Abstract
The strength of self-peptide–major histocompatibility complex (MHC) recognition dictates naïve CD8+ T cell homeostasis, but its effect on foreign antigen reactivity is controversial. As CD5 expression correlates with self-recognition, we studied CD5lo and CD5hi naïve CD8+ T cells. Gene expression characteristics suggested CD5hi cells were better poised for reactivity and differentiation compared to the CD5lo population, and we found that the CD5hi pool exhibited more efficient clonal recruitment and expansion, as well as enhanced reactivity to inflammatory cues, during recognition of foreign antigen. Yet foreign peptide–MHC recognition was similar for both subsets. Thus, CD8+ T cells with higher self-reactivity dominate the immune response against foreign antigens, with implications for T cell repertoire diversity and autoimmunity.
The nature of the TCR interaction with foreign peptide–MHC (pMHC) complexes dictates the response magnitude and differentiation characteristics of antigen specific T cells1–4. In addition studies suggest TCR interactions with self-pMHC also impact the naïve T cell response to foreign-pMHC5–11. Thymic positive selection and naïve T cell homeostasis require low affinity TCR recognition of self-pMHC ligands12–16, but there is controversy about how such interactions affect the subsequent response to foreign-pMHC: published studies argue self-pMHC recognition enhances6 or diminishes7 the response to foreign antigens, or selectively impairs sensitivity to low-affinity foreign ligands14. However, those reports investigated the impact of self-pMHC withdrawal rather than studying how the degree of self-pMHC sensitivity influences the T cell response to foreign-pMHC.
Homeostatic TCR interactions with self-pMHC are thought to be of very low affinity and involve recognition of multiple self-peptides by an individual T cell clone, precluding direct assessment of self-pMHC recognition characteristics in the polyclonal T cell pool. However, differences in the expression of the cell surface protein CD5 have proven to be a valuable surrogate for the strength of the TCR-self-pMHC interactions14,17–21. CD5 expression on naïve T cells accurately predicts basal TCR signaling intensity and the capacity of T cells to rapidly engage key TCR signaling pathways9–11, and correlates with the ability of naïve CD8+ T cells to respond to homeostatic cues22–26. However, the underlying basis for the distinct response characteristics of naïve CD5lo and CD5hi populations is unclear, as is the impact of these differences on reactivity toward foreign-pMHC.
Recent studies used CD5 expression on naïve CD4+ T cells to correlate the strength of self-pMHC interaction with foreign-pMHC reactivity9–11. In one study, analysis of TCR transgenic mice suggested a direct correlation between the abundance of cell surface CD5 and the ability to bind cognate foreign-pMHC tetramers9, suggesting TCR affinity for self-pMHC predicts the affinity for foreign-pMHC. Those authors observed more vigorous responses by CD5hi than CD5lo naïve CD4+ T cells toward foreign-pMHC. Another report failed to observe any correlation between CD5 expression and TCR affinity for foreign-pMHC ligands, however, and found that CD5lo T cells expanded more efficiently than CD5hi cells during the primary response to foreign antigen10,11. Hence, whether and how CD5 expression predicts the capacity of naïve T cells to bind to and/or respond toward foreign-pMHC ligands is unclear.
Here, we report that CD5hi and CD5lo naïve CD8+ T cells differ in gene expression characteristics and that the CD5hi population manifests improved clonal recruitment and expansion in response to foreign-pMHC. These response differences did not correlate with the strength of the TCR interaction with foreign-pMHC, but CD5hi naïve CD8+ T cells showed superior utilization of in vivo inflammatory signals. Our data suggest pre-determined heterogeneity among naïve T cells dictates their capacity to respond to foreign antigens, with consequences for diversity of the functional T cell repertoire. Moreover, the finding that T cells with strong reactivity toward self-pMHC dominate the foreign-pMHC response has implications for outgrowth of autoreactive T cells.
Results
Distinct phenotype of CD5hi and CD5lo CD8+ T cells
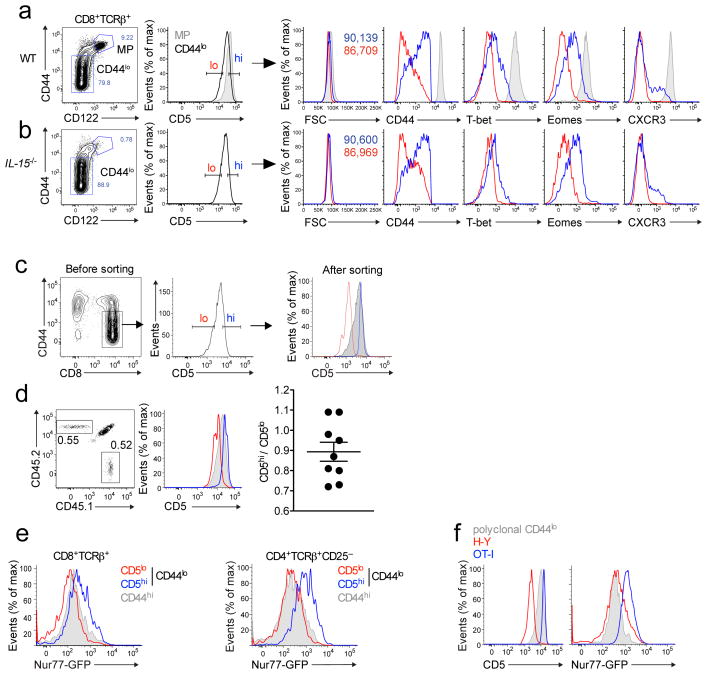
We first examined phenotypic differences between naïve (CD44loCD122lo) CD5lo and CD5hi CD8+ T cells. Extending previous work24,26,27 CD5hi cells were slightly larger, had elevated expression of CD44 and modestly increased interleukin 2Rβ (CD122) and IL-7Rα (CD127) expression, but slightly lower TCR, CD8+ and CD62L expression compared to the CD5lo population (Fig. 1a, Supplementary Fig. 1a–c). The CD5hi naïve CD8+ T cell population also showed elevated expression of T-bet and eomesodermin (transcription factors associated with activated CD8+ T cell differentiation28) and a subset of CD5hi cells expressed the chemokine receptor CXCR3 (Fig. 1a). The phenotypic characteristics of CD5hi naïve CD8+ T cells had some similarities to memory CD8+ T cells. However, the frequency and phenotype of CD5hi naïve CD8+ T cells was similar in IL-15-deficient mice, which lack typical CD8+ memory T cells29 (Fig. 1b and Supplementary Fig. 1b,c). Hence, the CD5hi naïve CD8+ T cell population neither derives from nor depends on memory-phenotype CD8+ T cells.
Figure 1. CD5 expression by naïve CD8+ T cells identifies stable populations with unique phenotypic traits.
Flow cytometry of cells combined from spleen and lymph nodes of wild-type (a) or Il15−/− (b) mice were stained for CD44 and CXCR3 and transcription factors T-bet and Eomes. Data were gated on naive (CD44lo CD122lo) CD8+ T cells in the lowest 20% (red) and highest 20% (blue) with respect to CD5 expression. Memory phenotype (MP) cells (CD44hiCD122hi) are indicated as gray shaded histograms. Data represent expression of indicated molecules and forward scatter (as a measure of cell size). (c,d) Naïve CD8+ T cells were sorted on CD5 expression as indicated and congenic populations co-transferred into normal recipient mice, which were analyzed 4–8 weeks later without immunization. Representative data are shown for the histograms (c,d); d, right, summary of compiled data (each symbol represents an individual mouse). (e,f) Flow cytometry of CD4+ CD25– and CD8+ T cells from Nur77gfp transgenic mice. T cells were gated on the lower or upper CD5 expression and analyzed for GFP expression (e). MP cells (gray) are included for comparison. In (f) flow cytometry of HY or OT-I TCR transgenic CD8+ T cells, gated on the CD44lo population, for CD5 and Nur77gfp expression as compared to polyclonal naïve CD8+ T cells (gray). Data in (a,c,d) are representative of 4 independent experiments (a n=10; c,d n=9), in (b,f) 2 independent experiments (b n=4; f n=6–7) and in (e) 3–4 independent experiments (n=6–11).
To determine whether the CD5hi and CD5lo populations are stable, we sorted polyclonal naïve CD8+ T cells into CD5hi and CD5lo populations (reflecting the upper and lower 20% of CD5 distribution, respectively), and congenically distinct cell populations were co-transferred into normal recipients. Both transferred populations maintained distinct CD5 expression and persisted for at least 8 weeks, indicating equivalent steady state survival (similar to studies on naïve CD4+ T cells9) (Fig. 1c,d and data not shown). The majority of donor cells maintained a naïve phenotype, though a fraction of CD5hi cells converted to CD44hi phenotype (Supplementary Fig. 1d), consistent with their enhanced response to homeostatic cues22–26.
Biochemical approaches indicate a correlation between CD5 abundance and the degree of basal TCR signaling5,9,11, however such methods cannot permit assessment of TCR signal strength in individual cells. Hence we examined Nur77gfp transgenic reporter mice, in which green fluorescent protein (GFP) expression provides a sensitive readout of TCR signaling30. For CD8+ and CD4+ naïve T cell subsets, CD5hi cells showed increased GFP expression compared to the CD5lo population (Fig. 1e), and this correlation held for Nur77gfp expression in H-Y and OT-I TCR transgenic CD8+ T cells (which reflect CD5lo and CD5hi populations, respectively)9,22,25 (Fig. 1f). Thus, CD5hi and CD5lo naïve CD8+ T cells are distinct, stable populations, with CD5hi cells displaying characteristics of cells that undergo more intense or frequent TCR interactions with self-pMHC.
Distinct transcriptional profiles of CD5hi and CD5lo CD8+ T cells
We next conducted gene expression analysis on polyclonal CD5hi and CD5lo naïve CD8+ T cells. In total, 57 unique genes were significantly changed by at least 2-fold (47 upregulated, 10 downregulated) in CD5hi relative to CD5lo naïve CD8+ T cells (Table 1). Among genes upregulated in CD5hi cells were those for the transcription factors Eomes, T-bet, Helios and Id3, many of which play a key role in activated T cell differentiation28, and molecules associated with trafficking and adhesion of effector T cells (CXCR3, XCL1 and CD44). Conversely, the kinase Itk (which can serve as a negative regulator of T-bet31,32) was downregulated in CD5hi cells.
Table 1. Major gene expression differences between sorted CD5hi and CD5lo naïve polyclonal CD8+ T cells.
