Abstract
In this essay, we discuss new insights into the wide-ranging impacts of mammalian transposable elements (TE) on gene expression and function. Nearly half of each mammalian genome is comprised of these mobile, repetitive elements. While most TEs are ancient relics, certain classes can move from one chromosomal location to another even now. Indeed, striking recent data show that extensive transposition occurs not only in the germline over evolutionary time, but also in developing somatic tissues and particular human cancers. While occasional germline TE insertions may contribute to genetic variation, many other, similar TEs appear to have little or no impact on neighboring genes. However, the effects of somatic insertions on gene expression and function remain almost completely unknown. We present a conceptual framework to understand how the ages, allele frequencies, molecular structures and especially the genomic context of mammalian TEs each can influence their various possible functional consequences.
Keywords: gene expression, gene regulation, retrotransposon, transposition, transcriptional variation
Introduction
Since their discovery in maize more than six decades ago by Barbara McClintock, transposons have been identified in virtually every eukaryotic genome that has been analyzed, including metazoans all the way to mammals [1]. These mobile genetic elements make up almost 50% of the human genome [2] and slightly less in the mouse [3]. A recent, probabilistic reannotation of genome sequences indicated that much more, i.e. up to approximately 70%, of mammalian genomes may be derived from transposons [4].
Both DNA transposons and RNA transposons (called retrotransposons) are abundantly represented in mammalian genomes [2,3]. The mechanisms by which they can move are highly divergent. DNA transposons move from one genomic location to another by a “cut and paste” process, whereby a transposase excises and reinserts them elsewhere. Retrotransposons are transcribed and the resulting RNA intermediates are copied by reverse transcriptase into cDNAs that are either synthesized directly at new target sites or inserted en bloc, as intact new elements, into new genomic locations [1].
In contrast to retrotransposons, DNA transposons are not actively mobilized in most mammalian genomes, except for particular classes that appear to have been mobilized more recently in bats [5-7]. Retrotransposons comprise a much more substantial fraction of each mammalian genome that has been sequenced. They include autonomous L1 (LINE, Long Interspersed Element) and non-autonomous SINE (Short Interspersed Element) retrotransposons. In primates including humans, another active, non-autonomous family has been identified, called SVA elements [8-10]. Various categories of endogenous retroviruses (ERVs) also have been defined, of which several classes are still actively mobilized in mouse, rat and perhaps human [11-15].
In discovering transposons and finding that they are mobile genetic elements, McClintock distinguished them from unmoving genes in the chromosomes. Significantly, she also termed transposons “controlling elements” because of their regulatory influences on neighboring gene expression and function, particularly in the development of various maize somatic tissues.
Given their huge numbers and widespread distribution in the mammalian genome, we hypothesize that many TE insertions have become fixed evolutionary relics in mammals that are biologically inert, with no detectable impacts on gene regulation or function. Ancient TEs thus have survived ongoing directional selection. Occasional TEs may have been exapted and exert positive effects, but very few would have deleterious impacts on gene function. By contrast, younger or de novo integrants that may have been mobilized in somatic tissues of a single individual may exert a wide range of effects on gene expression, from highly deleterious transcriptional dysregulation to neutral or rare positive consequences. The biological impacts of recently integrated elements are difficult to assess experimentally, since they may differ between tissues. Moreover, their allele frequencies may be substantially less than 50% in a chimeric population of cells.
Germline vs. Somatic mobilization
Germline transposition contributes to variation between individuals and within populations
Initial evidence demonstrating that human retrotransposons can move autonomously from one genomic location to another came from careful analysis of a disease caused by a new insertion. The first de novo L1 retrotransposon insertions to be identified occurred in two unrelated patients with hemophilia A. They caused insertional mutagenesis by integrating directly into the same exon of the factor VIII gene, thereby disrupting its open reading frame [16]. Subsequent investigations have revealed an ever-expanding list of approximately 100 individual cases of various human diseases attributable to new transposon insertions, either in the germline or somatic tissues [17-19]. By definition, germline insertions are present in the genomes of virtually all cells in offspring, but absent from the orthologous target site in both parents. They occur de novo in the germline cell lineage extending from the zygote to gametogonia, gametocytes and gametes.
Significant germline transposition in mammals has continued over evolutionary time up to the present, resulting in ongoing formation of insertion/deletion (indel) variants that are present or absent in individuals, populations and related species. In non-placental mammals such as the opossum, various classes of retrotransposons each have contributed to relatively high percentages of the overall genome [20,21]. In the mouse, several classes of transposable elements including L1, SINEs and various families of ERVs including intracisternal A particles (IAP), early transposons (ETn) and mammalian apparent LTR retrotransposons (MaLR) all have mobilized very recently. Resulting TE indels now are present in distinct genomic locations, distinguishing between closely related lineages [11,12,22-24]. De novo retrotransposon insertions even distinguish mice on the same genetic background that were separated merely ∼30 generations ago from the reference genome [25]. Similarly, human populations and individuals are delineated by numerous, polymorphic genomic integrants, present in some and absent from others [26,27]. A recent analysis of 1,000 Genomes data cataloged thousands of individual TE integrant polymorphisms in roughly 200 individuals from three major human populations. A detailed comparison of such transposon variants with single nucleotide polymorphisms (SNPs) revealed that insertion rates differ among populations [28]. However, a study of previously unreported L1 integrants in a mother/father/child family trio identified no de novo insertions in the child [29].
Mobilization rates of germline transposition seem to be comparatively greater in the mouse (per generation) than in human [12,23,25,29]. In addition, particular mouse lineages and human populations may support more frequent mobilization of certain TE classes. Nevertheless, precise rates have not been determined, and a detailed molecular basis for such differences in retrotransposition frequencies has not been established.
Germline transposition may help drive speciation
Over long evolutionary times, retention of exapted or exonized TEs may increase divergence and help drive speciation. For example, a human endogenous defective retrovirus element, HERV-W, has been incorporated into the gene encoding syncytin in human and other primates, thereby contributing to placental morphogenesis [30]. More recent analysis revealed biases toward exonized TEs at the 5′ ends of coding sequences in mouse and human, and differences between Alu and other TE families [31]. Moreover, germline transmission of retrotransposed pseudogenes also can contribute to functional differences between species. For example, a retrotransposed cyclophilin A pseudogene, mobilized into the TRIM5 locus, contributes to resistance to human immunodeficiency virus (HIV) in the owl monkey by driving fusion transcripts [32].
Endogenous transposition in somatic cells
In contrast with germline TE insertions, somatic insertions are not meiotically heritable from one generation to the next, although they can be mitotically transmitted within an individual (Fig. 1). Somatic insertions theoretically can occur in any cell type(s) aside from the gametes. In a few well-documented examples of somatic mobilization, new L1 retrotransposon insertions were identified as mosaics in various non-cancerous tissues of mouse and human, i.e. they were absent from the orthologous loci of at least some cells (Fig. 1). These results implied that L1 elements can mobilize during early embryonic development [33-36].
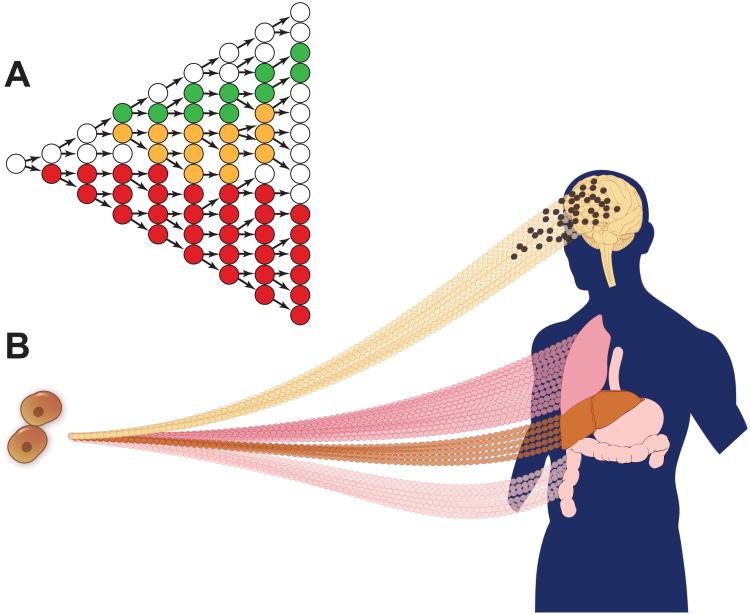
Figure 1. Somatic transposition leads to tissue-specific genetic variation.
