Abstract
The optimal medium for cardiac differentiation of adult primitive cells remains to be established. We quantitatively compared the efficacy of IGF-1, dynorphin B, insulin, oxytocin, bFGF, and TGF-β1 in inducing cardiomyogenic differentiation. Adult mouse skeletal muscle-derived Sca1+/CD45-/c-kit-/Thy-1+ (SM+) and Sca1-/CD45-/c-kit-/Thy-1+ (SM−) cells were cultured in basic medium (BM; DMEM, FBS, IGF-1, dynorphin B) alone and BM supplemented with insulin, oxytocin, bFGF, or TGF-β1. Cardiac differentiation was evaluated by the expression of cardiac-specific markers at the mRNA (qRT-PCR) and protein (immunocytochemistry) levels. BM+TGF-β1 upregulated mRNA expression of Nkx2.5 and GATA-4 after 4 days and Myl2 after 9 days. After 30 days, BM+TGF-β1 induced the greatest extent of cardiac differentiation (by morphology and expression of cardiac markers) in SM− cells. We conclude that TGF-β1 enhances cardiomyogenic differentiation in skeletal muscle-derived adult primitive cells. This strategy may be utilized to induce cardiac differentiation as well as to examine the cardiomyogenic potential of adult tissue-derived stem/progenitor cells.
Keywords: adult stem cell, cardiac differentiation, culture medium, skeletal muscle, growth factor
Introduction
Results from numerous studies in animals indicate that therapy with adult stem/progenitor cells can ameliorate left ventricular (LV) remodeling and improve LV function after myocardial infarction [9, 18, 25]. Based on this phenomenological evidence, several clinical trials of cell therapy for cardiac repair have been completed or are in progress [1]. Although controversial, differentiation of transplanted cells into cells of cardiac lineages has been proposed as one of the mechanisms underlying these beneficial effects. Therefore, it is crucial to determine the cardiomyogenic potential of transplanted cells in vitro prior to transplantation in vivo. In this regard, it is important to formulate culture conditions that can predictably induce cardiomyocytic differentiation in candidate cells for cardiac repair in vivo. However, the impact of different growth factors/cytokines on the cardiomyogenic differentiation of adult stem/progenitors in vitro has not been systematically investigated.
We have previously reported that specific subsets of adult skeletal muscle-derived primitive cells can undergo cardiomyocytic differentiation in vitro [35]. In this study, we quantitatively examined the ability of six specific growth factors/cytokines in five combinations to induce cardiomyocytic differentiation in two subsets (Sca-1+/CD45-/c-kit-/Thy-1+ [SM+] cells and Sca-1-/CD45-/c-kit-/Thy-1+ [SM−] cells) of skeletal muscle-derived adult primitive cells in vitro. These factors (insulin-like growth factor [IGF]-1 [5], dynorphin B [32], insulin [29], oxytocin [22], basic fibroblast growth factor [bFGF] [28], and transforming growth [TGF]-β1 [6, 21] were selected because of their known involvement in cardiac development and/ or their ability to induce cardiac differentiation in various stem cell lines. To our knowledge, this is the first systematic comparison of the ability of different growth factors and cytokines to induce cardiomyocytic differentiation in adult primitive cells in vitro in a controlled setting.
Materials and methods
The investigation conforms to the principles of laboratory animal care according to the Guide for the Care and Use of Laboratory Animals (Department of Health and Human Services, NIH Publication No. 86-23, revised 1996).
Animals
Adult C57BL/6 mice (age 6–10 weeks, body wt 20– 25 g) were used. Mice were bred and maintained at Indiana University according to protocols approved by the laboratory animal research facility (LARC) of the Indiana University School of Medicine. Each experimental group of donor C57BL/6 mice was obtained from the same litter.
Isolation of skeletal muscle-derived primitive cells
Cells were isolated according to previously published methods [35]. Briefly, following euthanasia, the hind limbs were removed and placed in Dulbecco’s Modified Eagle Medium (DMEM, Invitrogen). Adipose tissue and large vasculature were removed carefully with fine forceps and discarded. Skeletal muscle (sartorius, quadriceps, adductors, soleus, and gastrocnemius muscles) was carefully dissected, excised from tendons, and placed in 15 ml of fresh medium. Muscle tissue was triturated by repeated gentle pipetting and centrifuged at 500 rpm for 10 min. The pelleted muscle tissue was digested in collagenase I (220 U/ml, Worthington Biochemical) and Dispase (33 U/ml, BD Biosciences) in 50 ml of DMEM for 45 min at 37°C with gentle agitation [35]. Cells were then filtered through a 40-µm cell strainer into a sterile 50-ml conical tube. Viability was assessed by Trypan blue (Sigma) staining. Approximately 4 × 106 cells were obtained from each donor mouse.
Flow cytometric cell sorting
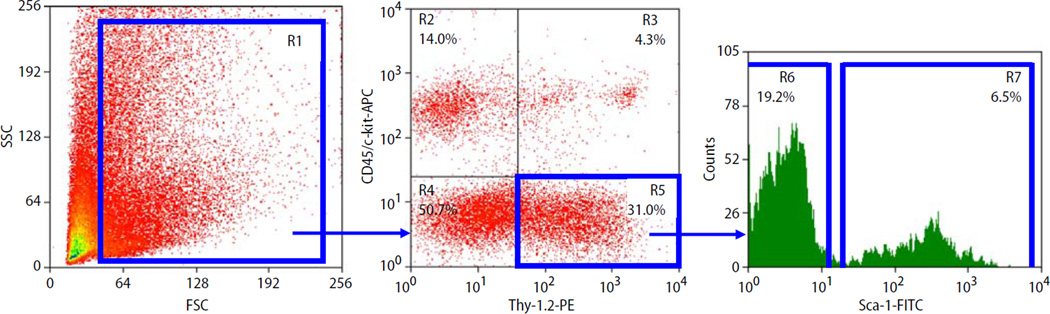
Freshly isolated cells were pelleted in PBS containing 1% FBS (HyClone), and FITC-conjugated monoclonal antimouse Sca-1 (E13–161.7, BD Pharmingen), allophycocyanin (APC)-conjugated monoclonal antimouse CD45 and c-kit (30-F11 and 2B8, respectively, BD Pharmingen), and biotin-conjugated antimouse Thy-1.2 (53-2.1, BD Pharmingen) primary antibodies were added simultaneously. PE-conjugated streptavidin (Caltag Laboratories) was added subsequently. All staining was performed at 4°C for 20 min, and cells were washed with PBS supplemented with 1% FBS after staining. Flow cytometric cell sorting was performed using a MoFlo (Dako) and a FACS Vantage SE [35]. The sorting strategy is summarized in Fig. 1. Sorted cells were collected into tubes containing 2 ml DMEM with 10% FBS. Isolated cells were reanalyzed immediately after sorting to determine purity of sorting. The viability of sorted cells always exceeded 90%.
Fig. 1.