Gene expression analysis was performed as described in Methods, and genes that showed statistically significant (P < 0.05), greater than 2-fold changes between the populations are indicated. Duplicates result from multiple probe sets for the same gene.
| Gene Symbol | Fold change | P-value | |
|---|---|---|---|
| UP in CD5hi | |||
| 1 | A430093F15Rik | 7.16 | 0.0170 |
| 2 | Endod1 | 5.79 | 0.0233 |
| 3 | Cxcr3 | 5.60 | 0.0217 |
| 4 | A530021J07Rik | 5.48 | 0.0033 |
| 5 | Ly6C1 | 5.27 | 0.0200 |
| 6 | Tbx21 (T-bet) | 4.96 | 0.0019 |
| A530021J07Rik | 4.74 | 0.0113 | |
| A530021J07Rik | 3.66 | 0.0144 | |
| 7 | Ndrg1 | 3.42 | 0.0224 |
| 8 | Eomes | 3.41 | 0.0255 |
| 9 | Ighv14–2 | 3.28 | 0.0059 |
| 10 | Cobll1 | 3.11 | 0.0033 |
| 11 | Ms4a4c | 3.08 | 0.0172 |
| 12 | Reck | 3.02 | 0.0201 |
| 13 | Itih5 | 3.00 | 0.0391 |
| 14 | Phactr2 | 2.97 | 0.0431 |
| 15 | Bcat1 | 2.91 | 0.0122 |
| 16 | Cldn10 | 2.88 | 0.0039 |
| 17 | 9230110F15Rik | 2.85 | 0.0293 |
| 18 | Serf1 | 2.76 | 0.0314 |
| 19 | Ptgfrn | 2.72 | 0.0458 |
| 20 | Xcl1 | 2.70 | 0.0173 |
| Eomes | 2.65 | 0.0361 | |
| 21 | Plac8 | 2.60 | 0.0137 |
| 22 | Rrm2 | 2.58 | 0.0286 |
| 23 | Fahd1 | 2.52 | 0.0457 |
| 24 | Mcart6 | 2.43 | 0.0019 |
| Ms4a4c | 2.41 | 0.0179 | |
| 25 | Ikzf2 (Helios) | 2.40 | 0.0273 |
| 26 | Xdh | 2.40 | 0.0031 |
| 27 | BB557941 | 2.40 | 0.0484 |
| 28 | Cd200 | 2.27 | 0.0288 |
| 29 | Anxa2 | 2.26 | 0.0213 |
| Ndrg1 | 2.23 | 0.0047 | |
| 30 | Gsto1 | 2.21 | 0.0197 |
| 31 | Cd5 | 2.20 | 0.0034 |
| 32 | Ptpn4 | 2.20 | 0.0291 |
| 33 | Chst11 | 2.17 | 0.0097 |
| 34 | Armcx4 | 2.15 | 0.0115 |
| 35 | Top2a | 2.15 | 0.0321 |
| 36 | Hopx | 2.14 | 0.0143 |
| Ndrg1 | 2.12 | 0.0008 | |
| 37 | Il10 | 2.10 | 0.0249 |
| 38 | Stmn1 | 2.09 | 0.0357 |
| 39 | Mrpl35 | 2.09 | 0.0337 |
| 40 | Lilrb3 & Pira | 2.08 | 0.0021 |
| 41 | Coro2a | 2.07 | 0.0034 |
| 42 | Cd44 | 2.06 | 0.0072 |
| 43 | Kctd15 | 2.03 | 0.0268 |
| 44 | Pogk | 2.03 | 0.0124 |
| 45 | Id3 | 2.02 | 0.0049 |
| 46 | Pck1 | 2.02 | 0.0151 |
| 47 | Aim1 | 2.02 | 0.0227 |
| Gene Symbol | Fold change | P-value | |
|---|---|---|---|
| DOWN in CD5hi | |||
| 1 | Dntt | 9.27 | 0.0110 |
| 2 | Slc6a19 | 4.18 | 0.0025 |
| Slc6a19 | 4.10 | 0.0143 | |
| 3 | Slc16a5 | 2.79 | 0.0257 |
| 4 | Ddc | 2.55 | 0.0035 |
| 5 | A130038J17Rik | 2.30 | 0.0175 |
| 6 | Grik4 | 2.20 | 0.0034 |
| 7 | Tmem154 | 2.10 | 0.0322 |
| 8 | 4930513N10Rik | 2.08 | 0.0453 |
| 9 | Tubb2a | 2.03 | 0.0200 |
| 10 | Itk | 2.01 | 0.0411 |
We further investigated the expression of XCL1, since it has been associated with efficient in vivo activation of CD8+ T cells (via enhancing T cell-dendritic cell colocalization)33. After brief in vitro stimulation of splenocytes, XCL1 protein expression was biased to a sub-population of CD5hi naïve CD8+ T cells (Fig. 2a,b; Supplementary Fig. 2a). Expression of CXCR3 and T-bet also marked a subset of CD5hi naïve CD8+ T cells (Fig. 2b)(Supplementary Fig. 2a). However, although memory phenotype (CD44hi) CD8+ T cells typically co-expressed these proteins, there was little coordinated expression in the naïve CD5hi pool (Supplementary Fig. 2b,c), indicating considerable heterogeneity within the CD5hi naïve CD8+ T cell population.
Figure 2. Naïve CD5lo and CD5hi CD8+ T cells have distinct gene-expression characteristics.
(a,b) Expression of XCL1, CXCR3 and T-bet by naïve (CD44lo) CD8+ T cells in the highest 20% (CD5hi) or lowest 20% (CD5lo) with respect to CD5 expression and memory phenotype (CD44hi) CD8+ T cell populations. (a) Splenocytes were stimulated with PMA/ionomycin in the presence of brefeldin A and then stained intracellularly for XCL1. Grey histograms represent unstimulated cells, solid lines indicate stimulated cells. (b) Frequencies of indicated subsets expressing XCL1, CXCR3 or T-bet. Expression of XCL1 was determined following activation (as in (a)), while CXCR3 and T-bet expression was determined in unstimulated B6 or T-bet reporter mice50. (c) CD44lo CD8+ T cells were flow sorted on the lower/upper 20% of CD5 expression and mRNA from 3 individual sorts was isolated and analyzed using an Affymetrix gene array. Differences between CD5lo and CD5hi transcription were compared against gene clusters associated with stages of the in vivo CD8+ T cell response34. Histograms represent fold-change ratio for expression in CD5lo versus CD5hi cells of genes from clusters II, III and X, revealing preferential expression in the CD5hi population (indicated as a fold change ratio <1). Data in (a) are representative of 4 experiments (9 mice); data in (b) were compiled from: 4 experiments (n=9) for XCL1; 3 experiments (n=7) for T-bet; 5 experiments (n=13) for CXCR3. Data in (c) derive from analysis of gene array analysis from 3 independent cell sorting experiments. * indicates p<0.001
Most individual gene expression differences between CD5hi and CD5lo naïve CD8+ T cell populations were subtle (Table 1), hence we explored whether there were changes in expression of gene sets. For a focused comparison, we used a χ2 test to align differences in CD5hi and CD5lo transcription with a database generated by ImmGen Consortium (Immgen.org), which had comprehensively defined patterns of gene expression following activation and differentiation of CD8+ T cells34. In that earlier work, a temporal analysis of gene expression over the course of the immune response allowed for the characterization of 10 clusters of correlated gene expression34. We investigated how expression of genes in these clusters were regulated in the CD5hi and CD5lo naïve CD8+ T cell subsets. This analysis revealed that the CD5hi population expressed significantly higher proportion of genes that characterize two early stages of the CD8+ T cell response and are associated with preparation for cell cycle (Cluster II) and active cell cycle and division (Cluster III) (Fig. 2c, Table 2). A more moderate (but still highly significant) correlation with Cluster X, which defines genes expressed at late effector and memory stages (Fig. 2c, Table 2). Together, these data suggest the CD5hi population is better poised for initial activation, compared to the CD5lo population.
TABLE 2.
Alignment between gene set expression differences for CD5hi and CD5lo naïve CD8+ T cells, and Immgen gene set clusters for CD8 T cell responses.
Gene set clusters and designated characteristics are listed (from ref34). The columns labeled “CD5lo” and “CD5hi” indicate the number of genes from each cluster that were preferentially expressed in that subset. The column labeled “Proportion” indicates the fraction of those genes expressed in the CD5lo subset. The significance of differential expression was determined by χ2 test (see Methods), and are listed in the column labeled “P-values”.
| Immgen Cluster | Characteristics | CD5lo | CD5hi | P-value | Proportion |
|---|---|---|---|---|---|
| Cluster I | Initial cytokine or effector response | 12 | 22 | 0.086 | 0.352 |
| Cluster II | Preparation for cell division | 187 | 334 | 1.193 E-10 | 0.358 |
| Cluster III | Cell cycle & division | 93 | 187 | 1.936 E-08 | 0.332 |
| Cluster IV | Naive and late memory | 51 | 40 | 0.248 | 0.560 |
| Cluster V | Early effector, late memory | 54 | 72 | 0.108 | 0.428 |
| Cluster VI | Short-term effector and memory | 27 | 37 | 0.211 | 0.421 |
| Cluster VII | Memory precursor | 61 | 49 | 0.252 | 0.554 |
| Cluster VIII | Naive or late effector or memory | 129 | 138 | 0.581 | 0.483 |
| Cluster IX | Short-term effector or memory | 39 | 55 | 0.098 | 0.414 |
| Cluster X | Late effector or memory | 34 | 59 | 0.009 | 0.365 |
Enhanced expansion of CD5hi CD8+ T cells in response to infection
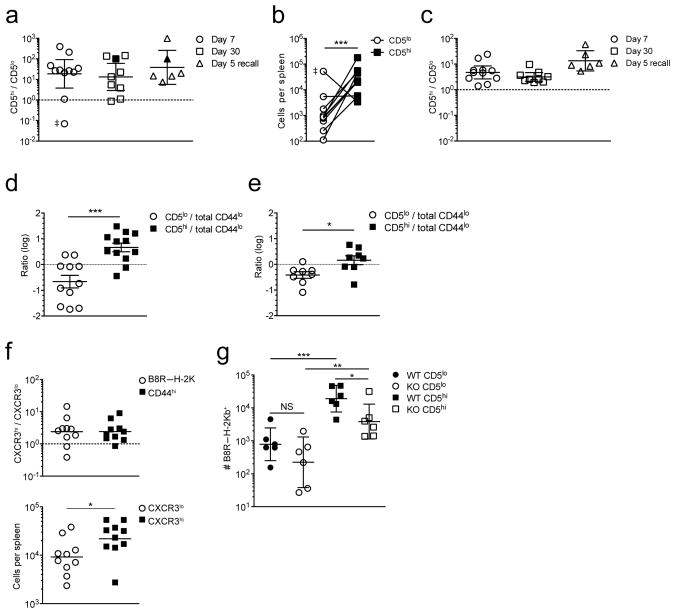
We next directly tested whether CD5hi and CD5lo naïve CD8+ T cells differ in their primary immune response against foreign antigen. In initial studies we assayed polyclonal CD8+ T cells specific for the H-2Kb restricted vaccinia virus epitope B8R20–27 (B8R), which are present at a frequency of ~1 per 1–2 × 104 CD8+ T cells in unimmunized C57BL/6 mice35. Naïve CD44lo CD8+ T cells were sorted by flow cytometry into congenically distinct CD5lo and CD5hi populations, and ~1.5 × 106 of each population co-transferred into recipients that were subsequently infected with LM-B8R, a recombinant attenuated Listeria monocytogenes expressing the B8R20–27 and the H-2Kb restricted ovalbumin peptide (OVA257–264: Ova). Assuming ~20% engraftment following adoptive transfer, this should seed ~20 B8R/Kb specific cells from each donor. At day 7 following infection, pMHC tetramer staining was used to identify responsive CD5hi and CD5lo donor cells, and the ratio (Fig. 3a) and absolute numbers (Fig. 3b) of each population was determined. In most cases, the CD5hi donor population dominated the response, on average accounting of ~95% of the B8R/Kb-specific population (Fig. 3a,b), although occasionally progeny of the CD5lo donors were more frequent (double dagger symbol in Fig. 3a,b). Tetramer binding may fail to identify all functionally responsive cells but similar results were obtained using peptide-induced interferon-γ (IFN-γ) production to identify antigen specific T cells (Supplementary Fig. 3a,b). At memory phase following priming and also during a recall response, the progeny of CD5hi donor cells maintained dominance over those from the CD5lo pool (Fig. 3a). The fact that this skewing was not exacerbated during the recall response indicates memory cells generated from CD5lo and CD5hi cells had similar re-expansion potential.