(A) Schematic depicting transmission of three distinct transposon integrants occurring at various stages of tissue development (red, yellow and green circles), resulting in mitotically inherited insertions occurring at different allele frequencies in cell populations. This schematic was inspired by [110]. (B) During both normal development and in tumor formation, somatic transposition can result in TE mosaicism (illustrated by black circles in developing brain and hot pink circles in colon). The developmental time point and tissue specificity of mobilization help to define the variable allele frequencies in cells and tissues, and the diverse possible functional impacts of such somatic transposition.
More recent studies revealed that significant endogenous transposition can occur frequently in somatic tissues without overt disease formation. For example, L1 retrotransposons marked with a reporter gene are mobilized in neuronal precursor cells, frequently targeting neuronal genes [37]. These findings, initially in rat and mouse neuronal progenitor cells, were later extended using quantitative PCR to detect somatic mobilization of endogenous L1 elements in human neuronal precursors [38]. Deep sequencing of genomic DNA from hippocampus and caudate nucleus from three normal adult human brains revealed tens of thousands of somatic indel polymorphisms caused by L1, Alu (SINE) and SVA retrotransposon insertions [39]. Interestingly, these insertions were enriched in particular protein-coding genes that tend to be expressed in the brain. A more recent study of individual human cortical and caudate neurons confirmed that somatic L1 retrotransposition events occur, albeit much more rarely than described above, i.e. at one insertion per 100 to 10,000 neural progenitors, resulting in very low levels of mosaicism (Fig. 1) [40].
Because they may be mobilized later in mammalian development, new somatic integrants are expected to exist in heterozygosity in individual cells, but at allele frequencies far less than 50% within a particular tissue or at the level of a whole organism (Fig. 1). Experimental measurement of these frequencies will help elucidate when transposition may occur during development of particular tissues [40].
Endogenous human retrotransposition also occurs in certain sporadic cancers, as demonstrated first by the identification of a somatic L1 insertion in the APC gene in a colorectal tumor [41]. Both targeted and genome-wide deep sequencing recently documented many additional examples of such somatic mobilization [14,42] (Fig. 1). Somatic TE integrants were identified in a fraction of tumors arising in certain tissues such as lung, colorectal, prostate and ovary but not others [14,42,43]. Roughly one-third of the new TE insertions landed within annotated genes.
Full-length L1 transcripts are expressed in human embryonic stem cells and in human induced pluripotent stem (iPS) cells [44]. Their expression is associated with decreased cytosine methylation in the genomic L1 promoter, located in the 5′ untranslated region (5′ UTR). Expression is associated with increased retrotransposition of engineered donor elements in human iPS cells [44]. By contrast, no endogenous retrotransposition was identified in three pluripotent mouse iPS cell lines analyzed by deep genomic sequencing [45]. Thus, divergent conclusions were formed from these studies, illustrating that TE transcription or engineered elements' mobilization may not necessarily reflect natural mobilization of endogenous retroelements. For example, engineered elements typically include an antiparallel reporter gene with a strong constitutive promoter and artificial intron [46]. Since experimentally marked TEs may have altered expression and mobilization, their utility as a model for native TEs may be obscured.
What host factors regulate mobilization of TEs?
Several classes of host factors recently have been shown to regulate not only the expression and mobilization of retrotransposition-competent TEs; they also may influence the impact of more ancient elements in bringing about genetic variation. Expression and mobilization of TEs are suppressed by DNA methylation and other host regulatory factors [47]. In the mouse, reduced genomic DNA methylation is linked to dramatic increases in transposition of particular TE classes, e.g. ERV (IAP) elements, thereby contributing to T cell lymphomas [48]. Interestingly, somatic ERV integrants recurrently were recovered from a single target gene, Notch1, and can initiate aberrant, downstream fusion transcripts in the tumors [48].
A histone methyltransferase, Setdb1, acts independently from DNA methyltransferases to repress certain families of ERVs in the mouse genome [49]. In knockout mice, those TEs are transcriptionally activated, although their resulting consequences on TE mobilization or the expression of neighboring genes remain unknown.
The methylcytosine binding protein MeCP2 has been shown to regulate expression and mobilization of L1 retrotransposons in somatic human cells [50] including brain cells. When MeCP2 is depleted, as in Rett syndrome patients, retrotransposition of both endogenous and of engineered L1 elements increases [51]. Similarly, disruption of ataxia telangiectasia mutated (ATM), a serine- threonine kinase involved in DNA damage repair pathways, results in increased somatic retrotransposition, both in an engineered model system and in brains from affected patients [52].
Additional cellular processes or proteins that inhibit endogenous retrotransposition include RNA interference [53], various APOBEC3 proteins [54], and heterogeneous nuclear ribonucleoprotein L [55]. However, in contrast to the epigenetic regulators outlined above, these factors inhibit retrotransposition after TE expression, so their impacts on TE-mediated genetic variation are secondary.
Wide-Ranging Biological Impacts of Transposons
The identification of sporadic diseases that are caused by new transposon insertions demonstrated that such integrants can affect gene expression and functions [56,57]. While many of the individual integrants disrupt coding exon sequences by direct insertional mutagenesis, others affect gene expression and structure by disrupting RNA splicing [58], introducing new promoter sequences and other mechanisms (Fig. 2).
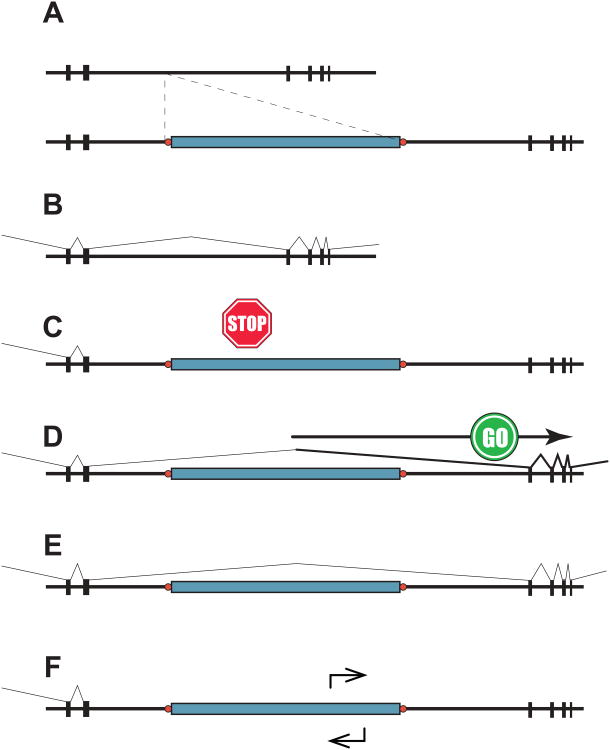
Figure 2. Variable effects of mammalian TEs on gene expression.
