Abstract
Demographic events and natural selection alter patterns of genetic variation within populations, and may play a substantial role in shaping the genetic architecture of complex phenotypes and disease. However, the joint impact of these basic evolutionary forces is often ignored in the assessment of statistical tests of association. Here, we provide a simulation-based framework for generating DNA sequences that incorporates selection and demography with flexible models for simulating phenotypic variation (sfs_coder). This tool also allows the user to perform locus-specific simulations by automatically querying annotated genomic functional elements and genetic maps. We demonstrate the effects of evolutionary forces on patterns of genetic variation by simulating recently inferred models of human selection and demography. We use these simulations to show that the demographic model and locus-specific features, such as the proportion of sites under selection, may have practical implications for estimating the statistical power of sequencing-based rare variant association tests. In particular, for some phenotype models, there may be higher power to detect rare variant associations in African populations compared to non-Africans, but power is considerably reduced in regions of the genome with rampant negative selection. Furthermore, we show that existing methods for simulating large samples based on resampling from a small set of observed haplotypes fail to recapitulate the distribution of rare variants in the presence of rapid population growth (as has been observed in several human populations).
Keywords: Rare variants, complex phenotypes, natural selection, demography, forward simulations, association tests
2 Introduction
Genome-wide association studies have identified many common loci that contribute to complex heritable phenotypes, but a large proportion of the heritability remains unexplained [Manolio et al., 2009; Witte, 2010]. Proposed sources of this missing heritability include rare variants, environmental interactions, structural variants, common variants of weak effect, and upward biases in the original estimates of heritability. Sequencing studies with large numbers of samples may offer new opportunities to find the unexplained heritability of complex phenotypes, especially rare causal variants as these sites were mostly absent from and very poorly tagged by the original array-based genotyping platforms.
Unfortunately, power to detect rare causal variants using single-marker statistical tests at the genome-wide scale is generally much lower than is desirable. As a result, researchers have proposed statistical methods to pool rare variants within a putatively causal locus and jointly test for the contribution of these variants to the phenotype [Hoffmann et al., 2010; Neale et al., 2011; Wu et al., 2011; Lee et al., 2012a]. While these methods all presuppose that low frequency causal sites have larger effects than high frequency causal sites, there are few mechanistic reasons for such a phenomenon other than the action of purifying selection restraining the frequencies of large effect mutations. Moreover, it has been argued that only those phenotypes with causal sites that are under selection will have a large fraction of heritability explained by rare variants [Simons et al., 2014]. Unfortunately, most rare variant association tools have not directly modeled selection on causal alleles or used simulations of selection to test their performance (but see King et al. [2010] and Price et al. [2010]). Since both natural selection and demography have strong effects on the frequency spectrum of variant sites, these evolutionary forces may have considerable impact on the performance of rare variant association tests [Zuk et al., 2014].
Demography and selection both impact genetic variation within populations, and population geneticists have developed a rich literature that models the effects of these forces on sampled DNA sequences (for reviews, see Emerson et al. [2001]; Nielsen [2005]). In general, changes in population size alter the probability of common ancestry between two sampled sequences per generation as the genealogical history is traced backwards in time. Meanwhile, selection acts to reduce the overall amount of genetic variation by shrinking the time to common ancestry, and in some cases also changes the shape of the genealogy. The height and shape of the genealogical tree relating sequenced chromosomes affect the total amount of variation in the samples and the frequencies of the variant sites.
However, only as the scale of sequencing experiments has increased in recent years has it become possible to apply models of selection and demography to real data sets. In particular, it is now possible to use statistical inference techniques to infer the parameters of demographic/selection models (e.g., population split times, growth rates, and the strength and rate of selection). Perhaps the most influential such model is known as the Poisson Random Field [Sawyer and Hartl, 1992], which has been used to infer both demographic events [Gutenkunst et al., 2009; Gravel et al., 2011; Tennessen et al., 2012] and selection [Bustamante et al., 2001; Williamson et al., 2005; Boyko et al., 2008; Torgerson et al., 2009].
As a result of these studies and others, we have rich information about the recent history of human continental groups and natural selection acting on human genomic elements such as conserved noncoding sequences and exons. In general, studies of human demography have found that human genetic variation is consistent with a population bottleneck as humans moved out of Africa and into Europe and Asia, and that in the recent past human populations have expanded rapidly. Studies of selection have found that most amino acid changes in proteins are weakly deleterious, and a substantial proportion of changes are strongly deleterious [Boyko et al., 2008]. Moreover, conserved noncoding elements have a qualitatively similar distribution of selective constraints with a lower mean strength of selection [Torgerson et al., 2009], but there exist ultra-conserved noncoding regions in the human genome with even stronger selective constraints than coding regions [Katzman et al., 2007].
There has been tremendous recent interest in both population genetic inference and association testing, and several studies connecting the fields have now been published (e.g., see Pritchard [2001]; Eyre-Walker [2010]; Maher et al. [2012]; Thornton et al. [2013]; Simons et al. [2014]; Lohmueller [2014]). Population genetics has direct implications for association studies, since the relationship between allele frequencies and effect sizes determines the power to detect causal sites. In particular, recent population growth and selection [Keinan and Clark, 2012; Nelson et al., 2012; Tennessen et al., 2012] have both increased the proportion of sites at low frequency and impacted the total number of segregating sites in a sample. Accounting for the impact of selection and demography on the frequency spectrum may be crucial to making sensible estimates of statistical power for association tests that pool putatively causal rare variants.
Perhaps the most widely used rare variant association test is the sequence kernel association test, or SKAT [Wu et al., 2011]. SKAT provides a regression-based framework for rare variant association testing, and has several advantages over tests that count the prevalence of rare variants in cases and controls (collectively known as burden tests, e.g. Cohen et al. [2004]; Morgenthaler and Thilly [2007]; Li and Leal [2008]). SKAT retains statistical power when rare variants have effects with opposing directions and provides the machinery for covariate adjustment. Furthermore, many burden tests can be treated as special cases of SKAT [Wu et al., 2011]. For these reasons, we focus on SKAT in this article (specifically SKAT-O, an optimized version of SKAT [Lee et al., 2012a,b]).
Here, we introduce a simulation tool that incorporates recently inferred population genetic models of natural selection and demography, and accounts for the inferred functional elements and local recombination rate of any desired locus in the human genome. We demonstrate how local genomic features can impact patterns of variation within sampled DNA sequences, and show that accounting for these patterns may have practical implications for rare variant association test power calculations under some phenotype models. We also consider the impact of linked selection on patterns of genetic variation and discuss the simulation of phenotypes under models with selection.
3 Materials and Methods
3.1 sfs_coder: A Python-based interface to SFS_CODE
We built a Python-based front-end to the forward simulator SFS_CODE, which we have named sfs_coder. sfs_coder is designed to allow users to simulate human DNA sequences using inferred demographic histories and human selection models. A typical workflow in sfs_coder consists of 1) importing the appropriate modules, 2) performing SFS_CODE simulations of the desired population genetic model and/or locus in the human genome, and 3) analyzing the output of the command or simulating phenotype data using the simulated genetic data. The post-processing analysis tools include locus-by-locus computation of π, Tajima's D, ZnS, Watterson's θ, Fay and Wu's H, and the site frequency spectrum. Each step can be accomplished with a few lines of code if the user simulates one of the models that we have included, but any acceptable SFS_CODE command can be called from sfs_coder. Furthermore, advanced Python users can use sfs_coder's object-oriented framework to write their own analysis tools beyond what we have provided.
