Abstract
Disorders of cobalamin deficiency are a heterogeneous group of disorders with at least 19 autosomal recessive-associated genes. Familial samples of an infant who died due to presumed cobalamin deficiency were referred for clinical exome sequencing. The patient died before obtaining a blood sample or skin biopsy, autopsy was declined, and DNA yielded from the newborn screening blood spot was insufficient for diagnostic testing. Whole-exome sequencing of the mother, father, and unaffected sister and tailored bioinformatics analysis was applied to search for mutations in underlying disorders with recessive inheritance. This approach identified alterations within two genes, each of which was carried by one parent. The mother carried a missense alteration in the MTR gene (c.3518C>T; p.P1173L) which was absent in the father and the sister. The father carried a translational frameshift alteration in the LMBRD1 gene (c.1056delG; p.L352Lfs*18) which was absent in the mother and present in the heterozygous state in the sister. These mutations in the MTR (MIM# 156570) and LMBRD1 (MIM# 612625) genes have been described in patients with disorders of cobalamin metabolism complementation groups cblG and cblF, respectively. The child’s clinical presentation and biochemical results demonstrated overlap with both cblG and cblF. Sanger sequencing using DNA from the infant’s blood spot confirmed the inheritance of the two alterations in compound heterozygous form. We present the first example of exome sequencing leading to a diagnosis in the absence of the affected patient. Furthermore, the data support the possibility for potential digenic inheritance associated with cobalamin deficiency.
Electronic supplementary material
The online version of this chapter (doi:10.1007/8904_2014_294) contains supplementary material, which is available to authorized users.
Introduction
The inborn errors of intracellular cobalamin metabolism are a group of clinically and genetically heterogeneous disorders inherited in an autosomal recessive manner and have been categorized based on complementation analysis and designated mut (MIM #251000), cblA-cblG (MIM #25110, 277400, 277410, 236270, 277380, 250940) (reviewed in Watkins and Rosenblatt 2011), CblJ (MIM #614857) (Coelho et al. 2012), and CblX (Yu et al. 2013). Cobalamin deficiency generally leads to an accumulation of methylmalonic acid and homocysteine in the blood and urine stemming from decreased activity of the enzymes methylmalonyl-CoA mutase and methionine synthase (Gulati et al. 1996, 1997; Gailus et al. 2010; Watkins and Rosenblatt 2011). Clinical symptoms generally include hematologic and neurologic defects (Watkins and Rosenblatt 2011). However, even within a single complementation class clinical symptoms and biochemical profile can be highly variable, which led to the need to distinguish the disorders based on the affected gene (Froese and Gravel 2010). Autosomal recessive mutations within at least 19 genes are associated with disorders of cobalamin metabolism. To our knowledge, there have been no reports of digenic inheritance (Sarafoglou and Hoffman 2009; Froese and Gravel 2010).
The number of patients diagnosed through research and clinical exome sequencing has increased exponentially since the inception of the technology in 2008 and the introduction of the clinical test in 2011 (NIH Health Consensus Conference Statement, 2001; Worthey et al. 2011; Dixon-Salazar et al. 2012; Need et al. 2012; Sailer et al. 2012; Hanchard et al. 2013; Shamseldin et al. 2013). Exome sequencing has been instrumental in discovering the pathogenic etiology in patients in whom traditional molecular methods were uninformative and is uniquely useful in overcoming limitations posed by traditional molecular diagnostic strategies in the identification of oligogenic findings. A recent meta-analysis of digenic inheritance publications points out that despite the capacity of high-throughput sequencing to discover digenic inheritance, strikingly few reports have been published (between 1 and 7 annually), and the rate of discovery has not increased over the last decade (Schaffer 2013). Often complicating the interpretation of genetic variants are factors such as reduced penetrance and variable expressivity, which has been, in part, attributed to a two-locus model (Strauch et al. 2003).
To our knowledge, there are no reports of exome sequencing leading to a diagnosis in a patient using only DNA from the affected individual’s family members. Herein, we describe this approach and the subsequent postmortem diagnosis of an affected neonate.
