 10.1111/j.1365-2559.1991.tb00229.x. [DOI] [PubMed] [Google Scholar]
- 18.Hammond ME, Hayes DF, Dowsett M, Allred DC, Hagerty KL, Badve S, Fitzgibbons PL, Francis G, Goldstein NS, Hayes M, Hicks DG, Lester S, Love R, Mangu PB, McShane L, Miller K, Osborne CK, Paik S, Perlmutter J, Rhodes A, Sasano H, Schwartz JN, Sweep FC, Taube S, Torlakovic EE, Valenstein P, Viale G, Visscher D, Wheeler T, Williams RB, Wittliff JL, Wolff AC. American Society of Clinical Oncology/College Of American Pathologists guideline recommendations for immunohistochemical testing of estrogen and progesterone receptors in breast cancer. J. Clin. Oncol. 2010;28:2784–2795. doi: 10.1200/JCO.2009.25.6529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wolff AC, Hammond ME, Schwartz JN, Hagerty KL, Allred DC, Cote RJ, Dowsett M, Fitzgibbons PL, Hanna WM, Langer A, McShane LM, Paik S, Pegram MD, Perez EA, Press MF, Rhodes A, Sturgeon C, Taube SE, Tubbs R, Vance GH, van de Vijver M, Wheeler TM, Hayes DF. American Society of Clinical Oncology/College of American Pathologists guideline recommendations for human epidermal growth factor receptor 2 testing in breast cancer. J. Clin. Oncol. 2007;25:118–145. doi: 10.1200/JCO.2006.09.2775. [DOI] [PubMed] [Google Scholar]
- 20.Won KY, Kim GY, Kim YW, Song JY, Lim SJ. Clinicopathologic correlation of beclin-1 and bcl-2 expression in human breast cancer. Hum Pathol. 2010;41:107–112. doi: 10.1016/j.humpath.2009.07.006. [DOI] [PubMed] [Google Scholar]
- 21.Goldhirsch A, Wood WC, Coates AS, Gelber RD, Thurlimann B, Senn HJ. Strategies for subtypes--dealing with the diversity of breast cancer: highlights of the St. Gallen International Expert Consensus on the Primary Therapy of Early Breast Cancer 2011. Ann Oncol. 2011;22:1736–1747. doi: 10.1093/annonc/mdr304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yoon JK, Kim do H, Koo JS. Implications of differences in expression of sarcosine metabolism-related proteins according to the molecular subtype of breast cancer. J Transl Med. 2014;12:149. doi: 10.1186/1479-5876-12-149. [DOI] [PMC free article] [PubMed] [Google Scholar]