In the next sections, we detail the models that are simulated in this paper, all of which are immediately accessible through our free software, except as noted. The software is available at sfscode.sourceforge.net and complete documentation is available at uricchio.github.io/sfs_coder.
3.2 Simulations of human demography and selection
We simulated human demography and selection for three human continental groups (African, Asian, and European) using the parameter estimates of previous studies. All of the demographic models we considered include discrete population size changes, population splits, and continuous migration [Schaffner et al., 2005]. Two of the models incorporate the above features with recent exponential growth in the European and Asian continental groups [Gutenkunst et al., 2009; Gravel et al., 2011], and the final model incorporates all of the above features with recent acceleration of the growth rate in the African and European continental groups [Tennessen et al., 2012]. The model of Schaffner et al. [2005] was simulated with the coalescent simulator cosi (http://www.broadinstitute.org/~sfs/cosi/) and is not included in sfs_coder, while the other models were simulated using the forward simulator SFSJCODE [Hernandez, 2008] and sfs_coder. The parameters of the population split times, migration rates, bottlenecks, and growth rates were obtained from the maximum likelihood estimates of the relevant publications [Gutenkunst et al., 2009; Gravel et al., 2011; Tennessen et al., 2012]. Model parameters were scaled as described in the SFS_CODE manual, available at sfscode.sourceforge.net. Sample SFS_CODE command lines for each of the different models are provided in the Appendix.
In all simulations of selection on coding regions, we used the distribution of selection coefficients on non-synonymous sites that was inferred by Boyko et al. [2008]. For conserved noncoding elements, we applied the distribution inferred by Torgerson et al. [2009]. Both are Γ-distributed, and parameters for the distributions are given in the Appendix.
To summarize the results of our simulations, we plot the cumulative site frequency spectrum (cSFS). The value of the cSFS at frequency x is defined as the proportion of variant sites below or equal to frequency x in our simulations. We also report the nucleotide diversity, π, for some of our simulations. π is defined as the mean number of pairwise differences per base pair between a random pair of chromosomes within the sample. The values of π that we report are the mean over a set of independent simulations.
3.3 Simulations of genomic elements
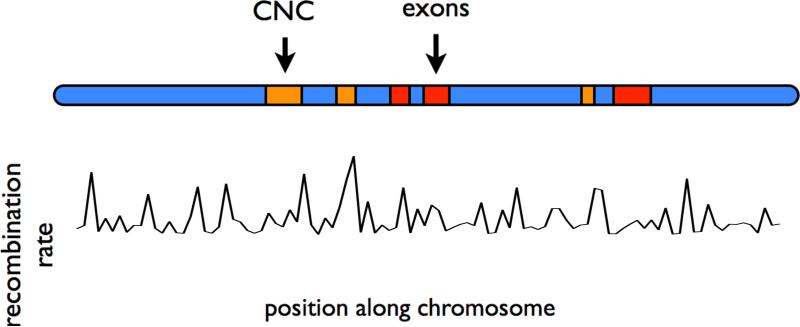
sfs_coder allows users to input the coordinates of a human genomic region and models the local genomic structure of this region. We model human genomic elements within sfs_coder as shown in Figure 1. The positions of exons were extracted from GENCODE v14 [Harrow et al., 2012]. The positions of conserved noncoding elements were inferred by Siepel et al. [2005], and recombination rates by International HapMap Consortium et al. [2007]. Data sources for each of these elements are included in sfs_coder.
Figure 1.
A model of human genomic sequences that incorporates selection on exons and conserved noncoding elements (with separate distributions of selection coefficients) and local recombination rates. Selection coefficients on exons and conserved noncoding elements were taken from Boyko et al. [2008] and Torgerson et al. [2009], respectively. The positions of the conserved noncoding elements were inferred by Siepel et al. [2005], and the recombination map was inferred by International HapMap Consortium et al. [2007]. (CNC: conserved noncoding).
Furthermore, we allow users to specify one of several recently inferred models of human demography, namely those of Gutenkunst et al. [2009], Gravel et al. [2011], and Tennessen et al. [2012], simultaneous to simulating complex genomic structure. Both the exonic regions and conserved noncoding regions are under selection in the simulations, with selection coefficients drawn from distributions that were inferred specifically for these regions by recent studies [Boyko et al., 2008; Torgerson et al., 2009].
3.4 Haplotype resampling with Hapgen2
Haplotype resampling methods provide an efficient mechanism for simulating large samples based on an existing reference panel, and constitute an alternative to forward simulations of DNA sequences. Such methods include Hapgen2 [Su et al., 2011] and others, and are often applied based on the deep catalog of variation represented by the HapMap project [International HapMap Consortium et al., 2007] or the 1000 Genomes Project [1000 Genomes Project Consortium et al., 2012]. Haplotype resampling has the demonstrated ability to recapitulate the haplotype and genetic variation of large samples when the population size remains constant, but it is not clear that they will perform well in cases of rapid population growth. Recent studies have suggested that the excess of rare variants associated with recent growth may only be detectable with very large sample sizes [Keinan and Clark, 2012; Tennessen et al., 2012].
We simulated thirty unlinked 10Mb regions of the human genome under the European demographic model of Tennessen et al. [2012] and sampled 104 chromosomes using SFS_CODE. Each 10Mb region was based on chr15:59200000-69200000, and incorporated natural selection on all exons and conserved noncoding regions, as well as the genetic map inferred by the HapMap project [International HapMap Consortium et al., 2007]. We randomly chose 100 chromosomes from this simulation to form a reference panel. We then used Hapgen2 to resample this reference panel up to a larger sample size of 103, 5 × 103, or 104 chromosomes and compared the distributions of derived allele frequencies (DAF) at each sample size to a random subsample of equivalent size from SFS_CODE simulations.
3.5 Choosing a region under strong linked selection
We performed simulations of a genomic region under strong linked selection using sfs_coder. To select a candidate region for these simulations, we computed the density of phastCons elements [Siepel et al., 2005] and total genetic distance separately in 1Mb sliding windows (10 kb sliding distance) across the human genome (hg19). We then took the intersect of those windows that were in the top 10% of the distribution of phastCons (phastConsElements46wayPlacental) density and the bottom 10% of the distribution of recombination distance (HapMapII GRCh37). From that intersection, we extracted windows that had a mean B value less than 25 (indicating very strong background selection [McVicker et al., 2009]). We found 931 transcripts (GENCODE version 14) that fell within these regions. We then picked chr3:50320000-50350000 for our simulations of selection and power since it had among the highest densities of transcripts within this set of regions. Our background selection simulations incorporate the 2 Mb surrounding this region (chr3:49335000-51335000). This region also contains 13 Genome-Wide Association Study loci in the NHGRI GWAS Catalog.
