Abstract
Hematopoiesis is maintained throughout life by self-renewing hematopoietic stem cells (HSCs) that differentiate to produce both myeloid and lymphoid cells. The NR4A family of orphan nuclear receptors, which regulates cell fate in many tissues, appears to play a key role in HSC proliferation and differentiation. Using a NR4A1GFP BAC transgenic reporter mouse we have investigated NR4A1 expression and its regulation in early hematopoiesis. We show that NR4A1 is most highly expressed in a subset of Lin−Sca-1+c-Kit+ CD48−CD150+ long-term (LT) HSCs, and its expression is tightly associated with HSC quiescence. We also show that NR4A1 expression in HSCs is induced by PGE2, a known enhancer of stem cell engraftment potential. Finally, we find that both NR4A1GFP+ and NR4A1GFP− HSCs successfully engraft primary and secondary irradiated hosts; however, NR4A1GFP+ HSCs are distinctly myeloid-biased. These results show that NR4A1 expression identifies a highly quiescent and distinct population of myeloid-biased LT-HSCs.
Keywords: hematopoiesis, hematopoietic stem cell, NR4A1 (Nur77), PGE2, myeloid bias, quiescence
Introduction
Hematopoietic stem cells (HSCs) maintain the blood system through a delicate balance of quiescence, self-renewal, and differentiation. Although both cell intrinsic and extrinsic factors contribute to preserving this balance, the precise regulatory mechanisms involved remain poorly understood. The most primitive HSCs, long-term (LT) HSCs, are thought to be the most quiescent and are defined by their ability to maintain the blood throughout the lifetime of an organism [1]. LT-HSCs, which are a subset of lineage−c-Kit+Sca-1+ (LSK) bone marrow cells have been identified by the differential expression of several surface proteins: LSKs with a CD150+CD48− phenotype (LT-HSC) [2], and LSKs with a CD34−CD135− [3, 4] have both been associated with LT-HSC function.
While the LT-HSC population defined by these phenotypic markers is enriched for cells with stem cell activity, this pool of cells nevertheless is functionally heterogeneous, underscoring one limitation of defining stem potential by surface phenotype alone. For example, LT-HSCs expressing the highest levels of CD150 appear to have more potent self-renewal ability and, interestingly, display a myeloid-skewed reconstitution pattern upon transplantation, while CD150− HSCs produce lymphoid-skewed reconstitution [5]. Similar LT-HSC heterogeneity was observed by Benz et al. who used single-cell transplants of LT-HSCs to identify three distinct patterns of HSC expansion and differentiation [6]. Another limitation of defining LT-HSC subpopulations strictly on the basis of surface phenotype is that the surface proteins are only indirectly linked to intracellular regulatory pathways and therefore provide limited information about a cell’s internal state.
Recent studies have suggested that the NR4A family of orphan nuclear hormone receptors may play a critical role in integrating the molecular pathways essential for normal HSC quiescence and regulated differentiation. NR4A1, in particular, is an early response gene whose expression levels are exquisitely sensitive to diverse external stimuli including physical stresses, growth factors, prostaglandins, inflammatory cytokines and neurotransmitters [7-10]. NR4A1’s activity is both transcriptionally and post-translationally controlled. It is active as a transcription factor in the nucleus and as a modifier of bcl-2 at the mitochondria and has been shown to regulate cell fate via influences on apoptosis, metabolism, and proliferation [11] [12-17].
Within the hematopoietic system, NR4A1 is expressed at high levels by a subset of mature monocytes (the “patrolling monocytes”) [18] and has been detected by PCR analysis in LSKs [19]. NR4A1’s potential role in regulating hematopoietic proliferation has been suggested by studies showing that mice deficient in two NR4A family members (NR4A1 and NR4A3) die of acute myeloid leukemia (AML) 2-4 weeks after birth [19]. In addition, mice bearing one copy of NR4A1 along with complete deletion of NR4A3, or neither NR4A1 allele and one copy of NR4A3, show increased hematopoietic stem and progenitor cell (HSPC) proliferation and go on to develop mixed myelodysplastic/myeloproliferative neoplasms over weeks and months (MDS/MPNs) [20]. Finally, NR4A1 has also been shown to stabilize and increase the transcriptional activity of HIF-1α[21, 22], which has been implicated in maintaining HSC quiescence and regulating HSC metabolism [23, 24].
Given these observations, we wondered whether NR4A1 might be expressed by a distinct subset of LSKs whose features may include quiescence. To directly address this possibility, we have examined hematopoietic stem and progenitor cells using a NR4A1GFP transgenic (Tg) reporter mouse [10] and show that NR4A1 expression specifies a distinct population of quiescent, myeloid-biased LT-HSCs. Moreover, we find that HSCs upregulate NR4A1 expression in response to in vitro treatment with PGE2, which has been shown to enhance HSC reconstitution potential [25, 26]. We propose that NR4A1 both senses and mediates hematopoietic biochemical pathways that link HSC quiescence, self-renewal and differentiation potential in vivo.
Materials and Methods
Mice
C57BL/6J wild-type mice were obtained from The Jackson Laboratory, Taconic Farms, or Charles River Laboratories International, Inc. NR4A1GFP BAC transgenic reporter mice were previously generated [27] and are available from The Jackson Laboratory (016617). The GFP-Cre fusion protein is located at the start codon of the NR4A1 gene in a BAC construct. NR4A1GFP mice are healthy and undergo normal hematopoiesis, indistinguishable from that of their non-transgenic littermates. B6.SJL mice were obtained from Taconic Farms, Inc. All mouse husbandry and experiments followed the guidelines of the Haverford College, Columbia University, and University of Pennsylvania Animal Care and Use Committees. Mice used were typically between 4 and 30 weeks of age and euthanized by CO2 inhalation.
Flow cytometry
Bone marrow cells were flushed from tibias, femurs, and also sometimes pelvic and humeral bones, using 1X Delbecco’s phosphate buffered saline without calcium or magnesium (DPBS, Gibco) supplemented with 0.1% fatty-acid free bovine serum albumin (BSA, Fisher Scientific). Red blood cells were lysed with 1X ammonium-chloride-potassium (ACK, Lonza) and cells were filtered through sterile nylon mesh (40 or 70μm, Becton Dickinson Falcon) to obtain single cell suspensions. Cells were maintained on ice when possible throughout all procedures. Bone marrow cells were enriched for lineage negative (Lin-) cells by incubating with lineage biotin antibody cocktail containing biotinylated antibodies against lineage markers (CD5, B220, Mac-1, GR-1, 7-4, and Ter119), followed by anti-biotin microbeads (Lineage Cell Depletion Kit, Mouse, Miltenyi Biotec). Lineage positive (Lin+) bone marrow cells were depleted using LS Columns (Miltenyi Biotec) and MidiMACs magnets (Miltenyi Biotec) according to the manufacturer’s instructions. To allow for dead cell exclusion during flow cytometric analysis, lineage negative cells were stained with LIVE/DEAD Aqua Dead cell stain kit according to manufacturer’s instructions (Life Technologies). Cells were then washed with PBS+0.1% BSA and further stained with specific combinations of fluorochrome conjugated anti-mouse antibodies: anti c-Kit-APC-eFluor780 (eBioscience); anti Sca-1-PerCP-Cy5.5 (Biolegend); anti CD150-PE-Cy7 (Biolegend); anti CD48-eFluor450 (eBioscience); APC-conjugated antibodies against lineage antigens Ter119, Mac-1, B220, Gr-1, and CD3 (Biolegend and eBioscience). All antibodies are listed in Table S1. Stained cells were analyzed using a FACSAriaII (Becton Dickinson) or a MACSQuant (Miltenyi Biotec). Cells were sorted using a FACS Aria II cell sorter (Becton Dickinson). All flow cytometry and FACS data were analyzed with FlowJo software (TreeStar Inc.).