Subjects and Methods
DNA from the mother, father, and sister of a patient with cobalamin deficiency were referred to Ambry Genetics (Aliso Viejo, CA) for clinical diagnostic exome sequencing (DES). The patient had previously died at 3.5 months before DNA could be collected. A newborn screening blood spot was available from the patient, providing sufficient DNA for Sanger sequencing confirmation of findings, but not for whole-exome sequencing due to limited DNA quantity and quality. The family was consented for DES and research. The patient was born to non-consanguineous parents after an unremarkable prenatal course. Though B12 levels were not measured, the mother was on a normal diet including meat during pregnancy and breast-feeding. At 27 4/7 weeks gestation, CBC was normal. The patient was apparently healthy for the first 2.5 months. Newborn screening was normal. Specifically, propionylcarnitine (C3) level at birth was 1.43 μmol/L (normal <7.0 μmol/L), propionylcarnitine/acetylcarnitine (C3/C2) was 0.06 μmol/L (normal <0.20 μmol/L), methylmalonylcarnitine (C4-DC) was 0.10 μmol/L (normal <1.00 μmol/L), and methionine was 0.08 μmol/L (normal <1.25 μmol/L). At 2.5 months, he had recurring apnea episodes and was diagnosed with reflux. He then presented with seizures 10 days later. MRI showed brain atrophy with volume decrease and sulcal enlargement. EEG was consistent with diffuse encephalopathy. He had microcephaly with height and weight at the 50% tile. Ammonia, lactate, and pyruvate were normal. Total homocysteine was elevated at 125 μmol/L (normal 4.0–15.4 μmol/L). Urine organic acids showed mild elevation of MMA (8, normal 0–5) and hydantoin-5-propionate. Urine amino acids revealed homocystinuria (204 μM/gCR) and plasma amino acids showed low methionine (3, normal 10–60). Serum MMA was 0.19 nmol/mL. Vitamin B12 was low at 173 pg/mL (211–946), but mom’s level was normal. RBC folate was 580 ng/mL. CSF amino acids showed normal methionine levels, and homocystine was not detected. CSF neurotransmitter studies were normal (5-methylterahydrofolate in CSF was 59 nmol/L). Blood smear was normal without megaloblastic anemia or cytopenia. He was placed on daily hydroxocobalamin IM injections, betaine, B6, folate, and carnitine. His homocysteine fell from 125.2 to 30.5 to 18.6. His methionine increased from 3 to 12 in the same period. RBC folate changed from 508 to 503. The patient was ventilated for nearly 2 weeks and failed extubation due to continued respiratory distress with paradoxical episodes of respiratory pauses. He subsequently died at 3.5 months.
Genomic deoxyribonucleic acid (gDNA) was isolated from whole blood from each parent and the sister. Samples were prepared using the SureSelect Target Enrichment System (Agilent Technologies, Santa Clara, CA) (Gnirke et al. 2009). Briefly, each DNA sample was sheared, blunt-end repaired, and adaptor-ligated using indexed adapters. Using solution-based hybridization with oligonucleotide probes, the coding exons and neighboring intronic sequence of the genome were enriched and the nontargeted sequences were washed away. The enriched exome libraries were then applied to the solid surface flow cell for clonal amplification and sequencing using paired-end, 100 bp cycle chemistry on the Illumina HiSeq 2000 (Illumina, San Diego, CA). Each sample was sequenced sufficiently to yield greater than 90% of the targeted region with at least 10x base-pair coverage and greater than 85% of targeted bases with quality score of Q30 or higher, which translates to an expected base-calling error rate of 1:1,000 or an expected base-calling accuracy of 99.9%.
Initial data processing and base calling, including extraction of cluster intensities, was done using RTA 1.12.4 (HiSeq Control Software 1.4.5). Sequence quality filtering script was executed with the Illumina CASAVA software (ver 1.8.2, Illumina, Hayward, CA). Data yield (Mbases), %PF (pass filter), # of reads, % of raw clusters per lane, and quality scores were examined in Demultiplex_Stats.htm file. The sequence data were aligned to the reference human genome (GRCh37) and variant calls were generated using CASAVA and PINDEL (Ye et al. 2009). Exons plus at least two bases into the 5′ and 3′ ends of all the introns were analyzed. Data analysis focused on nonsense variants, small insertions and deletions, canonical splice site alterations, or non-synonymous missense alterations. The human gene mutation database (HGMD) (Strauch et al. 2003), the single nucleotide polymorphism database (dbSNP) (Sherry et al. 2001), 1,000 Genomes (1000, Genomes Project Consortium, 2010), HapMap data (International HapMap C 2003), and online search engines (e.g., PubMed) were used to search for previously described gene mutations and polymorphisms. The filtering pipeline protects all variants annotated within the HGMD (Stenson et al. 2009) and/or the online Mendelian inheritance in man (OMIM) databases. Stepwise filtering included the removal of common SNPs, intergenic and 3′/5′ UTR variants, non-splice-related intronic variants, and lastly synonymous variants. Variants were then filtered further based on family history and possible inheritance models. Data are annotated with the ambry variant analyzer tool (AVA), including nucleotide and amino acid conservation, biochemical nature of amino acid substitutions, population frequency (ESP and 1,000 genomes), and predicted functional impact (including PolyPhen (Gitiaux et al. 2013) and SIFT (Grohmann et al. 2004) in silico prediction tools). Each candidate mutation was assessed by a molecular geneticist to identify the most likely causative mutation(s). Multiple sequence alignments were viewed using IGV (integrative genomics viewer) software (Robinson et al. 2011).