Shown here are models of possible genomic and transcriptional variation due to mammalian transposon integrants. (A) Schematic of a gene without a TE insertion (top) and with a polymorphic TE insertion (bottom, indicated by blue rectangle). Black vertical rectangles, gene exons; horizontal black lines, genomic sequences including introns; red circles, target site duplications. (B – E) Effects of TE insertions on gene expression. (B) Typical gene expression with RNA splicing betwPeen exons, removing introns in the absence of a transposon insertion. (C) Premature transcriptional termination (Stop) triggered by an intronic TE acting at long genomic distances [24,82]. (D) TE-mediated transcriptional activation or upregulation (Go), due to internal promoter or enhancer activity. (E) Intergenic TEs may incorporate enhancer or silencer activities that variably affect gene expression upstream and/or downstream. These effects can help TEs influence transcriptional regulatory networks. (F) We hypothesize that many TE integrants may have no detectable effect on gene expression, depending on TE age and structure, tissue-specific differences and genomic context. See text for further details.
TEs can introduce promoter activities and initiate novel transcripts
A recently appreciated, functional impact of pre-existing or ancient retroelements in interspersed genomic locations consists of their ability to initiate fusion transcripts [59]. While resulting fusion transcripts might only rarely encode intact proteins, they also can add to the transcriptome as long noncoding RNAs. A provocative recent analysis of mouse and human transcriptomes suggested that between 6 and 30% of all transcripts expressed in a range of tissues are initiated from transposable elements [60]. This study suggested that proximal transposon integrants can function as alternative gene promoters and initiate expression of noncoding transcripts. A more recent study of transcription start sites, based on cap-analysis of gene expression (CAGE) clusters, revealed that about 18% of transcription start sites overlap with repetitive elements, including many overlapping with L1, SINE, LTR and other elements [61]. These data highlighted that cell lineage-specificity is a central characteristic of the transposon-initiated transcripts.
An aberrantly activated, individual ERV integrant upstream of a myeloid-specific proto-oncogene, colony-stimulating factor 1 receptor (CSF1R), initiates expression of that gene in certain human lymphomas. The resulting, aberrantly expressed gene contributes to lymphoma pathogenesis [62]. In another example, an L1 insertion in the c-Met proto-oncogene initiates expression of L1-Met fusion transcripts that are associated with downregulated canonical Met expression [63]. The mechanism by which such fusion transcripts might decrease native gene expression is unclear. Many additional examples of fusion transcripts initiated from transposable elements in mouse and man have been described, including antisense transcripts [12,59,64,65].
Promoters within transposable element integrants can initiate variable transcription of adjacent (or more distant) genes. Perhaps the best-characterized examples of such variably expressed transcripts are provided by mouse ERVs within or near the agouti viable yellow (Avy) and axin fused (AxinFu) genes. In those cases, promoters in the ERVs' long terminal repeats, up to ∼100 kb distant, can act as gene-specific, alternative promoters, initiating fusion transcripts that encode most or all of the canonical open reading frames [66,67]. Human retrotransposons also can add to and alter the cellular transcriptome [59,68].
Small RNAs can be transcribed abundantly from TE templates. These include both Dicer-dependent, small interfering RNAs, microRNAs [69] and Dicer-independent, Piwi-associated RNAs. They appear to play various roles in regulating both the TEs themselves as well as other cellular transcripts in various stages of development and in the germline [53,70,71]. Recent analysis of a disease-causing locus, linked with infantile encephalopathy, revealed a point mutation in a primate-specific L1 retrotransposon which results in aberrant formation of a small RNA-like hairpin template [72]. Thus, TEs can serve as templates for expression of noncoding RNAs that play significant roles in human brain development. In some cases, mutated small RNAs can be pathogenic.
Additional genomic features introduced by new TE insertions
In addition to introducing promoter activities, mammalian transposons also contain other genetic features that can affect gene expression. These include enhancer elements, boundary elements, transcription splice sites, termination (polyadenylation signal) sequences and microRNA binding sites [57].
DNase hypersensitivity sites can serve as markers of regulatory DNA sequences. Their recent mapping demonstrated that they are localized frequently within certain individual TEs in a cell type-specific fashion [73]. These results corroborate cell-specific transcription of many TEs [61], strongly suggesting that transposons can act as enhancers. The most highly enriched DNase hypersensitivity sites are located in long terminal repeats of ERV elements [73]. Approximately 10 - 20% of conserved, non-exonic gene regulatory sequences were deposited by TEs [74,75].
Alternative splice sites can be introduced directly by TE insertions. For example, a long terminal repeat of an ERV-K element in the human Leptin receptor triggers alternative splicing and encodes 3′ end of alternative transcripts [76]. Mammalian L1 retrotransposons contain numerous splice donor and acceptor sites, which can disrupt normal gene expression and contribute to transcriptional variation [77,78].
Annotated RefSeq transcripts frequently can contain a retrotransposon incorporated into their 3′ UTR, potentially providing a target for microRNA binding which may result in reduced expression of such transcripts or encoded proteins [69]. TEs can incorporate transcriptional polyadenylation (termination) sequences into intragenic regions [79], e.g. in the 3′ UTR of genes [80].
We recently described a previously uncharacterized form of transcriptional disruption, triggered by intronic ERV elements in the mouse, that can act at considerable genomic distances up to >12.5 kb upstream [24]. As part of the study, we mapped thousands of polymorphic ERV insertions in widely divergent mouse strains. While premature transcriptional termination was strongly associated with a small but significant fraction of intronic ERVs located nearby, a large majority of such TEs may not trigger significant transcriptional disruption in any tissue or developmental timepoint [24].
Premature transcriptional termination attributed to ERVs has been identified at genes including CDK5 regulatory subunit associated protein 1 (Cdk5Rap1, also called Cabp) [81], solute carrier family 15 [H+/peptide transporter], member 2 (Slc15a2), polymerase [RNA] I polypeptide A (Polr1a, also called Rpo1-4), spondin 1 (Spon1) and up to perhaps 100 other genes [24,82]. In these cases, intronic ERVs trigger alternative RNA splicing, resulting in expression of intronic sequences and usage of cryptic existing polyadenylation signals. In heterozygous animals, the parent of origin of the ERVs is associated with levels of non-terminated transcripts. When the ERV is paternally inherited, non-terminated transcripts are reduced and terminated transcripts are increased. By contrast, when the ERV is maternally inherited, the non-terminated transcripts are significantly increased [24].
SINEs also can affect transcripts in several distinct ways. Intronic Alu elements are relatively enriched adjacent to alternatively spliced exons, when compared with constitutively spliced exons. Pairs of oppositely oriented elements can form duplexes that can trigger A-to-I editing by specific double-stranded RNA adenosine deaminases, thereby affecting the nuclear retention and alternative splicing of resulting transcripts [83]. Moreover, a form of mRNA degradation appears to be mediated by intermolecular interactions between long noncoding transcripts containing Alu sequences that are partially complementary to Alu sequences in the 3′ untranslated region of transcripts. This process is mediated by Staufen1, a double-stranded RNA binding protein in mouse and human [84,85]. Alu sequences in the 3′ UTRs of transcripts also can provide targets for various microRNAs [86].
TEs can influence transcriptional networks
Substantial evidence indicates that retrotransposons have introduced or propagated various tissue-specific gene regulatory elements in mouse [87], human [88] and other mammalian genomes [89]. Many of the conserved, non-exonic gene regulatory sequences introduced by TEs are strongly enriched near genes with developmental and transcriptional regulatory roles [74,75].
Exapted TE fragments have helped form regulatory networks involving various transcription factors. For example, binding sites for the CCCTC binding factor, CTCF, have been linked to the expansion of mouse SINE B2 elements, which dispersed them in up to 10,000 locations [87]. A comparative study of CTCF binding sites across multiple mammalian genomes revealed that a large majority of new, non-conserved binding sites likely were mobilized by more recent, species-specific SINE integrants, because they are embedded within SINE retrotransposon sequences. In addition, more highly conserved CTCF binding sites, present at orthologous locations across many species, appear to have been mobilized by ancient SINEs [89]. These sites can divide large-scale topological or chromatin interaction domains from one another, thereby serving as insulators [90].