Figure 3. Naïve CD5lo and CD5hi CD8+ T cell precursors differ in their capacity to respond to foreign antigen.
CD44lo CD8+ T cells in the lowest or highest 20% for CD5 expression were sorted and 1.25–1.5 × 106 each of congenically distinct CD5lo and CD5hi cells co-transferred into CD45.1/CD45.2 host animals, which were infected the next day with LM-B8R. B8R/Kb–specific cells were isolated from the spleen by B8R/Kb tetramer enrichment at the indicated time points. (a) shows the ratio of donor CD5hi and CD5lo B8R/Kb-specific cells at days 7 and 30 after primary LM-B8R infection or 5 days after re-challenge of memory recipients with virulent LM-B8R, while (b) shows total donor cells number for the d7 timepoint. Each dot is a host animal. Filled symbols in (a) indicate mice where a B8R/Kb-specific response was not detected for CD5lo donor cells – the CD5hi/CD5lo ratio was arbitrarily set to 100. The double dagger symbol (‡) indicates an outlier: exclusion of this group did not alter the statistical significance shown in (b). (c) The ratio of bulk LM-specific donor CD5hi/CD5lo CD8+ T cells from the experiments shown in (a). Data in (a,c) are shown as the geometric mean ± 95% confidence interval. (d,e) Total CD8+ CD44lo cells were sorted and combined with congenically distinct CD44lo CD5lo or CD44lo CD5hi populations and 1.25 – 1.5 × 106 of each population co-transferred into congenic recipients which were infected with LM-B8R 1d later. The B8R/Kb-specific (d) and bulk LM-specific (e) responses were determined 7d after infection. The graphs indicate mean ± SEM. (f) CD5hi naïve CD8+ T cells (CD44lo cells in the upper 20% for CD5 expression) were sorted into the lowest or highest 30% for CXCR3 expression. 1–2×106 of congenically distinct CXCR3lo and CXCR3hi CD5hi cells were co-transferred into CD45.1/45.2 donors, and infected 1d later with LM-B8R. B8R/Kb-specific cells were isolated from the spleen 7d after infection by tetramer enrichment. The upper panel shows the ratio of cells derived from CXCR3hi versus CXCR3lo donor cells in the B8R/Kb-specific and bulk LM-B8R responsive populations. The lower panel shows the number of B8R/Kb–specific CXCR3hi and CXCR3lo donor cells isolated. The geometric mean is indicated. (g) Mixed bone marrow chimeras were generated using congenic wild-type and Cd25 (IL-2Rα)-deficient bone marrow. Following T cell reconstitution, CD44lo CD5lo and CD5hi populations were sorted and congenically distinct combinations of WT and KO cells co-transferred into congenic recipients that were infected with LM-B8R 1d later. At d7 following infection, the spleen was recovered and the number of B8R/Kb–specific donor cells enumerated. Data show the geometric mean ± 95% confidence interval. In all figures, each symbol indicates data from a single animal. Data in (a-c) are compiled from four experiments at day 7 (n = 11), 3 experiments at day 30 (n = 9), and 2 experiments for day 5 recall (n = 6); (d) shows data compiled from 3 experiments (n = 11–12 mice), (e) from two experiments (n = 8); (f) is compiled from 3 experiments (n=10); (g) is compiled from 3 individual experiments (n = 6). For this figure, statistical analysis is indicated as follows: *** for p < 0.001; ** for p < 0.01; * for p < 0.05.
The dominance of CD5hi naïve CD8+ T cell responses was not unique to B8R/Kb specific T cells or to Listeria infection: The OVA/Kb specific response induced by LM-B8R infection and the gp33/Db specific response induced by lymphocytic choriomeningitis virus (LCMV) infection were also biased to the CD5hi donor cells (Supplementary Fig. 3c,d). Beyond individual antigen specificities, the bulk pathogen-specific response – identified as donor CD8+ T cells that had acquired an antigen-experienced CD44hi, CD8lo, CD11ahi phenotype – also showed an advantage for the CD5hi donor pool, albeit less pronounced than observed for individual pMHC specific responses (Fig. 3c, Supplementary Fig. 3e).
The preferential expansion of the CD5hi donor population did not reflect greater intrinsic capacity of these cells for TCR-induced proliferation, as CD5lo and CD5hi naïve CD8+ T cells proliferated similarly upon in vitro stimulation with anti-CD3 plus anti-CD28 (Supplementary Fig. 3f), consistent with earlier studies9,20,24. The enhanced CD5hi T cell B8R/Kb-specific response was also seen when using Rag-1−/− recipient mice, ruling out a required contribution of host T or B lymphocytes (Supplementary Fig. 3g).
Since naïve CD5hi cells express intermediate amounts of CD44, it was formally possible that some memory-phenotype T cells had contaminated the CD5hi donor population. Yet when CD5hi and CD5lo donor populations were sorted to have equally low CD44 expression, the CD5hi donor population still dominated the response to LM-B8R (Supplementary Fig. 3h), arguing against memory cell contamination in our studies. The differential response of CD5hi and CD5lo naïve CD8+ T cells suggested that reactivity improves proportionally to increasing CD5 expression. To explore this, we measured the response to infection of CD5lo and CD5hi populations relative to total naïve CD8+ T cell pool (containing the full spectrum of CD5 expression). The CD5lo population expanded less than bulk naïve CD8+ T cells, while the CD5hi cells proliferated more (Fig. 3d,e), suggesting that CD5lo and CD5hi populations represent the extremes of a continuum in reactivity to foreign antigen.
Given that the CD5hi population is heterogeneous (Fig. 2b, Supplementary Fig. 2c), it was possible that a small subset within this pool was responsible for their superior antigen-specific responses. We investigated this hypothesis by subdividing the naïve CD5hi population based on CXCR3 expression, since this chemokine receptor has been associated with enhanced in vivo antigen detection by memory CD8+ T cells36,37. We sorted congenic populations of naïve CD5hi cells into CXCR3lo and CXCR3hi populations, and tested their response toward LM-B8R infection using the co-transfer model described above. We observed that expansion of the CXCR3hi CD5hi population was significantly greater than that of the CXCR3lo CD5hi subset, in both the B8R/Kb specific and bulk CD44hi responder populations (Fig. 3f). However, these differences were of lower magnitude than those between CD5hi and CD5lo populations (compare Figs. 3A,B and F), arguing against the hypothesis that a small subset of CD5hi cells accounts for all the enhanced reactivity of this population.
Aside from TCR signals, CD8+ T cell responses are strongly influenced by cytokines. Naïve CD5hi CD8+ T cells are more reactive to γc cytokines24,26 and possess enhanced ability to produce IL-2 following TCR stimulation11, which might induce autocrine IL-2R signaling. Also, the capacity of inflammatory cues to augment the magnitude of CD8+ T cell responses involves sustained upregulation of CD25 (ref38). To test whether CD25 expression impacts the differential response of CD5lo and CD5hi naïve CD8+ T cells, we assessed reactivity of CD25-deficient polyclonal CD8+ T cells, generated in mixed bone marrow chimeras, to avoid the lymphoproliferation and autoimmunity that occurs in CD25-deficient mice39. The distribution of CD5 expression was similar in wild-type and Cd25−/− resting naïve CD8+ T cells (Supplementary Fig. 3i). As expected, the B8R/Kb-specific wild-type CD5hi population expanded more than wild-type CD5lo cells (Fig. 3g) but, while CD25 deficiency did not have a statistically significant effect on the response by B8R/Kb-specific CD5lo cells, the response by Cd25−/− CD5hi cells was modestly but significantly impaired (Fig. 3g). While these data suggest the CD5hi pool may be more reliant on IL-2 responsiveness, the responses of CD5hi and CD5lo cells were not normalized by CD25 deficiency, indicating that elevated IL-2 sensitivity cannot fully account for the differences between these populations.
Together these data indicate that the antigen specific response to pathogens is dominated by CD5hi naïve CD8+ T cells.
