Abstract
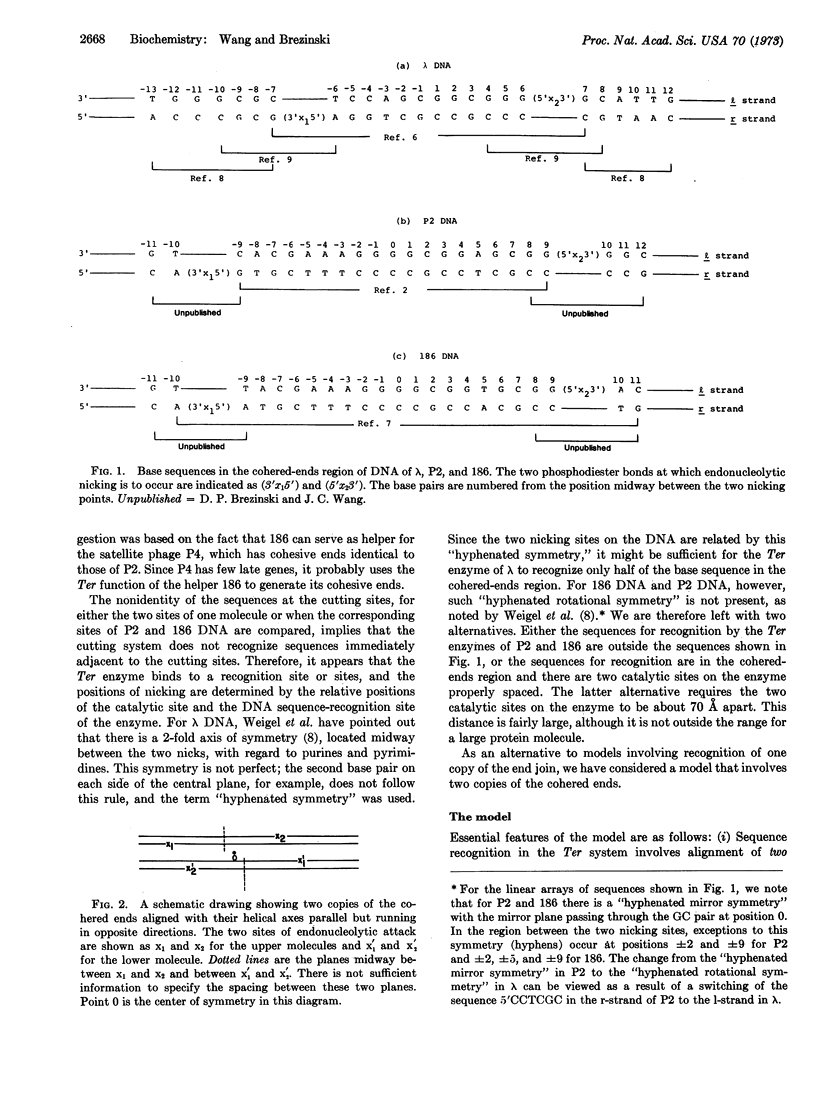
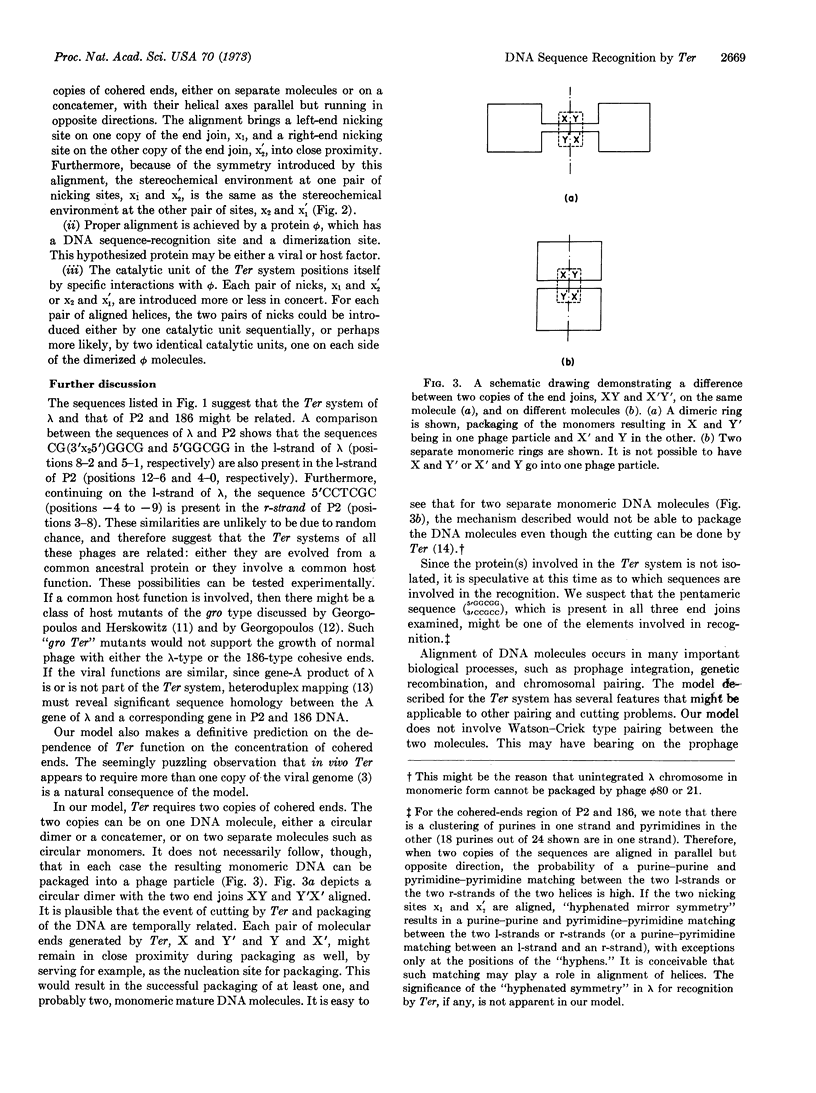
Based on the 3′- and 5′-terminal sequences of DNA of phage λ, P2, and 186, a model is proposed for recognition of DNA sequences by enzymes responsible for generation of cohesive ends. Two copies of the cohered ends, either on separate molecules or on a concatemer, are aligned with their helical axes parallel but running in opposite directions. The nicking system is dimeric, with each of the two monomers carrying identical sequence-recognition sites. Two pairs of nicks are introduced into the two aligned DNA molecules by the nicking system. The applicability of this model to other biological processes, such as integration of a viral genome into a host genome and the cutting of concatemeric T7 DNA, is discussed.
Keywords: cohesive ends, sequence recognition, DNA alignment
Full text
PDF

Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Brezinski D. P., Wang J. C. The 3'-terminal nucleotide sequences of lambda DNA. Biochem Biophys Res Commun. 1973 Jan 23;50(2):398–404. doi: 10.1016/0006-291x(73)90854-1. [DOI] [PubMed] [Google Scholar]
- Davis R. W., Parkinson J. S. Deletion mutants of bacteriophage lambda. 3. Physical structure of att-phi. J Mol Biol. 1971 Mar 14;56(2):403–423. doi: 10.1016/0022-2836(71)90473-6. [DOI] [PubMed] [Google Scholar]