Another transcriptional regulatory network involving p53 binding sites and responsive elements also has been linked to TEs, i.e. primate-specific Alu elements [91], ERVs [92] and the 5′ UTR promoter sequences of L1 retrotransposons [93]. In the latter case, p53 can activate expression of L1 elements.
TEs can serve as targets of variable epigenetic regulation
TEs comprise some of the main targets of variable epigenetic regulation [47,94]. Bioinformatics screens for “meta-stable” epialleles, focused on promoter activity and euchromatic histone modifications, identified numerous loci that display variable DNA methylation and associated, variable levels of transcription [95]. Most of these epialleles, whose expression differences are inherited transgenerationally but are variably established, appear to involve endogenous mouse transposons [96,97].
In cultured human embryonic carcinoma cells, engineered L1 integrants undergo variable de novo epigenetic silencing mediated by changes in histone acetylation, although the impact of such epigenetic variation on neighboring gene expression or function has remained unclear [98]. Spreading of heterochromatin adjacent to polymorphic retrotransposon integrants has been described in the mouse, occasionally affecting expression of certain genes including B3galtl [99]. Genes with CpG islands that are hypermethylated frequently in human cancers are depleted in SINE and L1 retrotransposons near their transcription start sites, suggesting that TEs may play an important role in affecting epigenetic regulation at genes linked to tumor formation [100].
Evolutionary implications of endogenous transposons
The many forms of functional variation attributable to active transposition have engendered considerable speculation about the evolutionary implications of such mobilization [69,101]. As somatic L1 retrotransposition may contribute to mosaicism in brain cells, leading to increased neuronal diversity and resulting functional variation, such mobilization may confer a positive advantage upon individual hosts [102,103]. However, the idea that most somatic retrotransposition events may be neutral or indeed are deleterious to individual hosts is highly likely. For example, recent evidence indicates that active endogenous transposition may occur early in the development of certain human cancers [14,42,43]. Thus, in typical circumstances, somatic TE mobilization is unlikely to be beneficial to the host.
The evolutionary consequences of TE insertions must also be considered from a population genetics perspective. Mobilization of TEs that is deleterious for an individual may be beneficial for the broader population [103].
Identifying the Determinants of TE-Mediated Variation
Despite numerous diverse functional consequences attributed to transposon integrants, which we emphasize are repetitive, virtually identical elements interspersed across genomes, we note that many or most mammalian germline TE insertions appear unlikely to affect nearby gene transcription or functions (Fig. 3). In addition, most of the TEs that do affect neighboring gene expression seem to exert their effects in a limited, tissue-specific fashion.
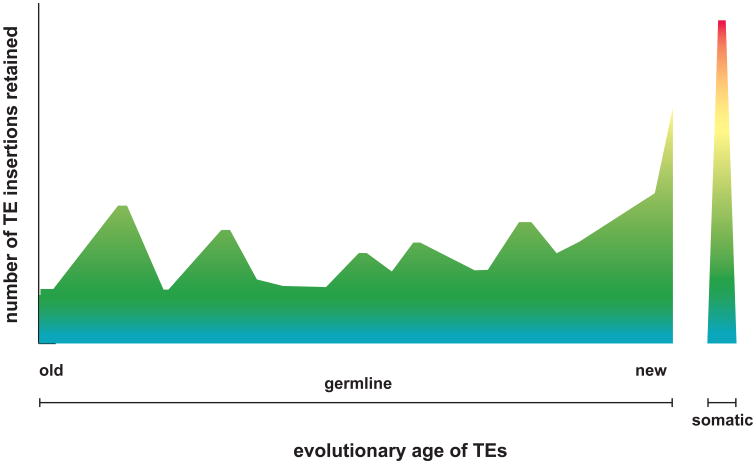
Figure 3. Variable impacts of transposon insertions accumulated over time.
This schematic depicts hypothetical genomic counts (y-axis) and functional impacts (color scale) of TE insertions retained over evolutionary time, ranging from “old” (left) to “new” (right) integrants. The newest TE integrants are generated in somatic tissues (right). We hypothesize that their variable functional impacts (blue, neutral impact; red, high or deleterious impact) are influenced by the duration and strength of directional selection against deleterious insertions. In this way, surviving “older” insertions would be expected to have accumulated more mutations, to be fixed at a population level, and to exert minimal negative impacts on gene expression and function (green, blue, see Fig. 2F). By contrast, some young TE insertions, including somatic insertions, may disrupt gene expression more strongly (yellow, orange, red). Their allele frequencies would be significantly less than 100%. See text for further details.
Strong evidence supporting the idea that many or most germline TEs do not uniformly or broadly affect genes was provided recently by the ENCODE project. A large majority of the DNase-hypersensitivity sites that mapped within TEs are hypersensitive to DNase treatments in a cell line-specific or tissue-specific fashion [73], rather than on a constitutive basis. Thus, despite the fact that transposons are redundantly represented as repetitive elements in many genomic locations, TE-mediated effects are not exerted uniformly or globally, but rather are idiosyncratic and contextual.
Another illustration of these points is provided by highly divergent inbred mouse strains, which have many thousands of polymorphic transposon insertions widely dispersed across their genomes [11,12,23,24,104]. Despite extensive genomic variation due to TE polymorphisms, the divergent mouse lineages all are viable, fertile and lack overt diseases. Their transcriptomes are more closely related on a tissue-specific basis than on a lineage-specific basis [105,106].
What genomic features could influence the variable biological impacts of TEs?
We propose that the ages, allele frequencies, molecular structures, and particularly the genomic context of individual mammalian TE integrants each can help to determine their wide-ranging possible functional impacts, ranging from no detectable effect to various large effects. In addition to affecting transcriptional splicing or termination in cis [77,79,80], intronic TE insertions may affect transcription of overlapping transcription units by disrupting read-through transcriptional processivity [107]. Effects in trans may include heterozygous TE insertions affecting gene expression from the homologous chromosome [24], and TE sequences in enhancer elements affecting expression quantitative trait loci (eQTL) in embryonic stem cells [108].
We suggest that the evolutionary times at which TEs first integrated would be inversely related to their remaining functional impact, reflecting the duration and strength of directional selection acting against their deleterious effects (Fig. 3). In this way, retained, older insertions would be expected to have accumulated more mutations, to be fixed at a population level, and to exert minimal negative impacts on gene expression and function. In relatively rare countervailing cases, they may have undergone exaptation or may exert other salutatory effects [103]. By contrast, younger TE insertions, including polymorphisms at a population level or even somatic integrants within a tissue or an individual, might be able to disrupt gene expression more strongly. There would be insufficient time for negative selection to mimimize their impacts by removing alleles that are deleterious. These younger insertions would be expected to be relatively free of mutations. Since they would not have undergone fixation in the population, their allele frequencies would be significantly less than 100%, so their potential impacts upon the population would be expected to be limited as well.
We also propose that the molecular structures of TE integrants themselves are another fundamentally important, albeit poorly understood, determinant of their impacts on gene expression and function. Mammalian TEs can be broadly classified as containing long terminal repeats or not. Integrants possessing long terminal repeats frequently have strong internal promoter activities, as demonstrated by DNase hypersensitivity experiments and analysis of transcription start sites, assessed recently in a variety of human cell lines [61,73]. Other insertions by non-LTR retrotransposons such as L1 elements frequently are 5′ truncated, so different innate features will be present or absent in various integrants. For example, both a sense and an antisense promoter are present in the 5′ untranslated region of human L1s, but these are absent from most integrants, which instead are truncated [46]. A striking difference in the length distributions of L1 integrants in human cancers vs. in the germline was recently reported [14], implying that the functional impacts of severely truncated integrants (observed in cancers) might be significantly less than those of longer elements (mobilized in the germline). SINE elements include both RNA polymerase III promoter sites [109] and CTCF binding sites [87,88], which can help shape their impacts on gene expression.