Representative dot-plots show the expression (in percentage) of Sca-1 (FITC), CD45 (APC), c-kit (APC), and Thy-1.2 (PE) in skeletal muscle-derived cells. Blue lines indicate regions included into the sorting logic. From region 1 (R1), CD45-/c-kit- cells positive for Thy-1.2 are identified in region 5 (R5). Based on Sca-1 expression, cells from R5 are divided into Sca-1 +/CD45-/c-kit-/Thy-1 + (SM+) cells in region 7 (R7) and Sca-1-/CD45-/c-kit-/Thy-1+ (SM−) cells in region 6 (R6). The percentages in R6 and R7 relate to the total number of cells in R1. FSC forward scatter characteristics, SSC side scatter characteristics
Cell culture
Freshly isolated SM+ and SM− cells were seeded at 5,000 cells/plate in gelatin-coated 22-mm diameter plates. The basic medium (BM) consisted of DMEM supplemented with 10% FBS, IGF-1 (25 nM [2]; R&D Research), and dynorphin B (1 µM [32]; NeoMPS). IGF-1 and dynorphin B, both of which have been shown to induce cardiac differentiation [2, 32], were added to the BM to synergistically augment the overall cardiomyogenic effects of these media. To determine the impact of specific supplements on cardiomyogenic differentiation, cells were cultured in BM alone and BM supplemented with insulin (10 µg/ ml [3]; Sigma), oxytocin (0.1 µM [22]; Becham California), bFGF (20 ng/ml [23, 30]; R&D Research), or TGF-β1 (2.5 ng/ml [6]; R&D Research). These concentrations were based on our careful review of the literature. We specifically avoided using a higher dose of TGF-β1 to obviate the induction of osteoblastic features [14]. Half of the medium was changed twice a week and cells were allowed to differentiate for 30 days.
Phenotypic analysis
Cells in each of 10 groups were evaluated on a daily basis under light microscopy and images were acquired using a digital camera (MRC5, Carl Zeiss, Inc., Thornwood, NY) and the Axiovision software (Carl Zeiss) to document changes in cellular morphology.
Assessment of mRNA expression by real-time qRT-PCR
For the analysis of mRNA expression of markers of cardiac differentiation (Nkx2.5, GATA-4, and Myl2), total mRNA was isolated from freshly isolated cells SM+ and SM− cells and after 4 and 9 days of culture (RNeasy, Qiagen), and reverse-transcribed (TaqMan, Applied Biosystems). Quantitative assessment of mRNA expression of all of the genes of interest and β2-microglobulin was performed by real-time qRT-PCR using an ABI PRISM® 7000 System [35].
All of the primer sequences are provided in supplemental Table 1. Primers were designed with the Primer Express software. A 25-µl reaction mixture containing 12.5 µl of SYBR Green PCR Master Mix and 10 ng of forward and reverse primers was used. The threshold cycle (Ct), i.e., the cycle number at which the amount of amplified gene of interest reached a fixed threshold, was subsequently determined. Relative quantitation of mRNA expression was calculated with the comparative Ct method. The relative quantitative value of target, normalized to an endogenous control β2-microglobulin gene and relative to a calibrator, was expressed as 2−ΔΔCt (−fold difference), where ΔCt = (Ct of target genes [Nkx2.5, GATA-4, and Myl2]) – (Ct of endogenous control gene [β2-microglobulin]), and ΔΔCt = (ΔCt of samples for target gene) – (ΔCt of calibrator for the target gene). To avoid the possibility of amplifying contaminating DNA (1) all of the primers for real-time RT-PCR were designed to contain an intron sequence for specific cDNA amplification; (2) reactions were performed with appropriate negative controls (template-free controls); (3) a uniform amplification of the products was rechecked by analyzing the melting curves of the amplified products (dissociation graphs); and (4) the melting temperature (Tm) was 57–60°C with the probe Tm at least 10°C higher than the primer Tm [35]. Three independent experiments were performed for each set of genes.
Immunocytochemistry
The expression of cardiac-specific transcription factors (cTFs) and structural proteins (cSPs) was examined by immunocytochemical staining after 30 days of differentiation [35]. Briefly, cells were fixed (4% paraformaldehyde), permeabilized (0.2% Triton X-100), blocked (10% donkey serum) and incubated with primary antibodies against Nkx2.5 (Santa Cruz), GATA-4 (Santa Cruz), cardiac myosin heavy chain (cMyHC, Novus Biologicals), α-sarcomeric actin (Sigma), and a panel of primary antibodies against specific markers of noncardiac lineages (as described below) at a concentration of 1:100–1:200 for 16 h at 4°C. Following washing with PBS, appropriate fluorochrome-conjugated secondary antibodies (Jackson Immunoresearch) were added at a concentration of 1:100. Following staining with DAPI, cells were washed with PBS and a coverslip was applied for microscopic examination. Appropriate negative and specific positive controls were employed for each antigen to ensure the specificity of staining.
Quantitative assessment of cardiomyocytic differentiation
All images were acquired using a LSM 510 (Zeiss) confocal microscope. For quantitative assessment of the expression of cardiac-specific antigens, 500 cells/plate on average were systematically evaluated in multiple fields from all four quarters of the plate [35]. As previously reported [12, 33, 35], cardiac-specific transcription factors (Nkx2.5 [12, 35] and GATA-4 [33, 35]) were noted both in cytoplasm and in the nucleus. Cells with nuclear staining with or without cytoplasmic staining were counted as positives. For the purpose of exclusion, we also stained cells for markers specific for several noncardiac lineages (skeletal muscle [myogenin and Myf5]; neuron [nestin and GFAP]; fibroblast [PDGFR-α and -β]; chondrocyte [collagens I and II]; epithelial cell [cytokeratin 17]; macrophage [CD68]; and others) [35]. This rigorous approach enabled us to specifically identify cells with a cardiomyocytic phenotype. Cells that were positive for cardiac-specific markers and negative for noncardiac lineage markers were counted as positive, and the total number of cells in each field was calculated based on nuclear staining with DAPI. The percentage of cells differentiating into cardiomyocytes was calculated as the number of positive cells divided by the total number of nuclei in the field [35]. These experiments were performed in triplicate.
Statistical analysis
Data are mean ± SEM. The percentage of cells with a cardiac phenotype and quantitative mRNA data (-fold changes in mRNA levels) for cardiac-specific markers was analyzed with one-way or two-way (time and group) ANOVA. If the ANOVA showed an overall difference, post hoc contrasts were performed with Student’s t tests for unpaired data, and the resulting probability values were adjusted according to the Bonferroni correction (SPSS). A P < 0.05 was considered statistically significant.
Results
Isolated skeletal muscle-derived primitive cells
Skeletal muscle-derived primitive cells were isolated by FACS. Based on the expression of Sca-1, CD45, c-kit, and Thy-1, cells were sorted into Sca-1+/CD45-/ c-kit-/Thy-1+ (SM+) and Sca-1-/CD45-/c-kit-/Thy-1 + (SM−) populations according to the protocol summarized in Fig. 1.
