Abstract
The transcription factor grainyhead-like 2 (GRHL2) is evolutionarily conserved in many different species, and is involved in morphogenesis, epithelial differentiation, and the control of the epithelial-mesenchymal transition. It has also recently been implicated in carcinogenesis, but its role in this remains controversial. Expression of GRHL2 has not previously been reported in cervical cancer, so the present study aimed to characterize GRHL2 expression in cervical cancer-derived cell lines (CCCLs) and cervical tissues with different grades of lesions. Microarray analysis found that the expression of 58 genes was down-regulated in CCCLs compared to HaCaT cells (non-tumorigenic human epithelial cell line). The expression of eight of these genes was validated by quantitative real-time PCR (qPCR), and GRHL2 was found to be the most down-regulated. Western blot assays corroborated that GRHL2 protein levels were strongly down-regulated in CCCLs. Cervical cells from women without cervical lesions were shown to express GRHL2, while immunohistochemistry found that positivity to GRHL2 decreased in cervical cancer tissues. In conclusion, a loss or strong reduction in GRHL2 expression appears to be a characteristic of cervical cancer, suggesting that GRHL2 down-regulation is a necessary step during cervical carcinogenesis. However, further studies are needed to delineate the role of GRHL2 in cervical cancer and during malignant progression.
Keywords: GRHL2, grainyhead-like 2, cervical cancer, epithelial-mesenchymal transition, carcinogenesis
Introduction
Cervical cancer is both one of the most common cancers and one of the principal causes of mortality in women worldwide (http://globocan.iarc.fr). Its main etiological factor is human papillomavirus infection [1]. Following the initial viral infection of proliferating cells in the basal layer of the cervical epithelium, infected cells proliferate and expand laterally. Viral gene expression is very tightly regulated [2], with the expression of capsid genes being activated as epithelial cells begin to mature and emigrate to suprabasal layers. The papillomavirus life cycle is time- and differentiation-dependent of the epithelial host cell, although the cellular factors involved in these processes remain unknown [3,4].
In this context, grainyhead-like 2 (GRHL2), a member of the grainyhead subfamily of transcription factors, plays an important role in regulating epithelial cell differentiation [5-7] and has recently been implicated in carcinogenesis [8,9]. Moreover, GHRL2 has been suggested to control the epithelial-mesenchymal transition (EMT), with loss of the epithelial phenotype appearing to be an essential characteristic of tumor progression [10-12]. Little is known about the expression of GRHL2 in cancer, although it was reported to be significantly down-regulated at the messenger (m)RNA and protein level in gastric cancer [8], and higher expression of GRHL2 was observed in oral squamous cell carcinoma [13] and hepatocellular carcinoma [9]. Interestingly, dual roles of this transcription factor were reported in breast cancer [14]. Thus, the role of GRHL2 is still controversial, with evidence for it being both a tumor suppressor gene [8,11,12] and an oncogene [9,13-17].
To the best of our knowledge there are no published reports of GRHL2 expression in cervical cancer. Therefore, in view of the important and controversial evidence previously reported from different epithelial cancers, we set out to characterize the expression of GRHL2 in cervical cancer in the present study.
Materials and methods
Cell culture
Cervical cancer-derived cell lines HeLa, SiHa, and C-33A, and the non-tumorigenic human epithelial cell line HaCaT were obtained from the German Cancer Research Center (DKFZ, Heidelberg, Germany). Cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum, penicillin (100 U/ml), and streptomycin (100 μg/ml), at 37°C with an atmosphere of 5% CO2 and 90% relative humidity. All products mentioned before were acquired from GIBCO®, Life Technologies Corporation, Carlsbad, California.
RNA extraction
A total of 3-5 × 106 cells of each cell line were seeded on p100 culture dishes, cultured in DMEM for 24 h at 37°C, then washed with cold phosphate buffered saline. RNA extraction was performed using the RNA Mini Kit PureLinkTM (Ambion®, Life Technologies Corporation) according to the manufacturer’s instructions. RNA samples were stored at -80°C until use.