Cell cycle analysis
Lin− cells were incubated for 45 min at 37°C, 5% CO2 in 1X PBS+0.1% BSA + 6μg /ml Hoechst 33342 (Invitrogen) and 5μM Fumitremorgin C (Sigma Aldrich) to prevent Hoechst dye efflux [28]. Pyronin Y (Sigma Aldrich, 0.08-0.16μg /ml) was added and cells were incubated for an additional 15 minutes. Cells were washed and surface stained as described previously. Propidium Iodide (PI, 0.02 μg /ml) was used for dead cell exclusion.
5-Fluorouracil treatment
NR4A1GFP transgenic and littermate control mice received a single intraperitoneal injection of 60 or 150mg/kg 5-FU in PBS or an equivalent volume of sterile 1X PBS alone. Forty-two hours later, mice were euthanized by CO2 inhalation and bone marrow was harvested and lineage depleted. Lin-cells were stained for flow cytometric analysis as described previously. LIVE/DEAD Aqua Dead cell stain (Life Technologies) was used for dead cell exclusion.
Bone marrow transplantation
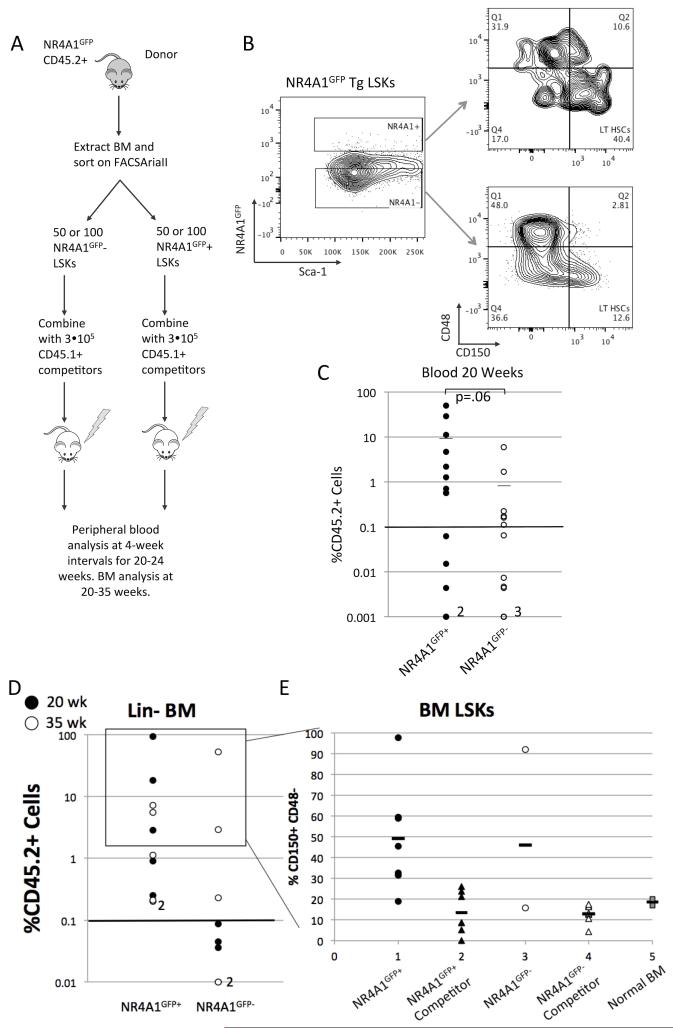
CD45.1+ B6.SJL mice were irradiated with a cesium-137 irradiator in two equal doses of 500 rads separated by at least 30 minutes. Lineage-depleted bone marrow from CD45.2+ NR4A1GFP mice was stained with the following fluorochrome conjugated antibodies: anti Sca-1-PE, anti cKit-APC, biotinylated antibodies against lineage antigens (Ter119, Mac-1, CD3, B220, Gr-1, CD3, CD4, CD8) and streptavidin-PE-Texas Red. NR4A1GFP+ and NR4A1GFP− Lineage-Sca-1+c-Kit+ (LSKs) cells were sorted using a FACSAria II (Becton Dickinson). One hundred or fifty NR4A1GFP+ or NR4A1GFP− LSK cells were injected into lethally irradiated B6.SJL recipient mice, along with 3 ×105 B6.SJL whole bone marrow competitor cells. Peripheral blood was collected every 4 weeks over 20-24 weeks to assess blood chimerism and lineage distribution over time. Red blood cells were lysed using 1X ACK (Lonza) and stained with LIVE/DEAD Aqua Dead cell stain kit (Life Technologies) for dead cell exclusion and the following antibody surface stain: anti CD45.1-PE, anti CD45.2-APC, anti Mac-1-PerCP, anti Gr-1-APC-Cy7, anti CD19-BV421, anti CD3-biotin, anti CD4-biotin, anti CD8-biotin with streptavidin-PE-Cy7. Recipient mice were euthanized at indicated times. Harvested bone marrow was lineage depleted and Lin-cells were surface stained for flow cytometric analysis as described previously. Thirty-five weeks after primary engraftment, the bone marrow of selected primary recipient mice (3 NR4A1GFP+ recipients and 3 NR4A1GFP− recipients) was harvested and 2.5 × 107 whole bone marrow cells were transplanted from each mouse into a corresponding irradiated secondary B6.SJL recipient. The blood of the secondary recipients was analyzed every 4 to 6 weeks as described above.
Short-term cell culture
NR4A1GFP transgenic lineage-depleted bone marrow was incubated for two hours at 37°C, 5% CO2 in PBS+0.1% fatty-acid-free BSA and supplemented with either 1ng/ml SCF (Peprotech), 1ng/ml IFNγ (Peprotech and R & D Systems), 10ng/ml G-CSF (Peprotech), 50ng/ml IGF-1, or 1μM PGE2 (Sigma Aldrich). After incubation, cells were washed and stained for flow cytometric analyses.
Statistical Analyses
P-values were calculated using unpaired, one-tailed student’s t-test.