3.6 Simulations of phenotypes and the power of SKAT-O
We followed Wu et al. [2011] in simulating phenotypes and testing the power of the rare variant association method SKAT-O. Effect sizes β(x) of causal variants were taken as β(x) = –0.4 log10(x), where x is the minor allele frequency. Thus, lower frequency sites have larger effect sizes. Among variants under 3% frequency, 5% were taken to be causal. Phenotypes, Y, of each sampled individual were then generated as
| (1) |
where X1 is a standard normal covariate, X2 is a dichotomous covariate that takes the value 0 with probability 0.5 and the value 1 otherwise, and ε is a standard normal random variable (not taken as a covariate). β is the vector of effect sizes and G represents the genotypes. This phenotype model is included in sfs_coder, as are the models of Eyre-Walker [2010] and Simons et al. [2014], which map selection coefficients, rather than allele frequencies, to effect sizes. Our software also allows the user to set the desired genetic variance explained by the causal sequence for each of the models.
For each of the demographic models considered in our power calculations, we generated 250 independent simulations with the relevant sample size and selective constraint for the human genomic locus at chromosome 3, hg19 positions 50320000-50350000. For each simulation, we randomly selected causal loci and generated phenotypes as described above. We resampled causal sites from each genetic simulation 4 times, for a total of 103 simulations of phenotypes for each demographic model. We then ran SKAT-O and computed the fraction of results with p-values under 10–6.
Depending on the exact sequencing experiment performed, the number of statistical tests might range from approximately 2 × 104 (all genes) to 105 (all 30 kb sequences in the genome), so Bonferroni corrected significance thresholds may range from 5 × 10–7 to 2.5 × 10–6. Here we have chosen 10–6, but we emphasize that the trends in power as a function of sample size, selection, and demography are not dependent on this choice.
We obtained the SKAT R-package from http://www.hsph.harvard.edu/skat/download/.
4 Results
4.1 Demography, sample size, and selection impact rare variants
The results of rare variant association tests are contingent on the joint distribution of variant frequencies and effect sizes. In this section, we examine the effects of selection and demography on the simulated frequency spectrum through simulations.
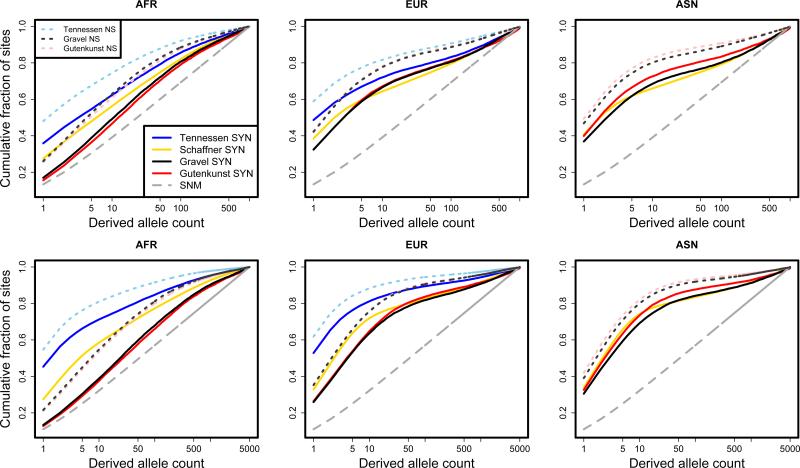
We simulated human demography and selection under several previously inferred models [Schaffner et al., 2005; Gutenkunst et al., 2009; Gravel et al., 2011; Tennessen et al., 2012]. In Figure 2, we plot the simulated cumulative site frequency spectra of each of the three demographic models that we considered. Consistent with population genetic theory [Keinan and Clark, 2012] (and the data that was used to infer the models), exponential or two-phase exponential growth results in a large excess of rare variants relative to a constant population size model (gray dashed line). Furthermore, when sample size is large (5 × 103 chromosomes, lower panels), the two-phase model of exponential growth [Tennessen et al., 2012] generates many more low frequency variants than the other growth models.
Figure 2.
Simulated cumulative site frequency spectra in three human continental groups under several recently inferred demographic models. Sample size is 103 chromosomes in the top panels and 5 × 103 chromosomes in the lower panels. Note that the model of Tennessen et al. [2012] did not infer demography of the Asian continental group, so we do not plot a curve for this population. However, the Asian continental group is included in the Tennessen et al. [2012] simulations. Each curve was calculated using 103 independent simulations of 5 × 103 base pairs each. The gray dashed curve represents the analytical expectation based on the standard neutral model and is not the result of simulations. (SNM: standard neutral model, AFR: African population, EUR: European population, ASN: Asian population, NS: non-synonymous, SYN: synonymous).
Our simulations also included natural selection on non-synonymous sites [Boyko et al., 2008]. Negative selection tends to constrain variants to lower frequencies, so the site frequency spectra of non-synonymous sites are shifted towards the left (Figure 2, dotted lines). Note that we do not include non-synonymous sites for the model of Schaffner et al. [2005] in Figure 2 because the coalescent simulator cosi does not allow for the introduction of natural selection.
4.2 Haplotype resampling under-estimates the number of rare variants in large samples
As an alternative to forward simulations, investigators might opt to resample haplotypes from a previously sequenced sample. In this section, we test whether a haplotype resampling method (also known as a “sideways” simulation, [Chen et al., 2014]) is able to recapitulate the extent of rare variation expected in large samples when based on a modest reference panel of 100 chromosomes in the context of rapid population growth.
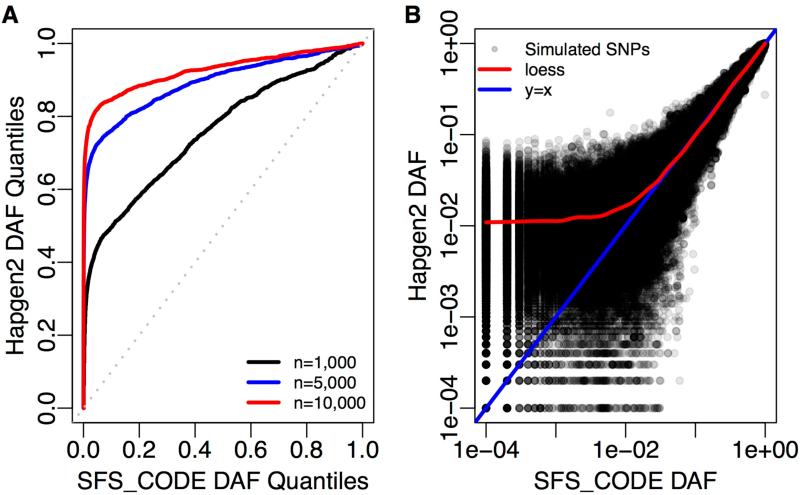
We find that under the demographic model of Tennessen et al. [2012], Hapgen2 [Su et al., 2011] does a poor job of recapitulating the extent of rare variation expected at large sample sizes. In Figure 3A, we show a quantile-quantile (QQ) plot of the DAF distribution inferred from Hapgen2 versus the DAF distribution expected by SFS_CODE under the Tennessen et al. [2012] European demographic model. If Hapgen2 were able to recapitulate the underlying DAF distribution Figure 3A would follow the diagonal dotted line. However, we find that as the sample size increases, the extent to which Hapgen2 underestimates the fraction of rare variants increases (indicated by curves deviating above the diagonal). In Figure 3B we look closely at the expected (based on SFS_CODE) and inferred (based on Hapgen2) frequencies of each SNP observed in the sample of 104 chromosomes using a scatter plot. We do not expect points to fall along the diagonal in this case because of the resampling procedure, but we would expect the points to be symmetrically distributed about the diagonal (blue curve). Instead, we find that Hapgen2 DAF frequencies are skewed toward higher frequencies for rare variants. This is demonstrated using a loess smoothing (red curve). The loess curve shows that the Hapgen2 DAF may be strongly biased by the reference panel size.