Microarrays
RNA from CCCLs was analyzed by expression microarrays using the Gene Atlas Personal Microarray System (Affymetrix, Santa Clara, California). Briefly, total RNA was isolated from freshly seeded cells after 24 h of culture. The RNA integrity was assayed using the 2100 Bionalyzer (Agilent, Santa Clara, California) and 250 ng RNA from each sample was converted into double-stranded complementary (c)DNA using the WT Expression Kit (Ambion®, Life Technologies Corporation). This was then used to generate multiple copies of biotinylated cRNA by in vitro transcription with the GeneChip® WT Terminal Labeling and Controls Kit (Affymetrix). Biotinylated cRNA (10 μg) of each sample was hybridized to the Human Gene 1.1 ST Array Strip (Affymetrix) for 16 h at 43°C in the GeneAtlas Hybridization Station. After hybridization, microarray strips were washed and stained in the GeneAtlas Fluidics Station. Stained microarrays were scanned with the Imaging Station of the GeneAtlas System and image analysis and probe data extraction were carried out using Affymetrix GeneAtlas Software. The probe level data were summarized and normalized into probeset level data using the RMA algorithm before statistical testing; these calculations were performed using the Affymetrix expression console v. 1.1 (Affymetrix). Normalized data were analyzed using geWorkbench 2.4 software [18], and CCCL data were compared with that of HaCaT cells.
QPCR
Total RNA was reverse-transcribed into single-stranded cDNA using the SuperScript™ III First-Strand Synthesis System primed with oligo(dT) (Invitrogen™, Life Technologies Corporation). Gene expression levels were performed with 2.0 LightCycler technology using the LightCycler FastStar DNA Master PLUS SYBR Green I kit (Roche Applied Science, Penzberg, Germany). Analysis of gene expression was performed with LightCycler 4.1 software. The reference genes ACTB, GADPH, and RPL32 were used to determine the relative quantification of target genes by the ΔΔCp method. To analyze ΔΔCp, at least two independent experiments with duplicates were conducted. These data were normalized independently with the three reference genes and analyzed relative to HaCaT cells (set as 1). Relative expression was also calculated as ΔCp by subtracting the target gene Cp minus the reference gene Cp from the same sample. This type of analysis is normally used to avoid external calibrators and to take only intrinsic references from each sample into account. Notably, ΔCp is inversely proportional to the expression of the target gene.
Western blotting
Cells were lysed with radioimmunoprecipitation assay (RIPA) buffer by sonication (15 pulses, 90% amp). Total extracts were incubated for 30 min at 4°C and subsequently centrifuged at 17,000 g for 5 min at 4°C. Protein concentrations were determined using the Bio-Rad DC Protein kit (Bio-Rad Laboratories, Hercules, California) and 50 μg of the extracts were electrophoresed using 8% sodium dodecyl sulfate polyacrylamide gel electrophoresis. These gels were transferred onto polyvinylidene difluoride membranes (Merck Millipore, Billerica, Massachusetts) and incubated with 5% western blocking reagent to block nonspecific binding. Primary antibodies antiRHL2 (cat. no. HPA004820, Atlas Antibodies, Stockholm, Sweden; cat. no. Ab8631, Abcam, Cambridge, United Kingdom), or anti-β-actin (cat. no. sc-1616, Santa Cruz Biotechnology, Santa Cruz, California) were incubated overnight at 4°C, and specific secondary antibodies (cat. no. sc-2005, sc-2004 and sc2020, Santa Cruz Biotechnology) were incubated for 1 h at room temperature, followed by chemiluminescent detection (cat. no. WBKLs0100, Millipore) with ChemiDoc XRS (Bio-Rad Laboratories). Densitometric analysis was carried out to determine the intensity of GRHL2 expression relative to that of β-actin using Scion Image software (Scion Corporation, Frederick, Maryland).
Immunohistochemistry
Biopsies diagnosed as CIN 1, CIN 3 and CC were used to create tissue microarrays (TMA) by punching representative tissues for each sample with a needle to obtain 1 mm tissue cores, fixing in formalin, and transferring to a recipient paraffin block using a Tissue Microarray ATA 100 Chemicon (Temecula, California). TMA sections of 4 μm were sectioned using a Leica RM 2125 microtome (Wetzlar, Germany) and transferred to positively charged slides (Biocare Medical, Concord, California) [19]. Inmunodetection was performed automatically using the Ventana BenchMark System (Ventana Medical Systems, Mannheim, Germany) with an anti-GRHL2 antibody (cat. no. HPA004820, Atlas Antibodies) and Ultraview Universal DAB detection kit (cat. no. 760-500, Ventana Medical Systems). GRHL2 expression was evaluated by a pathologist and classified according to the level of positivity as negative, low, medium, or high.
Statistical analysis
All experiments were performed in duplicate and repeated independently at least twice. The results are expressed as means ± SD. For differences between groups, analysis was performed using an unpaired, two tailed Student’s t-test and significance at P < 0.05.
Results
Genes were differentially expressed between non-tumorigenic HaCaT cells and cervical cancer-derived cell lines
To identify genes with possible functions in the initiation, development, or maintenance of cervical cancer, we extracted mRNA from three CCCLs, HeLa, SiHa, and C-33A, and the keratinocyte non-tumorigenic cell line HaCaT as a control. We then compared the expression levels of comon genes using microrray analysis and identified six that were up-regulated and 58 that were down-regulated in the tumorigenic cell lines (Figure 1). The complete lists of differentially regulated genes are shown in Supplemental Data.