Nonstandard bioinformatics filtering was performed in order to search for gene alterations which may have been transmitted to the deceased son. Specifically, analysis was performed to search for mutations in recessive genes carried by both the mother and father and not present in the homozygous or compound heterozygous state in the sister, as well as for X-linked recessive genes carried by mother. This analysis included all captured genes, including clinically novel genes. Additionally, a manual review of the known cobalamin deficiency genes was performed including the following genes: ABCD4, AMN, CD320, CUBN, FTCD, GIF, HCFC1, LMBRD1, MCEE, MMAA, MMAB, MMACHC, MMADHC, MTHFR, MTR, MTRR, MUT, PCFT, TCN1, and TCN2.
Identified candidate alterations were confirmed using automated fluorescence dideoxy sequencing. Co-segregation analysis was performed using each available family member. Amplification primers were designed using PrimerZ (Guenther et al. 2009). PCR primers were tagged with established sequencing primers on the 5′ end. Sequencing was performed on an ABI3730 (Life Technologies, Carlsbad, CA) using standard procedures.
Several months subsequent to exome sequencing, we were notified of the availability of Guthrie blood spot card from the patient and isolated gDNA for genotyping to confirm the presence of identified mutations.
Results
Exome sequencing of the mother, father, and sister resulted in an average of 16.7 Gb of sequence per sample (Fig. 1a; Table 1). Mean coverage of captured regions was 141x per sample, with 89% covered with at least 10x coverage and an average of 89% base call quality of Q30 or greater. Stepwise filtering for the removal of SNPs, intergenic and 3′/5′ UTR variants, non-splice-related intronic variants, and synonymous variants resulted in ~111,000 variants per sample (Table 2). Nonstandard bioinformatics filtering was performed in order to search for gene alterations which may have been transmitted to the deceased neonate but not to his unaffected sister. Five alterations met these criteria. Four were within two genes with described autosomal recessive inheritance and the fifth by the mother on the X chromosome. Of these three genes none related to the patient’s phenotype. Therefore, all of the autosomal recessive genes with alterations carried by the father (58) and mother (51) were analyzed for relation to the patient’s phenotype. Among these, two notable genes (two alterations) with potential clinical relevance were identified: (1) a missense alteration in the MTR gene (c.3518C>T; p.P1173L) present in the mother and (2) a single nucleotide deletion predicting a translational frameshift alteration in the LMBRD1 gene (c.1056delG; p.L352Lfs*18) present in the heterozygous state in the father. Automated fluorescence dideoxy sequencing using DNA from the infant’s blood spot confirmed the inheritance of each of the two alterations in heterozygous state (Fig. 1b). Furthermore, familial co-segregation analysis revealed that the MTR alteration was absent in the father and sister and the LMBRD1 alteration was absent in the mother and present in the heterozygous state in the sister (Table 3).
Fig. 1.
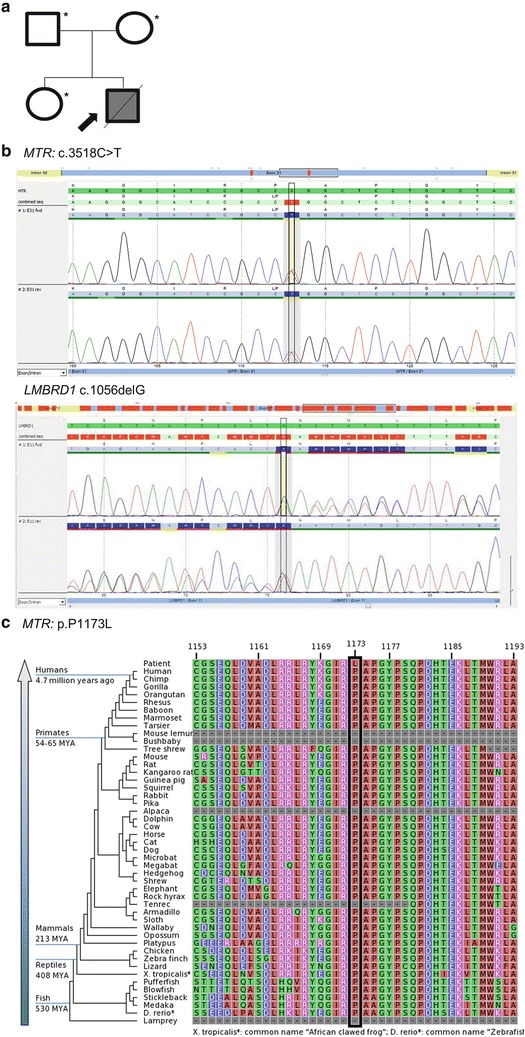
Pedigree and MTR c.3518C>T (p.P1173L) and LMBRD1 c.1056delG (p.L352fsX18) mutations. (a) Familial pedigree. Shaded shapes indicate affected individuals. Asterisk (*) indicates whole-exome sequencing performed. (b) An electropherogram of the MTR c.3518C>T (p.P1173L) and LMBRD1 c.1056delG (p.L352fsX18) alterations in the proband. (c) Sequence conservation plots at the MTR p.P1173L mutated site amino acid position across different species
Table 1.