Figure 3.
Generating large samples by haplotype resampling results in a deficiency of rare variants in the presence of population growth. For this plot we simulated 104 chromosomes from thirty unlinked 10Mb regions of the human genome (300Mb total) in SFS_CODE under the Tennessen et al. [2012] demographic model discussed in the text. We then used 100 chromosomes as a reference panel in Hapgen2 to generate a much larger sample (as indicated in the legend). (A) shows a quantile-quantile plot of the derived allele frequencies (DAF) for chromosomes simulated using Hapgen2 versus the expected DAF distribution from SFS_CODE (with sample sizes indicated in the legend). (B) shows a scatter plot of the frequencies of each SNP inferred by Hapgen2 compared to SFS_CODE for a sample of size 104 chromosomes. Both figures demonstrate that Hapgen2 fails to recapitulate the extent of rare variation expected under rapid population growth, particularly for large sample sizes.
4.3 Power estimates may be impacted by local genomic context and demography
Forward simulations allow investigators to model the effects of demography and selection on sampled DNA sequences [Peng et al., 2014]. Since recombination and natural selection jointly impact the number of segregating sites and the proportion of sites at low frequency, it may be important to accurately account for these features when performing power calculations. Moreover, in the case of a targeted resequencing study, it is desirable to model the genomic architecture of the target locus directly.
We tested the performance of the method SKAT-O with our simulations of demography and selection [Lee et al., 2012a]. We simulated 30 kb of sequence from chromosome 3 (hg19 coordinates 50320000-50350000, which is a region under strong selection, see Methods), under two different demographic models [Gutenkunst et al., 2009; Tennessen et al., 2012], with and without selection on coding and conserved noncoding elements. Selection coefficients were drawn from the distributions inferred by Boyko et al. [2008] for coding regions and Torgerson et al. [2009] for conserved noncoding regions. We also ran simulations where the entire 30 kb region was treated as a single gene (i.e., ignoring the local structure of conserved elements and allowing selection on all non-synonymous sites).
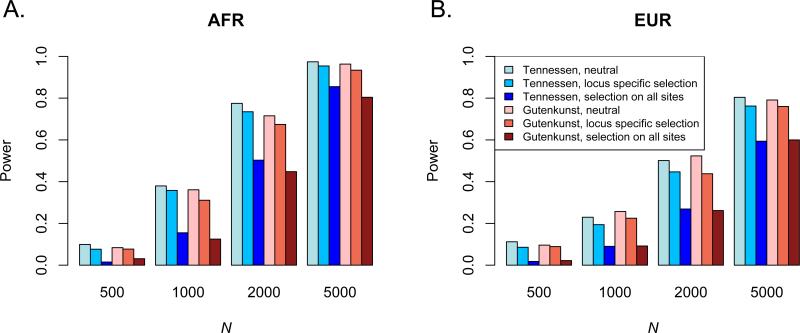
Following Wu et al. [2011], we generated phenotypes by allowing 5% of the sampled variants under 3% frequency to be causal (see Methods). We ran SKAT-O on the phenotypes and genotypes from the African and European continental groups and computed the fraction of simulations with p-values under 10–6. Selection impacts power by increasing the fraction of sites at low frequency and decreasing the overall level of genetic variation. The net result is a loss of power. When the entire locus is under selection, genetic variation is pushed to even lower levels and the site frequency spectrum is further shifted to rare alleles, which substantially reduces power. Note that we have not forced the genetic variance explained by the test sequence to be the same between the neutral and selected models, but this is precisely the point of this experiment. Because of reduced levels of overall variation, regions under direct selection must have larger effect sizes in order to explain an equal proportion of the genetic variance as neutral regions and reach the same power as neutral sequences.
In general, we observe higher power in the African continental group for this phenotype model (Figure 4, with > 80% power for a sample size of 5 × 103), which is in line with the overall higher level of genetic diversity within Africans (Figure 5). We also observe a subtle difference in power between the demographic models of Tennessen et al. [2012] (which includes rapid growth of the African population) and Gutenkunst et al. [2009] (which includes only an ancient expansion in the African population) in the African population.
Figure 4.
Power of SKAT-O at the α = 10–6 level for two different models of demography with and without selection, for different sample sizes N. We applied both a locus specific model of selection (and a model that treats the entire 30kb locus as a single gene. All results are for a region on chromosome 3, hgl9 coordinates 50320000-50350000.
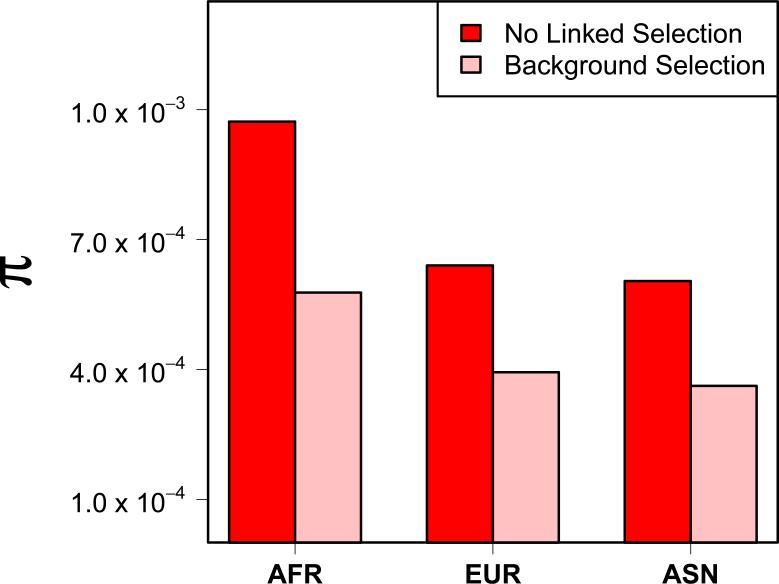
Figure 5.
We calculated the pairwise nucleotide diversity π for for the locus at chr3:50320000-50350000, for simulations with and without background selection and the demographic model of Gutenkunst et al. [2009]. The simulations without background selection simulated only this locus, while the background selection simulations also included 1 Mb of flanking sequence on either side of the locus of interest. Note, sites in the flanking 1Mb are not included in the diversity calculations.