Figure 1.
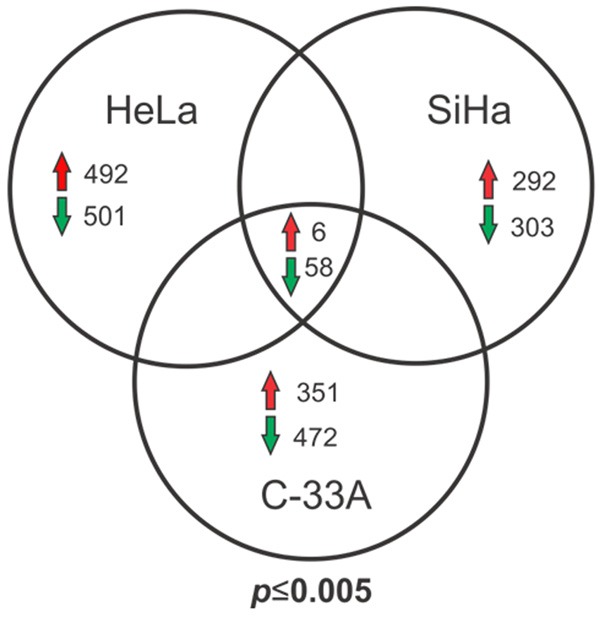
Venn diagram of differentially expressed genes in CCCLs versus HaCaT cells determined using microarray analysis. Comparison of genes differentially expressed between CCCLs (HeLa, SiHa, and C-33A) with respect to the keratinocyte-derived non-tumorigenic cell line HaCaT. Common up- or down-regulated genes in the three tumorigenic cell lines vs. HaCaT are shown. Statistically significant (P ≤ 0.005).
Validation of selected down-regulated genes in CCCLs by quantitative real-time qPCR
Based on the results obtained by microarray analysis, we selected eight of the 58 genes found to be down-regulated and analyzed their expression by qPCR, taking into account any relationship with neoplastic processes (Figure 2A). qPCR validated the down-regulation of the eight selected genes in CCCLs compared with HaCaT cells (Figure 2B). Using three different reference genes (ACTB, GAPDH, and RPL32) for normalization and HaCaT cells for calibration (value set as 1), relative expression values ranged from 0.015-0.000002, with GRHL2 found to be the most down-regulated gene in CCCLs.
Figure 2.
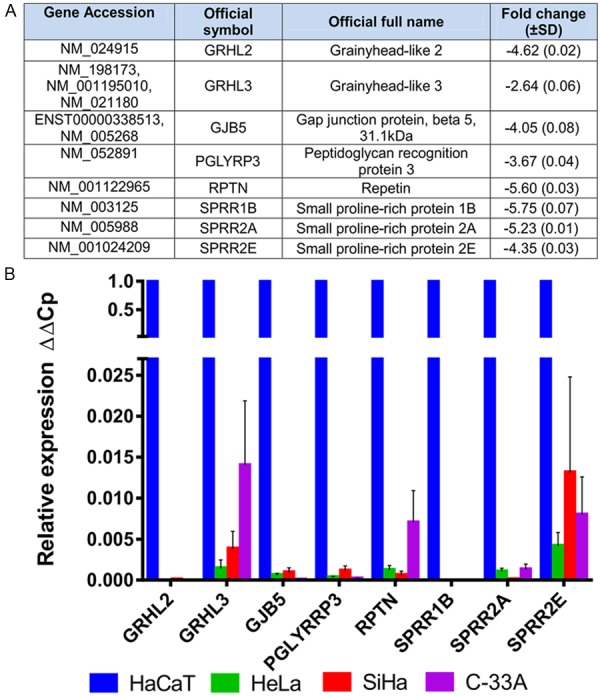
Down-regulated genes in CCCLs vs. HaCaT cells. A. Group of selected genes from microarray expression assays. The fold change value shown is a mean of the three CCCLs, HeLa, SiHa, and C-33A; B. Individual relative expression (ΔΔCp) of each selected gene as determined by qPCR in HeLa, SiHa, and C-33A. The values obtained were normalized to three reference genes, ACTB, GAPDH, and RPL32, and determined relative to HaCaT cells (set as 1). Means ± SD from at least three independent experiments are shown.