Distinct clonal responses by CD5hi and CD5lo CD8+ T cells
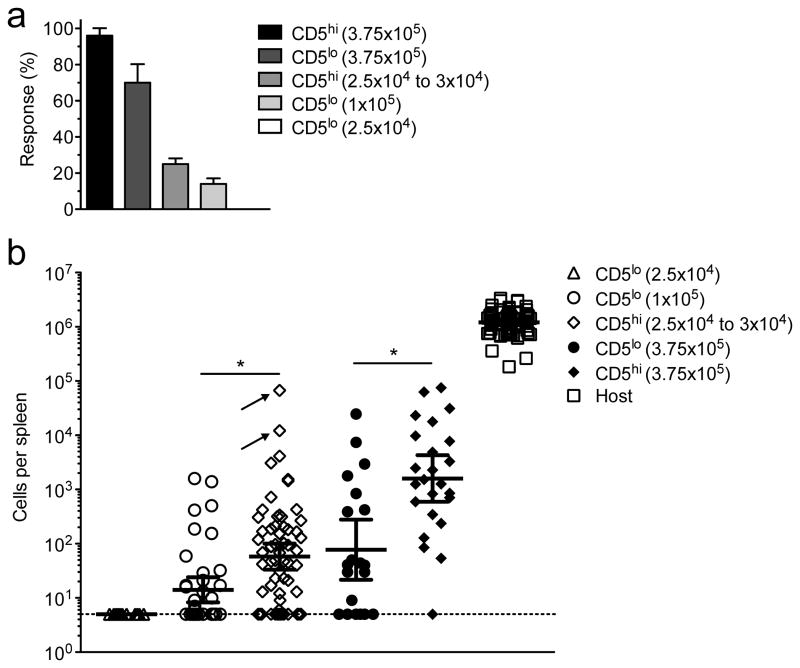
Our analysis of bulk naive CD8+ T cell responses could not determine whether the differential expansion of CD5hi and CD5lo naïve CD8+ T cell populations reflected enhanced responses by all antigen reactive CD5hi cells, or dominance by a small number of CD5hi CD8+ T cell clones. This is relevant because expansion characteristics of individual antigen specific naïve T cells can vary considerably2,4,40– 42. Accordingly, we reduced the number of adoptively transferred naïve CD44lo CD5hi or CD5lo polyclonal naïve CD8+ T cells to 25–30 × 103 cells. Based on the frequency of B8R/Kb specific precursors35, 20% engraftment would seed ~1 B8R/Kb specific donor CD8+ T cell per 3–5 donor cell cohorts, giving an average predicted response rate of ~27.5%. To increase the efficiency of detecting a clonal response, we used simultaneous transfer of up to 8 congenically distinct donor populations into a single recipient, as described by others2,4,40 (Supplementary Fig. 4). Using CD5hi naïve CD8+ T cells, ~ 24% (46/188) of transfers led to a B8R/Kb specific response, which was not significantly different from the predicted frequency (Fig. 4a) and consistent with studies using naïve OT-I T cells40. In contrast, adoptive transfer of 25 × 103 CD5lo cells led to no detectable B8R/Kb specific donor responses (0/40), significantly below the predicted rate (see legend to Fig. 4a). Increasing the input of CD5lo donor cells to 100 × 103 led to detectable responses, but only in 14% (18/125) of transfers (Fig. 4a). This response rate suggests that less than 1/7th the expected number of CD5lo CD8+ T cell precursors were capable of mounting a detectable response. Examining non-clonal responses by 375 × 103 CD5hi or CD5lo donor cells showed that >95% of CD5hi (23/24) but only 70% of donor CD5lo populations (14/20) mounted a response (Fig. 4b), further demonstrating the reduced response rate in the CD5lo pool.
Figure 4. Analysis of clonal naïve CD5lo and CD5hi CD8+ T cell responses to infection.
The indicated number of sorted CD5lo or CD5hi cells was transferred into congenic recipients and the B8R/Kb-specific donor-derived response was determined in the spleen 7 days after infection with LM-B8R. (The congenic marker scheme used is illustrated in Fig. S4).
(a) Response rate for the indicated number of transferred donor cells for which a B8R/Kb-specific response was observed above the limit of detection (5 cells). Error bars are calculated standard deviation. Statistical analysis using a one-sided binomial test showed that the response rate for 2.5×104 and 1×105 CD5lo cells (0/40 and 18/125, respectively) were significantly different from that expected (25% and 100%, respectively) (p<0.0001 for both) while the response rate for 2.5–3.0 × 104 CD5hi cells was not significantly different from average expected rate (~27.5%) (p=0.2). (b) Numbers of B8R/Kb-specific CD8+ T cells derived from each donor population, shown as the geometric mean ± 95% confidence interval. Each symbol represents cell numbers from an individual donor cohort. Donor populations that did not mount a detectable response are shown on the limit of detection line. Two very large clonal responses are indicated by arrows. Host responses are shown for comparison. Statistical significance was assessed only on responses above the limit of detection, using the Mann-Whitney test on log-transformed values. The numbers of transferred cohorts for each condition is given in the text, and the data derive from 2–4 separate experiments. The symbol * indicates p<0.05.
Furthermore, the mean clonal expansion magnitude (“burst size”) of the responding CD5lo population (from the 100 × 103 cell transfer) was significantly smaller than that of CD5hi cells (25–30 × 103 cell transfer)(Fig. 4b). It was also notable that the two largest clonal responses were seen for cells derived from CD5hi precursors, and were 10–100 fold greater than the largest CD5lo clonal response (arrows in Fig. 4B): Modeling the outcome if all the measured CD5hi and CD5lo clonal responses had occurred in a single animal, those two clones would account for nearly 80% of the B8R/Kb specific population (data not shown).
Hence, clonal analysis revealed two ways in which the CD5hi and CD5lo T cell responses differ: First, the CD5hi population displayed a markedly greater response rate. Second, even among cells that did engage in the B8R/Kb specific response, the average burst size of the CD5lo pool was reduced compared to CD5hi responders. Together, these differences can account for much of the expansion advantage of the CD5hipool.
Efficient recruitment of CD5hi CD8+ T cells into the immune response
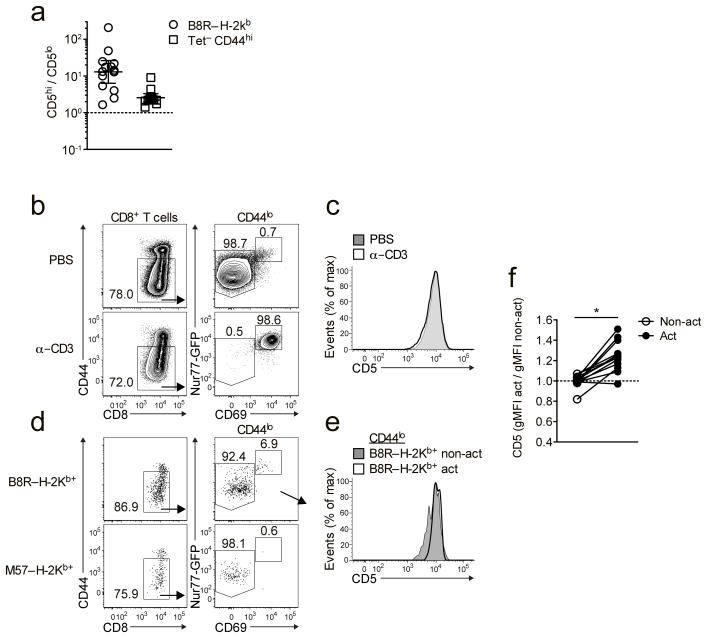
The increased clonal recruitment of CD5hi versus CD5lo cells might reflect preferential initial activation of CD5hi cells, or similar initial response by both populations, followed by improved proliferation/survival of the CD5hi population. The superior response by CD5hi cells was already apparent at days 3–4 of the in vivo response to LM-B8R (Fig. 5a), hence we next investigated whether CD5hi cells were preferentially activated during the initial response to infection. This was not feasible using adoptive transfer of polyclonal cells, and to determine the response of endogenous CD5hi and CD5lo cells it was first necessary to test whether CD5 expression changes during short term in vivo activation. Nur77gfp mice were injected with anti-CD3 i.v., and 5h later T cell activation was determined by induction of CD69 and Nur77gfp. Despite robust activation, naïve CD8+ T cells showed no change in CD5 expression (Fig. 5b,c), indicating that CD5hi and CD5lo naïve populations could still be distinguished. Next, Nur77gfp mice were infected with LM-B8R and 5h later splenic CD8+ T cells specific for B8R/Kb and for an irrelevant antigen (M57/Kb with the murine cytomegalovirus (MCMV) epitope, M57) were enriched using MHC class I tetramer capture. Following LM-B8R infection, activated naïve CD8+ T cells were evident among the B8R/Kb–specific population but not in the control M57/Kb-specific population (Fig. 5d), and the activated B8R/Kb–specific population was enriched for CD5hi cells (Fig. 5e,f). These data suggest that initial recruitment and/or activation favors the CD5hi naïve CD8+ T cell pool during the response to foreign antigen.
Figure 5. Early recruitment of CD5lo and CD5hi cells into the immune response after infection.
(a) Congenically distinct CD44lo CD5lo and CD5hi cells were sorted and co-transferred into congenic recipients as in Fig. 3. Donor-derived B8R/Kb-specific responses were analyzed from the spleen on day 3 or 4 after infection with LM-B8R. Data are shown as the geometric mean ± 95% confidence interval. (b,c) Nur77gfp transgenic mice were injected i.v. with 50 μg anti-CD3 or PBS and spleens harvested 5 hrs later. Representative data is shown for expression of Nur77GFP and CD69 (b) and CD5 levels (c) on CD44lo CD8+ T cells. (d-f) Nur77gfp transgenic mice were infected with 1 × 107 CFU LM-B8R and spleens harvested 5h later. B8R/Kb- and M57/Kb-specific CD8+ T cells were isolated by tetramer based enrichment. (d) Nur77gfp and CD69 expression was assessed on B8R/Kb- and M57/Kb-specific CD8+ T cells, and (e) B8R/Kb-specific cells with activated (Nur77gfp-hi, CD69+) and non-activated (Nur77gfp-lo, CD69−) phenotype assessed for CD5 expression. (d,e) are representative data. (f) shows normalized geometric mean fluorescence intensity (gMFI) for CD5 expression on non-activated and activated B8R/Kb specific naïve CD8+ T cells. (a) shows data compiled from a total of 3 experiments (1 at day 3; n=5, 2 at day 4; n=9); Data in (b,c) are from 2 separate experiments (n=3); (d–f) show representative or compiled data from 3 experiments (n=13) for B8R/Kb–specific cells and 2 experiments (n=9) for M57/Kb–specific cells. * indicates p < 0.01.
CD5lo and CD5hi cells show similar foreign pMHC binding characteristics
Some studies suggest CD5 expression on naïve TCR transgenic CD4+ T cells correlates with the TCR affinity for foreign-pMHC ligands, indicated by increased pMHC tetramer labeling of CD5hi versus CD5lo clones9. However, we found comparable pMHC tetramer staining intensities were observed on CD5hi and CD5lo naïve CD8+ T cell populations isolated by tetramer enrichment from unimmunized mice, (Fig. 6a), suggesting similar capacities for foreign-pMHC ligand binding. Furthermore, B8R/Kb tetramer geometric mean fluorescence intensity (gMFI) was not significantly different on effector cells derived from clonal CD5lo and CD5hi responses revealed that intensity did not significantly differ for antigen-specific progeny of CD5lo versus CD5hi clones, whereas the burst size of CD5hi clones was significantly higher than that of CD5lo clones (Fig. 6b). Thus, we observed minimal correlation between pMHC tetramer-staining intensity and either CD5 expression or clonal expansion characteristics of specific CD8+ T cells.
Figure 6. CD5hi and CD5lo cells show similar TCR binding to cognate antigen.