Impact of media composition on cell morphology during culture
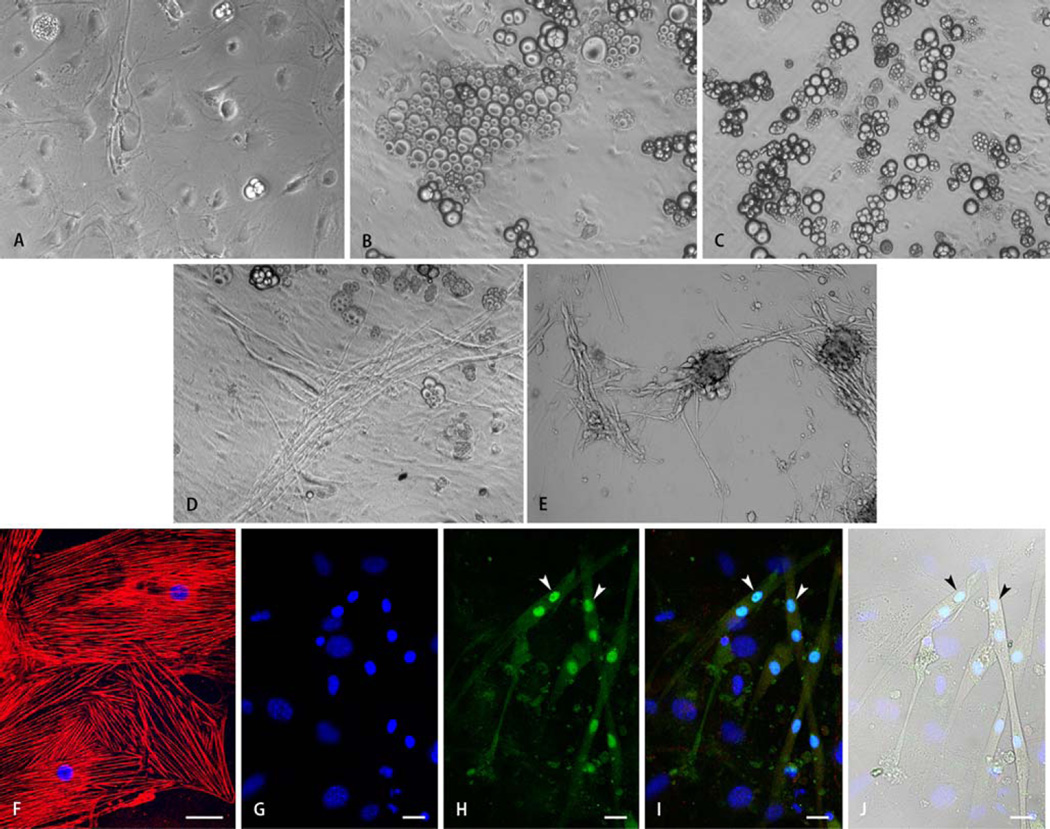
Within the first 48 h after plating, both SM+ and SM− cells adhered to the gelatin-coated surface. During culture, SM+ cells cultured in all five different media exhibited greater morphological heterogeneity compared with SM− cells (Fig. 2a – e). During the 2nd–4th weeks, SM+ cells cultured in BM alone gradually differentiated into a combination of large flat cells with epithelial morphology and positive immunostaining for cytokeratin 17 (Fig. 2f), isolated spindle-shaped cells, oval cells with endothelial morphology, and scattered cellular aggregates with an adipocytic phenotype (Fig. 2a). Addition of insulin (Fig. 2b) or oxytocin (Fig. 2c) to the BM resulted in markedly greater differentiation into adipocytes. Although BM+bFGF (Fig. 2d) and BM+TGF-β1 (Fig. 2e) induced differentiation into varied phenotypes, the frequency of spindle-shaped cells was less compared with SM− cells in respective media. In addition, SM+ cells cultured in BM+bFGF (Fig. 2d) and BM+TGF−β1 (Fig. 2e) exhibited greater predisposition to differentiate into myotubes (Fig. 2g – j). Overall, SM+ cells exhibited greater propensity to differentiate into an adipose phenotype in all media.
Fig. 2.
Representative light microscopic images of SM+ cells after 30 days of differentiation demonstrating the effects of culture media composition on cellular morphology. Panels a–c show the heterogeneous (epithelial, endothelial, adipose, and others) morphology of SM+ cells cultured in basic medium (BM) containing IGF-1 and dynorphin B (a), and BM supplemented with insulin (b), or oxytocin (c). Exposure of SM+ cells to insulin or oxytocin increased differentiation into an adipose phenotype. Supplementation of BM with bFGF (d) or TGF-β1 (e) resulted in enhanced myotube formation from SM+ cells. Morphological observations were confirmed via immunostaining (f–j). Panel f shows differentiation of SM+ cells cultured in BM supplemented with insulin into epithelial cells positive for the epithelial cytoskeletal marker cytokeratin 17 (red). Panels g–j show representative images of SM+ cells stained for the cardiac-specific marker cardiac myosin heavy chain (i,j red), and skeletal muscle-specific transcription factor myogenin (h–j, green). Nuclei are stained with DAPI (f,g,i,j, blue). Myogenin expression was localized to nuclei (H,l, exemplified by arrowheads). Nuclei expressing myogenin (h–j, exemplified by arrowheads) belonged to cells negative for cardiac marker in red fluorescence (h,j) and exhibited morphology characteristic of myotubes (j). Scale bar = 40 µm (f) or 20 µm (g–j)
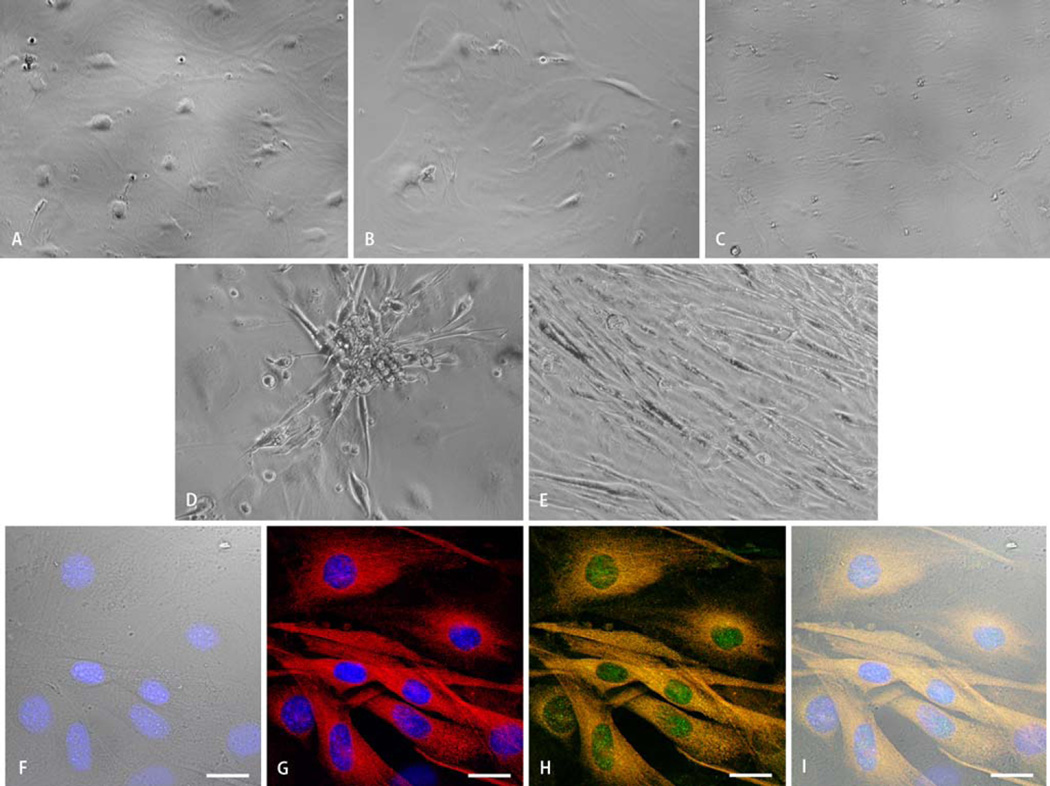
Compared with SM+ cells, SM− cells cultured in all five media combinations exhibited greater morphological homogeneity (Fig. 3a – e). SM− cells cultured in BM alone differentiated into cells with epithelial and endothelial phenotypes with isolated spindle-shaped cells (Fig. 3a). SM− cells in BM+insulin (Fig. 3b) and BM+oxytocin (Fig. 3c) differentiated into similar combinatorial phenotypes with predominant epithelial cells. In contrast, a greater fraction of SM− cells cultured in BM+bFGF (Fig. 3d) and BM+TGF–β1 (Fig. 3e) showed predominantly spindle-shaped morphology. However, a fraction of these SM− cells cultured in BM+bFGF gradually acquired the phenotype characteristic of myotubes, while the mononucleate spindle-shaped SM− cells in BM+TGF–β1 predominantly remained isolated with formation of occasional small clusters. Spontaneous rhythmic contractions were observed in both single cells and clusters of small cells in BM+TGF-β1 medium.