GRHL2 expression is remarkably decreased at the mRNA and protein level in CCCLs
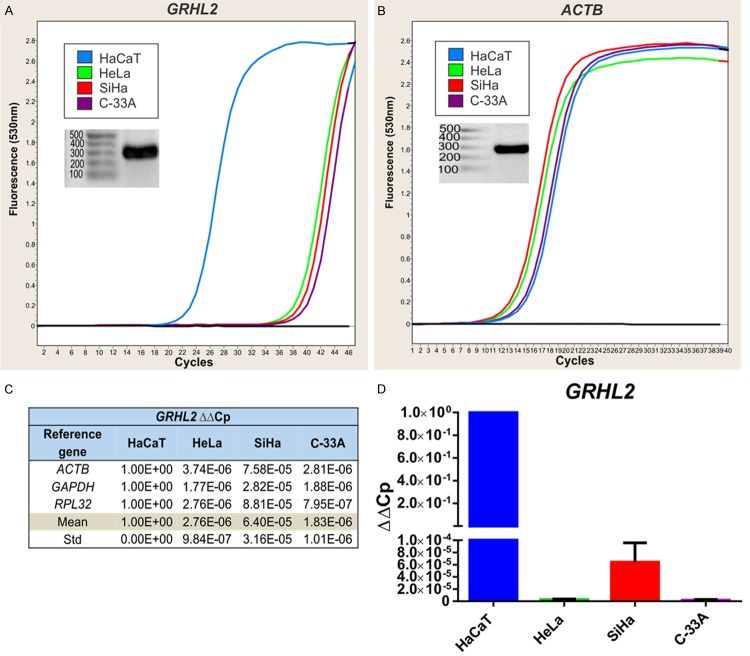
In the CCCLs analyzed, two members of the Grainyhead-like family of genes (GRHL2 and GRHL3) were found to be down-regulated, particularly GRHL2. Because the functions of these genes are closely interrelated and involved in mammalian organogenesis [20,21] and because GRHL2 has recently been shown to play a role in gastric and breast carcinogenesis [8,14], we focused further on its analysis. We observed a notable difference in GRHL2 expression (> 10 cycles) between the CCCLs and HaCaT cells when comparing amplification curves (Figure 3A). This difference was not observed for the reference genes, e.g. ACTB (Figure 3B).
Figure 3.
GRHL2 expression as determined by qPCR. Amplification curves obtained from GRHL2 (A) and the reference gene ACTB (B) in HaCaT (blue), HeLa (green), SiHa (red), and C-33A (purple). PCR products visualized on agarose gels are also shown. GRHL2 relative expression is shown as ΔΔCp, and the values presented were normalized to each reference gene (ACTB, GAPDH and RPL32) and determined relative to HaCaT (C). The mean ± SD of all cell lines are displayed in a graph (D).
To confirm specific amplification of primer pairs, the expected molecular weights of all qPCR products were observed on agarose gels. The relative expression of GRHL2 in each CCCL was determined using the ΔΔCp method, and data are shown in Figure 3C and 3D. These results indicated that GRHL2 was expressed at negligible levels in CCCLs compared with HaCaT cells.
To corroborate GRHL2 mRNA levels, protein expression was also determined. Western blotting with two different anti-GRHL2 antibodies showed that a protein of expected molecular weight was expressed only in HaCaT cells (Figure 4A, 4B). Densitometric analysis revealed a 6-fold increase in GRHL2 expression in HaCaT versus HeLa, SiHa, and C-33A cells, particularly using the Atlas antibody (Figure 4, right panels).
Figure 4.
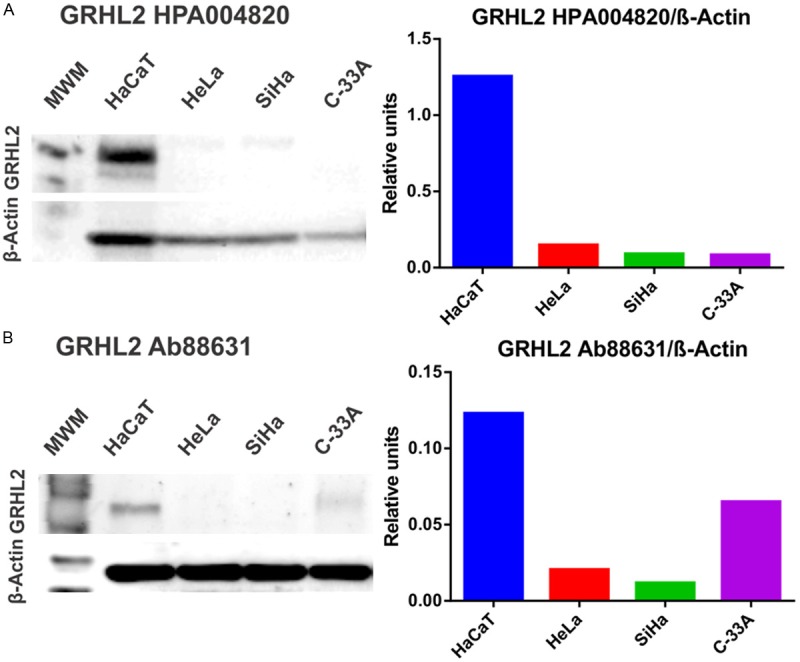
Detection of GRHL2 protein expression in HaCaT, HeLa, SiHa, and C-33A using western blotting. GRHL2 expression was measured using two independent antibodies, HPA004820-Atlas antibodies (A, left) and Ab88631-Abcam (B, left). β-actin was used as a protein loading control. Densitometric analysis expressing the ratio GRHL2/β-actin is also shown (A and B, right panels). MWM: Molecular weight marker.