(a) B8R/Kb and M57/Kb specific CD8+ T cells were enriched from unimmunized mice and CD44lo precursors were gated on the lower and upper 20% of CD5 expression. Tetramer staining intensity (gMFI) was determined for both groups and expressed as a ratio, to normalize between experiments. (b) B8R/Kb tetramer staining (gMFI) and burst size of expanded clonal populations derived from CD5lo and CD5hi donor cells at d7 following LM-B8R infection. Data derive from transfers of 1×105 CD5lo and 2.5–3.0 × 104 CD5hi cells (see Fig. 4). Tetramer gMFI on donor cells was normalized to tetramer gMFI on host B8R/Kb-specific cells from the same mice, to yield relative fluorescence intensity (RFI). (c) Spleen and lymph node cells from unimmunized mice were subjected to tetramer enrichment. CD5 expression as gMFI on B8R/Kb-specific and M57/Kb-specific CD44lo CD8+ T cell populations was compared to normalized CD5 expression on bulk CD44lo CD8+ T cells. (d) CD5 expression levels on indicated MHC class I-restricted TCR transgenic CD8+ T cells. The histograms shows representative data while the bar graph shows CD5 relative fluorescence intensity (RFI), CD5 staining on polyclonal B6 CD44lo CD8+ T cells being set at “100”. (e) MHC class I tetramer binding to TCR transgenic cells after staining with the indicated range of tetramer concentrations. RFI reflects the geometric MFI of tetramer staining normalized to maximum intensity, which was set at “100”. Similar data were obtained in a second experiment. (f) In vitro activation of TCR transgenic CD8+ T cells. T cells were incubated with titrated doses of peptide for 6 hrs and CD69 expression was assessed by flow cytometry. Data are normalized to maximum CD69 expression. Similar data were obtained in a repeat experiment. (g) Expression of Kb or Db on RMA-S cells following incubation with indicated doses of the peptides recognized by studied TCR transgenic T cells. RFI reflects the MFI of Class I MHC molecule staining, normalized to maximum intensity, which was set at “100”. Data are representative of three experiments. (h) OT-I bone marrow chimeras were generated by transferring OT-I bone marrow (RAG+) into irradiated WT or β2m−/− hosts. Flow plots showing CD5 expression is representative of 4 separate sets of chimeras. Congenically distinct CD44lo OT-I CD8+ T cells were enriched from each chimera source and 103 cells of each population co-transferred into congenic recipients that were untreated or infected 1-3d later with attenuated ΔActA LM-OVA. (i) Donor cell engraftment (“take”) was determined uninfected recipients, and compared to ratios of donor-derived populations at 7d and 12d after LM-OVA infection. Data are shown as mean ± SEM.
In (a) data are compiled from 4 experiments (n = 14 for B8R/Kb, n = 9 for M57/Kb); (b) data are from 18 (CD5lo) and 46 (CD5hi) clonal responses from at least 3 experiments. * indicates p<0.05. Statistical significance was not changed by exclusion of the outlier (marked as ‡); In (c) the data is compiled from 6 experiments (n = 21) for B8R/Kb-specific cells and two experiments (n = 8) for M57/Kb-specific cells. **p < 0.001. In (d) data are compiled from 2–3 experiments (n=4–6); (e,f,g) are representative of 2–3 independent experiments; (h,i) Data are representative or compiled from 4 independent sets of chimeras.
It was also possible that foreign antigen specific T cells are selectively under-represented in the CD5lo pool. We did observe modest, but in some cases significant, skewing to higher CD5 expression within the foreign-pMHC tetramer binding naïve CD8+ T cell pool (Fig. 6c), and accordingly there were slightly more B8R/Kb specific cells in sorted CD5hi versus CD5lo populations (Supplementary Fig. 5). However, such skewing only contributed an average ~1.5-fold increase in antigen-specific precursors within the CD5hi population, relative to CD5lo cells, which could not explain the larger differences in clonal recruitment or population expansion of antigen specific CD5hi versus CD5lo cells (Figs. 3,4).
To avoid potential artifacts from the tetramer enrichment protocol, we analyzed four TCR transgenic lines which differ in CD5 surface expression, following the order H-Y < F5 < P14 < OT-I22,23,26 (Fig. 6d). All the TCR transgenic strains bound cognate pMHC tetramers with similar efficiency in dose titration (Fig. 6e) indicating that, in contrast to studies with CD4+ TCR-transgenic T cells9, CD5lo versus CD5hi expression did not predict the strength of foreign-pMHC ligand binding to TCR-transgenic CD8+ T cells. Interpreting tetramer staining may be complicated by the finding that CD8 and TCR expression are reduced on naïve CD5hi versus CD5lo CD8+ T cells (Supplementary Fig. 1a), and CD8 contributes to Class I pMHC tetramer binding43. Hence we also tested reactivity (as CD69 induction) of CD5hi and CD5lo TCR transgenic T cells to their cognate foreign ligands in dose titration. Although differences in antigen sensitivity were seen, they did not correlate with CD5 expression levels (Fig. 6f): for example CD5hi OT-I and CD5lo F5 CD8+ T cells showed similar antigen sensitivity. Instead, dose sensitivity corresponded with peptide binding to the relevant MHC molecules (Fig. 6g). Hence, these data indicate that CD5 expression predicted neither tetramer binding nor in vitro antigen sensitivity of naïve CD8+ T cells.
An expectation from our findings would be that CD5hi and CD5lo naïve CD8+ T cells with identical TCRs would display distinct response characteristics. This hypothesis was supported by earlier studies using TCR transgenic CD8+ T cells sorted into CD5hi and CD5lo pools24, but as CD5 levels are typically determined during thymic development, we sought to manipulate positive selection to produce cells with distinct CD5 expression levels. Bone marrow chimeras were generated using OT-I TCR transgenic donor marrow to reconstitute wild-type or β2m−/− hosts – in the latter, positive selection is mediated by hematopoietic cells, resulting in generation of OT-I cells with lower CD5lo expression (Fig. 6H and data not shown). When assessed for their response to LM-OVA infection, CD5hi OT-I expanded ~3–4-fold greater than the CD5lo OT-I population (Fig. 6I) indicating that CD5 expression levels correlated with the magnitude of the immune response, even when TCR specificity was normalized. In aggregate, our data suggest that the advantage of CD5hi over CD5lo naïve CD8+ T cells in their response to foreign antigen cannot be explained by differences in precursor frequency or avidity for foreign pMHC ligands.
Naïve CD5hi cells utilize inflammatory signals during the response to antigen
Besides TCR signals, the magnitude of the CD8+ T cell response is influenced by inflammatory cues44,45, hence we next tested the impact of inflammation on the response of CD5hi and CD5lo naïve CD8+ T cells. Since CD8+ T cell expansion is reduced in the absence of innate cues46,47 this system was not suitable for analysis of rare antigen-specific polyclonal CD8+ T cells, hence we used H-Y and OT-I TCR transgenic T cells models as examples of CD5lo and CD5hi clones, respectively. Low numbers of naïve H-Y and OT-I CD8+ T cells from were transferred into congenic hosts and stimulated by injection of dendritic cells (DCs) loaded with cognate peptides, with or without co-infection using wild-type ΔactA LM, which expresses no stimulatory antigens for either TCR transgenic, as a source of inflammatory stimulation. OVA peptide was used to stimulate OT-I cells, while H-Y T cells were stimulated with C2A, a variant of the Smcy peptide that enhances HY TCR recognition without altering MHC binding48 (Supplementary Fig. 6a–c), to minimize differences in ligand dose sensitivity of H-Y and OT-I T clones.
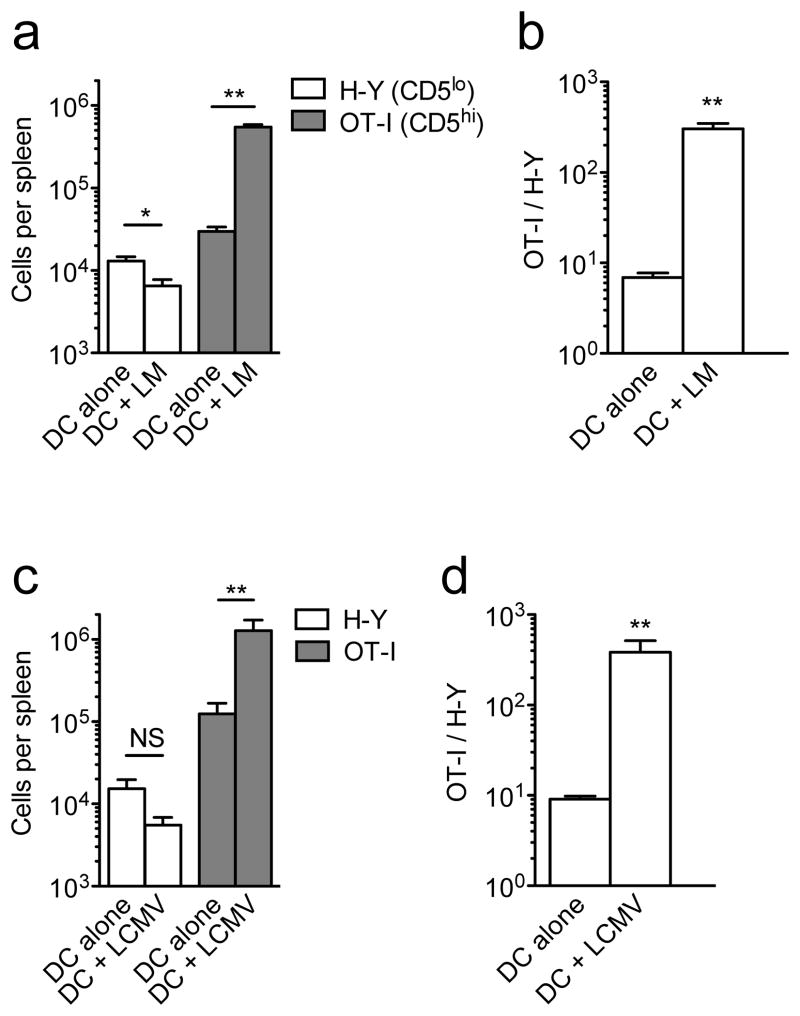
As expected, antigen-bearing DCs alone provoked modest responses by both H-Y and OT-I cells (Fig. 7a) and, when corrected for donor cell engraftment, there was moderately increased expansion of the OT-I versus H-Y pool (Fig. 7b). LM co-infection enhanced expansion by the OT-I population, as anticipated from earlier studies38,49, but did not increase H-Y T cell expanasion, and in fact caused a slight reduction in cell numbers (Fig. 7a). Accordingly, LM co-infection greatly increased the difference between HY and OT-I population sizes (Fig. 7b). Similar effects were seen using LCMV co-infection (Fig. 7c,d) and preliminary studies using co-administration of the TLR9 agonist CpG yielded comparable results (Supplementary Fig. 6d). These data suggest that, while the CD5hi clone OT-I responds to pro-inflammatory signals with enhanced expansion, this pathway is not operative for CD5lo H-Y CD8+ T cells. The pro-inflammatory cytokines IL-12 and Type-I IFN act as a “3rd signals” to promote CD8+ T cell responses46,47, but preliminary in vitro experiments did not suggest differences in the responses of HY and OT-I T cells to those cytokines (data not shown), indicating a more complex basis for the altered response. Nevertheless, our data suggest qualitative differences in the response of CD5hi versus CD5lo CD8+ T cells when foreign antigen stimulation is delivered in the context of innate immune cues in vivo.
Figure 7. CD5hi cells preferentially utilize pro-inflammatory signals during expansion.