We propose that the most significant factor that defines the impact of TEs on gene expression and biological function is very likely to be their genomic context, i.e. the collection of neighboring features including promoters, tissue-specific transcription units, enhancers, boundary elements, splice sites, polyadenylation sites, and others that can interact with each TE integrant. Since new TE insertions located at interspersed locations are themselves virtually identical, one might expect their effects in triggering genetic variation would be comparable. However, their impacts are wide-ranging [24,104]. Many TEs are likely to lack pre-existing, weak polyadenylation or alternative splicing signals nearby, which if present could have been co-opted by them in cis or in trans [24]. Others may be located in densely silenced, constitutive heterochromatin, or away from actively transcribed genes, and therefore would lack any detectable functional impact.
The genomic distribution of newly integrated TEs can reflect mechanism-based target specificities that may define their resulting genomic milieus and, in turn, their impacts on gene expression. Their orientation in the genome, relative to gene transcription units, is another important contextual factor that determines their functional consequences. Sense-oriented TEs typically can affect gene expression more strongly, by mechanisms that are not fully characterized. Because their genetic impact can be deleterious, such insertions undergo directional selection. Older, residual elements have survived such ongoing selection, resulting in the removal of the more deleterious insertions, so the genomic distribution of the surviving elements would be biased more extensively toward antisense TEs, particularly as the elements age [11,12,24,104].
The distance between TEs and other genomic features also is likely to be an important determinant that modulates their impact. Agouti is variably expressed upon epigenetic regulation at an ERV approximately 100 kb distant {Duhl, 1994 #4425}. However, the extent to which such chromosomal distances influence the TEs' effects on gene regulation is unknown.
Many further experiments will be required to characterize the determinants of wide-ranging TE-mediated impacts on gene expression, structure and function. For example, it is very likely that targeted, experimental manipulation of particular elements already known to disrupt gene transcription, along with others that have no discernable impacts, will shed substantial light on some of the factors that shape these variable effects by TEs. Ideally, such studies will be performed in a variety of organisms in vivo and in vitro. They will need to include experimental disruption of TE integrant structures and their genomic locations. Insights will also be gained from studies of the genetic impact of engineered agents such as lentiviruses, used as delivery vehicles in gene therapy experiments.
Conclusions
TEs are actively mobilized in mammalian genomes. Striking recent findings have documented TE movement not only in the germline, but also in developing somatic tissues and in certain cancers. In this essay, we have discussed new insights into some of the variable biological impacts that endogenous mammalian TEs can exert in wide-ranging genomic, cellular and organismal contexts, i.e. by affecting and disrupting gene expression, regulation, structure and function in various ways. The basis for this extensive variability in the functional consequences of mammalian transposition is not well established. Notably, many transposon insertions appear to have little or no impact on neighboring genes. Such “inert” insertions include many ancient, evolutionarily established elements that have persisted despite negative selection. Other TE insertions with little functional consequence probably include somatic insertions occurring at low allele frequencies, short or promoter-less elements, and integrants that landed far from active transcription units or in constitutively heterochromatic genomic locations. In some cases, ancient elements might have been co-opted for more positive, beneficial uses, including roles in transcriptional regulatory networks, by the host through exaptation. By contrast, many TEs may be functionally deleterious, including those that mutagenized exons or transcriptional regulatory regions directly, those with active promoter, splice site or terminator features, those that landed near native genetic regulatory elements, and those that are strong targets for epigenetic regulation. Notably, the effects of somatic insertions on gene expression and function remain almost completely unknown. We conclude that the ages, allele frequencies, molecular structures and most importantly the genomic context of transposed mammalian integrants all are important factors in defining their various possible functional impacts in mammalian genomes.
Supplementary Material
Acknowledgments
We thank Anthony Baker (Medical Visuals, Health Sciences Library, Ohio State) for expert help preparing illustrations, and members of the Symer laboratory for discussions.
Footnotes
We declare no conflicts of interest associated with this manuscript.
References
- 1.Symer DE, Boeke JD. An everlasting war dance between retrotransposons and their metazoan hosts. In: Kurth RBN, editor. Retroviruses: Molecular BIology, Genomics and Pathogenesis. Norwich, U.K.: Caister Academic Press; 2010. pp. 1–33. [Google Scholar]
- 2.Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 3.Waterston RH, Lindblad-Toh K, Birney E, Rogers J, Abril JF, et al. Initial sequencing and comparative analysis of the mouse genome. Nature. 2002;420:520–62. doi: 10.1038/nature01262. [DOI] [PubMed] [Google Scholar]
- 4.de Koning AP, Gu W, Castoe TA, Batzer MA, Pollock DD. Repetitive elements may comprise over two-thirds of the human genome. PLoS genetics. 2011;7:e1002384. doi: 10.1371/journal.pgen.1002384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Feschotte C, Pritham EJ. DNA transposons and the evolution of eukaryotic genomes. Annu Rev Genet. 2007;41:331–68. doi: 10.1146/annurev.genet.40.110405.090448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ray DA, Feschotte C, Pagan HJ, Smith JD, Pritham EJ, et al. Multiple waves of recent DNA transposon activity in the bat, Myotis lucifugus. Genome Res. 2008;18:717–28. doi: 10.1101/gr.071886.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pagan HJ, Macas J, Novak P, McCulloch ES, Stevens RD, et al. Survey sequencing reveals elevated DNA transposon activity, novel elements, and variation in repetitive landscapes among vesper bats. Genome Biol Evol. 2012;4:575–85. doi: 10.1093/gbe/evs038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cordaux R, Batzer MA. The impact of retrotransposons on human genome evolution. Nat Rev Genet. 2009;10:691–703. doi: 10.1038/nrg2640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Prufer K, Munch K, Hellmann I, Akagi K, Miller JR, et al. The bonobo genome compared with the chimpanzee and human genomes. Nature. 2012;486:527–31. doi: 10.1038/nature11128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hancks DC, Goodier JL, Mandal PK, Cheung LE, Kazazian HH., Jr Retrotransposition of marked SVA elements by human L1s in cultured cells. Hum Mol Genet. 2011;20:3386–400. doi: 10.1093/hmg/ddr245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang Y, Maksakova IA, Gagnier L, van de Lagemaat LN, Mager DL. Genome-wide assessments reveal extremely high levels of polymorphism of two active families of mouse endogenous retroviral elements. PLoS Genet. 2008;4:e1000007. doi: 10.1371/journal.pgen.1000007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Akagi K, Li J, Stephens RM, Volfovsky N, Symer DE. Extensive variation between inbred mouse strains due to endogenous L1 retrotransposition. Genome Res. 2008;18:869–80. doi: 10.1101/gr.075770.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang Y, Liska F, Gosele C, Sedova L, Kren V, et al. A novel active endogenous retrovirus family contributes to genome variability in rat inbred strains. Genome research. 2010;20:19–27. doi: 10.1101/gr.100073.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee E, Iskow R, Yang L, Gokcumen O, Haseley P, et al. Landscape of somatic retrotransposition in human cancers. Science. 2012;337:967–71. doi: 10.1126/science.1222077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jern P, Coffin JM. Effects of retroviruses on host genome function. Annual review of genetics. 2008;42:709–32. doi: 10.1146/annurev.genet.42.110807.091501. [DOI] [PubMed] [Google Scholar]