Fig. 3.
Representative light microscopic images of SM− cells after 30 days of differentiation demonstrating the effects of culture media composition on cellular morphology. Panels a–c show the predominant epithelial morphology of SM-cells cultured in basic medium (BM) containing IGF-1 and dynorphin B (a), and BM supplemented with insulin (b), or oxytocin (c). Scattered cells with adipose morphology were noted throughout the culture plates. Panels d and e show the increased frequency of spindle-shaped cells when SM− cells were exposed to BM supplemented with bFGF (d) or TGF-β1 (e). Cardiomyogenic differentiation was verified via immunostaining. Panels f–i show representative confocal microscopic images of SM− cells after 30 days of differentiation in BM supplemented with TGF-β1. Mononucleate spindle-shaped cells are identified in the transmission image (f). Cardiac differentiation is evidenced by positivity for cardiac-specific transcription factor (Nkx2.5h–i, green) and structural protein (cardiac myosin heavy chain, g–i, red). Panels h–i identify cells positive for both cardiac-specific transcription factor (in green) and structural protein (in red) resulting in yellow fluorescence. Nuclei are stained with DAPI (f,g,i, blue). Scale bar = 20 µm
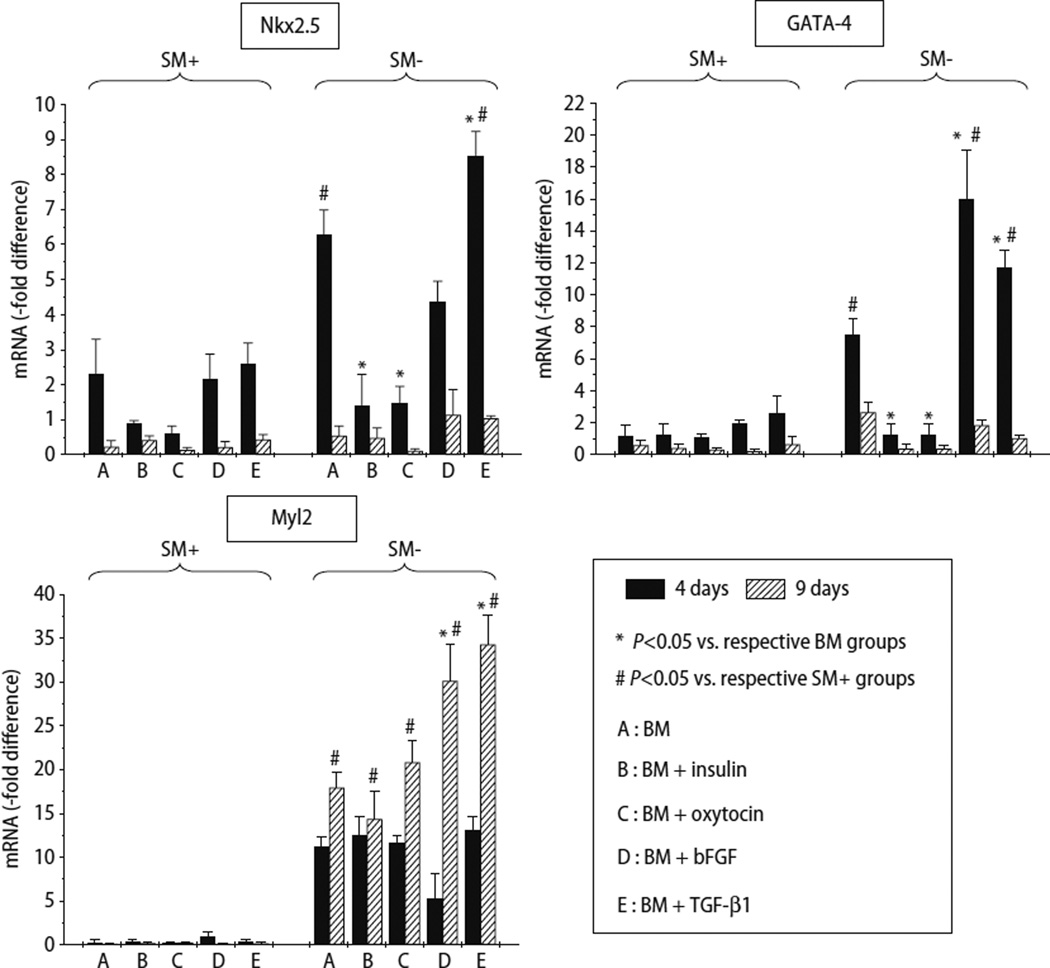
Analysis of mRNA expression of cardiac markers
The expression of Nkx2.5 was greater in all media at 4 days of culture as compared with 9 days, and greater in SM− cells compared with SM+ cells (Fig. 4). In SM− cells, the expression of Nkx2.5 was greatest in cells cultured in BM+TGF–β1, and to a lesser extent in BM alone and BM+bFGF (Fig. 4). The expression of GATA-4 followed a similar pattern except that the greatest expression was in SM− cells cultured in BM+bFGF, followed by BM+TGF-β1, and BM alone (Fig. 4). Compared with SM+ cells, Myl2 expression was uniformly greater in SM− cells, particularly in presence of bFGF and TGF-β1. Importantly, Myl2 expression was greater after 9 days compared with levels after 4 days of culture. These observations are consistent with the paradigm that the expression of transcription factors would logically precede the expression of structural proteins during lineage commitment. Although SM+ cells expressed low levels of Myl2 (a structural protein) at 4 and 9 days, the expression of Nkx2.5 and GATA-4 (transcription factors) was clearly detectable in SM+ cells at these time-points (Fig. 4). Cell type-specific differences in the time-course of antigen expression may be responsible for the differential expression levels of Myl2 in SM− and SM+ cells at these earlier time-points.