Congenically distinct CD44lo H-Y and OT-I TCR transgenic CD8+ T cells were co-transferred into congenic recipients. Mice were co-immunized the next day with single peptide-pulsed matured DCs, with or without collateral infection with ΔactA−/ − LM (a,b) or LCMV Armstrong (c,d). (a,c) Show total numbers of splenic H-Y and OT-I CD8+ T cells 7d post-immunization with antigen pulsed DC, with or without indicated infections, while (b,d) shows ratios of OT-I/H-Y normalized to the “take” ratio observed in unimmunized animals 1d after adoptive transfer. All graphs show mean ± SEM. Data in (a,b) are compiled from 3 independent experiments (n=10); data in (c.d) are compiled from 2 experiments (n=6 for DC alone and n=7 for DC+LCMV). * = p<0.01; ** = p <0.001.
Discussion
Our data show that naïve CD8+ T cells with heightened recognition of self-pMHC ligands display enhanced reactivity toward foreign pMHC antigens. We confirmed and extended the utility of CD5 expression as a measure of the strength of self ligand encounter – showing that CD5hi cells exhibited increased expression of the Nur77-GFP reporter (a surrogate for TCR signaling) and changes in gene expression indicative of enhanced response sensitivity. Comparison of the in vivo response to foreign antigen revealed multiple steps at which the CD5hi population of naïve CD8+ T cells manifest an advantage over their CD5lo counterparts: initial activation and response rates were more efficient, the clonal burst size greater, and sensitivity to inflammatory cues enhanced. On the other hand, we did not observe a consistent difference in the capacity of polyclonal or TCR transgenic CD5hi versus CD5lo cells to bind to foreign pMHC tetramers, nor did TCR transgenic models suggest a difference in foreign pMHC response sensitivity. Taken together, our studies support a model in which the differences between CD5hi and CD5lo naïve CD8+ T cells are established prior to encounter with foreign antigen, and that numerous properties of the CD5hi population make their responses more efficient and competitive.
Our data differ from two elegant reports that used CD5 expression to characterize heterogeneity in the naïve CD4 T cell response. While one study found that CD5hi cells had enhanced TCR engagement with foreign pMHC ligands and superior response to antigen in vivo9, another reported that CD5lo and CD5hi cells had similar engagement with pMHC ligands and that CD5lo cells showed greater in vivo expansion than their CD5hi counterparts10,11. Although we observed some skewing in the size of the foreign pMHC tetramer binding population in favor of the CD5hi pool, this effect was modest and average tetramer binding intensity was similar for antigen specific CD5lo and CD5hi cells. Hence, our data and others10,11 argue against the conceptually complex model that the structural capacity to bind foreign pMHC ligands is dictated by T cell sensitivity toward self-pMHC molecules. These discrepancies may reflect distinct properties of CD4 and CD8+ T cells (as discussed9), although this argument does not pertain to the divergent conclusions reached with studies on CD4 T cells9,11. In any case, our findings reinforce the concept that, at least for naïve CD8+ T cells, the distinct responses of the CD5hi and CD5lo population likely reflects pre-existing, intrinsic properties of the cells, rather than arising from differences in foreign antigen perception.
Contrasting with our findings and others9, some reports found that CD5lo cells expanded more effectively than CD5hi cells, despite similar foreign ligand recognition properties10,11. Significantly, recent studies argued that CD5hi naïve CD4+ T cells exhibited a strong response to TCR stimulation, but this led to increased susceptibility for activation-driven cell death induced by IL-211. Our data suggest that optimal expansion of activated CD5hi naïve CD8+ T cells was dependent on CD25 expression, and enhanced IL-2 sensitivity (through improved CD122 signaling) was reported in the landmark studies of Cho et al. on of homeostasis of CD5hi naïve CD8+ T cells24. Potentially, enhanced IL-2 sensitivity is a boon to responding CD5hi CD8+ T cells, yet may be detrimental to CD5hi CD4 T cells (at least in some situations) by making them more vulnerable to induced cell death. It is also worth noting that the range of CD5 expression levels (and associated basal TCR signaling) is greater in naïve CD4 T cells compared to naïve CD8+ T cells9–11, perhaps indicative of distinct functional thresholds between the subsets.
Our studies build on considerable work that suggested CD5 levels correlate with TCR engagement by self-pMHC9–11,18–21. Our work defines the properties of the CD5hi population prior to antigen encounter, and mechanisms with which these cells outcompete other naïve CD8+ T cell populations during an active immune response. It is unclear whether CD5 itself contributes to the distinct function of CD5hi versus CD5lo cells - recent studies using Cd5−/− mice do not support that concept9,11, although this does not negate the value of CD5 expression level as a marker. As we show here, the CD5hi population differs from CD5lo cells in their expression of several genes. However, even within the CD5hi pool there is heterogeneity in T-bet, CXCR3 and induced XCL1 expression – hence there may be other features of CD5hi cells that better correlate with their improved functional prowess. We found a modest but significant advantage of CXCR3hi CD5hi over CXCR3lo CD5hi populations, suggesting CXCR3 expression may be a core feature of the optimal foreign antigen reactivity by CD5hi naïve CD8+ T cells.
Together, these findings suggest that naïve CD8+ T cells with the greatest level of self-reactivity are the most efficiently recruited into the foreign-pMHC specific response. Since sensitivity to TCR signals may change following naïve T cell activation, it is possible that progeny of some CD5hi clones could exhibit overt self-reactivity following activation, with significance for the induction of autoimmune disease following response to infection. At the same time, our findings leave open the question of why the CD5lo pool is maintained in the naïve CD8+ T cell repertoire. CD5lo cells are relatively resistant to deprivation of IL-726, making it possible that these cells are efficiently maintained during naïve T cell competition for homeostatic cytokines. Alternatively, CD5lo naïve CD8+ T cells may show superior responses to pathogens in certain situations: as shown for naïve CD4+ T cells heightened initial reactivity may accompany increased sensitivity to cell death10,11. Whether some immune responses favor the CD5lo population of naïve CD8+ T cells awaits further investigation.
METHODS
Mice
We purchased 6- to 12-week-old female C57BL/6 and B6.SJL mice from the National Cancer Institute. For adoptive cell transfer recipients, we used F1 CD45.1/2 females generated from C57BL/6J (Jackson Laboratories) crossed with B6.SJL (NCI) mice. Il15−/− and TCR-transgenic P14 mice51 were kind gifts from D. Masopust (University of Minnesota, Minneapolis, MN). P14 and OT-I52 and were maintained on a C57BL/6N and B6.PL (Thy-1.1) backgrounds. HY TCR transgenic mice were maintained on a Rag2−/− background (apart from initial cell surface phenotype studies, in which cells from female Rag2+/+ HY mice were analyzed with the T3.70 monoclonal antibody to identify HY-specific CD8+ T cells). F5 Rag1−/− mice were a kind gift of L. Cauley (University of Connecticut) and Cd25−/− mice were obtained from Jackson Labs. The Nur77gfp transgenic reporter mice have been previously described30, and were maintained on a C57BL/6N background. T-bet–ZsGreen reporter mice50 were initially obtained from J. Zhu (US National Institute of Allergy and Infectious Diseases, Bethesda, MD), and maintained on the C57BL/6N background. All mice were maintained in SPF conditions, and all mouse protocols were approved by the University of Minnesota Institutional Animal Care and Use Committee. No samples/animals were excluded from the analysis. The investigators were not blinded to group allocations or assessment.
Bacterial and Viral Infections
ΔActA attenuated LM (DP-L1942)53 and ΔActA LM-OVA were provided by J. Harty (University of Iowa, Iowa City, IA) and LM-B8R (both virulent and ΔActA), which contains both the Kb-restricted CD8+ epitopes B8R20–27 and OVA257–264 was a kind gift of R. Kedl (National Jewish Medical Research Center, University of Colorado, Denver, CO). LCMV Armstrong was a gift of D. Masopust. LM was grown in tryptic soy broth containing 50 μg/mL streptomycin to an OD600 of ~0.1. For primary infection with attenuated LM-B8R, 3 × 106 CFU were injected intravenously (i.v.). For secondary infections with virulent LM-B8R, mice were injected with 1 ×106 CFU i.v. In experiments where wild-type ΔActA LM was used to induce inflammation, 3–6 × 106 CFU bacteria were mixed with peptide-pulsed DCs and co-injected i.v. For LCMV infections, mice were injected with 2 × 105 PFU intraperitoneally.
Dendritic Cell Immunizations
Splenic DCs were prepared as previously described49. Briefly, to generate splenic DCs, mice were injected s.c. with 5 × 106 B16 cells expressing Flt3L (provided by M. Prlic and M. Bevan, University of Washington, Seattle, via J. Harty, University of Iowa). When tumors were palpable (5 × 5 mm), mice were injected with 2 μg LPS i.v. to mature the DCs and spleens were harvested ~16 h later. Following digestion with collagenase D for 20 min at 37°C, RBCs were lysed and splenocytes were resuspended in media comprised of 2 parts complete RPMI, 1 part B16-Flt3L-conditioned complete RPMI, 50 ng/mL GM-CSF, and 2 μM peptide. The C2A mutant of the SMCY peptide54 was used to stimulate HY CD8+ T cells, while OVA257–264 was used for stimulation of OT-I. Splenocytes were pulsed with peptide for 2 h at 37 °C, thoroughly washed, and DCs purified using Miltenyi CD11c microbeads. Mice were co-injected i.v. with 1 × 106 DCs pulsed with each peptide, with co-administration of LM, LCMV or CpG as indicated.
Flow Cytometry
Cells were stained with the following antibodies from eBioscience or BD Biosciences unless otherwise noted: CD4 (RM4-5), CD8+ (53–6.7), CD5 (53–7.3), CD27 (LG.7F9), CD44 (IM7), CD45.1 (A20), CD45.2 (104), Thy1.1 (HIS51 or OX-7), Thy1.2 (53–2.1), CD62L (MEL-14), CD69 (H1.2F3), CD122 (TM-b1), CD127 (A7R34), TCRβ (H57-597), CXCR3 (CXCR3-173), IFN-γ (XMG1.2), MHC class II (M5/114.15.2), and F4/80 (BM8). The B8R/Kb and OVA/Kb tetramers were generated as previously described43. The MCMV M57/Kb, LCMV gp33/Db, influenza NP68/Db, and HY SMCY/Db tetramers were provided by the NIH Tetramer Facility. For intracellular staining of transcription factors, cells were fixed and permeabilized with Foxp3 Fixation and Permeabilization Buffers (eBioscience) and stained with antibodies to T-bet (4B10) and Eomesodermin (Dan11mag) in Permeabilization Solution. Data was collected on LSR-II or Fortessa flow cytometers (BD Biosciences) and data were analyzed by using FlowJo analysis software (Tree Star).