Figure 4 shows that the effect of selection on power for this particular phenotype model ranges from ≈ 5 – 50% depending on the sample size and demographic model. However, we emphasize that these results should not be interpreted as general effects of human selection and demography on statistical power, but rather a reflection of a specific phenotype model commonly used in the literature [Wu et al., 2011]. In general, the effects of recent selection and demography on power may be more or less severe depending on sample size and the relationship between effect sizes and allele frequency, which is still a matter of some debate. One of the main advantages of our simulation method over other methods for estimating power is that the selection strength operating on every variant in the simulation is known. In two of the phenotype models included in sfs_coder, we use selection coefficients rather than allele frequencies to draw effect sizes when simulating phenotypes. In the Discussion section we further deliberate on the simulation of phenotypes. Here we have examined direct selection, but linked selection may also play a role in altering patterns of variation and affecting power calculations, as we address in the next section.
4.4 The impact of linked selection
Patterns of genetic diversity at neutral sites can be altered via physical linkage to sites under selection [Smith and Haigh, 1974; Charlesworth et al., 1993]. The effects of linked negative selection (known as “background selection”) on variation in humans have been well studied [McVicker et al., 2009]. Sites that are closely linked to deleterious variants are also prohibited from increasing in frequency, resulting in a local decrease in genetic diversity. The mean number of variants segregating at such loci is lower than in regions that are unlinked to selected loci. Background selection also has an effect on the shape of the site frequency spectrum (i.e., the proportion of variant sites at a given frequency) [Zeng and Charlesworth, 2011; Nicolaisen and Desai, 2013], but this effect is often subtle.
We ran 250 simulations incorporating the demographic model of Gutenkunst et al. [2009] for a 2Mb region that is centered on the 30 kb region considered in Figure 4 (chr3:50320000-50350000). This locus is expected to have among the strongest effects of background selection in the human genome under the model of McVicker et al. [2009]. All inferred exons and conserved noncoding elements within this region were simulated. In Figure 5, we show the mean observed nucleotide diversity π for these simulations as compared to simulations we used for Figure 4, which did not include the flanking sequences (but did incorporate selection on the 30 Kb central region). The overall level of diversity is strongly reduced in the background selection simulations. Additionally, there is a very small shift in the site frequency spectrum towards rare variants in the background selection simulations (data not shown).
We used these background selection simulations to compute power of SKAT-O with the same phenotype model that we considered in Figure 4. For a sample size of 2,000 individuals, power in Europeans drops from 44% for simulations without background selection to 38% when background selection is included, and power in Africans drops from 67% to 57%, due to the overall reduction in genetic diversity. Note that this reduction in overall diversity also implies a reduction in variance explained by the test sequence under the phenotype model of Wu et al. [2011], as discussed in the previous section.
5 Discussion
Simulations play an increasingly prominent role in statistical and population genetics because they can be used to generate DNA sequence data under models that are too complex to handle analytically. In statistical genetics, simulations have been used to generate sequence data under various demographic scenarios and to assess the performance of various statistical tests of association in the presence of complex demographics (e.g., see Wu et al. [2011]). However, natural selection also impacts patterns of genetic variation and thus might influence conclusions about statistical power.
Here, we introduced a new simulation tool (sfs_coder) that allows users to model human demography, selection, recombination, and genomic elements (exons and conserved noncoding sequences) for any locus in the human genome. Both selection and demography alter the frequency spectrum of variant sites and the number of variant sites within a sample of sequenced chromosomes. Jointly modeling these evolutionary forces may prove to be an important step forward for the use of simulations in assessing the performance of tests of statistical association [King et al., 2010], and indeed we showed here that natural selection and demography can impact the power of rare variant association tests under some phenotype models.
As an alternative to forward simulations of DNA sequences, researchers have also proposed “sideways” simulations [Chen et al., 2014]. These simulations use a reference panel of sequenced (or genotyped) chromosomes to generate new chromosomes under the model of Li and Stephens [2003]. The advantage of these methods is that they rely on observed genetic data, so the impact of natural selection and demography on the genealogy and the sampled genetic diversity are present in the reference panel and do not need to be directly modeled. However, here we showed that these methods do not recapitulate expected patterns of variation for rare variants when the population has experienced recent growth and the size of the reference panel is much smaller than the desired sample size.
While it is straightforward to use forward simulation tools such as sfs_coder to model the effects of selection and demography on DNA sequences, an important caveat of forward simulation is model misspec-ification. Here, we have tapped into the deep population genetics literature to incorporate recently inferred models, but these models should not be considered absolute truth. In particular, in future studies with larger sample sizes, considerable refinement could be made in the estimates of recent human growth as well as the strength of selective constraint. For example, there remains some debate about the rate of recent human growth and its influence on DNA sequences. One study of neutral genomic regions did not find support for recent acceleration in the growth rate [Gazave et al., 2014], in contrast to Tennessen et al. [2012]. Furthermore, more diverse annotations of genomic elements than we have considered here may allow for more precise distributions of selection coefficients for each locus in the genome.
Another important consideration when performing forward simulations is the choice of simulation parameters, especially sequence length and population size. Larger sequence lengths and population sizes can dramatically increase the computation time of forward simulations, so it is advantageous to pick these variables to be as small as possible [Hoggart et al., 2007]. However, we showed here that ignoring the impact of linked negative selection can alter patterns of simulated diversity and potentially affect power calculations. Furthermore, for some evolutionary models the simulated population size can also bias simulated patterns of genetic diversity if it is chosen to be too small [Uricchio and Hernandez, 2014]. Some further work is needed to explore appropriate choices of sequence length and population size for forward simulations of background selection and demography, and we have left these choices up to the end-user in our software sfs_coder.
Applications of simulations abound in the genetics literature, but one use that is of particular interest to statistical geneticists is the estimation of statistical power. For rare variant association tests (or any test of association that pools putatively causal variants), power is a function of the joint distribution of allele frequencies and effect sizes. Here, we showed that power is higher in Africans as compared to non-Africans for a simple phenotype model that asserts a logarithmic increase in effect size as allele frequency decreases. A side-effect of this phenotype model is that populations with more genetic variation have a greater proportion of variance explained by the test sequence. For a real phenotype, this may or may not hold, since it is not necessarily true that 1) effect sizes will be the same in two different populations or 2) the environmental variance is the same in two different populations. Exactly how the joint distribution of effect sizes and allele frequencies differ between populations may depend intimately on the action of selection on causal sites in each population and recent demographic history.
Generally, the distribution of effect sizes is not known, so arbitrary distributions have been proposed in previous work (and were applied here). These distributions assign larger effect sizes to rare variants than common causal alleles, but they do not necessarily have an obvious interpretation in terms of the strength and rate of selection in the human genome. Rare variants are not likely to contribute substantially to the variance observed in complex phenotypes unless causal sites are under selection [Simons et al., 2014], so the interpretability of power studies could be improved substantially by performing assessments with genetic models that include selection. In future studies it will be advantageous to simulate phenotypes where the effect sizes are modeled directly based on the selection coefficients of the causal sites, reminiscent of the work of Eyre-Walker [2010]. Tools such as sfs_coder should enable this work as human geneticists push further into the age of deep sequencing, large sample sizes, and complex genetic architectures.