Fig. 4.
Quantitative assessment of mRNA expression by qRT-PCR of cardiac-specific markers in SM+ and SM− cells after 4 and 9 days of culture in 5 different media. mRNA level is expressed as -fold change compared with freshly isolated unfractionated skeletal muscle-derived cells. Three independent experiments were performed. Data are mean ± SEM
Impact of media composition on cardiomyogenic differentiation
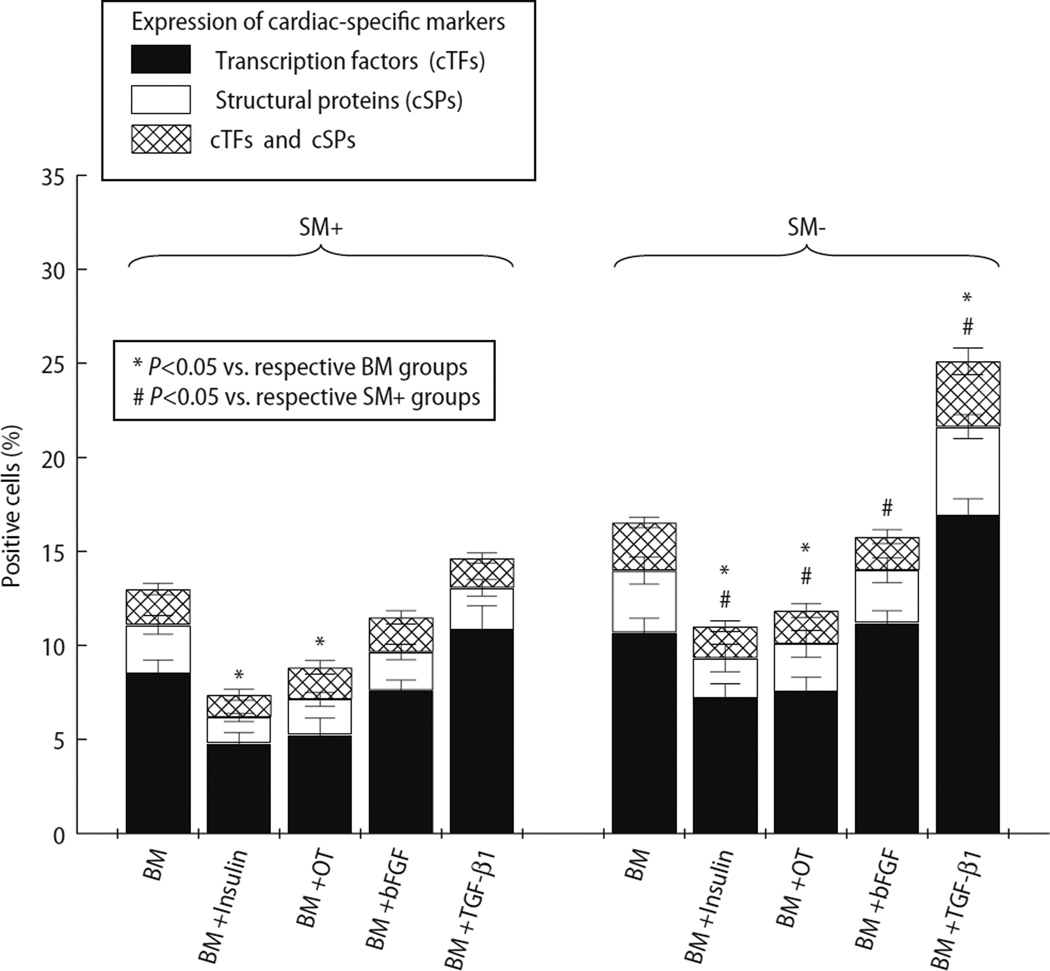
Freshly isolated SM+ and SM− cells were allowed to differentiate in five different media combinations. After 30 days, cells were immunostained for cardiac-specific transcription factors (Nkx2.5 and GATA-4) and cardiac-specific structural proteins (cMyHC and α-sarcomeric actin). Fig. 3f – i demonstrates cardiomyocytic differentiation of SM− cells cultured in medium containing BM+TGF-β1. After 30 days, BM alone, BM+insulin, BM+oxytocin, BM+bFGF, and BM+TGF-β1 induced cardiac differentiation in 11.1 ± 1.0%, 6.2 ± 0.4%, 7.1 ± 1.0%, 9.7 ± 0.8%, and 13.1 ± 1.5% of SM+ cells, respectively (Fig. 5). In cultured SM− cells, BM alone, BM+insulin, BM+oxytocin, BM+bFGF, and BM+TGF-β1 induced cardiac differentiation in 14.0 ± 1.3%, 9.3 ± 1.0%, 10.1 ± 0.2%, 14.0 ± 1.1% and 21.6 ± 1.6% cells, respectively (Fig. 5). Addition of insulin or oxytocin to BM significantly attenuated cardiac differentiation. BM+TGF-β1 was most effective in inducing cardiomyogenic differentiation of SM− cells. Compared with insulin, oxytocin, and bFGF, addition of TGF-β1 also resulted in greater cardiac differentiation of SM+ cells. These findings are consistent with data from mRNA analysis, which showed that addition of TGF-β1 enhanced the expression of both cardiac-specific transcription factors and structural proteins. In addition, we observed spontaneous rhythmic contractions in SM− cell-derived cardiomyocytic cells (supplemental Video 1) confirming their cardiomyocytic nature. Overall, compared with SM+ cells, SM− cells exhibited greater predisposition to undergo cardiomyocytic differentiation (Fig. 5). The myotube differentiation rate was not determined because cells were cultured in cardiomyogenic media, and because these myotubes frequently fused to form syntitia, thereby precluding an accurate cell count.
Fig. 5.
Percentage of cells positive for cardiac-specific markers (Y axis), demonstrating the effects of different media (X axis) on cardiomyogenic differentiation of SM+ and SM− cells. Cells positive for cardiac transcription factors (cTFs) and/or structural proteins (cSPs) were counted and expressed as a percentage of total cells. The percentage of cells positive for cTFs is represented by solid bars, cSPs by white bars, and cells positive for both by cross-hatched bars. Cardiomyocytic differentiation was noted in both SM+ and SM− populations. However, compared with SM+ cells, SM− cells exhibited greater cardiomyogenic potential in all media. Supplementation of BM with TGF-β1 resulted in cardiomyocytic differentiation in the greatest fraction of cells in both groups
Discussion
In order to improve the outcomes of cell therapy for cardiac repair, it is imperative to critically evaluate the ability of the candidate cell to differentiate into cardiac lineages in vitro. However, little consensus exists regarding the most effective medium composition for inducing cardiac differentiation in vitro. Our results demonstrate that: (1) the addition of TGF-β1 to medium containing IGF-1 and dynorphin B significantly enhances cardiomyogenic differentiation of adult skeletal muscle-derived primitive cells; (2) the addition of insulin or oxytocin reduces cardiac differentiation and promotes adipocytic differentiation; and (3) SM− cells exhibit greater predisposition to differentiate into a cardiomyocytic phenotype, indicating that this specific cell type is inherently more amenable to cardiac differentiation. These results have considerable implications for the selection of media for in vitro experiments aimed at identifying the most suitable cell population for cardiac repair in vivo.