XCL1 expression assay
Peripheral lymphocytes were stained with anti-XCL1 mAb (MTAC-2)55 that was kindly provided by R. Kroczek (Robert Koch-Institute, Berlin, Germany). To examine XCL1 production, bulk splenocytes were stimulated to PMA/ionomycin for 3–5 h at 37°C in the presence of Brefeldin A. Cells were stained for cell surface markers then fixed and permeabilized with BD Cytofix/Cytoperm or eBiosciences Foxp3/transcription factor fixation/permeabilization solutions, prior to intracellular staining for XCL1.
Cell Sorting and Adoptive Transfer
For adoptive transfer experiments, spleens and lymph nodes from C57BL/6 (CD45.2/2) and B6.SJL (CD45.1/1) mice digested with collagenase D (Roche) and negatively enriched for CD8+ T cells using Miltenyi enrichment antibody cocktail and beads. Cells were then stained with anti-CD8, CD5, and CD44 and CD8+CD44lo cells (i.e. excluding CD44hi cells) were sorted on the lower or upper 20% of CD5 expression using a BD FACSAria I. In some studies, the CD5hi CD44lo population was further gated on the lower or upper 30% of CXCR3 prior to sorting. Approximately 1.25–1.5 × 106 each of congenically mismatched CD5lo and CD5hi cells were co-transferred into CD45.1/2 recipients and infected with LM-B8R in the next day. For recall experiments, CD5lo/hi recipients that had been infected with ΔActA LM-B8R >40 days previous were challenged with virulent LM-B8R. Varied combinations of congenic backgrounds for donor and host animals in transfer studies.
In experiments where we transferred single B8R/Kb-specific CD8+ T cell clones, CD8+ T cells were negative enriched from the spleens and lymph node cells of 4 to 8 congenically distinct donors by using different combinations of CD45.1/2 and CD90.1/2 (Supplementary Fig. 6). Equal numbers of CD8+ T cells from each congenic donor group were mixed, stained with anti-CD8, CD5, and CD44 and sorted for naïve CD8+ T cells in the lower or upper 20% for CD5 expression. The indicated number of CD5lo/hi cells for each congenic group was then transferred into congenic recipients. Mice were infected 1–2 days post-transfer with attenuated LM-B8R, and the response to B8R/Kb was assessed 7 days later. Background staining for congenic markers was very low (1 event or less, data not shown), and we set our limit of detection at ≥3 flow cytometric events in the antigen specific population, which equates to ~5 total B8R/Kb specific CD8+ T cells
In adoptive transfer experiments using TCR transgenic CD8+ T cells, CD44lo Thy-1.1 OT-I cells (RAG+/+ or RAG-1−/−) were enriched by negative selection as previously described56. Female Rag-2−/− HY CD8+ T cells, which are all CD44lo, were negatively enriched using Miltenyi beads. Mixtures containing 1000 each of the OT-I and HY populations were co-transferred i.v. into B6.SJL mice and these recipients were immunized 1 day later. To assess the “take”, 2 × 105 cells from the same mixture of OT-I and HY cells was transferred into recipients, cells from these mice were then analyzed by flow cytometry the day of immunization. Similar “take” ratios were observed when animals receiving 1000 OT-I and HY T cells were enriched using magnetic beads on the day of immunization (data not shown).
MHC Class I Tetramer Enrichment
To analyze CD8+ T cell antigen-specific precursors or CD5lo/hi donor responses following infection, MHC class I tetramer enrichment was used as previously described35. Briefly, spleen and lymph nodes (for analyzing precursors) or spleen only (LM infection) were digested with collagenase D. Cells were labeled with PE- or APC-conjugated tetramers and enriched over magnetic columns using anti-PE or APC magnetic beads (Miltenyi). A small portion of the enriched fraction was added to AccuCheck counting beads (Invitrogen) to accurately back-calculate total numbers. Tetramer-enriched fractions were then stained with additional extracellular antibodies and fixed with paraformaldehyde prior to analysis by flow cytometry.
Mixed Bone-Marrow Chimeras
We generated mixed bone-marrow chimeras by mixing T cell-depleted bone marrow from congenic strains and injecting 5–10 × 106 cells into lethally irradiated (1000 rads) host animals. For chimeras with WT and Cd25−/− bone marrow, roughly equal numbers of cells from CD45.1/2+ WT and CD45.2+ Cd25−/− mice (6–8 weeks of age) were injected into CD45.1+ WT hosts. For OT-I chimeras, Thy-disparate OT-I (RAG+) bone marrow was injected into congenically distinct WT or β2m−/− recipients. Cells from chimeras were used >10 weeks after transplant.
In vitro Stimulation
To assess CD8+ T cell activation, 2×104 purified CD44lo CD8+ TCR transgenic cells were incubated at 37°C with 1–2×106 splenic antigen presenting cells in 96-well round-bottom plates with titrated doses of cognate peptide: OT-I with Kb/OVA257–264 (SIINFEKL), P14 with Db/gp33–41 (KAVYNFATC), F5 with Db/NP366–374 (ASNENMDAM), HY with Db/Smcy (KCSRNRQYL) or Db/C2A (KASRNRQYL). Cells were stimulated for 6 h and then stained for CD69 expression.
RMA-S MHC Class I Stabilization Assay
RMA-S cells were cultured in RPMI containing 10% FCS at 30°C with 5% CO2 overnight. In a 96-well round-bottom plate 1 × 105 RMA-S cells were incubated with titrated doses of peptide for 1 h and then the plate was moved to a 37 °C CO2 incubator for 3 h. Cells were then stained for stable surface class I molecules using H-2Kb (Y3) or H-2Db (28.14.8) antibodies.
Gene Transcription Analysis
Naïve CD44lo CD8+ T cells from spleens and lymph nodes were flow sorted on the lower and upper 20% of CD5 expression as described above. For each sample ≥1 × 106 cells were used for RNA extraction using a RNeasy microkit (Qiagen). RNA was used to generate biotinylated cRNA using the MessageAmpIII RNA Amplification kit (Ambion) following the manufacturer’s recommendations. Samples were hybridized to Affymetrix murine 430 2.0 gene chips at the BioMedical Genomics Center (University of Minnesota) following standard procedures. RNA samples from three independent sorts were analyzed. Gene expression analysis that led to Table 1 used Genespring software: Data were MAS5 normalized and filtered for present/absent calls in at least one group, and for stastically significant (P < 0.05) fold change of >2.0. For enrichment analysis (Table 2), cluster genes expressed by either CD5hi or CD5lo cells were determined to be any genes with a Fold Change (FC)>0. Significance was determined by χ2 where equal distribution was taken as the null hypothesis. Histograms show fold change within the CD5hi versus CD5lo comparison, binned as indicated, for genes within the indicated clusters.
Statistics
Unless indicated otherwise in the figure legend, a two-tailed, unpaired Student’s t test was performed on log-transformed data using Prism (GraphPad Software). When making multiple comparisons, one-way ANOVA with Dunnett’s Multiple Comparison post-test was used. Sample sizes were chosen based on previous experience and similar studies. In the clonal analysis shown in Fig. 4, the data distribution was not normal or lognormal, and the non-parametric Mann-Whitney test was applied. The P values are indicated with asterisks, defined in each figure legend.
Supplementary Material
Acknowledgments
We thank the University of Minnesota Flow Core personnel for flow cytometry support and cell sorting. We thank M. Jenkins and M. Mescher for critical review of the manuscript, G. Stritesky for insight on analysis of Nur77 mice, J. Ding and S. Peery for excellent technical assistance with mice and members of the Jamequist laboratory provided valuable discussions throughout. We also thank R. Kroczek (Robert Koch-Institute, Berlin, Germany) for generously providing conjugated MTAC-2 antibody. The authors declare no financial interests. This work was supported by NIH award R37 AI-38903 (S.C.J.) and the Irvington Institute Fellowship Program of the Cancer Research Institute (R.B.F.). Research reported in this publication was also supported by NIH grant P30 CA77598 utilizing the Biostatistics and Bioinformatics Core shared resource of the Masonic Cancer Center, University of Minnesota and by the by the National Center for Advancing Translational Sciences of the National Institutes of Health Award Number UL1TR000114. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Accession codes. GEO: microarray data, GSE62142.
AUTHOR CONTRIBUTIONS
R.B.F. and S.C.J. designed the experimental approaches; R.B.F. and S.E.H. conducted experiments; Y.X. and K.A.H. provided mouse strains and bone marrow chimeras; J.A.B. and A.W.G. analyzed gene expression data; R.B.F. and S.C.J. wrote and edited the manuscript.