Results
NR4A1 is preferentially expressed by CD150+CD48− LSK cells in the BM
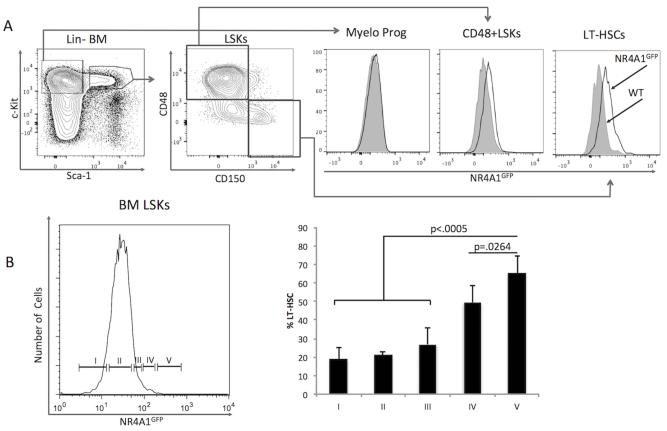
To determine if NR4A1 gene expression might be developmentally regulated during early bone marrow hematopoiesis, we examined NR4A1GFP expression levels in hematopoietic stem and progenitor cell subpopulations using our NR4A1GFP BAC transgenic reporter mouse, in which GFP expression is driven by the NR4A1 promoter [10]. Among Lin- bone marrow cells, NR4A1GFP expression is elevated in the cKit+Sca-1+ (LSK) compartment. Within the LSK population, NR4A1GFP is preferentially expressed in the most immature CD150+CD48− LT-HSC (LT-HSC) subset (Fig. 1A). Reciprocally, partitioning LSKs by their level of NR4A1 expression showed that cells expressing higher levels of NR4A1 were progressively enriched in LT-HSCs (Fig. 1B). Within this LT-HSC compartment, we observed a direct correlation between NR4A1 and CD150 expression levels (Fig. S1). NR4A1GFP was not detected in committed myeloid progenitors (Lin−Sca-1−c-Kit+) (Fig. 1A). Of note, NR4A1 expression by LSKs is considerably lower than that seen among patrolling monocytes [18], yet consistently higher than other mature, Lin+ cells (Fig. S2).
Fig. 1. LT-HSCs express the highest levels of NR4A1.
Lineage-depleted (Lin−) NR4A1GFP transgenic (Tg) reporter mouse bone marrow (BM) cells were isolated, stained and analyzed by flow cytometry. Non-Tg littermates were used as controls (grey). (A) Relative NR4A1GFP expression levels in hematopoietic subpopulations: Myelo Prog, myeloid-committed progenitors (Lin−Sca-1−c-Kit+); LSKs, Lin−Sca-1+c-Kit+ cells; LT-HSCs, long-term hematopoietic stem cells. (B) Proportional contribution of LT-HSC compartment (CD150+CD48−) to LSKs with increasing NR4A1GFP expression levels. Data are from four independent experiments, each using 1-2 NR4A1GFP Tg mice and 1-2 non-Tg littermate control mice. P-values were calculated using unpaired, one-tailed student’s t-test.
NR4A1GFP+ HSCs are quiescent
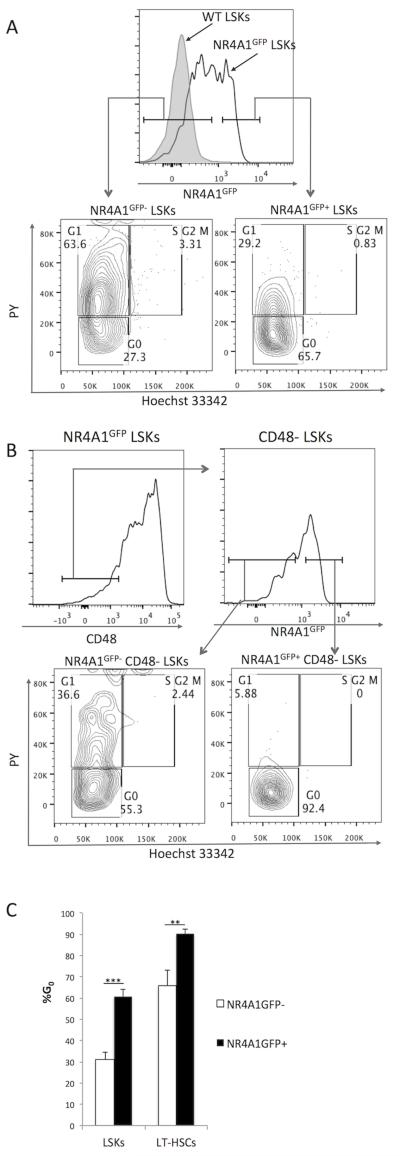
This pattern of NR4A1GFP expression suggested that NR4A1 expression might be associated with functional attributes of immature HSCs, including HSC quiescence. To address this directly, we assessed the cell cycle status of NR4A1GFP+ vs. NR4A1GFP− LSKs, staining Lin− BM cells from NR4A1GFP mice with Hoechst 33342 and Pyronin Y. We found that the majority of NR4A1GFP+ LSKs (61% +/−3.5) were in G0, as compared to only 30% (+/−3.5) of NR4A1GFP− LSKs (Fig. 2A,C). The association between NR4A1GFP expression and quiescence was also evident among LT-HSCs. Among CD48− LSKs, only 65% (+/−7.3) of NR4A1GFP− cells in G0, while almost all (90% +/−2.1) of NR4A1GFP+ cells were in G0 (Fig. 2B, C). Together, these data indicate that NR4A1GFP+ LSKs and LT-HSCs are markedly quiescent.
Fig. 2. NR4A1 expression identifies quiescent LSKs.
Lineage− bone marrow (Lin− BM) cells from NR4A1GFP mice were isolated and stained with Hoechst33342 and Pyronin Y (PY) for cell cycle analysis. (A) Comparison of cell cycle status of NR4A1GFP+ vs. NR4A1GFP− LSKs. (B) Comparison of cell cycle status of CD48− NR4A1GFP+ vs. CD48− NR4A1GFP− LSKs. Results are representative of three independent experiments, each of which analyzed 2-3 NR4A1GFP mice and one non-transgenic littermate control and/or one B6 WT control (Note: The Hoechst and PY staining protocol requires a 60-minute 37°C incubation, which increases baseline NR4A1GFP expression (Fig. S3)). (C) Percentage of NR4A1GFP+ vs. NR4A1GFP− LSKs and CD48-LT-HSCs found in G0 based on Hoechst and PY staining. Results are measurements from four different NR4A1GFP transgenic reporter mice. **p=.0007, ***p<.0001 (unpaired student’s t-test).