Members of the TGF-β superfamily play important roles in cardiac development during embryogenesis [27, 31] as well as in various cardiac pathologies [24, 34]. TGF-β1 has been shown to induce cardiac differentiation in vitro in embryonic explants and stem cells [6, 21] as well as in adult bone marrow-derived cells [14]. Our results show that compared with other combinations, the BM+TGF-β1 combination is most effective in inducing cardiomyocytic differentiation of SM− cells in vitro, an effect that was verified by morphological examination as well as expression of cardiac-specific transcription factors and structural proteins at the mRNA and protein levels. TGF-β1 enhanced the expression of Nkx2.5 and GATA-4 at 4 days of culture, which was followed by increased expression of structural proteins (Myl2 at day 9, and cMyHC and α-sarcomeric actin at later stages). Following TGF-β1 receptor binding, the receptor-regulated Smads (R-Smads: Smad2 and Smad3) are phosphorylated and form a complex with the common-mediator Smad (Co-Smad: Smad4) that migrates to the nucleus and binds to specific DNA sequences leading to transcriptional activation [31]. Importantly, the regulatory region of the Nkx2.5 gene, an early [17] and essential [19] transcription factor for cardiomyogenesis, contains several Smad [16] as well as GATA [7] binding sites. Notwithstanding the complex and poorly understood interaction between Nkx2.5, GATA-4, and other factors during cardiac development, it is plausible that TGF-β1-induced cardiac differentiation is triggered via Smad activation of Nkx2.5 and GATA-4, which is followed by the expression of cardiac structural proteins.
In a recent study by Li et al. [15], TGF-β1 induced differentiation of bone marrow-derived CD117+ cells into immature cardiomyocytes in vitro. Importantly, in a previous study [14], transplantation of TGF-β1-primed CD117+ cells into infarcted hearts resulted in myocyte regeneration and improvement in LV function. In our study, while a rigorous assessment of the cardiomyocyte phenotype via a combinatorial approach (morphology, transcription factor, structural protein, spontaneous contraction) enabled an accurate assessment of the comparative efficacy of different growth factors in cardiac differentiation induction, whether these cardiomyocytic cells achieved the status of mature cardiomyocytes in vitro remained unclear. Therefore, to address this issue of maturity from a translational standpoint, future studies will be necessary to investigate whether TGF-β1-pre-differentiated SM− cells can improve LV structure and function in vivo following transplantation into the infarcted heart.
bFGF (FGF-2) also induced expression of Nkx2.5 and GATA-4; however, immunocytochemical quantitation revealed significantly less cardiac differentiation. Although bFGF is expressed in the developing heart [28], its role on cardiac specification vs. proliferation is unclear [21, 29]. In the study by Hidai et al. [10], FGF-2 suppressed the expression of retinoic acid-induced GATA-4 expression, and promoted skeletal muscle differentiation in embryonal P19 cells. Interestingly, inhibition of FGFR-1, which serves as a receptor for both FGF-1 and FGF-2, was able to block the expression of GATA-4 without affecting α-MyHC expression. In the study by Barron et al. [4], transient exposure to FGF-2 with continuous BMP-2 presence was able to induce a contractile phenotype in non-precardiac mesoderm. However, longer exposure to FGF-2 was necessary to achieve Nkx2.5 induction. Our results are consistent with this dichotomy between the induction of cardiac transcription factors vs. structural proteins by bFGF, and its role in skeletal myogenesis. Further studies will be necessary to determine the signaling machinery activated by bFGF in adult primitive cells.
IGF-1 receptor is expressed in the developing heart [5] and IGF-1 has been shown to induce cardiac differentiation of transplanted embryonic stem cells [13]. Similarly, the P19 embryonal cells express prodynorphin, and dynorphin B can induce cardiac differentiation in these cells [32]. In our study, although BM (containing IGF-1 and dynorphin B) alone upregulated the expression of Nkx2.5 and GATA-4 in SM− cells, cardiomyocytic differentiation was considerably less than that achieved with the addition of TGF-β1. These differences from previous reports could be accounted for by the differences in substrates, embryonic cells vs. adult primitive cells.
The addition of insulin to BM negatively impacted the expression of cardiac markers. In addition, insulin induced marked adipocytic differentiation. In previous studies, insulin has been shown to induce contractility in embryonic explants [29] and promote cardiac differentiation in embryonic neural precursors [3]. Similar observations have been made in adult bone marrow-derived mesenchymal stem cells [26]. It is plausible that adding insulin to a medium containing IGF-1 and dynorphin B negatively impacted cardiac differentiation. Similarly, the addition of oxytocin also reduced cardiac differentiation compared with BM alone and promoted an adipocytic phenotype. Oxytocin is expressed in the developing heart [11], and the evidence supporting a role of oxytocin in cardiac differentiation comes primarily from studies that used the embryonal P19 cells [8, 22]. Interestingly, amongst adult cells, the cardiomyogenic properties of oxytocin has been documented in Sca-1+ cardiac progenitors [20]. However, in our study, the effects of oxytocin in Sca-1+ (SM+) and Sca-1-(SM−) cells were similar. Thus, the role of oxytocin in cardiac differentiation of non-cardiac adult primitive cells remains to be established.
The current results also expand our previous observations regarding the impact of Sca-1 expression on cardiac differentiation of CD45-/c-kit-/Thy-1 + adult skeletal muscle-derived cells [35]. Compared with SM− cells, SM+ cells consistently exhibited lower levels of cardiac-specific gene and protein expression in response to the various growth factors used, indicating that the negative influence of Sca-1+ expression on cardiac differentiation of skeletal muscle-derived CD45-/c-kit-/Thy-1+ primitive cells [35] is independent of the culture medium. The greater cardiomyocytic differentiation of SM− cells vs. SM+ cells with the same growth factors implies cell type-specific differences. Therefore, these results also underscore the importance of carefully selecting the candidate cell for cardiac repair.
Our study has several limitations. First, we did not demonstrate calcium transients in differentiated cells. Instead, we documented spontaneous rhythmic contractions in cardiomyocytic cells in culture (supplemental Video 1). Second, the cardiomyogenic effects of insulin and oxytocin might have been influenced by the presence of IGF-1 and dynorphin B in the BM. However, both IGF-1 and dynorphin B are known to induce cardiomyogenesis [2, 32], and their presence was therefore more likely to synergistically augment the cardiomyogenic effects in all arms. Since the composition of BM was constant in all groups, our comparison remains valid. Finally, we did not fully assess whether these SM− cell-derived cardiomyocytes in vitro attained features consistent with mature adult cardiomyocytes, a question that may perhaps be addressed better in a more conducive milieu in vivo following transplantation into the infarcted heart.