HiSeq sequencing run metrics
| Father | Mother | Sister | Average | |
|---|---|---|---|---|
| Yield (Gb) | 14.722 | 17.446 | 18.036 | 16.735 |
| Quality | ||||
| % of >= Q30 bases (PF) | 88.715 | 88.945 | 88.995 | 88.89 |
| Mean quality score (PF) | 35.115 | 35.195 | 35.205 | 35.17 |
| Base coverage | ||||
| %Base_1x (%) | 94.03 | 94.24 | 94.05 | 94.11 |
| %Base_4x (%) | 91.75 | 92.21 | 92.07 | 92.01 |
| %Base_5x (%) | 90.72 | 91.31 | 91.18 | 91.07 |
| %Base_10x (%) | 89.31 | 90.05 | 89.96 | 89.77 |
| %Base_20x (%) | 85.22 | 86.44 | 86.43 | 86.03 |
| %Base_50x (%) | 73.65 | 76.95 | 77.26 | 75.95 |
| %Base_100x (%) | 50.60 | 58.85 | 59.74 | 56.40 |
| Mean_coverage (%) | 121.46 | 148.73 | 153.09 | 141.09 |
The bold values signify the average of the three individual's (father, mother, sister), values
Table 2.
Bioinformatics stepwise variant filtering
| Stepwise filteringa | Father | Mother | Sister | Average | |
|---|---|---|---|---|---|
| No. of variants in coding regionsb | 110,636 | 108,825 | 112,270 | 110,577 | |
| No. post-removal of intergenic and 3′/5′ UTR variants | 82,300 | 81,626 | 83,470 | 82,465 | |
| No. post-removal of non-splice-related intronicc variants | 21,855 | 21,963 | 22,319 | 22,046 | |
| No. post-removal of synonymous variants | 11,631 | 11,622 | 11,874 | 11,709 | |
| (A) Autosomal recessived | 2 genes (4 alterations) | ||||
| No. of genes in (A) related to phenotype | 0 genes (0 alterations) | ||||
| (B) X-linked recessivee | 1 gene (1 alteration) | ||||
| No. of genes in (B) related to phenotype | 0 genes (0 alterations) | ||||
| (C) Autosomal recessive carrier variantsf | 58 | 51 | N/A | N/A | |
| No. of genes in (C) related to phenotype | 2 genes (2 alterations) | ||||
The bold values signify the average of the three individual's (father, mother, sister), values
aStepwise filtering protects variants annotated within the human gene mutation database (HGMD) and/or the online Mendelian inheritance in man (OMIM) databases
bVariants refer to single nucleotide alterations, insertions, deletions, and indels with at least 10x base-pair coverage
cIntronic refers to >3 bp into the introns
dMother and father carrier, daughter negative or carrier
eMother carrier
fMother and father
Table 3.
Familial co-segregation analysis results
Special consideration was given to the known cobalamin metabolism genes which had almost complete base-pair coverage at ≥10x (>99%), with exceptions noted in some exons in AMN, CD320, FTDC, MMAB, and HCFC1 (Supplementary Table 1). No candidate alterations were identified, other than the described MTR and LMBRD1 alterations.
The P1173L (c.3518C>T) alteration is located in exon 31 of the MTR gene (NM_000254) (OMIM), is predicted to be deleterious by PolyPhen (Adzhubei et al. 2010) and SIFT (Ng and Henikoff 2006) in silico analyses, and is located at highly conserved amino acid position (Fig. 1c). Based on data from the NHLBI Exome Sequencing Project (ESP), the T-allele has an overall frequency of approximately 0.04% (4/10,744) total alleles.