- Kaiser A. D., Wu R. Structure and function of DNA cohesive ends. Cold Spring Harb Symp Quant Biol. 1968;33:729–734. doi: 10.1101/sqb.1968.033.01.083. [DOI] [PubMed] [Google Scholar]
- Mousset S., Thomas R. Ter, a function which generates the ends of the mature lambda chromosome. Nature. 1969 Jan 18;221(5177):242–244. doi: 10.1038/221242a0. [DOI] [PubMed] [Google Scholar]
- Padmanabhan R., Wu R. Nucleotide sequence analysis of DNA. IV. Complete nucleotide sequence of the left-hand cohesive end of coliphage 186 DNA. J Mol Biol. 1972 Apr 14;65(3):447–467. doi: 10.1016/0022-2836(72)90201-x. [DOI] [PubMed] [Google Scholar]
- Szpirer J., Brachet P. Relations physiologiques entre les phages tempérés lambda et phi80. Mol Gen Genet. 1970;108(1):78–92. doi: 10.1007/BF00343187. [DOI] [PubMed] [Google Scholar]
- Wang J. C., Kaiser A. D. Evidence that the cohesive ends of mature lambda DNA are generated by the gene A product. Nat New Biol. 1973 Jan 3;241(105):16–17. doi: 10.1038/newbio241016a0. [DOI] [PubMed] [Google Scholar]
- Weigel P. H., Englund P. T., Murray K., Old R. W. The 3'-terminal nucleotide sequences of bacteriophage lambda DNA. Proc Natl Acad Sci U S A. 1973 Apr;70(4):1151–1155. doi: 10.1073/pnas.70.4.1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu R., Taylor E. Nucleotide sequence analysis of DNA. II. Complete nucleotide sequence of the cohesive ends of bacteriophage lambda DNA. J Mol Biol. 1971 May 14;57(3):491–511. doi: 10.1016/0022-2836(71)90105-7. [DOI] [PubMed] [Google Scholar]