In conclusion, our results indicate that TGF-β1 enhances cardiomyogenic differentiation of skeletal muscle-derived adult primitive cells. This influence was achieved via the upregulation of Nkx2.5 and GATA-4 expression. These results may have considerable implications for the formulation of optimal medium for the induction of cardiac differentiation in vitro, and selecting the candidate cell population for therapeutic cardiac repair in vivo.
Supplementary Material
Acknowledgments
We gratefully acknowledge Barbara Turgeon for expert secretarial assistance. This study was supported in part by NIH grants R01 HL-72410, HL-55757, HL-68088, HL-70897, HL-76794, HL-78825, and R21 HL-89737.
Footnotes
Electronic supplementary material: The online version of this article (doi:10.1007/S00395-008-0729-9) contains supplementary material, which is available to authorized users.
Contributor Information
Ahmed Abdel-Latif, Division of Cardiology, University of Louisville, 550 S. Jackson St., ACB, 3rd floor, Louisville (KY) 40292, USA.
Ewa K. Zuba-Surma, Division of Cardiology, University of Louisville, 550 S. Jackson St., ACB, 3rd floor, Louisville (KY) 40292, USA.
Jamie Case, Dept. of Medicine, Herman B Wells Center for Pediatric Research, Indiana University School of Medicine, Indianapolis (IN), USA.
Sumit Tiwari, Division of Cardiology, University of Louisville, 550 S. Jackson St., ACB, 3rd floor, Louisville (KY) 40292, USA.
Greg Hunt, Division of Cardiology, University of Louisville, 550 S. Jackson St., ACB, 3rd floor, Louisville (KY) 40292, USA.
Smita Ranjan, Division of Cardiology, University of Louisville, 550 S. Jackson St., ACB, 3rd floor, Louisville (KY) 40292, USA.
Robert J. Vincent, Division of Cardiology, University of Louisville, 550 S. Jackson St., ACB, 3rd floor, Louisville (KY) 40292, USA.
Edward F. Srour, Dept. of Microbiology/Immunology, Herman B Wells Center for Pediatric Research, Indiana University School of Medicine, Indianapolis (IN), USA.
Roberto Bolli, Division of Cardiology, University of Louisville, 550 S. Jackson St., ACB, 3rd floor, Louisville (KY) 40292, USA.
Buddhadeb Dawn, Division of Cardiology, University of Louisville, 550 S. Jackson St., ACB, 3rd floor, Louisville (KY) 40292, USA.
References
- 1.Abdel-Latif A, Bolli R, Tleyjeh IM, Montori VM, Perin EC, Hornung CA, Zuba-Surma EK, Al-Mallah M, Dawn B. Adult bone marrow-derived cells for cardiac repair: a systematic review and meta-analysis. Arch Intern Med. 2007;167:989–997. doi: 10.1001/archinte.167.10.989. [DOI] [PubMed] [Google Scholar]
- 2.Antin PB, Yatskievych T, Dominguez JL, Chieffi P. Regulation of avian precardiac mesoderm development by insulin and insulin-like growth factors. J Cell Physiol. 1996;168:42–50. doi: 10.1002/(SICI)1097-4652(199607)168:1<42::AID-JCP6>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- 3.Bani-Yaghoub M, Kendall SE, Moore DP, Bellum S, Cowling RA, Nikopoulos GN, Kubu CJ, Vary C, Verdi JM. Insulin acts as a myogenic differentiation signal for neural stem cells with multilineage differentiation potential. Development. 2004;131:4287–4298. doi: 10.1242/dev.01295. [DOI] [PubMed] [Google Scholar]
- 4.Barron M, Gao M, Lough J. Requirement for BMP and FGF signaling during cardiogenic induction in non-precardiac mesoderm is specific, transient, and cooperative. Dev Dyn. 2000;218:383–393. doi: 10.1002/(SICI)1097-0177(200006)218:2<383::AID-DVDY11>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- 5.Bassas L, Lesniak MA, Serrano J, Roth J, de Pablo F. Developmental regulation of insulin and type I insulin-like growth factor receptors and absence of type II receptors in chicken embryo tissues. Diabetes. 1988;37:637–644. doi: 10.2337/diab.37.5.637. [DOI] [PubMed] [Google Scholar]
- 6.Behfar A, Zingman LV, Hodgson DM, Rauzier JM, Kane GC, Terzic A, Puceat M. Stem cell differentiation requires a paracrine pathway in the heart. Faseb J. 2002;16:1558–1566. doi: 10.1096/fj.02-0072com. [DOI] [PubMed] [Google Scholar]
- 7.Brown CO3rd, Chi X, Garcia-Gras E, Shirai M, Feng XH, Schwartz RJ. The cardiac determination factor, Nkx2-5, is activated by mutual cofactors GATA-4 and Smad1/4 via a novel upstream enhancer. J Biol Chem. 2004;279:10659–10669. doi: 10.1074/jbc.M301648200. [DOI] [PubMed] [Google Scholar]
- 8.Danalache BA, Paquin J, Donghao W, Grygorczyk R, Moore JC, Mummery CL, Gutkowska J, Jankowski M. Nitric oxide signaling in oxytocin-mediated cardiomyogenesis. Stem Cells. 2007;25:679–688. doi: 10.1634/stemcells.2005-0610. [DOI] [PubMed] [Google Scholar]
- 9.Dawn B, Bolli R. Adult bone marrow-derived cells: regenerative potential, plasticity, and tissue commitment. Basic Res Cardiol. 2005;100:494–503. doi: 10.1007/s00395-005-0552-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hidai C, Masako O, Ikeda H, Nagashima H, Matsuoka R, Quertermous T, Kasanuki H, Kokubun S, Kawana M. FGF-1 enhanced cardiogenesis in differentiating embryonal carcinoma cell cultures, which was opposite to the effect of FGF-2. J Mol Cell Cardiol. 2003;35:421–425. doi: 10.1016/s0022-2828(03)00019-1. [DOI] [PubMed] [Google Scholar]
- 11.Jankowski M, Danalache B, Wang D, Bhat P, Hajjar F, Marcinkiewicz M, Paquin J, McCann SM, Gutkowska J. Oxytocin in cardiac ontogeny. Proc Natl Acad Sci USA. 2004;101:13074–13079. doi: 10.1073/pnas.0405324101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kodama H, Hirotani T, Suzuki Y, Ogawa S, Yamazaki K. Cardiomyogenic differentiation in cardiac myxoma expressing lineage-specific transcription factors. Am J Pathol. 2002;161:381–389. doi: 10.1016/S0002-9440(10)64193-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kofidis T, de Bruin JL, Yamane T, Balsam LB, Lebl DR, Swijnenburg RJ, Tanaka M, Weissman IL, Robbins RC. Insulin-like growth factor promotes engraftment, differentiation, and functional improvement after transfer of embryonic stem cells for myocardial restoration. Stem Cells. 2004;22:1239–1245. doi: 10.1634/stemcells.2004-0127. [DOI] [PubMed] [Google Scholar]