The c.1056delG (p.L352Lfs*18) alteration is located in exon 11 of the LMBRD1 gene and results from the deletion of 1 nucleotide, causing a translational frameshift with a predicted alternate stop codon.
Discussion
Herein, whole-exome sequencing and nonstandard bioinformatics analysis of the parents and sister of a deceased infant with cobalamin deficiency identified alterations within well-described genes associated with cobalamin deficiency, of which each parent was a carrier. Sanger sequencing using DNA from the infant’s blood spot confirmed the presence of both alterations in compound heterozygous form. To our knowledge, these results represent both the first example of the molecular diagnosis in the absence of the affected patient as well as the first example of digenic inheritance associated with cobalamin deficiency.
Mutations in the MTR and LMBRD1 genes have been described in patients with disorders of cobalamin metabolism. Specifically, MTR gene alterations are associated with homocystinuria/hyperhomocysteinemia, and LMBRD1 gene alterations are observed in patients with methylmalonic acidemia/aciduria and hyperhomocysteinemia/homocystinuria (Watkins and Rosenblatt 2011). c.3518C>T (p.P1173L) is the most common alteration observed in the MTR gene, with a frequency of about 40% (16/38 chromosomes) of patients with cblG deficiency (Watkins et al. 2002). LMBRD1 c.1056delG (p.L352Lfs*18) is the most frequently reported alteration, found to be present in 75% (18/24 chromosomes) of one cohort of 12 patients with cblF deficiency (Rutsch et al. 2009, 2011).
The identified alterations are likely founder mutations among individuals of European ancestry, consistent with the family’s reported ancestry (Watkins et al. 2002; Rutsch et al. 2011). MTR and LMBRD1 are two of the several proteins essential to cobalamin metabolism (reviewed in Sarafoglou and Hoffman 2009; Watkins and Rosenblatt 2011). MTR is a cobalamin-dependent enzyme that catalyzes a methyl transfer reaction from methyltetrahydrofolate to homocysteine to form methionine and tetrahyrofolate (Gulati et al. 1997). Mutations in the MTR gene lead to the cblG complementation class of the cobalamin metabolism disorders (Gulati et al. 1996; Watkins et al. 2002). Patients with cblG typically present in the first year of life with elevated homocysteine, decreased methionine, seizures, and cerebral atrophy (Sarafoglou and Hoffman 2009). Other clinical findings in patients with cblG include decreased S-adenosylmethionine, lethargy, feeding difficulties, vomiting, abnormal tonus, mental retardation, failure to thrive, blindness, ataxia, delayed myelination, and megaloblastic anemia (Sarafoglou and Hoffman 2009). LMBRD1 is a lysosomal membrane protein thought to be involved in lysosomal export of cobalamin in which alteration leads to the cblF complementation class of the cobalamin metabolism disorders (Rutsch et al. 2009, 2011; Gailus et al. 2010). Cells from cblF patients accumulate large amounts of free cobalamin within the lysosomes, but there is a deficiency of both cobalamin coenzyme derivatives and decreased activity of MMA mutase and methionine sythetase (reviewed in Watkins and Rosenblatt, 2011). Immortalized fibroblasts from patients with cblF show restored cobalamin coenzyme synthesis and function upon transfection with an LMBRD1 wild-type complementary DNA (cDNA) construct (Rutsch et al. 2011). Typical clinical findings in patients with cblF are evident by the first year of life and include increased homocystine and MMA, encephalopathy, poor head growth, decreased cobalamin levels, mental retardation, ataxia, and megaloblastic anemia and/or pancytopenia (Sarafoglou and Hoffman 2009).
The proband’s clinical phenotype did not fit any of the well-characterized cobalamin metabolism disorders but had overlapping features. His elevated homocysteine and low methionine are seen in both cblF and cblG, while the elevated MMA and low cobalamin levels are seen with cblF but not typically with cblG. He did not have the megaloblastic anemia that is observed in both cblF and cblG. However, the phenotype produced from digenic inheritance is often distinct from that observed in either of the contributing genes (e.g., Helwig et al. 1995; Van Goethem et al. 2003; Yang et al. 2009). In fact, a recent review on digenic inheritance cautions medical geneticists to consider digenic inheritance as a possible mechanism for patients with novel phenotypes (Schaffer 2013). Further, the clinical features observed among patients with LMBRD1 are widely variable (Froese and Gravel 2010; Watkins and Rosenblatt 2011), and specifically patients with the c.1056delG alteration in the compound heterozygous or homozygous state show considerable phenotypic variability ranging from death in infancy to asymptomatic long-term survival (Rutsch et al. 2009).