- 16.Kazazian HH, Jr, Wong C, Youssoufian H, Scott AF, Phillips DG, et al. Haemophilia A resulting from de novo insertion of L1 sequences represents a novel mechanism for mutation in man. Nature. 1988;332:164–6. doi: 10.1038/332164a0. [DOI] [PubMed] [Google Scholar]
- 17.Chen JM, Ferec C, Cooper DN. LINE-1 Endonuclease-Dependent Retrotranspositional Events Causing Human Genetic Disease: Mutation Detection Bias and Multiple Mechanisms of Target Gene Disruption. J Biomed Biotechnol. 2006;2006:56182. doi: 10.1155/JBB/2006/56182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Levin HL, Moran JV. Dynamic interactions between transposable elements and their hosts. Nature reviews Genetics. 2011;12:615–27. doi: 10.1038/nrg3030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hancks DC, Kazazian HH., Jr Active human retrotransposons: variation and disease. Current opinion in genetics & development. 2012;22:191–203. doi: 10.1016/j.gde.2012.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gentles AJ, Wakefield MJ, Kohany O, Gu W, Batzer MA, et al. Evolutionary dynamics of transposable elements in the short-tailed opossum Monodelphis domestica. Genome Res. 2007;17:992–1004. doi: 10.1101/gr.6070707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mikkelsen TS, Wakefield MJ, Aken B, Amemiya CT, Chang JL, et al. Genome of the marsupial Monodelphis domestica reveals innovation in non-coding sequences. Nature. 2007;447:167–77. doi: 10.1038/nature05805. [DOI] [PubMed] [Google Scholar]
- 22.Horie K, Saito ES, Keng VW, Ikeda R, Ishihara H, et al. Retrotransposons influence the mouse transcriptome: implication for the divergence of genetic traits. Genetics. 2007;176:815–27. doi: 10.1534/genetics.107.071647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Akagi K, Stephens RM, Li J, Evdokimov E, Kuehn MR, et al. MouseIndelDB: a database integrating genomic indel polymorphisms that distinguish mouse strains. Nucleic Acids Res. 2010;38:D600–6. doi: 10.1093/nar/gkp1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li J, Akagi K, Hu Y, Trivett AL, Hlynialuk CJ, et al. Mouse endogenous retroviruses can trigger premature transcriptional termination at a distance. Genome research. 2012;22:870–84. doi: 10.1101/gr.130740.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Quinlan AR, Clark RA, Sokolova S, Leibowitz ML, Zhang Y, et al. Genome-wide mapping and assembly of structural variant breakpoints in the mouse genome. Genome Res. 2010;20:623–35. doi: 10.1101/gr.102970.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xing J, Zhang Y, Han K, Salem AH, Sen SK, et al. Mobile elements create structural variation: analysis of a complete human genome. Genome Res. 2009;19:1516–26. doi: 10.1101/gr.091827.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ewing AD, Kazazian HH., Jr Whole-genome resequencing allows detection of many rare LINE-1 insertion alleles in humans. Genome Res. 2011;21:985–90. doi: 10.1101/gr.114777.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stewart C, Kural D, Stromberg MP, Walker JA, Konkel MK, et al. A comprehensive map of mobile element insertion polymorphisms in humans. PLoS genetics. 2011;7:e1002236. doi: 10.1371/journal.pgen.1002236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rouchka E, Montoya-Durango DE, Stribinskis V, Ramos K, Kalbfleisch T. Assessment of genetic variation for the LINE-1 retrotransposon from next generation sequence data. BMC bioinformatics. 2010;11(Suppl 9):S12. doi: 10.1186/1471-2105-11-S9-S12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mi S, Lee X, Li X, Veldman GM, Finnerty H, et al. Syncytin is a captive retroviral envelope protein involved in human placental morphogenesis. Nature. 2000;403:785–9. doi: 10.1038/35001608. [DOI] [PubMed] [Google Scholar]
- 31.Sela N, Mersch B, Hotz-Wagenblatt A, Ast G. Characteristics of transposable element exonization within human and mouse. PLoS One. 2010;5:e10907. doi: 10.1371/journal.pone.0010907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sayah DM, Sokolskaja E, Berthoux L, Luban J. Cyclophilin A retrotransposition into TRIM5 explains owl monkey resistance to HIV-1. Nature. 2004;430:569–73. doi: 10.1038/nature02777. [DOI] [PubMed] [Google Scholar]
- 33.Kazazian HH. Retrotransposon insertions in germ cells and somatic cells. Dev Biol (Basel) 2001;106:307–13. discussion 13-4, 17-29. [PubMed] [Google Scholar]
- 34.Prak ET, Dodson AW, Farkash EA, Kazazian HH., Jr Tracking an embryonic L1 retrotransposition event. Proc Natl Acad Sci U S A. 2003;100:1832–7. doi: 10.1073/pnas.0337627100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.van den Hurk JA, Meij IC, Seleme MC, Kano H, Nikopoulos K, et al. L1 retrotransposition can occur early in human embryonic development. Hum Mol Genet. 2007;16:1587–92. doi: 10.1093/hmg/ddm108. [DOI] [PubMed] [Google Scholar]
- 36.Kano H, Godoy I, Courtney C, Vetter MR, Gerton GL, et al. L1 retrotransposition occurs mainly in embryogenesis and creates somatic mosaicism. Genes Dev. 2009;23:1303–12. doi: 10.1101/gad.1803909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Muotri AR, Chu VT, Marchetto MC, Deng W, Moran JV, et al. Somatic mosaicism in neuronal precursor cells mediated by L1 retrotransposition. Nature. 2005;435:903–10. doi: 10.1038/nature03663. [DOI] [PubMed] [Google Scholar]
- 38.Coufal NG, Garcia-Perez JL, Peng GE, Yeo GW, Mu Y, et al. L1 retrotransposition in human neural progenitor cells. Nature. 2009;460:1127–31. doi: 10.1038/nature08248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Baillie JK, Barnett MW, Upton KR, Gerhardt DJ, Richmond TA, et al. Somatic retrotransposition alters the genetic landscape of the human brain. Nature. 2011;479:534–7. doi: 10.1038/nature10531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Evrony GD, Cai X, Lee E, Hills LB, Elhosary PC, et al. Single-neuron sequencing analysis of l1 retrotransposition and somatic mutation in the human brain. Cell. 2012;151:483–96. doi: 10.1016/j.cell.2012.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Miki Y, Nishisho I, Horii A, Miyoshi Y, Utsunomiya J, et al. Disruption of the APC gene by a retrotransposal insertion of L1 sequence in a colon cancer. Cancer Res. 1992;52:643–5. [PubMed] [Google Scholar]
- 42.Iskow RC, McCabe MT, Mills RE, Torene S, Pittard WS, et al. Natural mutagenesis of human genomes by endogenous retrotransposons. Cell. 2010;141:1253–61. doi: 10.1016/j.cell.2010.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Solyom S, Ewing AD, Rahrmann EP, Doucet T, Nelson HH, et al. Extensive somatic L1 retrotransposition in colorectal tumors. Genome research. 2012;22:2328–38. doi: 10.1101/gr.145235.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wissing S, Munoz-Lopez M, Macia A, Yang Z, Montano M, et al. Reprogramming somatic cells into iPS cells activates LINE-1 retroelement mobility. Human molecular genetics. 2012;21:208–18. doi: 10.1093/hmg/ddr455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Quinlan AR, Boland MJ, Leibowitz ML, Shumilina S, Pehrson SM, et al. Genome sequencing of mouse induced pluripotent stem cells reveals retroelement stability and infrequent DNA rearrangement during reprogramming. Cell Stem Cell. 2011;9:366–73. doi: 10.1016/j.stem.2011.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Symer DE, Connelly C, Szak ST, Caputo EM, Cost GJ, et al. Human L1 retrotransposition is associated with genetic instability in vivo. Cell. 2002;110:327–38. doi: 10.1016/s0092-8674(02)00839-5. [DOI] [PubMed] [Google Scholar]