5-Fluorouracil treatment in vivo enriches NR4A1GFP+ HSCs
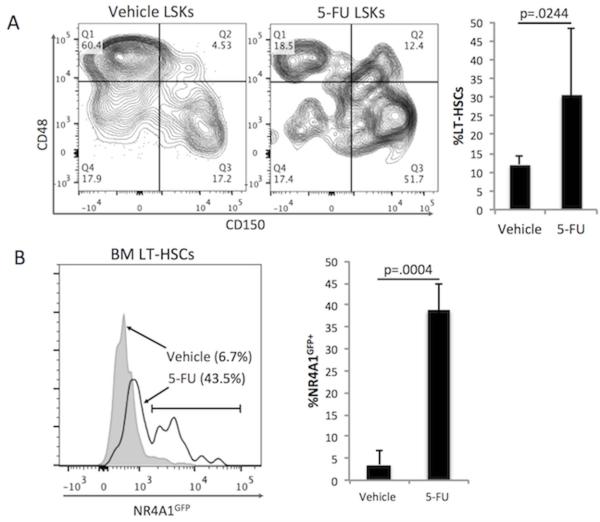
Based on our observations that NR4A1 expression is a feature of non-cycling HSCs, we predicted that NR4A1GFP+ HSCs would be resistant to cell-cycle selective killing. To test this prediction, NR4A1GFP Tg mice were treated with the cell cycle selective chemotherapeutic agent 5-fluorouracil (5-FU), and bone marrow was collected and analyzed after 48 hours. FACS analysis revealed that the BM of 5-FU-treated mice was enriched for LT-HSCs (Fig. 3A). In addition, NR4A1GFP+ cells were enriched within the LT-HSC compartment: an average 39% of 5-FU treated LT-HSCs were NR4A1GFP+ while only 3.6% of vehicle-treated LT-HSCs were NR4A1GFP+ (Fig. 3B). Consistent with our cell cycle results in Figure 2, these data show that 5-FU preferentially spared NR4A1GFP+ LSKs.
Fig. 3. NR4A1GFP+ LSKs are preserved after treatment with 5-Florouracil.
NR4A1GFP Tg reporter mice were treated with 5-fluorouracil (5-FU) or an equivalent volume of PBS via intraperitoneal injection. Mice were euthanized 42h after treatment, and Lin− BM cells were isolated, stained and analyzed by FACS. (A) CD150 and CD48 expression levels of LSK cells from 5-FU treated and control mice (left). The bar graph (right) shows the average proportion of LT-HSCs among LSKs after 5-FU or vehicle treatment (data are from two independent experiments with a total of three 5-FU-treated NR4A1GFP transgenic (Tg) mice; three vehicle-treated Tg mice; two 5-FU-treated littermate control (LMC) mice; two vehicle-treated LMC mice). (B) NR4A1GFP expression of LT-HSCs (CD48−CD150+ LSKs) from 5-FU and vehicle treated mice (left). The bar graph (right) shows the proportion of NR4A1GFP+ LT-HSCs from the three 5-FU-treated Tg mice and the three vehicle-treated Tg mice. P-values were calculated using unpaired, one-tailed student’s t-test.
PGE2 upregulates NR4A1 expression in LSKs
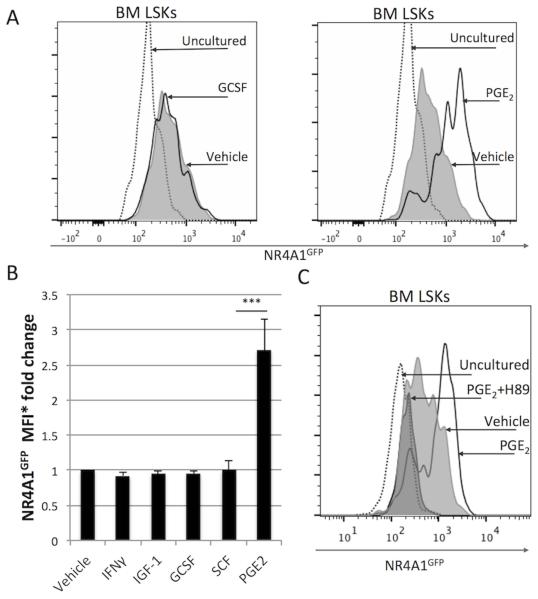
Because the HSC niche is thought to provide factors that maintain HSC quiescence, we hypothesized that the expression of NR4A1 might be directly induced by regulatory molecules produced by bone marrow cells. To test this prediction, we incubated NR4A1GFP Lin− bone marrow cells in vitro for two hours in the presence of selected molecules known to play a role in stem cell proliferation and/or differentiation, including SCF, IFNγ, G-CSF, IGF-1, and PGE2. Culturing HSCs at 37°C increased NR4A1GFP expression to some extent, particularly in the presence of serum. The addition of select polypeptide cytokines did not enhance NR4A1GFP expression; however, the lipid-derived prostaglandin PGE2 potently induced NR4A1GFP expression above that induced by culture alone, increasing expression 3-fold within 90-120’ (Fig. 4A, B and S3). PGE2’s ability to upregulate NR4A1GFP in LSKs was abrogated upon addition of the PKA inhibitor H89 (Fig. 4C), indicating that PGE2 upregulates NR4A1GFP in a PKA-dependent manner. These results, together with Hoggatt et al.’s observations that in vitro PGE2 treatment increases HSC potency [25, 26], suggest that NR4A1 is one molecular indicator of stem cell activity.
Fig. 4. PGE2 upregulates NR4A1 expression in HSCs.
Lin− bone marrow cells were isolated from NR4A1GFP reporter mice and cultured for 2h with either: SCF (1ng/ml), IFNγ (100ng/ml), G-CSF (10ng/ml), IGF-1 (50ng/ml) or PGE2 (1μM). After culture, cells were washed and stained for flow cytometry analysis. (A) NR4A1GFP expression in LSKs after short-term culture with G-CSF (left) or PGE2 (right) versus vehicle-treated and uncultured controls. (B) Change in NR4A1GFP median fluorescence intensity (MFI) in LSKs after short-term culture with factors indicated versus vehicle-treated and uncultured controls. (C) PGE2 induction of NR4A1GFP in LSKs is dependent on protein kinase A (PKA). Lin− bone marrow cells were isolated from NR4A1GFP reporter mice and cultured for 2h in 1μM PGE2 with or without PKA-inhibitor H89. Data are derived from 3 independent experiments, each performed with 1-2 NR4A1GFP transgenic (Tg) mice and 1-2 non-Tg littermate controls. *Median Fluorescence Intensity. ***p=.0003 (unpaired student’s t-test).