The unaffected mother, unaffected father, and unaffected sister do not carry both the MTR and LMBRD1 alterations which would have ruled out the possibility for a digenic affect. The alterations were present in the compound heterozygous state in the proband, reducing, but not eliminating, the likelihood for copy-number variation (CNV) on the corresponding allele of each gene. Gross deletions and duplications have not been reported in MTR, but a single report of a 6,785 bp deletion of exon 2 has been reported in LMBRD1 (Miousse et al. 2011). A gross deletion in LMBRD1 would either have occurred de novo in the proband or be inherited from a parent (most likely the mother). The mother and father’s next-gen sequencing data show comparable coverage within this gene compared with other genes, reducing the likelihood for the presence of deletion within the gene (~120X vs. mean = 134X ± 44.4). Due to insufficient DNA isolated from the blood spot of the deceased proband we are unable to entirely exclude this possibility. Whole- or partial-gene deletions including exon 11 of LMBRD1 or exon 31 or MTR are excluded on the basis of heterozygosity of the mutation call.
Although a digenic inheritance pattern has not been previously reported in a patient with cobalamin deficiency, it is well described in a growing number of inherited conditions including nonsyndromic hearing loss, Bardet–Biedl syndrome, retinitis pigmentosa, polycystic kidney disease, and facioscapulohumeral muscular dystrophy type 2 (Vockley 2011; Schaffer 2013). Common to syndromes in which digenic inheritance has been described are variable expressivity, reduced penetrance, pleiotrpy, and genetic heterogeneity (reviewed in Vockley 2011), all of which are observed in cobalamin deficiency disorders. The data herein suggest that digenic inheritance may represent a novel molecular mechanism for disorders of cobalamin deficiency. It should be noted, however, that we could not completely rule out an alteration occurring in a dominant de novo manner, a novel gene alteration, or alterations not detectable by exome sequencing (including copy number variation). Biochemical analysis and in vitro transfection studies are needed to further define the impact of digenic haploinsufficiency on this metabolic pathway. This unique case highlights the power of whole-exome sequencing as a diagnostic tool even in the absence of a sample from the affected individual and suggests a novel mechanism and inheritance pattern for disorders of cobalamin metabolism.
Electronic Supplementary Material
Cobalamin - Text & Tables -revised with track changes.docx (DOCX 153 KB)
Acknowledgments
We are grateful to the family of the patient for their participation.
Compliance with Ethics Guidelines
Synopsis
In this report, we simultaneously demonstrate the power of new molecular technologies to diagnose a deceased patient while uncovering the first potential example of digenic inheritance associated with cobalamin deficiency.
Conflict of Interest
Kelly Gonzalez, Xiang Li, Hsiao-Mei Lu, Hong Lu, Elizabeth Chao, and Wenqi Zeng are employed and receive a salary from Ambry Genetics. Exome sequencing is among the commercially available tests. Joan Pellegrino and Ryan Miller declare that they have no conflict of interest.
Informed Consent
All procedures followed were in accordance with the ethical standards of the responsible committee on human experimentation (institutional and national) and with the Helsinki Declaration of 1975, as revised in 2000 (5). Informed consent was obtained from all patients for being included in the study. Additional informed consent was obtained from all patients for which identifying information is included in this article.
Animal Rights
This article does not contain any studies with animal subjects performed by any of the authors.
Details of the Contributions of Individual Authors
Kelly Gonzalez was involved in the study’s conception and design, analysis and interpretation of data, and drafting the article and is the guarantor for the manuscript. Xiang Li, Hsiao-Mei Lu, and Hong Lu were involved in analysis and interpretation of data and critical review and revision of drafts for important intellectual content. Joan Pellegrino and Ryan Miller were involved in drafting the article, analysis and interpretation of data, and critical review and revision of drafts for important intellectual content. Elizabeth Chao and Wenqi Zeng were involved in the study’s conception and design, analysis and interpretation of data, and critical review and revision of drafts for important intellectual content.
Electronic-Database Information
Accession numbers and URLs for data in this article are as follows:
ESP [Internet]: Exome variant server, NHLBI GO exome sequencing project (ESP), Seattle, WA. http://evs.gs.washington.edu/EVS/. Accessed Feb 2013.
OMIM [Internet]: Online Mendelian inheritance in man, OMIM®. McKusick-Nathans Institute of Genetic Medicine, Johns Hopkins University, Baltimore, MD. World Wide Web. http://omim.org/. Accessed 2 June 2013.