GRHL2 is expressed in cervical samples without lesions
Although HaCaT cells are non-tumorigenic, they are an immortal cell line so have genomic instabilities. Therefore, to better characterize normal expression levels of GRHL2 in keratinocytes, we next determined GRHL2 mRNA expression levels in cervical epithelial samples obtained by cervical cytobrush of women without cervical lesions (as visualized by colposcopy).
A total of 15 control cervical samples were collected for RNA extraction and retrotranscription to measure GRHL2 expression using qPCR. ΔCp was then calculated using ACTB, GAPDH, and RPL32 as references genes. As shown in Figure 5, the average ΔCp for HaCaT cells was 9.2±0.8, which was very similar to that for cervical control samples (9.1±2.7), and much lower than that for CCCLs (21.1±5.3), representing a difference of at least 10 cycles. To achieve a general perspective of GRHL2 expression in cervical samples vs. HaCaT cells and CCCLs, ΔCps obtained from independent assays are displayed on a box and whisker plot (Figure 5). Cervical control samples were found to express very similar levels of GRHL2 to HaCaT cells, whereas CCCLs showed a strongly diminished expression pattern (P < 0.001).
Figure 5.
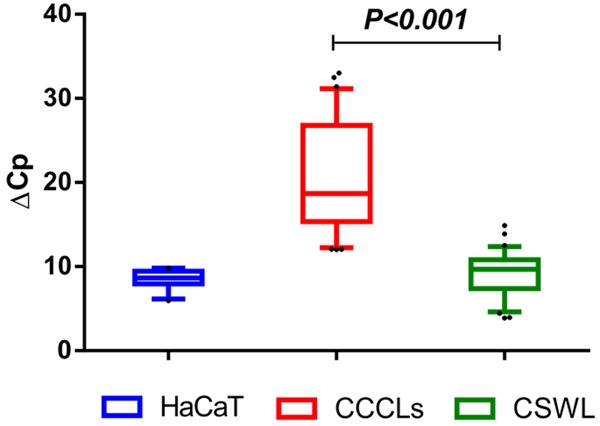
GRHL2 expression at the mRNA level in cervical samples without lesions. Box plot showing relative expression levels of GRHL2 as measured by qPCR in 15 cervical samples obtained from women without cervical lesions (CSWL). Relative expression is shown as ΔCp values (Cp of GRHL2 minus Cp of reference gene from each sample). ACTB, GAPDH, and RPL32 were used as reference genes. ΔCp values from HaCaT and CCCLs from all experiments are also included for comparison. The graph shows medians (dark lines), 5-95 percentile limits (boxes), interquartile ranges (whiskers), and outliers (points). *Student’s t-test, two-tailed P-value < 0.001.
Decreased expression of GRHL2 in cervical cancer tissues
To corroborate the low expression level of GRHL2 observed in CCCLs, tissues diagnosed as cervical intraepithelial neoplasia grade 1 (CIN 1), cervical intraepithelial neoplasia grade 3 (CIN 3), as well as cervical cancer (CC), were assembled in tissue arrays to detect GRHL2 using immunohistochemistry.
The GRHL2 signal was evaluated by a pathologist, and representative images of the assays performed in CC and precursor lesions (CIN 1 and CIN 3) are displayed in Figure 6A. A summary of the results is shown in Figure 6B, and it was clear that GRHL2 expression in cervical tissue samples diminished with increasing malignancy. This result is consistent with the strongly diminished GRHL2 expression observed in CCCLs (Figure 4).
Figure 6.