NR4A1GFP+ LSKs reconstitute long-term hematopoiesis in irradiated hosts
Given that NR4A1GFP+ LSKs display features of quiescent long-term stem cells, and that NR4A1GFP expression increases upon PGE2 treatment, we predicted that NR4A1 expression identifies HSCs with stem cell repopulating activity. To test this prediction, we assessed the ability of NR4A1GFP+ versus NR4A1GFP− LSKs to reconstitute the blood system following competitive bone marrow transplant into irradiated recipient mice (Fig 5A). We used fluorescence-activated cell sorting (FACS) to isolate 50 or 100 NR4A1GFP+ or NR4A1GFP− CD45.2+ LSKs and injected them into lethally irradiated CD45.1+ B6.SJL recipient mice, along with 3×105 B6.SJL whole bone marrow competitor cells (Fig 5B). Recipient mice were bled every 4 weeks to assess blood chimerism and lineage distribution over time. Recipient mice were euthanized at the end of the experiment and their bone marrow was extracted, lineage depleted and stained for flow cytometry analysis. NR4A1GFP+ LSKs contributed to a significant proportion (>0.1%) of blood cells in 9 out of 14 recipients, while NR4A1GFP− LSKs contributed to a significant proportion of cells in 6 out of 13 recipients (p=.36, unpaired student’s t-test). Donor chimerism in peripheral blood at 20 weeks averaged 7% in mice receiving NR4A1GFP+ LSKs versus 0.6% in mice receiving NR4A1GFP− LSKs (p= 0.06, unpaired student’s t-test) (Fig. 5C). At the end of the engraftment experiment (20 or 35 weeks), bone marrow from surviving mice was analyzed via flow cytometry. Of these mice, NR4A1GFP+ LSKs engrafted the bone marrow of all recipients (10/10), while NR4A1GFP− LSKs engrafted only 3/8 mice (p = 0.0014) (Fig. 5D). Both NR4A1GFP− and NR4A1GFP+ donor cells gave rise to LSKs that were enriched in CD48−CD150+ LT-HSCs. Competitor LSKs in the same recipient mice showed proportions of CD48−CD150+ LT-HSCs that averaged 6.3% of LSKs, similar to their representation in wild type, untransplanted LSK populations (Fig. 5E). Taken together these data suggest that both pools of LSKs include long term repopulating stem cells, although NR4A1GFP+ LSKs may be more enriched for long-term stem cell activity.
Fig. 5. NR4A1GFP+ expression identifies long-term repopulating cells.
(A) Experimental design of BM transplant of NR4A1GFP+ vs. NR4A1GFP− LSKs, as described in materials and methods. (B) Representative CD48 and CD150 profiles of FACS sorted NR4A1GFP+ and NR4A1GFP− LSK populations. (C) Peripheral blood chimerism at 20 weeks from surviving recipients of NR4A1GFP+ LSKs (14 mice) and NR4A1GFP− LSKs (13 mice). Data are pooled from two independent engraftment experiments using a total of 14 NR4A1GFP transgenic donor mice, 2 B6.SJL mice for competitor cells. Two NR4A1GFP+ recipients and 3 NR4A1GFP− recipients had chimerism below .001% as indicated. 9/14 NR4A1GFP+ recipients had greater than 0.1% chimerism in the blood and 6/13 NR4A1GFP− recipients had greater than 0.1% chimerism (p=.362, unpaired student’s t-test). Average CD45.2% chimerism in the blood was 0.64+/−1.7% for NR4A1GFP− recipients and 7.1+/−14.5 for NR4A1GFP+ recipients (p=0.06 unpaired student’s t-test). (D) CD45.2+ percent chimerism in lineage-depleted BM from recipients of NR4A1GFP+ or NR4A1GFP− LSKs. Filled circles represent mice that were euthanized after 20wks post-injection and open circles represent mice from an independent engraftment experiment that were euthanized after 35 weeks post-injection. 10/10 NR4A1GFP+ recipients had greater than 0.1% chimerism in the BM and 3/8 NR4A1GFP− recipients had greater than 0.1% engraftment. (E) Proportion of donor-derived and competitor-derived LSKs with a LT-HSC phenotype (LSK CD150+CD48−) from the BM of mice that received NR4A1GFP+ LSKs or NR4A1GFP− LSKs, or from the BM of non-irradiated, non-transplanted NR4A1GFP mice (“Normal BM”). The box surrounding data points in D indicates those mice from which it was possible to obtain sufficient numbers of CD45.2+ LSKs for the analysis in part E.
NR4A1 expression in LSKs identifies a myeloid-biased LT-HSC
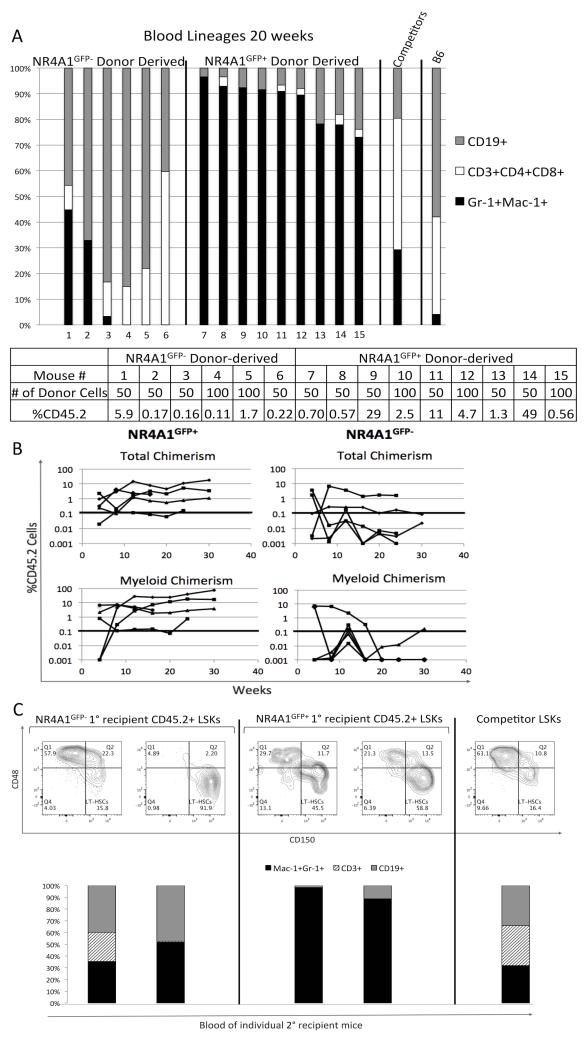
Recent studies suggest that the LT-HSC compartment is heterogeneous in terms of myeloid versus lymphoid lineage potential [29-32]. To examine the lineage distribution of cells derived from NR4A1GFP+ or NR4A1GFP− LSKs, we determined the expression of lineage-associated surface molecules Gr-1, Mac-1, CD4/CD3/CD8, and CD19 on CD45.2+ circulating mature blood cells from recipient mice at 20 weeks post-transplant. Strikingly, we found that donor-derived cells in mice transplanted with NR4A1GFP+ LSKs were markedly skewed toward the myeloid lineage (Figure 6A); the average lymphoid to myeloid ratio (ρ) was 0.27, indicating that they are myeloid biased [29]. On the other hand, NR4A1GFP− LSKs that successfully contributed to hematopoiesis after 20 weeks generated predominantly T and B lymphocytes. NR4A1GFP+ LSKs produced a %myeloid of 86.9%+/−8.3, while NR4A1GFP− LSKs produced a %myeloid of 13.5+/−20 (p<.0001, unpaired student’s t-test. See Fig 6A legend for definition of %myeloid).
Fig. 6. NR4A1GFP+ LSKs are myeloid-biased.
Kinetic analyses showed that the differences in the ability of NR4A1GFP+ versus NR4A1GFP− LSKs to reconstitute myeloid populations were evident as early as 8 weeks after transplantation. The proportion of CD45.2+ myeloid cells in these NR4A1GFP+ recipients increased over 12 weeks and remained high through 30 weeks, while the proportion of CD45.2+ myeloid cells in NR4A1GFP− recipients was negligible by 20 weeks after transplantation (Fig. 6B).