RefSeq: NCBI (2002) The NCBI handbook. National Library of Medicine (US), National Center for Biotechnology Information, Bethesda (MD). Chapter 18, The reference sequence (RefSeq) project.
Footnotes
Competing interests: None declared
Contributor Information
Kelly D. Farwell Gonzalez, Email: kgonzalez@ambrygen.com
Collaborators: Johannes Zschocke and K Michael Gibson
References
- 1000 Genomes Project Consortium. Abecasis GR, Altshuler DL, Auton A, et al. A map of human genome variation from population-scale sequencing. Nature. 2010;467(7319):1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adzhubei IA, Schmidt S, Peshkin L, et al. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7(4):248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coelho D, Kim JC, Miousse IR, et al. Mutations in ABCD4 cause a new inborn error of vitamin B12 metabolism. Nat Genet. 2012;44(10):1152–1155. doi: 10.1038/ng.2386. [DOI] [PubMed] [Google Scholar]
- Dixon-Salazar TJ, Silhavy JL, Udpa N, et al. Exome sequencing can improve diagnosis and alter patient management. Sci Trans Med. 2012;4(138):138ra178. doi: 10.1126/scitranslmed.3003544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Froese DS, Gravel RA. Genetic disorders of vitamin B(1)(2) metabolism: eight complementation groups–eight genes. Expert Rev Mol Med. 2010;12:e37. doi: 10.1017/S1462399410001651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gailus S, Hohne W, Gasnier B, Nurnberg P, Fowler B, Rutsch F. Insights into lysosomal cobalamin trafficking: lessons learned from cblF disease. J Mol Med. 2010;88(5):459–466. doi: 10.1007/s00109-010-0601-x. [DOI] [PubMed] [Google Scholar]
- Gitiaux C, Bergounioux J, Magen M, et al. Diaphragmatic weakness with progressive sensory and motor polyneuropathy: case report of a neonatal IGHMBP2-related neuropathy. J Child Neurol. 2013;28(6):787–790. doi: 10.1177/0883073812450209. [DOI] [PubMed] [Google Scholar]
- Gnirke A, Melnikov A, Maguire J, et al. Solution hybrid selection with ultra-long oligonucleotides for massively parallel targeted sequencing. Nat Biotech. 2009;27(2):182–189. doi: 10.1038/nbt.1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grohmann K, Rossoll W, Kobsar I, et al. Characterization of Ighmbp2 in motor neurons and implications for the pathomechanism in a mouse model of human spinal muscular atrophy with respiratory distress type 1 (SMARD1) Hum Mol Genet. 2004;13(18):2031–2042. doi: 10.1093/hmg/ddh222. [DOI] [PubMed] [Google Scholar]
- Guenther UP, Handoko L, Laggerbauer B, et al. IGHMBP2 is a ribosome-associated helicase inactive in the neuromuscular disorder distal SMA type 1 (DSMA1) Hum Mol Genet. 2009;18(7):1288–1300. doi: 10.1093/hmg/ddp028. [DOI] [PubMed] [Google Scholar]
- Gulati S, Baker P, Li YN, et al. Defects in human methionine synthase in cblG patients. Hum Mol Genet. 1996;5(12):1859–1865. doi: 10.1093/hmg/5.12.1859. [DOI] [PubMed] [Google Scholar]
- Gulati S, Chen Z, Brody LC, Rosenblatt DS, Banerjee R. Defects in auxiliary redox proteins lead to functional methionine synthase deficiency. J Biol Chem. 1997;272(31):19171–19175. doi: 10.1074/jbc.272.31.19171. [DOI] [PubMed] [Google Scholar]
- Hanchard NA, Murdock DR, Magoulas PL, et al. Exploring the utility of whole-exome sequencing as a diagnostic tool in a child with atypical episodic muscle weakness. Clin Genet. 2013;83(5):457–461. doi: 10.1111/j.1399-0004.2012.01951.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helwig U, Imai K, Schmahl W, et al. Interaction between undulated and patch leads to an extreme form of spina bifida in double-mutant mice. Nat Genet. 1995;11(1):60–63. doi: 10.1038/ng0995-60. [DOI] [PubMed] [Google Scholar]
- International HapMap C The international HapMap project. Nature. 2003;426(6968):789–796. doi: 10.1038/nature02168. [DOI] [PubMed] [Google Scholar]
- Miousse IR, Watkins D, Rosenblatt DS. Novel splice site mutations and a large deletion in three patients with the cblF inborn error of vitamin B12 metabolism. Mol Genet Metabol. 2011;102(4):505–507. doi: 10.1016/j.ymgme.2011.01.002. [DOI] [PubMed] [Google Scholar]