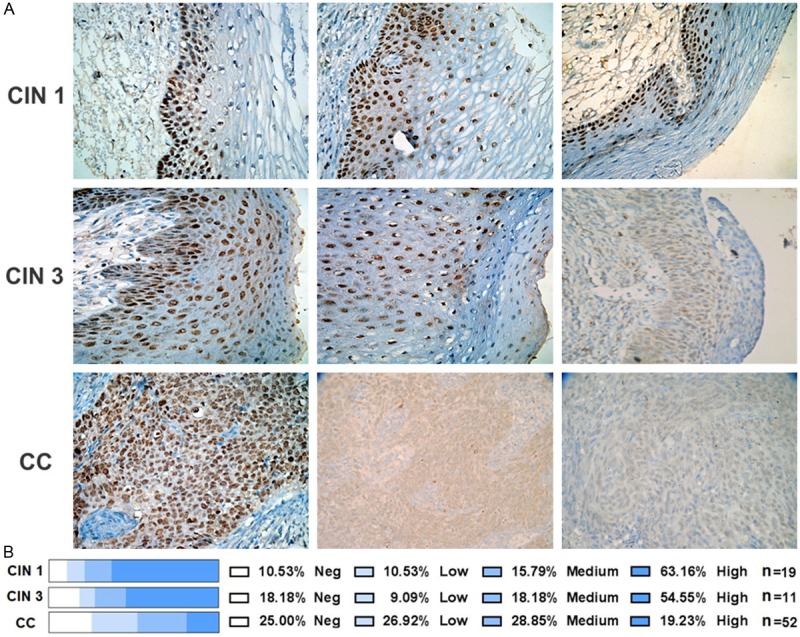
Immunodetection of GRHL2 in precursor and cervical cancer tissues. Cervical tissues microarrays (including CIN 1, CIN 3, and CC biopsies) were used to detect GRHL2 by immunohistochemistry. An anti-GRHL2 antibody (HPA004820-Atlas antibodies) was utilized as the primary antibody. Representative images from each histological diagnosis are shown at a magnification of ×40 (A). GRHL2 expression was evaluated by the pathologist and classified according to the degree of positivity: negative, low, medium, or high. Percentages of the tissues found with the different grades of GRHL2 positivity are included (B). The biopsies evaluated for each pathological diagnosis (n) are also included.
Discussion
We used microarray expression analysis to reveal the differential expression of genes in the three CCCLs tested compared to HaCaT cells, of which GRHL2 was strongly down-regulated in the tumorigenic cell lines (Figure 2). The functions of GRHL2 are mainly related to epithelial behavior such as morphogenesis, cell polarity, differentiation, and wound healing [5-7,22-25]. Although diminished expression of GRHL2 has been reported in other malignant cells including human gastric carcinoma cell lines [8] and breast cancer-derived cell lines, to our knowledge no previous reports have investigated GRHL2 expression in cervical cancer. Our study is therefore the first to demonstrate, both at the mRNA and protein level, that GRHL2 is strongly down-regulated in CCCLs.
Previous studies of breast cancer cells with an epithelial phenotype revealed that they expressed higher levels of GRHL2 [14,26]. In this respect, the functional role of GRHL2 as a proto-oncogene or tumor suppressor gene remains controversial in breast carcinogenesis [10-12,14,15]. Additionally, the regulatory system that governs the expression of this transcription factor (GRHL2/ZEB1/miR-200) is not fully understood.
Similar levels of GRHL2 as observed by qPCR in HaCaT cells were also seen in normal epithelial cervical cells (Figure 5), indicating that the cell line model used in our study did not misrepresent the tissue-specific level of GRHL2 in healthy human cervical samples. This result is in agreement with previous reports of different epithelial cells such as those of the mucociliary airway epithelium and lung epithelial cells, where GRHL2 regulates processes related to morphogenesis, differentiation, and barrier functions through its transcriptional up-regulation of genes related to tight and adherent junctions [6,7,24,25].
Immunohistochemistry showed that GRHL2 expression diminished in cervical tissues in line with an increase in malignancy (Figure 6). This supports the findings of Xiang and co-workers [8], who reported weaker GRHL2 expression in gastric cancer versus normal tissue. Similarly, Werner et al. [14] observed reduced GRHL2 expression at the invasive front of primary breast tumors, while decreased GRHL2 expression levels were also reported in the claudin-low molecular subclass of breast tumors and clear cell renal carcinomas [11]. Reduced expression of GRHL2 in breast tumors has been associated with an EMT phenotype, increased cell survival, and migratory and invasive behaviour, which is reminiscent of a tumor suppressor gene [11,12]. Conversely, high GRHL2 expression has been related to a poor prognosis for breast cancer metastasis [10,11,15,17], although we are currently exploring the hypothesis that high expression of GRHL2 indicates a loss of function of this protein.