- 47.Yoder JA, Walsh CP, Bestor TH. Cytosine methylation and the ecology of intragenomic parasites. Trends Genet. 1997;13:335–40. doi: 10.1016/s0168-9525(97)01181-5. [DOI] [PubMed] [Google Scholar]
- 48.Howard G, Eiges R, Gaudet F, Jaenisch R, Eden A. Activation and transposition of endogenous retroviral elements in hypomethylation induced tumors in mice. Oncogene. 2008;27:404–8. doi: 10.1038/sj.onc.1210631. [DOI] [PubMed] [Google Scholar]
- 49.Matsui T, Leung D, Miyashita H, Maksakova IA, Miyachi H, et al. Proviral silencing in embryonic stem cells requires the histone methyltransferase ESET. Nature. 2010;464:927–31. doi: 10.1038/nature08858. [DOI] [PubMed] [Google Scholar]
- 50.Yu F, Zingler N, Schumann G, Stratling WH. Methyl-CpG-binding protein 2 represses LINE-1 expression and retrotransposition but not Alu transcription. Nucleic acids research. 2001;29:4493–501. doi: 10.1093/nar/29.21.4493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Muotri AR, Marchetto MC, Coufal NG, Oefner R, Yeo G, et al. L1 retrotransposition in neurons is modulated by MeCP2. Nature. 2010;468:443–6. doi: 10.1038/nature09544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Coufal NG, Garcia-Perez JL, Peng GE, Marchetto MC, Muotri AR, et al. Ataxia telangiectasia mutated (ATM) modulates long interspersed element-1 (L1) retrotransposition in human neural stem cells. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:20382–7. doi: 10.1073/pnas.1100273108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yang N, Kazazian HH., Jr L1 retrotransposition is suppressed by endogenously encoded small interfering RNAs in human cultured cells. Nat Struct Mol Biol. 2006;13:763–71. doi: 10.1038/nsmb1141. [DOI] [PubMed] [Google Scholar]
- 54.Beauregard A, Curcio MJ, Belfort M. The Take and Give Between Retrotransposable Elements and their Hosts. Annu Rev Genet. 2008;42:587–617. doi: 10.1146/annurev.genet.42.110807.091549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Peddigari S, Li PW, Rabe JL, Martin SL. hnRNPL and nucleolin bind LINE-1 RNA and function as host factors to modulate retrotransposition. Nucleic acids research. 2012 doi: 10.1093/nar/gks1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kazazian HH., Jr Mobile elements and disease. Curr Opin Genet Dev. 1998;8:343–50. doi: 10.1016/s0959-437x(98)80092-0. [DOI] [PubMed] [Google Scholar]
- 57.Rebollo R, Romanish MT, Mager DL. Transposable Elements: An Abundant and Natural Source of Regulatory Sequences for Host Genes. Annual review of genetics. 2012 doi: 10.1146/annurev-genet-110711-155621. [DOI] [PubMed] [Google Scholar]
- 58.Chen J, Rattner A, Nathans J. Effects of L1 retrotransposon insertion on transcript processing, localization and accumulation: lessons from the retinal degeneration 7 mouse and implications for the genomic ecology of L1 elements. Hum Mol Genet. 2006;15:2146–56. doi: 10.1093/hmg/ddl138. [DOI] [PubMed] [Google Scholar]
- 59.Rangwala SH, Zhang L, Kazazian HH., Jr Many LINE1 elements contribute to the transcriptome of human somatic cells. Genome Biol. 2009;10:R100. doi: 10.1186/gb-2009-10-9-r100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Faulkner GJ, Kimura Y, Daub CO, Wani S, Plessy C, et al. The regulated retrotransposon transcriptome of mammalian cells. Nat Genet. 2009;41:563–71. doi: 10.1038/ng.368. [DOI] [PubMed] [Google Scholar]
- 61.Djebali S, Davis CA, Merkel A, Dobin A, Lassmann T, et al. Landscape of transcription in human cells. Nature. 2012;489:101–8. doi: 10.1038/nature11233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lamprecht B, Walter K, Kreher S, Kumar R, Hummel M, et al. Derepression of an endogenous long terminal repeat activates the CSF1R proto-oncogene in human lymphoma. Nature medicine. 2010;16:571–9. doi: 10.1038/nm.2129. 1p following 9. [DOI] [PubMed] [Google Scholar]
- 63.Weber B, Kimhi S, Howard G, Eden A, Lyko F. Demethylation of a LINE-1 antisense promoter in the cMet locus impairs Met signalling through induction of illegitimate transcription. Oncogene. 2010;29:5775–84. doi: 10.1038/onc.2010.227. [DOI] [PubMed] [Google Scholar]
- 64.Matlik K, Redik K, Speek M. L1 antisense promoter drives tissue-specific transcription of human genes. J Biomed Biotechnol. 2006;2006:71753. doi: 10.1155/JBB/2006/71753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kim DS, Hahn Y. Human-specific antisense transcripts induced by the insertion of transposable element. Int J Mol Med. 2010;26:151–7. [PubMed] [Google Scholar]
- 66.Duhl DM, Vrieling H, Miller KA, Wolff GL, Barsh GS. Neomorphic agouti mutations in obese yellow mice. Nature genetics. 1994;8:59–65. doi: 10.1038/ng0994-59. [DOI] [PubMed] [Google Scholar]
- 67.Rakyan VK, Chong S, Champ ME, Cuthbert PC, Morgan HD, et al. Transgenerational inheritance of epigenetic states at the murine Axin(Fu) allele occurs after maternal and paternal transmission. Proc Natl Acad Sci U S A. 2003;100:2538–43. doi: 10.1073/pnas.0436776100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Perot P, Mugnier N, Montgiraud C, Gimenez J, Jaillard M, et al. Microarray-based sketches of the HERV transcriptome landscape. PLoS One. 2012;7:e40194. doi: 10.1371/journal.pone.0040194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Feschotte C. Transposable elements and the evolution of regulatory networks. Nature reviews Genetics. 2008;9:397–405. doi: 10.1038/nrg2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lee KJ, Conley AB, Lunyak VV, Jordan IK. Do human transposable element small RNAs serve primarily as genome defenders or genome regulators? Mobile genetic elements. 2012;2:19–25. doi: 10.4161/mge.19031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Aravin AA, Hannon GJ, Brennecke J. The Piwi-piRNA pathway provides an adaptive defense in the transposon arms race. Science. 2007;318:761–4. doi: 10.1126/science.1146484. [DOI] [PubMed] [Google Scholar]
- 72.Cartault F, Munier P, Benko E, Desguerre I, Hanein S, et al. Mutation in a primate-conserved retrotransposon reveals a noncoding RNA as a mediator of infantile encephalopathy. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:4980–5. doi: 10.1073/pnas.1111596109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Thurman RE, Rynes E, Humbert R, Vierstra J, Maurano MT, et al. The accessible chromatin landscape of the human genome. Nature. 2012;489:75–82. doi: 10.1038/nature11232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lowe CB, Bejerano G, Haussler D. Thousands of human mobile element fragments undergo strong purifying selection near developmental genes. Proc Natl Acad Sci U S A. 2007;104:8005–10. doi: 10.1073/pnas.0611223104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lowe CB, Haussler D. 29 mammalian genomes reveal novel exaptations of mobile elements for likely regulatory functions in the human genome. PLoS One. 2012;7:e43128. doi: 10.1371/journal.pone.0043128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kapitonov VV, Jurka J. The long terminal repeat of an endogenous retrovirus induces alternative splicing and encodes an additional carboxy-terminal sequence in the human leptin receptor. J Mol Evol. 1999;48:248–51. doi: 10.1007/pl00013153. [DOI] [PubMed] [Google Scholar]