Acknowledgments
This work was partially supported by the National Institutes of Health (grants 1R01HG007644 to R.D.H and 1R01CA088164 to J.S.W.) and a Sloan Foundation Research Fellowship (to R.D.H.). L.H.U. was supported by an Achievement Rewards for College Scientists fellowship and R.T. was supported by National Science Foundation Graduate Research fellowship 1144247. We thank M. Cyrus Maher for providing a useful script.
7 Appendix
We ran used the default parameter settings of the model “bestfit”, included in the cosi distribution, to simulate the model of Schaffner et al. [2005].
The following are representative command lines for our SFS_CODE simulations.
Gutenkunst et al. [2009]: sfs.code 3 10 -N 7000 -n 50 50 0 -A -L 1 100 -t 0.001 -r 0.001 -TS 0.219178 0 1 -TS 0.544658 1 2 -TE 0.60274 -Td 0 P 0 1.68493 -Td 0.219178 P 1 0.170732 -Td 0.544658 P 1 0.47619 -Tg 0.544658 P 1 58.4 -Td 0.544658 P 2 0.242857 -Tg 0.544658 P 2 80.3 -Tm 0.219178 P 0 1 6.15 -Tm 0.219178 P 1 0 0.5 -Tm 0.544658 L 0.738 0.4674 0.06 0.192 0.01938 0.09792
Gravel et al. [2011]: sfs.code 3 10 -N 7000 -n 50 50 0 -A -L 1 100 -t 0.001 -r 0.001 -TS 0.265753 0 1 -TS 0.342466 1 2 -TE 0.405479 -Td 0 P 0 1.982738 -Td 0.265753 P 1 0.128575 -Td 0.342466 P 1 0.554541 -Tg 0.342466 P 1 55.48 -Td 0.342466 P 2 0.29554 -Tg 0.342466 P 2 70.08 -Tm 0.265753 P 0 1 4.3422 -Tm 0.265753 P 1 0 0.5583 -Tm 0.342466 L 0.7237 0.225794 0.09305 0.115754 0.00858 0.03421
Tennessen et al. [2012]: sfs.code 3 10 -N 7000 -n 50 50 0 -A -L 1 100 -t 0.001 -r 0.001 -TS 0.265753 0 1 -TS 0.342466 1 2 -TE 0.405479 -Td 0 P 0 1.982738 -Td 0.265753 P 1 0.128575 -Td 0.342466 P 1 0.554541 -Tg 0.342466 P 1 44.822 -Td 0.342466 P 2 0.29554 -Tg 0.342466 P 2 70.08 -Tm 0.265753 P 0 1 4.3422 -Tm 0.265753 P 1 0 0.5583 -Tm 0.342466 L 0.7237 0.225794 0.09305 0.115754 0.00858 0.03421 -Tg 0.391465 P 0 242.36 -Tg 0.391465 P 1 284.7
We rescaled the ancestral population size to N = 5 × 103 (as opposed to 7 × 103 above) for computational feasibility for our simulations. We applied the distribution of selection coefficients of Boyko et al. [2008] to coding loci, which is given by the flag -W 2 0 0 0 0.184 0.00040244. For conserved noncoding elements, we applied the distribution of Torgerson et al. [2009], which is given by -W 2 0 0 0 0.0415 0.0015625. See the SFS_CODE manual at sfscode.sourceforge.net for more information.
References
- 1000 Genomes Project Consortium. Abecasis GR, Auton A, Brooks LD, DePristo MA, Durbin RM, Handsaker RE, Kang HM, Marth GT, McVean GA. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491(7422):56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyko AR, Williamson SH, Indap AR, Degenhardt JD, Hernandez RD, Lohmueller KE, Adams MD, Schmidt S, Sninsky JJ, Sunyaev SR, White TJ, Nielsen R, Clark AG, Bustamante CD. Assessing the evolutionary impact of amino acid mutations in the human genome. PLoS genetics. 2008;4(5):e1000083. doi: 10.1371/journal.pgen.1000083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustamante CD, Wakeley J, Sawyer S, Hartl DL. Directional selection and the site-frequency spectrum. Genetics. 2001;159(4):1779–1788. doi: 10.1093/genetics/159.4.1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth B, Morgan MT, Charlesworth D. The effect of deleterious mutations on neutral molecular variation. Genetics. 1993;134(4):1289–1303. doi: 10.1093/genetics/134.4.1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H-S, Hutter CM, Mechanic LE, Amos CI, Bafna V, Hauser ER, Hernandez RD, Li C, Liberles DA, McAllister K, Moore JH, Paltoo DN, Papanicolaou GJ, Peng B, Ritchie MD, Rosenfeld G, Witte JS, Gillanders EM, J. FE. Genetic simulation tools for post-genome wide association studies of complex diseases. Genetic Epidemiology. 2014 doi: 10.1002/gepi.21870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen JC, Kiss RS, Pertsemlidis A, Marcel YL, McPherson R, Hobbs HH. Multiple rare alleles contribute to low plasma levels of hdl cholesterol. Science (New York, N.Y.) 2004;305(5685):869–872. doi: 10.1126/science.1099870. [DOI] [PubMed] [Google Scholar]
- Emerson BC, Paradis E, Thebaud C. Revealing the demographic histories of species using DNA sequences. TREE. 2001;16:707–716. [Google Scholar]
- Eyre-Walker A. Evolution in health and medicine sackler colloquium: Genetic architecture of a complex trait and its implications for fitness and genome-wide association studies. Proceedings of the National Academy of Sciences of the United States of America. 2010;107(Suppl 1):1752–1756. doi: 10.1073/pnas.0906182107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gazave E, Ma L, Chang D, Coventry A, Gao F, Muzny D, Boerwinkle E, Gibbs RA, Sing CF, Clark AG, Keinan A. Neutral genomic regions refine models of recent rapid human population growth. Proceedings of the National Academy of Sciences of the United States of America. 2014;111(2):757–762. doi: 10.1073/pnas.1310398110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gravel S, Henn BM, Gutenkunst RN, Indap AR, Marth GT, Clark AG, Yu F, Gibbs RA, 1000 Genomes Project. Bustamante CD. Demographic history and rare allele sharing among human populations. Proceedings of the National Academy of Sciences of the United States of America. 2011;108(29):11983–11988. doi: 10.1073/pnas.1019276108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutenkunst RN, Hernandez RD, Williamson SH, Bustamante CD. Inferring the joint demographic history of multiple populations from multidimensional snp frequency data. PLoS genetics. 2009;5(10):e1000695. doi: 10.1371/journal.pgen.1000695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrow J, Frankish A, Gonzalez JM, Tapanari E, Diekhans M, Kokocinski F, Aken BL, Barrell D, Zadissa A, Searle S, Barnes I, Bignell A, Boychenko V, Hunt T, Kay M, Mukherjee G, Rajan J, Despacio-Reyes G, Saunders G, Steward C, Harte R, Lin M, Howald C, Tanzer A, Derrien T, Chrast J, Walters N, Balasubramanian S, Pei B, Tress M, Rodriguez JM, Ezkurdia I, van Baren J, Brent M, Haussler D, Kellis M, Valencia A, Reymond A, Gerstein M, Guigó R, Hubbard TJ. Gencode: the reference human genome annotation for the encode project. Genome research. 2012;22(9):1760–1774. doi: 10.1101/gr.135350.