Taken together, the findings from the present study strongly indicate that loss or diminished GRHL2 expression is characteristic of cervical carcinogenesis, suggesting a tumor suppressor function for GRHL2 in this model. However, functional studies are needed to delineate its role during the malignant progression of cervical cancer cells.
Acknowledgements
LAT-R, LA-R, and MR-S are grateful for a scholarship from Consejo Nacional de Ciencia y tecnología-México. This work was supported by Fondo de Investigación en Salud-Instituto Mexicano del Seguro Social (FIS/IMSS/PROT/G12/1149).
Disclosure of conflict of interest
None.
Supporting Information
References
- 1.Psyrri A, DiMaio D. Human papillomavirus in cervical and head-and-neck cancer. Nat Clin Pract Oncol. 2008;5:24–31. doi: 10.1038/ncponc0984. [DOI] [PubMed] [Google Scholar]
- 2.zur Hausen H. Papillomaviruses and cancer: from basic studies to clinical application. Nat Rev Cancer. 2002;2:342–350. doi: 10.1038/nrc798. [DOI] [PubMed] [Google Scholar]
- 3.Stubenrauch F, Laimins L. Human papillomavirus life cycle: active and latent phases. Semin Cancer Biol. 1999;9:379–386. doi: 10.1006/scbi.1999.0141. [DOI] [PubMed] [Google Scholar]
- 4.Bernard HU. Regulatory elements in the viral genome. Virology. 2013;445:197–204. doi: 10.1016/j.virol.2013.04.035. [DOI] [PubMed] [Google Scholar]
- 5.Boglev Y, Wilanowski T, Caddy J, Parekh V, Auden A, Darido C, Hislop NR, Cangkrama M, Ting SB, Jane SM. The unique and cooperative roles of the Grainy head-like transcription factors in epidermal development reflect unexpected target gene specificity. Dev Biol. 2011;349:512–522. doi: 10.1016/j.ydbio.2010.11.011. [DOI] [PubMed] [Google Scholar]
- 6.Werth M, Walentin K, Aue A, Schonheit J, Wuebken A, Pode-Shakked N, Vilianovitch L, Erdmann B, Dekel B, Bader M, Barasch J, Rosenbauer F, Luft FC, Schmidt-Ott KM. The transcription factor grainyhead-like 2 regulates the molecular composition of the epithelial apical junctional complex. Development. 2010;137:3835–3845. doi: 10.1242/dev.055483. [DOI] [PubMed] [Google Scholar]
- 7.Senga K, Mostov KE, Mitaka T, Miyajima A, Tanimizu N. Grainyhead-like 2 regulates epithelial morphogenesis by establishing functional tight junctions through the organization of a molecular network among claudin3, claudin4, and Rab25. Mol Biol Cell. 2012;23:2845–2855. doi: 10.1091/mbc.E12-02-0097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xiang J, Fu X, Ran W, Chen X, Hang Z, Mao H, Wang Z. Expression and role of grainyhead-like 2 in gastric cancer. Med Oncol. 2013;30:714. doi: 10.1007/s12032-013-0714-5. [DOI] [PubMed] [Google Scholar]
- 9.Tanaka Y, Kanai F, Tada M, Tateishi R, Sanada M, Nannya Y, Ohta M, Asaoka Y, Seto M, Shiina S, Yoshida H, Kawabe T, Yokosuka O, Ogawa S, Omata M. Gain of GRHL2 is associated with early recurrence of hepatocellular carcinoma. J Hepatol. 2008;49:746–757. doi: 10.1016/j.jhep.2008.06.019. [DOI] [PubMed] [Google Scholar]
- 10.Xiang X, Deng Z, Zhuang X, Ju S, Mu J, Jiang H, Zhang L, Yan J, Miller D, Zhang HG. Grhl2 determines the epithelial phenotype of breast cancers and promotes tumor progression. PLoS One. 2012;7:e50781. doi: 10.1371/journal.pone.0050781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cieply B, Riley PT, Pifer PM, Widmeyer J, Addison JB, Ivanov AV, Denvir J, Frisch SM. Suppression of the epithelial-mesenchymal transition by Grainyhead-like-2. Cancer Res. 2012;72:2440–2453. doi: 10.1158/0008-5472.CAN-11-4038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cieply B, Farris J, Denvir J, Ford HL, Frisch SM. Epithelial-mesenchymal transition and tumor suppression are controlled by a reciprocal feedback loop between ZEB1 and Grainyhead-like-2. Cancer Res. 2013;73:6299–6309. doi: 10.1158/0008-5472.CAN-12-4082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kang X, Chen W, Kim RH, Kang MK, Park NH. Regulation of the hTERT promoter activity by MSH2, the hnRNPs K and D, and GRHL2 in human oral squamous cell carcinoma cells. Oncogene. 