To determine if myeloid bias persists among HSCs derived from NR4A1GFP+ LSKs we performed secondary engraftments using bone marrow from primary recipients of NR4A1GFP+ and NR4A1GFP− LSKs. Fourteen weeks after engraftment, we found that two of three secondary recipients in each group revealed blood chimerism, confirming that both NR4A1GFP+ and NR4A1GFP− LSKs include cells with long-term repopulating potential. Notably, peripheral blood cells derived from CD45.2+ NR4A1GFP+ LSKs maintained a myeloid bias (Fig. 6C). Together, these findings suggest that NR4A1GFP+ LSKs are enriched for a myeloid-biased, long-term repopulating HSC.
Discussion
In this study, we show that NR4A1 expression identifies a subpopulation of quiescent long term HSCs. We show that PGE2, which improves hematopoietic stem and progenitor cell engraftment [25, 26, 33], induces NR4A1 expression in HSCs and progenitors. Our bone marrow transplant experiments show that both NR4A1GFP− and NR4A1GFP+ LSKs successfully engraft irradiated primary and secondary hosts, although NR4A1GFP+ LSKs may include a higher proportion of long-term repopulating cells. Unlike NR4A1GFP− LSKs, NR4A1GFP+ LSKs are markedly myeloid-biased after transplantation.
The myeloid bias of NR4A1GFP+ HSCs, coupled with the observation that NR4A1GFP+ HSC-derived LSKs have long-term repopulating ability, suggest that NR4A1GFP+ HSCs represent a distinct subpopulation of myeloid-biased LT-HSCs. These data raise the possibility that NR4A1GFP+ HSCs undergo an alternate developmental pathway such as the “myeloid by-pass” model proposed by Yamamoto et al., in which particular HSCs differentiate directly into self-renewing myeloid-restricted repopulating progenitors (MyRPs) [34]. These data, combined with indications that NR4A1GFP−-derived cells are lymphoid skewed, support the notion of distinct Ly-bi and My-bi subsets of LT-HSCs. Weksberg et al. find that Ly-bi, Hoechst side-population (SP)-CD150− HSCs cycle more actively, while My-bi, SP-CD150+ HSCs, like NR4A1GFP+ HSCs, are more quiescent [5]. Together, these observations suggest that NR4A1 expression may be used to differentiate between My-bi and Ly-bi subpopulations of HSCs.
NR4A1 transcription is rapidly induced by external stimuli, so its expression may report features of HSC microenvironments. NR4A1GFP expression may therefore reflect residence in a particular niche that maintains quiescence and fosters myeloid bias. We found that PGE2 upregulates NR4A1, suggesting that cellular components of the stem cell niche including PGE2-secreting osteoblasts [35] or endothelial cells may help to maintain NR4A1 levels and thereby maintain HSC potency [36]. Interestingly, one recently published report suggests that PGE2’s impact on HSC potency is mediated by stabilization of Hypoxia-Inducible Factor-1α (HIF-1α) [37]. Since studies in diverse cell types indicate that NR4A1 and HIF-1α can reciprocally stabilize one another’s activity [21, 22], and both NR4A1 and HIF-1α have been shown to regulate genes associated with glycolysis [13, 38-40], our results suggest the possibility that NR4A1 may be part of a transcriptional network that regulates HSC stem cell function by influencing metabolic activity.
That NR4A1 expression is regulated in a PKA dependent manner is consistent with a number of observations in other tissues showing that NR4A1 expression is induced by several receptor ligand interactions that activate the PKA/CREB pathway, including β-epinephrine [7, 41, 42]. Our preliminary observations suggest that, indeed, epinephrine also upregulates NR4A1 in HSCs and indicate that PGE2 may be only one of several molecules that can induce NR4A1 expression in the bone marrow niche. This may explain the increase in NR4A1 expression that we see when culturing cells in medium that includes serum, which likely contains diverse range of stimulating molecules.
How might we understand these observations in light of the data showing that the absence of NR4A1 and its family members rapidly leads to acute myeloid leukemia? Our data showing an association between NR4A1 expression and quiescence among myeloid biased stem cells suggests that it is part of network that provides a check on proliferative activity of immature myeloid precursors. In its absence, and the absence of family members that share and/or complement its activity, precursors may be free to divide without adequate regulation.
In summary, expression of the orphan nuclear hormone receptor NR4A1 specifies a quiescent, myeloid-biased LT-HSC. NR4A1 expression in HSCs is inducible by PGE2 in a PKA dependent manner. NR4A1GFP+ LSKs maintain myeloid-biased blood production through primary and secondary engraftment, suggesting that NR4A1GFP+ HSCs represent a functionally distinct long-term repopulating subpopulation. Their NR4A1GFP− HSC counterparts also include cells with long-term repopulating potential, but do not exhibit myeloid bias. Rather, NR4A1GFP− HSC these cells repopulate with more lymphoid skewing in primary recipients, and maintain their lineage repopulating proportions in secondary transplants, as well. Together with the elegant genetic knockout experiments showing that NR4A family members regulate myeloid leukemia development [19, 20] these data suggest that NR4A1 is one of a network of regulators of quiescence and reconstitution potential, integrating key aspects of HSC biology. Further studies of this family of orphan nuclear hormone receptors in HSCs may reveal significant insights into the integration of quiescence, metabolism, and lineage differentiation.
Supplementary Material
Fig. S1 NR4A1 expression levels correlate with CD150 expression among LSKs. Lineage-depleted (Lin−) NR4A1GFP Transgenic (Tg) reporter mouse bone marrow (BM) cells were isolated, stained and analyzed by flow cytometry. LSKs: Lin−Sca-1+c-Kit+ cells; LT-HSCs: long-term hematopoietic stem cells.
Fig. S2 Comparison of NR4A1GFP expression among bone marrow subpopulations. The cell population with highest expression of NR4A1GFP expression among Lin+ bone marrow cells (dotted line) corresponds to patrolling monocytes (data not shown). LSKs express low levels of NR4A1. NR4A1GFP expression in non-Tg littermate control Lin+ bone marrow is shown in grey. Data representative of more than five independent experiments. For further data on NR4A1GFP expression in patrolling monocytes, see Hanna et al. [18].
Fig. S3 NR4A1GFP expression increases during cell culture. Lineage-depleted BM was cultured at 37°C in PBS+0.1% BSA or PBS+10% FCSs for two hours and NR4A1GFP expression was compared to cells kept 4°C in PBS+0.1% BSA for 2 hours. Data are representative of more than five independent experiments.
Acknowledgements
We thank George Neusch for his tireless patience, thoughtful discussion and flow cytometry assistance; Alevtina Domashenko for sharing her laboratory techniques and her attention to detail; Jonathan Maltzman for valuable discussion and advice; and Michelle Nguyen-McCarty, for her help with cell purification techniques.
This material is based upon work supported by the National Science Foundation under grant NSF-RUI #IOS-1157986, and funding from HHMI, Haverford College, and grants from the NIH and NCI (2R01CA090833, 5P30CA013696). PSK and JH were supported by a grant from the NIH (1R01HL110806).