COMPETING FINANCIAL INTERESTS
None
References
- 1.Zehn D, Lee SY, Bevan MJ. Complete but curtailed T-cell response to very low-affinity antigen. Nature. 2009;458:211–214. doi: 10.1038/nature07657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tubo NJ, et al. Single Naive CD4(+) T Cells from a Diverse Repertoire Produce Different Effector Cell Types during Infection. Cell. 2013;153:785–796. doi: 10.1016/j.cell.2013.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fazilleau N, McHeyzer-Williams LJ, Rosen H, McHeyzer-Williams MG. The function of follicular helper T cells is regulated by the strength of T cell antigen receptor binding. Nature immunology. 2009;10:375–384. doi: 10.1038/ni.1704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Plumlee CR, Sheridan BS, Cicek BB, Lefrancois L. Environmental cues dictate the fate of individual CD8(+) T cells responding to infection. Immunity. 2013;39:347–356. doi: 10.1016/j.immuni.2013.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dorfman JR, Stefanova I, Yasutomo K, Germain RN. CD4+ T cell survival is not directly linked to self-MHC-induced TCR signaling. Nature immunology. 2000;1:329–335. doi: 10.1038/79783. [DOI] [PubMed] [Google Scholar]
- 6.Stefanova I, Dorfman JR, Germain RN. Self-recognition promotes the foreign antigen sensitivity of naive T lymphocytes. Nature. 2002;420:429–434. doi: 10.1038/nature01146. [DOI] [PubMed] [Google Scholar]
- 7.Bhandoola A, et al. Peripheral expression of self-MHC-II influences the reactivity and self-tolerance of mature CD4(+) T cells: evidence from a lymphopenic T cell model. Immunity. 2002;17:425–436. doi: 10.1016/s1074-7613(02)00417-x. [DOI] [PubMed] [Google Scholar]
- 8.Hochweller K, et al. Dendritic cells control T cell tonic signaling required for responsiveness to foreign antigen. Proc Natl Acad Sci U S A. 2010;107:5931–5936. doi: 10.1073/pnas.0911877107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mandl JN, Monteiro JP, Vrisekoop N, Germain RN. T cell-positive selection uses self-ligand binding strength to optimize repertoire recognition of foreign antigens. Immunity. 2013;38:263–274. doi: 10.1016/j.immuni.2012.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Weber KS, et al. Distinct CD4+ helper T cells involved in primary and secondary responses to infection. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:9511–9516. doi: 10.1073/pnas.1202408109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Persaud SP, Parker CR, Lo WL, Weber KS, Allen PM. Intrinsic CD4 T cell sensitivity and response to a pathogen are set and sustained by avidity for thymic and peripheral complexes of self peptide and MHC. Nature immunology. 2014 doi: 10.1038/ni.2822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Starr TK, Jameson SC, Hogquist KA. Positive and negative selection of T cells. Annu Rev Immunol. 2003;21:139–176. doi: 10.1146/annurev.immunol.21.120601.141107. [DOI] [PubMed] [Google Scholar]
- 13.Tanchot C, Lemonnier FA, Perarnau B, Freitas AA, Rocha B. Differential requirements for survival and proliferation of CD8 naive or memory T cells. Science. 1997;276:2057–2062. doi: 10.1126/science.276.5321.2057. [DOI] [PubMed] [Google Scholar]
- 14.Takada K, Jameson SC. Self-class I MHC molecules support survival of naive CD8 T cells, but depress their functional sensitivity through regulation of CD8 expression levels. J Exp Med. 2009;206:2253–2269. doi: 10.1084/jem.20082553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Takada K, Jameson SC. Naive T cell homeostasis: from awareness of space to a sense of place. Nat Rev Immunol. 2009;9:823–832. doi: 10.1038/nri2657. [DOI] [PubMed] [Google Scholar]
- 16.Sprent J, Surh CD. Normal T cell homeostasis: the conversion of naive cells into memory-phenotype cells. Nat Immunol. 2011;12:478–484. doi: 10.1038/ni.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tarakhovsky A, et al. A role for CD5 in TCR-mediated signal transduction and thymocyte selection. Science. 1995;269:535–537. doi: 10.1126/science.7542801. [DOI] [PubMed] [Google Scholar]
- 18.Azzam HS, et al. CD5 expression is developmentally regulated by T cell receptor (TCR) signals and TCR avidity. J Exp Med. 1998;188:2301–2311. doi: 10.1084/jem.188.12.2301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wong P, Barton GM, Forbush KA, Rudensky AY. Dynamic tuning of T cell reactivity by self-peptide-major histocompatibility complex ligands. J Exp Med. 2001;193:1179–1187. doi: 10.1084/jem.193.10.1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Smith K, et al. Sensory adaptation in naive peripheral CD4 T cells. J Exp Med. 2001;194:1253–1261. doi: 10.1084/jem.194.9.1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Seddon B, Zamoyska R. TCR signals mediated by Src family kinases are essential for the survival of naive T cells. J Immunol. 2002;169:2997–3005. doi: 10.4049/jimmunol.169.6.2997. [DOI] [PubMed] [Google Scholar]
- 22.Kieper WC, Burghardt JT, Surh CD. A role for TCR affinity in regulating naive T cell homeostasis. J Immunol. 2004;172:40–44. doi: 10.4049/jimmunol.172.1.40. [DOI] [PubMed] [Google Scholar]
- 23.Ge Q, Bai A, Jones B, Eisen HN, Chen J. Competition for self-peptide-MHC complexes and cytokines between naive and memory CD8+ T cells expressing the same or different T cell receptors. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:3041–3046. doi: 10.1073/pnas.0307339101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cho JH, Kim HO, Surh CD, Sprent J. T cell receptor-dependent regulation of lipid rafts controls naive CD8+ T cell homeostasis. Immunity. 2010;32:214–226. doi: 10.1016/j.immuni.2009.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Johnson LD, Jameson SC. Self-specific CD8+ T cells maintain a semi-naive state following lymphopenia-induced proliferation. J Immunol. 2010;184:5604–5611. doi: 10.4049/jimmunol.1000109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Palmer MJ, Mahajan VS, Chen J, Irvine DJ, Lauffenburger DA. Signaling thresholds govern heterogeneity in IL-7-receptor-mediated responses of naive CD8+ T cells. Immunology and cell biology. 2011;89:581–594. doi: 10.1038/icb.2011.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Park JH, et al. ‘Coreceptor tuning’: cytokine signals transcriptionally tailor CD8 coreceptor expression to the self-specificity of the TCR. Nat Immunol. 2007;8:1049–1059. doi: 10.1038/ni1512. [DOI] [PubMed] [Google Scholar]
- 28.Kaech SM, Cui W. Transcriptional control of effector and memory CD8+ T cell differentiation. Nature reviews. Immunology. 2012;12:749–761. doi: 10.1038/nri3307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Judge AD, Zhang X, Fujii H, Surh CD, Sprent J. Interleukin 15 controls both proliferation and survival of a subset of memory-phenotype CD8(+) T cells. J Exp Med. 2002;196:935–946. doi: 10.1084/jem.20020772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Moran AE, et al. T cell receptor signal strength in Treg and iNKT cell development demonstrated by a novel fluorescent reporter mouse. J Exp Med. 2011;208:1279–1289. doi: 10.1084/jem.20110308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chang JT, et al. Asymmetric Proteasome Segregation as a Mechanism for Unequal Partitioning of the Transcription Factor T-bet during T Lymphocyte Division. Immunity. 2011;34:492–504. doi: 10.1016/j.immuni.2011.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Miller AT, Wilcox HM, Lai Z, Berg LJ. Signaling through Itk promotes T helper 2 differentiation via negative regulation of T-bet. Immunity. 2004;21:67–80. doi: 10.1016/j.immuni.2004.06.009. [DOI] [PubMed] [Google Scholar]
- 33.Dorner BG, et al. Selective expression of the chemokine receptor XCR1 on cross-presenting dendritic cells determines cooperation with CD8+ T cells. Immunity. 2009;31:823–833. doi: 10.1016/j.immuni.2009.08.027. [DOI] [PubMed] [Google Scholar]
- 34.Best JA, et al. Transcriptional insights into the CD8(+) T cell response to infection and memory T cell formation. Nature immunology. 2013;14:404–412. doi: 10.1038/ni.2536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Haluszczak C, et al. The antigen-specific CD8+ T cell repertoire in unimmunized mice includes memory phenotype cells bearing markers of homeostatic expansion. J Exp Med. 2009;206:435–448. doi: 10.1084/jem.20081829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sung JH, et al. Chemokine guidance of central memory T cells is critical for antiviral recall responses in lymph nodes. Cell. 2012;150:1249–1263. doi: 10.1016/j.cell.2012.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kastenmuller W, et al. Peripheral prepositioning and local CXCL9 chemokine-mediated guidance orchestrate rapid memory CD8+ T cell responses in the lymph node. Immunity. 2013;38:502–513. doi: 10.1016/j.immuni.2012.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Starbeck-Miller GR, Xue HH, Harty JT. IL-12 and type I interferon prolong the division of activated CD8 T cells by maintaining high-affinity IL-2 signaling in vivo. J Exp Med. 2013;211:105–120. doi: 10.1084/jem.20130901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Malek TR, Castro I. Interleukin-2 receptor signaling: at the interface between tolerance and immunity. Immunity. 2010;33:153–165. doi: 10.1016/j.immuni.2010.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Buchholz VR, et al. Disparate individual fates compose robust CD8+ T cell immunity. Science. 2013;340:630–635. doi: 10.1126/science.1235454. [DOI] [PubMed] [Google Scholar]
- 41.Gerlach C, et al. Heterogeneous differentiation patterns of individual CD8+ T cells. Science. 2013;340:635–639. doi: 10.1126/science.1235487. [DOI] [PubMed] [Google Scholar]
- 42.van Heijst JW, et al. Recruitment of antigen-specific CD8+ T cells in response to infection is markedly efficient. Science. 2009;325:1265–1269. doi: 10.1126/science.1175455. [DOI] [PubMed] [Google Scholar]
- 43.Daniels MA, Jameson SC. Critical role for CD8 in T cell receptor binding and activation by peptide/major histocompatibility complex multimers. J Exp Med. 2000;191:335–346. doi: 10.1084/jem.191.2.335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Haring JS, Badovinac VP, Harty JT. Inflaming the CD8+ T cell response. Immunity. 2006;25:19–29. doi: 10.1016/j.immuni.2006.07.001. [DOI] [PubMed] [Google Scholar]
- 45.Cui W, Kaech SM. Generation of effector CD8+ T cells and their conversion to memory T cells. Immunological reviews. 2010;236:151–166. doi: 10.1111/j.1600-065X.2010.00926.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Badovinac VP, Messingham KA, Jabbari A, Haring JS, Harty JT. Accelerated CD8+ T-cell memory and prime-boost response after dendritic-cell vaccination. Nat Med. 2005;11:748–756. doi: 10.1038/nm1257. [DOI] [PubMed] [Google Scholar]
- 47.Curtsinger JM, Mescher MF. Inflammatory cytokines as a third signal for T cell activation. Curr Opin Immunol. 2010;22:333–340. doi: 10.1016/j.coi.2010.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Maile R, et al. Peripheral “CD8 tuning” dynamically modulates the size and responsiveness of an antigen-specific T cell pool in vivo. Journal of immunology. 2005;174:619–627. doi: 10.4049/jimmunol.174.2.619. [DOI] [PubMed] [Google Scholar]
- 49.Pham NL, Badovinac VP, Harty JT. A default pathway of memory CD8 T cell differentiation after dendritic cell immunization is deflected by encounter with inflammatory cytokines during antigen-driven proliferation. J Immunol. 2009;183:2337–2348. doi: 10.4049/jimmunol.0901203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhu J, et al. The transcription factor T-bet is induced by multiple pathways and prevents an endogenous Th2 cell program during Th1 cell responses. Immunity. 2012;37:660–673. doi: 10.1016/j.immuni.2012.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pircher H, Rohrer UH, Moskophidis D, Zinkernagel RM, Hengartner H. Lower receptor avidity required for thymic clonal deletion than for effector T-cell function. Nature. 1991;351:482–485. doi: 10.1038/351482a0. [DOI] [PubMed] [Google Scholar]
- 52.Hogquist KA, et al. T cell receptor antagonist peptides induce positive selection. Cell. 1994;76:17–27. doi: 10.1016/0092-8674(94)90169-4. [DOI] [PubMed] [Google Scholar]
- 53.Brundage RA, Smith GA, Camilli A, Theriot JA, Portnoy DA. Expression and phosphorylation of the Listeria monocytogenes ActA protein in mammalian cells. Proceedings of the National Academy of Sciences of the United States of America. 1993;90:11890–11894. doi: 10.1073/pnas.90.24.11890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Maile R, et al. Antigen-specific modulation of an immune response by in vivo administration of soluble MHC class I tetramers. Journal of immunology. 2001;167:3708–3714. doi: 10.4049/jimmunol.167.7.3708. [DOI] [PubMed] [Google Scholar]
- 55.Dorner BG, et al. MIP-1alpha, MIP-1beta, RANTES, and ATAC/lymphotactin function together with IFN-gamma as type 1 cytokines. Proc Natl Acad Sci U S A. 2002;99:6181–6186. doi: 10.1073/pnas.092141999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kieper WC, Prlic M, Schmidt CS, Mescher MF, Jameson SC. Il-12 enhances CD8 T cell homeostatic expansion. Journal of immunology. 2001;166:5515–5521. doi: 10.4049/jimmunol.166.9.5515. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.