- 77.Belancio VP, Hedges DJ, Deininger P. LINE-1 RNA splicing and influences on mammalian gene expression. Nucleic Acids Res. 2006;34:1512–21. doi: 10.1093/nar/gkl027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Belancio VP, Roy-Engel AM, Deininger P. The impact of multiple splice sites in human L1 elements. Gene. 2008;411:38–45. doi: 10.1016/j.gene.2007.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Roy-Engel AM, El-Sawy M, Farooq L, Odom GL, Perepelitsa-Belancio V, et al. Human retroelements may introduce intragenic polyadenylation signals. Cytogenet Genome Res. 2005;110:365–71. doi: 10.1159/000084968. [DOI] [PubMed] [Google Scholar]
- 80.Li Z, Mulligan MK, Wang X, Miles MF, Lu L, et al. A transposon in comt generates mRNA variants and causes widespread expression and behavioral differences among mice. PLoS One. 2010;5:e12181. doi: 10.1371/journal.pone.0012181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Druker R, Bruxner TJ, Lehrbach NJ, Whitelaw E. Complex patterns of transcription at the insertion site of a retrotransposon in the mouse. Nucleic Acids Res. 2004;32:5800–8. doi: 10.1093/nar/gkh914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Isbel L, Whitelaw E. Endogenous retroviruses in mammals: An emerging picture of how ERVs modify expression of adjacent genes. Bioessays. 2012 doi: 10.1002/bies.201200056. [DOI] [PubMed] [Google Scholar]
- 83.Lev-Maor G, Ram O, Kim E, Sela N, Goren A, et al. Intronic Alus influence alternative splicing. PLoS genetics. 2008;4:e1000204. doi: 10.1371/journal.pgen.1000204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Gong C, Kim YK, Woeller CF, Tang Y, Maquat LE. SMD and NMD are competitive pathways that contribute to myogenesis: effects on PAX3 and myogenin mRNAs. Genes & development. 2009;23:54–66. doi: 10.1101/gad.1717309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Gong C, Maquat LE. lncRNAs transactivate STAU1-mediated mRNA decay by duplexing with 3′ UTRs via Alu elements. Nature. 2011;470:284–8. doi: 10.1038/nature09701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Smalheiser NR, Torvik VI. Alu elements within human mRNAs are probable microRNA targets. Trends Genet. 2006;22:532–6. doi: 10.1016/j.tig.2006.08.007. [DOI] [PubMed] [Google Scholar]
- 87.Bourque G, Leong B, Vega VB, Chen X, Lee YL, et al. Evolution of the mammalian transcription factor binding repertoire via transposable elements. Genome Res. 2008;18:1752–62. doi: 10.1101/gr.080663.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kunarso G, Chia NY, Jeyakani J, Hwang C, Lu X, et al. Transposable elements have rewired the core regulatory network of human embryonic stem cells. Nat Genet. 2010;42:631–4. doi: 10.1038/ng.600. [DOI] [PubMed] [Google Scholar]
- 89.Schmidt D, Schwalie PC, Wilson MD, Ballester B, Goncalves A, et al. Waves of retrotransposon expansion remodel genome organization and CTCF binding in multiple mammalian lineages. Cell. 2012;148:335–48. doi: 10.1016/j.cell.2011.11.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Dixon JR, Selvaraj S, Yue F, Kim A, Li Y, et al. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature. 2012;485:376–80. doi: 10.1038/nature11082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Cui F, Sirotin MV, Zhurkin VB. Impact of Alu repeats on the evolution of human p53 binding sites. Biology direct. 2011;6:2. doi: 10.1186/1745-6150-6-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Wang T, Zeng J, Lowe CB, Sellers RG, Salama SR, et al. Species-specific endogenous retroviruses shape the transcriptional network of the human tumor suppressor protein p53. Proc Natl Acad Sci U S A. 2007;104:18613–8. doi: 10.1073/pnas.0703637104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Harris CR, Dewan A, Zupnick A, Normart R, Gabriel A, et al. p53 responsive elements in human retrotransposons. Oncogene. 2009;28:3857–65. doi: 10.1038/onc.2009.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Muotri AR, Marchetto MC, Coufal NG, Gage FH. The necessary junk: new functions for transposable elements. Hum Mol Genet. 2007;16(Spec No. 2):R159–67. doi: 10.1093/hmg/ddm196. [DOI] [PubMed] [Google Scholar]
- 95.Ekram MB, Kang K, Kim H, Kim J. Retrotransposons as a major source of epigenetic variations in the mammalian genome. Epigenetics: official journal of the DNA Methylation Society. 2012;7:370–82. doi: 10.4161/epi.19462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Daxinger L, Whitelaw E. Understanding transgenerational epigenetic inheritance via the gametes in mammals. Nat Rev Genet. 2012;13:153–62. doi: 10.1038/nrg3188. [DOI] [PubMed] [Google Scholar]
- 97.Cropley JE, Suter CM, Beckman KB, Martin DI. CpG methylation of a silent controlling element in the murine Avy allele is incomplete and unresponsive to methyl donor supplementation. PLoS One. 2010;5:e9055. doi: 10.1371/journal.pone.0009055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Garcia-Perez JL, Morell M, Scheys JO, Kulpa DA, Morell S, et al. Epigenetic silencing of engineered L1 retrotransposition events in human embryonic carcinoma cells. Nature. 2010;466:769–73. doi: 10.1038/nature09209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Rebollo R, Karimi MM, Bilenky M, Gagnier L, Miceli-Royer K, et al. Retrotransposon-induced heterochromatin spreading in the mouse revealed by insertional polymorphisms. PLoS Genet. 2011;7:e1002301. doi: 10.1371/journal.pgen.1002301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Estecio MR, Gallegos J, Vallot C, Castoro RJ, Chung W, et al. Genome architecture marked by retrotransposons modulates predisposition to DNA methylation in cancer. Genome research. 2010;20:1369–82. doi: 10.1101/gr.107318.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Belancio VP, Hedges DJ, Deininger P. Mammalian non-LTR retrotransposons: for better or worse, in sickness and in health. Genome Res. 2008;18:343–58. doi: 10.1101/gr.5558208. [DOI] [PubMed] [Google Scholar]
- 102.Singer T, McConnell MJ, Marchetto MC, Coufal NG, Gage FH. LINE-1 retrotransposons: mediators of somatic variation in neuronal genomes? Trends in neurosciences. 2010;33:345–54. doi: 10.1016/j.tins.2010.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Upton KR, Baillie JK, Faulkner GJ. Is somatic retrotransposition a parasitic or symbiotic phenomenon? Mobile genetic elements. 2011;1:279–82. doi: 10.4161/mge.18422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Nellaker C, Keane TM, Yalcin B, Wong K, Agam A, et al. The genomic landscape shaped by selection on transposable elements across 18 mouse strains. Genome biology. 2012;13:R45. doi: 10.1186/gb-2012-13-6-r45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Keane TM, Goodstadt L, Danecek P, White MA, Wong K, et al. Mouse genomic variation and its effect on phenotypes and gene regulation. Nature. 2011;477:289–94. doi: 10.1038/nature10413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Brawand D, Soumillon M, Necsulea A, Julien P, Csardi G, et al. The evolution of gene expression levels in mammalian organs. Nature. 2011;478:343–8. doi: 10.1038/nature10532. [DOI] [PubMed] [Google Scholar]
- 107.Han JS, Szak ST, Boeke JD. Transcriptional disruption by the L1 retrotransposon and implications for mammalian transcriptomes. Nature. 2004;429:268–74. doi: 10.1038/nature02536. [DOI] [PubMed] [Google Scholar]
- 108.Teng L, Firpi HA, Tan K. Enhancers in embryonic stem cells are enriched for transposable elements and genetic variations associated with cancers. Nucleic Acids Res. 2011;39:7371–9. doi: 10.1093/nar/gkr476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Borchert GM, Lanier W, Davidson BL. RNA polymerase III transcribes human microRNAs. Nat Struct Mol Biol. 2006;13:1097–101. doi: 10.1038/nsmb1167. [DOI] [PubMed] [Google Scholar]
- 110.Brosnan JA, Iacobuzio-Donahue CA. A new branch on the tree: next-generation sequencing in the study of cancer evolution. Seminars in cell & developmental biology. 2012;23:237–42. doi: 10.1016/j.semcdb.2011.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.