Funding:
This material is based upon work supported by the National Science Foundation under grant NSF-RUI #IOS-1157986 (JP), HHMI (JP), and grants from the NIH and NCI (2R01CA090833 (SE), 5P30CA013696 (SE), 1R01HL110806 (PK)).
Footnotes
Author Contributions:
Ruben H. Land, Anna K. Rayne, Ashley N. Vanderbeck, Trevor S. Barlowe, Shwetha Manjunath, Matthew Gross, Sophie Eiger, Nicole R. Cunningham, Jian Huang, Stephen Emerson, Jennifer Punt: Conception and design.
Ruben H. Land, Anna K. Rayne, Ashley N. Vanderbeck, Trevor S. Barlowe, Shwetha Manjunath, Matthew Gross, Sophie Eiger, Jian Huang: Collection and/or assembly of data
Peter Klein, Jian Huang: Provision of Study Materials
Ruben H. Land, Anna K. Rayne, Ashley N. Vanderbeck, Trevor S. Barlowe, Shwetha Manjunath, Matthew Gross, Sophie Eiger, Stephen Emerson, Jennifer Punt: Data analysis and interpretation.
Ruben H. Land, Ashley N. Vanderbeck, Stephen G. Emerson, Jennifer Punt: manuscript writing
Conflict-of-interest disclosure: All authors declare no competing financial interests.
References
- 1.Copley MR, Beer PA, Eaves CJ. Hematopoietic Stem Cell Heterogeneity Takes Center Stage. Cell Stem Cell. 2012;10:690–697. doi: 10.1016/j.stem.2012.05.006. [DOI] [PubMed] [Google Scholar]
- 2.Kiel MJ, Yilmaz OH, Iwashita T, et al. SLAM family receptors distinguish hematopoietic stem and progenitor cells and reveal endothelial niches for stem cells. Cell. 2005;121:1109–1121. doi: 10.1016/j.cell.2005.05.026. [DOI] [PubMed] [Google Scholar]
- 3.Osawa M, Hanada K, Hamada H, et al. Long-term lymphohematopoietic reconstitution by a single CD34-low/negative hematopoietic stem cell. Science. 1996;273:242–245. doi: 10.1126/science.273.5272.242. [DOI] [PubMed] [Google Scholar]
- 4.Sudo K, Ema H, Morita Y, et al. Age-associated characteristics of murine hematopoietic stem cells. J Exp Med. 2000;192:1273–1280. doi: 10.1084/jem.192.9.1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Weksberg DC, Chambers SM, Boles NC, et al. CD150-side population cells represent a functionally distinct population of long-term hematopoietic stem cells. Blood. 2008;111:2444–2451. doi: 10.1182/blood-2007-09-115006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Benz C, Copley MR, Kent DG, et al. Hematopoietic stem cell subtypes expand differentially during development and display distinct lymphopoietic programs. Cell Stem Cell. 2012;10:273–283. doi: 10.1016/j.stem.2012.02.007. [DOI] [PubMed] [Google Scholar]
- 7.Maxwell MA, Cleasby ME, Harding A, et al. Nur77 regulates lipolysis in skeletal muscle cells. Evidence for cross-talk between the beta-adrenergic and an orphan nuclear hormone receptor pathway. J Biol Chem. 2005;280:12573–12584. doi: 10.1074/jbc.M409580200. [DOI] [PubMed] [Google Scholar]
- 8.Maxwell MA, Muscat GE. The NR4A subgroup: immediate early response genes with pleiotropic physiological roles. Nucl Recept Signal. 2006;4:e002. doi: 10.1621/nrs.04002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pearen MA, Ryall JG, Maxwell MA, et al. The orphan nuclear receptor, NOR-1, is a target of beta-adrenergic signaling in skeletal muscle. Endocrinology. 2006;147:5217–5227. doi: 10.1210/en.2006-0447. [DOI] [PubMed] [Google Scholar]
- 10.Moran AE, Holzapfel KL, Xing Y, et al. T cell receptor signal strength in Treg and iNKT cell development demonstrated by a novel fluorescent reporter mouse. J Exp Med. 2011;208:1279–1289. doi: 10.1084/jem.20110308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li H, Kolluri SK, Gu J, et al. Cytochrome c release and apoptosis induced by mitochondrial targeting of nuclear orphan receptor TR3. Science. 2000;289:1159–1164. doi: 10.1126/science.289.5482.1159. [DOI] [PubMed] [Google Scholar]
- 12.Cunningham NR, Artim SC, Fornadel CM, et al. Immature CD4+CD8+ thymocytes and mature T cells regulate Nur77 distinctly in response to TCR stimulation. J Immunol. 2006;177:6660–6666. doi: 10.4049/jimmunol.177.10.6660. [DOI] [PubMed] [Google Scholar]
- 13.Chao LC, Wroblewski K, Zhang Z, et al. Insulin resistance and altered systemic glucose metabolism in mice lacking Nur77. Diabetes. 2009;58:2788–2796. doi: 10.2337/db09-0763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fassett MS, Jiang W, D’Alise AM, et al. Nuclear receptor Nr4a1 modulates both regulatory T-cell (Treg) differentiation and clonal deletion. Proc Natl Acad Sci U S A. 2012;109:3891–3896. doi: 10.1073/pnas.1200090109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rajpal A, Cho YA, Yelent B, et al. Transcriptional activation of known and novel apoptotic pathways by Nur77 orphan steroid receptor. EMBO J. 2003;22:6526–6536. doi: 10.1093/emboj/cdg620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liang B, Song X, Liu G, et al. Involvement of TR3/Nur77 translocation to the endoplasmic reticulum in ER stress-induced apoptosis. Exp Cell Res. 2007;313:2833–2844. doi: 10.1016/j.yexcr.2007.04.032. [DOI] [PubMed] [Google Scholar]
- 17.Thompson J, Winoto A. During negative selection, Nur77 family proteins translocate to mitochondria where they associate with Bcl-2 and expose its proapoptotic BH3 domain. J Exp Med. 2008;205:1029–1036. doi: 10.1084/jem.20080101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hanna RN, Carlin LM, Hubbeling HG, et al. The transcription factor NR4A1 (Nur77) controls bone marrow differentiation and the survival of Ly6C-monocytes. Nat Immunol. 2011;12:778–785. doi: 10.1038/ni.2063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mullican SE, Zhang S, Konopleva M, et al. Abrogation of nuclear receptors Nr4a3 and Nr4a1 leads to development of acute myeloid leukemia. Nat Med. 2007;13:730–735. doi: 10.1038/nm1579. [DOI] [PubMed] [Google Scholar]
- 20.Ramirez-Herrick AM, Mullican SE, Sheehan AM, et al. Reduced NR4A gene dosage leads to mixed myelodysplastic/myeloproliferative neoplasms in mice. Blood. 2011;117:2681–2690. doi: 10.1182/blood-2010-02-267906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim BY, Kim H, Cho EJ, et al. Nur77 upregulates HIF-alpha by inhibiting pVHL-mediated degradation. Exp Mol Med. 2008;40:71–83. doi: 10.3858/emm.2008.40.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yoo YG, Yeo MG, Kim DK, et al. Novel function of orphan nuclear receptor Nur77 in stabilizing hypoxia-inducible factor-1alpha. J Biol Chem. 2004;279:53365–53373. doi: 10.1074/jbc.M408554200. [DOI] [PubMed] [Google Scholar]