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez RD. A flexible forward simulator for populations subject to selection and demography. Bioinformatics (Oxford, England) 2008;24(23):2786–2787. doi: 10.1093/bioinformatics/btn522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann TJ, Marini NJ, Witte JS. Comprehensive approach to analyzing rare genetic variants. PloS one. 2010;5(11):e13584. doi: 10.1371/journal.pone.0013584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoggart Cf. J., Chadeau-Hyam M, Clark TG, Lampariello R, Whittaker JC, De Iorio M, Balding DJ. Sequence-level population simulations over large genomic regions. Genetics. 2007;177(3):1725–1731. doi: 10.1534/genetics.106.069088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International HapMap Consortium. Frazer KA, Ballinger DG, Cox DR, Hinds DA, Stuve LL, Gibbs RA, Belmont JW, Boudreau A, Hardenbol P, Leal SM, Pasternak S, Wheeler DA, Willis TD, Yu F, Yang H, Zeng C, Gao Y, Hu H, Hu W, Li C, Lin W, Liu S, Pan H, Tang X, Wang J, Wang W, Yu J, Zhang B, Zhang Q, Zhao H, Zhao H, Zhou J, Gabriel SB, Barry R, Blumenstiel B, Camargo A, Defelice M, Faggart M, Goyette M, Gupta S, Moore J, Nguyen H, Onofrio RC, Parkin M, Roy J, Stahl E, Winchester E, Ziaugra L, Altshuler D, Shen Y, Yao Z, Huang W, Chu X, He Y, Jin L, Liu Y, Shen Y, Sun W, Wang H, Wang Y, Wang Y, Xiong X, Xu L, Waye MMY, Tsui SKW, Xue H, Wong JT-F, Galver LM, Fan J-B, Gunderson K, Murray SS, Oliphant AR, Chee MS, Montpetit A, Chagnon F, Ferretti V, Leboeuf M, Olivier J-F, Phillips MS, Roumy S, Sallée C, Verner A, Hudson TJ, Kwok P-Y, Cai D, Koboldt DC, Miller RD, Pawlikowska L, Taillon-Miller P, Xiao M, Tsui L-C, Mak W, Song YQ, Tam PKH, Nakamura Y, Kawaguchi T, Kitamoto T, Morizono T, Nagashima A, Ohnishi Y, Sekine A, Tanaka T, Tsunoda T, Deloukas P, Bird CP, Delgado M, Dermitzakis ET, Gwilliam R, Hunt S, Morrison J, Powell D, Stranger BE, Whittaker P, Bentley DR, Daly MJ, de Bakker PIW, Barrett J, Chretien YR, Maller J, McCarroll S, Patterson N, Pe'er I, Price A, Purcell S, Richter DJ, Sabeti P, Saxena R, Schaffner SF, Sham PC, Varilly P, Stein LD, Krishnan L, Smith AV, Tello-Ruiz MK, Thorisson GA, Chakravarti A, Chen PE, Cutler DJ, Kashuk CS, Lin S, Abecasis GR, Guan W, Li Y, Munro HM, Qin ZS, Thomas DJ, McVean G, Auton A, Bottolo L, Cardin N, Eyheramendy S, Freeman C, Marchini J, Myers S, Spencer C, Stephens M, Donnelly P, Cardon LR, Clarke G, Evans DM, Morris AP, Weir BS, Mullikin JC, Sherry ST, Feolo M, Skol A, Zhang H, Matsuda I, Fukushima Y, Macer DR, Suda E, Rotimi CN, Adebamowo CA, Ajayi I, Aniagwu T, Marshall PA, Nkwodimmah C, Royal CDM, Leppert MF, Dixon M, Peiffer A, Qiu R, Kent A, Kato K, Niikawa N, Adewole IF, Knoppers BM, Foster MW, Clayton EW, Watkin J, Muzny D, Nazareth L, Sodergren E, Weinstock GM, Yakub I, Birren BW, Wilson RK, Fulton LL, Rogers J, Burton J, Carter NP, Clee CM, Griffiths M, Jones MC, McLay K, Plumb RW, Ross MT, Sims SK, Willey DL, Chen Z, Han H, Kang L, Godbout M, Wallenburg JC, L'Archevêque P, Bellemare G, Saeki K, Wang H, An D, Fu H, Li Q, Wang Z, Wang R, Holden AL, Brooks LD, McEwen JE, Guyer MS, Wang VO, Peterson JL, Shi M, Spiegel J, Sung LM, Zacharia LF, Collins FS, Kennedy K, Jamieson R, Stewart J. A second generation human haplotype map of over 3.1 million snps. Nature. 2007;449(7164):851–861. doi: 10.1038/nature06258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katzman S, Kern AD, Bejerano G, Fewell G, Fulton L, Wilson RK, Salama SR, Haussler D. Human genome ultraconserved elements are ultraselected. Science (New York, N.Y.) 2007;317(5840):915. doi: 10.1126/science.1142430. [DOI] [PubMed] [Google Scholar]
- Keinan A, Clark AG. Recent explosive human population growth has resulted in an excess of rare genetic variants. Science (New York, N.Y.) 2012;336(6082):740–743. doi: 10.1126/science.1217283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King CR, Rathouz PJ, Nicolae DL. An evolutionary framework for association testing in resequencing studies. PLoS genetics. 2010;6(11):e1001202. doi: 10.1371/journal.pgen.1001202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Emond MJ, Bamshad MJ, Barnes KC, Rieder MJ, Nickerson DA, NHLBI GO Exome Sequencing Project—ESP Lung Project Team. Christiani DC, Wurfel MM, Lin X. Optimal unified approach for rare-variant association testing with application to small-sample case-control whole-exome sequencing studies. American journal of human genetics. 2012a;91(2):224–237. doi: 10.1016/j.ajhg.2012.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Wu MC, Lin X. Optimal tests for rare variant effects in sequencing association studies. Biostatistics (Oxford, England) 2012b;13(4):762–775. doi: 10.1093/biostatistics/kxs014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Leal SM. Methods for detecting associations with rare variants for common diseases: application to analysis of sequence data. American journal of human genetics. 2008;83(3):311–321. doi: 10.1016/j.ajhg.2008.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li N, Stephens M. Modeling linkage disequilibrium and identifying recombination hotspots using single-nucleotide polymorphism data. Genetics. 2003;165(4):2213–2233. doi: 10.1093/genetics/165.4.2213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohmueller KE. The impact of population demography and selection on the genetic architecture of complex traits. PLoS genetics. 2014;10(5):e1004379. doi: 10.1371/journal.pgen.1004379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maher MC, Uricchio LH, Torgerson DG, Hernandez RD. Population genetics of rare variants and complex diseases. Human heredity. 2012;74(3-4):118–128. doi: 10.1159/000346826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manolio TA, Collins FS, Cox NJ, Goldstein DB, Hindorff LA, Hunter DJ, McCarthy MI, Ramos EM, Cardon LR, Chakravarti A, Cho JH, Guttmacher AE, Kong A, Kruglyak L, Mardis E, Rotimi CN, Slatkin M, Valle D, Whittemore AS, Boehnke M, Clark AG, Eichler EE, Gibson G, Haines JL, Mackay TFC, McCarroll SA, Visscher PM. Finding the missing heritability of complex diseases. Nature. 2009;461(7265):747–753. doi: 10.1038/nature08494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McVicker G, Gordon D, Davis C, Green P. Widespread genomic signatures of natural selection in hominid evolution. PLoS genetics. 2009;5(5):e1000471. doi: 10.1371/journal.pgen.1000471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgenthaler S, Thilly WG. A strategy to discover genes that carry multi-allelic or mono-allelic risk for common diseases: a cohort allelic sums test (cast). Mutation research. 2007;615(1-2):28–56. doi: 10.1016/j.mrfmmm.2006.09.003. [DOI] [PubMed] [Google Scholar]