2009;28:565–574. doi: 10.1038/onc.2008.404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Werner S, Frey S, Riethdorf S, Schulze C, Alawi M, Kling L, Vafaizadeh V, Sauter G, Terracciano L, Schumacher U, Pantel K, Assmann V. Dual roles of the transcription factor grainyhead-like 2 (GRHL2) in breast cancer. J Biol Chem. 2013;288:22993–23008. doi: 10.1074/jbc.M113.456293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yang X, Vasudevan P, Parekh V, Penev A, Cunningham JM. Bridging cancer biology with the clinic: relative expression of a GRHL2-mediated gene-set pair predicts breast cancer metastasis. PLoS One. 2013;8:e56195. doi: 10.1371/journal.pone.0056195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen W, Dong Q, Shin KH, Kim RH, Oh JE, Park NH, Kang MK. Grainyhead-like 2 enhances the human telomerase reverse transcriptase gene expression by inhibiting DNA methylation at the 5’-CpG island in normal human keratinocytes. J Biol Chem. 2010;285:40852–40863. doi: 10.1074/jbc.M110.103812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dompe N, Rivers CS, Li L, Cordes S, Schwickart M, Punnoose EA, Amler L, Seshagiri S, Tang J, Modrusan Z, Davis DP. A whole-genome RNAi screen identifies an 8q22 gene cluster that inhibits death receptor-mediated apoptosis. Proc Natl Acad Sci U S A. 2011;108:E943–951. doi: 10.1073/pnas.1100132108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Floratos A, Smith K, Ji Z, Watkinson J, Califano A. geWorkbench: an open source platform for integrative genomics. Bioinformatics. 2010;26:1779–1780. doi: 10.1093/bioinformatics/btq282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hidalgo A, Pina P, Guerrero G, Lazos M, Salcedo M. A simple method for the construction of small format tissue arrays. J Clin Pathol. 2003;56:144–146. doi: 10.1136/jcp.56.2.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wilanowski T, Tuckfield A, Cerruti L, O’Connell S, Saint R, Parekh V, Tao J, Cunningham JM, Jane SM. A highly conserved novel family of mammalian developmental transcription factors related to Drosophila grainyhead. Mech Dev. 2002;114:37–50. doi: 10.1016/s0925-4773(02)00046-1. [DOI] [PubMed] [Google Scholar]
- 21.Ting SB, Wilanowski T, Cerruti L, Zhao LL, Cunningham JM, Jane SM. The identification and characterization of human Sister-of-Mammalian Grainyhead (SOM) expands the grainyhead-like family of developmental transcription factors. Biochem J. 2003;370:953–962. doi: 10.1042/BJ20021476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Auden A, Caddy J, Wilanowski T, Ting SB, Cunningham JM, Jane SM. Spatial and temporal expression of the Grainyhead-like transcription factor family during murine development. Gene Expr Patterns. 2006;6:964–970. doi: 10.1016/j.modgep.2006.03.011. [DOI] [PubMed] [Google Scholar]
- 23.Pyrgaki C, Liu A, Niswander L. Grainyhead-like 2 regulates neural tube closure and adhesion molecule expression during neural fold fusion. Dev Biol. 2011;353:38–49. doi: 10.1016/j.ydbio.2011.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gao X, Vockley CM, Pauli F, Newberry KM, Xue Y, Randell SH, Reddy TE, Hogan BL. Evidence for multiple roles for grainyheadlike 2 in the establishment and maintenance of human mucociliary airway epithelium. Proc Natl Acad Sci U S A. 2013;110:9356–9361. doi: 10.1073/pnas.1307589110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Varma S, Cao Y, Tagne JB, Lakshminarayanan M, Li J, Friedman TB, Morell RJ, Warburton D, Kotton DN, Ramirez MI. The transcription factors Grainyhead-like 2 and NK2-homeobox 1 form a regulatory loop that coordinates lung epithelial cell morphogenesis and differentiation. J Biol Chem. 2012;287:37282–37295. doi: 10.1074/jbc.M112.408401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Neve RM, Chin K, Fridlyand J, Yeh J, Baehner FL, Fevr T, Clark L, Bayani N, Coppe JP, Tong F, Speed T, Spellman PT, DeVries S, Lapuk A, Wang NJ, Kuo WL, Stilwell JL, Pinkel D, Albertson DG, Waldman FM, McCormick F, Dickson RB, Johnson MD, Lippman M, Ethier S, Gazdar A, Gray JW. A collection of breast cancer cell lines for the study of functionally distinct cancer subtypes. Cancer Cell. 2006;10:515–527. doi: 10.1016/j.ccr.2006.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.