- 23.Eliasson P, Rehn M, Hammar P, et al. Hypoxia mediates low cell-cycle activity and increases the proportion of long-term-reconstituting hematopoietic stem cells during in vitro culture. Exp Hematol. 2010;38:301–310. e302. doi: 10.1016/j.exphem.2010.01.005. [DOI] [PubMed] [Google Scholar]
- 24.Takubo K, Goda N, Yamada W, et al. Regulation of the HIF-1alpha level is essential for hematopoietic stem cells. Cell Stem Cell. 2010;7:391–402. doi: 10.1016/j.stem.2010.06.020. [DOI] [PubMed] [Google Scholar]
- 25.Hoggatt J, Singh P, Sampath J, et al. Prostaglandin E2 enhances hematopoietic stem cell homing, survival, and proliferation. Blood. 2009;113:5444–5455. doi: 10.1182/blood-2009-01-201335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Goessling W, North TE, Loewer S, et al. Genetic interaction of PGE2 and Wnt signaling regulates developmental specification of stem cells and regeneration. Cell. 2009;136:1136–1147. doi: 10.1016/j.cell.2009.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moran AE, Holzapfel KL, Xing Y, et al. T cell receptor signal strength in Treg and iNKT cell development demonstrated by a novel fluorescent reporter mouse. J Exp Med. 2011;208:1279–1289. doi: 10.1084/jem.20110308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Scharenberg CW, Harkey MA, Torok-Storb B. The ABCG2 transporter is an efficient Hoechst 33342 efflux pump and is preferentially expressed by immature human hematopoietic progenitors. Blood. 2002;99:507–512. doi: 10.1182/blood.v99.2.507. [DOI] [PubMed] [Google Scholar]
- 29.Müller-Sieburg CE, Cho RH, Thoman M, et al. Deterministic regulation of hematopoietic stem cell self-renewal and differentiation. Blood. 2002;100:1302–1309. [PubMed] [Google Scholar]
- 30.Dykstra B, Kent D, Bowie M, et al. Long-term propagation of distinct hematopoietic differentiation programs in vivo. Cell Stem Cell. 2007;1:218–229. doi: 10.1016/j.stem.2007.05.015. [DOI] [PubMed] [Google Scholar]
- 31.Morita Y, Ema H, Nakauchi H. Heterogeneity and hierarchy within the most primitive hematopoietic stem cell compartment. Journal of Experimental Medicine. 2010;207:1173–1182. doi: 10.1084/jem.20091318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Oguro H, Ding L, Morrison SJ. SLAM Family Markers Resolve Functionally Distinct Subpopulations of Hematopoietic Stem Cells and Multipotent Progenitors. Cell Stem Cell. 2013;13:102–116. doi: 10.1016/j.stem.2013.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hoggatt J, Mohammad KS, Singh P, et al. Differential stem- and progenitor-cell trafficking by prostaglandin E2. Nature. 2013;495:365–369. doi: 10.1038/nature11929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yamamoto R, Morita Y, Ooehara J, et al. Clonal analysis unveils self-renewing lineage-restricted progenitors generated directly from hematopoietic stem cells. Cell. 2013;154:1112–1126. doi: 10.1016/j.cell.2013.08.007. [DOI] [PubMed] [Google Scholar]
- 35.Inada M, Matsumoto C, Uematsu S, et al. Membrane-bound prostaglandin E synthase-1-mediated prostaglandin E2 production by osteoblast plays a critical role in lipopolysaccharide-induced bone loss associated with inflammation. J Immunol. 2006;177:1879–1885. doi: 10.4049/jimmunol.177.3.1879. [DOI] [PubMed] [Google Scholar]
- 36.Wang Y, Krivtsov AV, Sinha AU, et al. The Wnt/beta-catenin pathway is required for the development of leukemia stem cells in AML. Science. 2010;327:1650–1653. doi: 10.1126/science.1186624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Speth JM, Hoggatt J, Singh P, et al. Pharmacologic increase in HIF1α enhances hematopoietic stem and progenitor homing and engraftment. Blood. 2014;123:203–207. doi: 10.1182/blood-2013-07-516336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Iyer NV, Kotch LE, Agani F, et al. Cellular and developmental control of O2 homeostasis by hypoxia-inducible factor 1 alpha. Genes Dev. 1998;12:149–162. doi: 10.1101/gad.12.2.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Simsek T, Kocabas F, Zheng J, et al. The distinct metabolic profile of hematopoietic stem cells reflects their location in a hypoxic niche. Cell Stem Cell. 2010;7:380–390. doi: 10.1016/j.stem.2010.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chao LC, Zhang Z, Pei L, et al. Nur77 coordinately regulates expression of genes linked to glucose metabolism in skeletal muscle. Mol Endocrinol. 2007;21:2152–2163. doi: 10.1210/me.2007-0169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kanzleiter T, Wilks D, Preston E, et al. Regulation of the nuclear hormone receptor nur77 in muscle: influence of exercise-activated pathways in vitro and obesity in vivo. Biochim Biophys Acta. 2009;1792:777–782. doi: 10.1016/j.bbadis.2009.05.002. [DOI] [PubMed] [Google Scholar]
- 42.Maira M, Martens C, Batsché E, et al. Dimer-specific potentiation of NGFI-B (Nur77) transcriptional activity by the protein kinase A pathway and AF-1-dependent coactivator recruitment. Mol Cell Biol. 2003;23:763–776. doi: 10.1128/MCB.23.3.763-776.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 NR4A1 expression levels correlate with CD150 expression among LSKs. Lineage-depleted (Lin−) NR4A1GFP Transgenic (Tg) reporter mouse bone marrow (BM) cells were isolated, stained and analyzed by flow cytometry. LSKs: Lin−Sca-1+c-Kit+ cells; LT-HSCs: long-term hematopoietic stem cells.
Fig. S2 Comparison of NR4A1GFP expression among bone marrow subpopulations. The cell population with highest expression of NR4A1GFP expression among Lin+ bone marrow cells (dotted line) corresponds to patrolling monocytes (data not shown). LSKs express low levels of NR4A1. NR4A1GFP expression in non-Tg littermate control Lin+ bone marrow is shown in grey. Data representative of more than five independent experiments. For further data on NR4A1GFP expression in patrolling monocytes, see Hanna et al. [18].
Fig. S3 NR4A1GFP expression increases during cell culture. Lineage-depleted BM was cultured at 37°C in PBS+0.1% BSA or PBS+10% FCSs for two hours and NR4A1GFP expression was compared to cells kept 4°C in PBS+0.1% BSA for 2 hours. Data are representative of more than five independent experiments.