- 14.Li TS, Hayashi M, Ito H, Furutani A, Murata T, Matsuzaki M, Hamano K. Regeneration of infarcted myocardium by intramyocardial implantation of ex vivo transforming growth factor-beta-preprogrammed bone marrow stem cells. Circulation. 2005;111:2438–2445. doi: 10.1161/01.CIR.0000167553.49133.81. [DOI] [PubMed] [Google Scholar]
- 15.Li TS, Komota T, Ohshima M, Qin SL, Kubo M, Ueda K, Hamano K. TGF-β induces the differentiation of bone marrow stem cells into immature cardiomyocytes. Biochem Biophys Res Commun. 2008;366:1074–1080. doi: 10.1016/j.bbrc.2007.12.095. [DOI] [PubMed] [Google Scholar]
- 16.Liberatore CM, Searcy-Schrick RD, Vincent EB, Yutzey KE. Nkx-2.5 gene induction in mice is mediated by a Smad consensus regulatory region. Dev Biol. 2002;244:243–256. doi: 10.1006/dbio.2002.0604. [DOI] [PubMed] [Google Scholar]
- 17.Lints TJ, Parsons LM, Hartley L, Lyons I, Harvey RP. Nkx-2.5: a novel murine homeobox gene expressed in early heart progenitor cells and their myogenic descendants. Development. 1993;119:419–431. doi: 10.1242/dev.119.2.419. [DOI] [PubMed] [Google Scholar]
- 18.Lyngbaek S, Schneider M, Hansen JL, Sheikh SP. Cardiac regeneration by resident stem and progenitor cells in the adult heart. Basic Res Cardiol. 2007;102:101–114. doi: 10.1007/s00395-007-0638-3. [DOI] [PubMed] [Google Scholar]
- 19.Lyons I, Parsons LM, Hartley L, Li R, Andrews JE, Robb L, Harvey RP. Myogenic and morphogenetic defects in the heart tubes of murine embryos lacking the homeo box gene Nkx2-5 . Genes Dev. 1995;9:1654–1666. doi: 10.1101/gad.9.13.1654. [DOI] [PubMed] [Google Scholar]
- 20.Matsuura K, Nagai T, Nishigaki N, Oyama T, Nishi J, Wada H, Sano M, Toko H, Akazawa H, Sato T, Nakaya H, Kasanuki H, Komuro I. Adult cardiac Sea-1-positive cells differentiate into beating cardiomyocytes. J Biol Chem. 2004;279:11384–11391. doi: 10.1074/jbc.M310822200. [DOI] [PubMed] [Google Scholar]
- 21.Muslin AJ, Williams LT. Well-defined growth factors promote cardiac development in axolotl mesodermal explants. Development. 1991;112:1095–1101. doi: 10.1242/dev.112.4.1095. [DOI] [PubMed] [Google Scholar]
- 22.Paquin J, Danalache BA, Jankowski M, McCann SM, Gutkowska J. Oxytocin induces differentiation of P19 embryonic stem cells to cardiomyocytes. Proc Natl Acad Sci USA. 2002;99:9550–9555. doi: 10.1073/pnas.152302499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rosenblatt-Velin N, Lepore MG, Cartoni C, Beermann F, Pedrazzini T. FGF-2 controls the differentiation of resident cardiac precursors into functional cardiomyocytes. J Clin Invest. 2005;115:1724–1733. doi: 10.1172/JCI23418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sakata Y, Chancey AL, Divakaran VG, Sekiguchi K, Sivasubramanian N, Mann DL. Transforming growth factorbeta receptor antagonism attenuates myocardial fibrosis in mice with cardiac-restricted overexpression of tumor necrosis factor. Basic Res Cardiol. 2008;103:60–68. doi: 10.1007/s00395-007-0689-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Segers VF, Lee RT. Stem-cell therapy for cardiac disease. Nature. 2008;451:937–942. doi: 10.1038/nature06800. [DOI] [PubMed] [Google Scholar]
- 26.Shim WS, Jiang S, Wong P, Tan J, Chua YL, Tan YS, Sin YK, Lim CH, Chua T, Teh M, Liu TC, Sim E. Ex vivo differentiation of human adult bone marrow stem cells into cardiomyocyte-like cells. Biochem Biophys Res Commun. 2004;324:481–488. doi: 10.1016/j.bbrc.2004.09.087. [DOI] [PubMed] [Google Scholar]
- 27.Solloway MJ, Harvey RP. Molecular pathways in myocardial development: a stem cell perspective. Cardiovasc Res. 2003;58:264–277. doi: 10.1016/s0008-6363(03)00286-4. [DOI] [PubMed] [Google Scholar]
- 28.Spirito P, Fu YM, Yu ZX, Epstein SE, Casscells W. Immunohistochemical localization of basic and acidic fibroblast growth factors in the developing rat heart. Circulation. 1991;84:322–332. doi: 10.1161/01.cir.84.1.322. [DOI] [PubMed] [Google Scholar]
- 29.Sugi Y, Lough J. Activin-A and FGF-2 mimic the inductive effects of anterior endoderm on terminal cardiac myogenesis in vitro. Dev Biol. 1995;168:567–574. doi: 10.1006/dbio.1995.1102. [DOI] [PubMed] [Google Scholar]
- 30.Ulloa-Montoya F, Verfaillie CM, Hu WS. Culture systems for pluripotent stem cells. J Biosci Bioeng. 2005;100:12–27. doi: 10.1263/jbb.100.12. [DOI] [PubMed] [Google Scholar]
- 31.Valdimarsdottir G, Mummery C. Functions of the TGF-β superfamily in human embryonic stem cells. Apmis. 2005;113:773–789. doi: 10.1111/j.1600-0463.2005.apm_3181.x. [DOI] [PubMed] [Google Scholar]
- 32.Ventura C, Maioli M. Opioid peptide gene expression primes cardiogenesis in embryonal pluripotent stem cells. Circ Res. 2000;87:189–194. doi: 10.1161/01.res.87.3.189. [DOI] [PubMed] [Google Scholar]
- 33.Winitsky SO, Gopal TV, Hassanzadeh S, Takahashi H, Gryder D, Rogawski MA, Takeda K, Yu ZX, Xu YH, Epstein ND. Adult murine skeletal muscle contains cells that can differentiate into beating cardiomyocytes in vitro. PLoS Biol. 2005;3:e87. doi: 10.1371/journal.pbio.0030087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wollert KC. Growth-differentiation factor-15 in cardiovascular disease: from bench to bedside, and back. Basic Res Cardiol. 2007;102:412–415. doi: 10.1007/s00395-007-0662-3. [DOI] [PubMed] [Google Scholar]
- 35.Zuba-Surma EK, Abdel-Latif A, Case J, Tiwari S, Hunt G, Kucia M, Vincent RJ, Ranjan S, Ratajczak MZ, Srour EF, Bolli R, Dawn B. Sca-1 expression is associated with decreased cardiomyogenic differentiation potential of skeletal muscle-derived adult primitive cells. J Mol Cell Cardiol. 2006;41:650–660. doi: 10.1016/j.yjmcc.2006.07.011. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.