- Neale BM, Rivas MA, Voight BF, Altshuler D, Devlin B, Orho-Melander M, Kathiresan S, Purcell SM, Roeder K, Daly MJ. Testing for an unusual distribution of rare variants. PLoS genetics. 2011;7(3):e1001322. doi: 10.1371/journal.pgen.1001322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson MR, Wegmann D, Ehm MG, Kessner D, St Jean P, Verzilli C, Shen J, Tang Z, Bacanu S-A, Fraser D, Warren L, Aponte J, Zawistowski M, Liu X, Zhang H, Zhang Y, Li J, Li Y, Li L, Woollard P, Topp S, Hall MD, Nangle K, Wang J, Abecasis G, Cardon LR, Zöllner S, Whittaker JC, Chissoe SL, Novembre J, Mooser V. An abundance of rare functional variants in 202 drug target genes sequenced in 14,002 people. Science (New York, N.Y.) 2012;337(6090):100–104. doi: 10.1126/science.1217876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolaisen LE, Desai MM. Distortions in genealogies due to purifying selection and recombination. Genetics. 2013;195(1):221–230. doi: 10.1534/genetics.113.152983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen R. Molecular signatures of natural selection. Annual review of genetics. 2005;39:197–218. doi: 10.1146/annurev.genet.39.073003.112420. [DOI] [PubMed] [Google Scholar]
- Peng B, Chen H, Mechanic L, Racine B, Clarke J, Gillanders E, Feuer E. Genetic data simulators and their applications: an overview. Genetic Epidemiology. 2014 doi: 10.1002/gepi.21876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price AL, Kryukov GV, de Bakker PIW, Purcell SM, Staples J, Wei L-J, Sunyaev SR. Pooled association tests for rare variants in exon-resequencing studies. American journal of human genetics. 2010;86(6):832–838. doi: 10.1016/j.ajhg.2010.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard JK. Are rare variants responsible for susceptibility to complex diseases? American journal of human genetics. 2001;69(1):124–137. doi: 10.1086/321272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawyer SA, Hartl DL. Population genetics of polymorphism and divergence. Genetics. 1992;132(4):1161–1176. doi: 10.1093/genetics/132.4.1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaffner SF, Foo C, Gabriel S, Reich D, Daly MJ, Altshuler D. Calibrating a coalescent simulation of human genome sequence variation. Genome research. 2005;15(11):1576–1583. doi: 10.1101/gr.3709305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siepel A, Bejerano G, Pedersen JS, Hinrichs AS, Hou M, Rosenbloom K, Clawson H, Spieth J, Hillier LW, Richards S, Weinstock GM, Wilson RK, Gibbs RA, Kent WJ, Miller W, Haussler D. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome research. 2005;15(8):1034–1050. doi: 10.1101/gr.3715005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simons YB, Turchin MC, Pritchard JK, Sella G. The deleterious mutation load is insensitive to recent population history. Nature genetics. 2014;46(3):220–224. doi: 10.1038/ng.2896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith JM, Haigh J. The hitch-hiking effect of a favourable gene. Genetical research. 1974;23(1):23–35. [PubMed] [Google Scholar]
- Su Z, Marchini J, Donnelly P. Hapgen2: simulation of multiple disease snps. Bioinformatics (Oxford, England) 2011;27(16):2304–2305. doi: 10.1093/bioinformatics/btr341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tennessen JA, Bigham AW, O'Connor TD, Fu W, Kenny EE, Gravel S, McGee S, Do R, Liu X, Jun G, Kang HM, Jordan D, Leal SM, Gabriel S, Rieder MJ, Abecasis G, Altshuler D, Nickerson DA, Boerwinkle E, Sunyaev S, Bustamante CD, Bamshad MJ, Akey JM, Broad GO, Seattle GO, NHLBI Exome Sequencing Project Evolution and functional impact of rare coding variation from deep sequencing of human exomes. Science (New York, N.Y.) 2012;337(6090):64–69. doi: 10.1126/science.1219240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornton KR, Foran AJ, Long AD. Properties and modeling of gwas when complex disease risk is due to non-complementing, deleterious mutations in genes of large effect. PLoS genetics. 2013;9(2):e1003258. doi: 10.1371/journal.pgen.1003258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torgerson DG, Boyko AR, Hernandez RD, Indap A, Hu X, White TJ, Sninsky JJ, Cargill M, Adams MD, Bustamante CD, Clark AG. Evolutionary processes acting on candidate cis-regulatory regions in humans inferred from patterns of polymorphism and divergence. PLoS genetics. 2009;5(8):e1000592. doi: 10.1371/journal.pgen.1000592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uricchio LH, Hernandez RD. Robust forward simulations of recurrent hitchhiking. Genetics. 2014;197(1):221–236. doi: 10.1534/genetics.113.156935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson SH, Hernandez R, Fledel-Alon A, Zhu L, Nielsen R, Bustamante CD. Simultaneous inference of selection and population growth from patterns of variation in the human genome. Proceedings of the National Academy of Sciences of the United States of America. 2005;102(22):7882–7887. doi: 10.1073/pnas.0502300102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witte JS. Genome-wide association studies and beyond. Annual review of public health. 2010;31:9–20. doi: 10.1146/annurev.publhealth.012809.103723. 4 p following 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu MC, Lee S, Cai T, Li Y, Boehnke M, Lin X. Rare-variant association testing for sequencing data with the sequence kernel association test. American journal of human genetics. 2011;89(1):82–93. doi: 10.1016/j.ajhg.2011.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng K, Charlesworth B. The joint effects of background selection and genetic recombination on local gene genealogies. Genetics. 2011;189(1):251–266. doi: 10.1534/genetics.111.130575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuk O, Schaffner SF, Samocha K, Do R, Hechter E, Kathiresan S, Daly MJ, Neale BM, Sunyaev SR, Lander ES. Searching for missing heritability: designing rare variant association studies. Proceedings of the National Academy of Sciences of the United States of America. 2014;111(4):E455–E464. doi: 10.1073/pnas.1322563111. [DOI] [PMC free article] [PubMed] [Google Scholar]