- National Institutes of Health Consensus Development Panel National institutes of health consensus development conference statement: phenylketonuria: screening and management, October 16–18, 2000. Pediatrics. 2001;108(4):972–982. doi: 10.1542/peds.108.4.972. [DOI] [PubMed] [Google Scholar]
- Need AC, Shashi V, Hitomi Y, et al. Clinical application of exome sequencing in undiagnosed genetic conditions. J Med Genet. 2012;49(6):353–361. doi: 10.1136/jmedgenet-2012-100819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng PC, Henikoff S. Predicting the effects of amino acid substitutions on protein function. Ann Rev Genom Hum Genet. 2006;7:61–80. doi: 10.1146/annurev.genom.7.080505.115630. [DOI] [PubMed] [Google Scholar]
- Robinson JT, Thorvaldsdottir H, Winckler W, et al. Integrative genomics viewer. Nat Biotech. 2011;29(1):24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutsch F, Gailus S, Miousse IR, et al. Identification of a putative lysosomal cobalamin exporter altered in the cblF defect of vitamin B12 metabolism. Nat Genet. 2009;41(2):234–239. doi: 10.1038/ng.294. [DOI] [PubMed] [Google Scholar]
- Rutsch F, Gailus S, Suormala T, Fowler B. LMBRD1: the gene for the cblF defect of vitamin B(1)(2) metabolism. J Inherit Metab Dis. 2011;34(1):121–126. doi: 10.1007/s10545-010-9083-9. [DOI] [PubMed] [Google Scholar]
- Sailer A, Scholz SW, Gibbs JR, et al. Exome sequencing in an SCA14 family demonstrates its utility in diagnosing heterogeneous diseases. Neurology. 2012;79(2):127–131. doi: 10.1212/WNL.0b013e31825f048e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarafoglou K, Hoffman G. Pediatric endocrinology and inborn errors of metabolism. New York: McGraw-Hill Medical; 2009. [Google Scholar]
- Schaffer AA. Digenic inheritance in medical genetics. J Med Genet. 2013;50(10):641–652. doi: 10.1136/jmedgenet-2013-101713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shamseldin HE, Swaid A, Alkuraya FS. Lifting the lid on unborn lethal Mendelian phenotypes through exome sequencing. Genet Med. 2013;15(4):307–309. doi: 10.1038/gim.2012.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherry ST, Ward MH, Kholodov M, et al. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 2001;29(1):308–311. doi: 10.1093/nar/29.1.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenson PD, Mort M, Ball EV, et al. The human gene mutation database: 2008 update. Genome Med. 2009;1(1):13. doi: 10.1186/gm13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strauch K, Fimmers R, Baur MP, Wienker TF. How to model a complex trait. 2. Analysis with two disease loci. Hum Hered. 2003;56(4):200–211. doi: 10.1159/000076394. [DOI] [PubMed] [Google Scholar]
- Van Goethem G, Lofgren A, Dermaut B, Ceuterick C, Martin JJ, Van Broeckhoven C. Digenic progressive external ophthalmoplegia in a sporadic patient: recessive mutations in POLG and C10orf2/Twinkle. Hum Mutat. 2003;22(2):175–176. doi: 10.1002/humu.10246. [DOI] [PubMed] [Google Scholar]
- Vockley J. Book digenic inheritance. Chichester: Wiley; 2011. Digenic inheritance. [Google Scholar]
- Watkins D, Rosenblatt DS. Inborn errors of cobalamin absorption and metabolism. Am J Med Genet C: Semin Med Genet. 2011;157(1):33–44. doi: 10.1002/ajmg.c.30288. [DOI] [PubMed] [Google Scholar]
- Watkins D, Ru M, Hwang HY, et al. Hyperhomocysteinemia due to methionine synthase deficiency, cblG: structure of the MTR gene, genotype diversity, and recognition of a common mutation, P1173L. Am J Hum Genet. 2002;71(1):143–153. doi: 10.1086/341354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worthey EA, Mayer AN, Syverson GD, et al. Making a definitive diagnosis: successful clinical application of whole exome sequencing in a child with intractable inflammatory bowel disease. Genet Med. 2011;13(3):255–262. doi: 10.1097/GIM.0b013e3182088158. [DOI] [PubMed] [Google Scholar]
- Yang T, Gurrola JG, 2nd, Wu H, et al. Mutations of KCNJ10 together with mutations of SLC26A4 cause digenic nonsyndromic hearing loss associated with enlarged vestibular aqueduct syndrome. Am J Hum Genet. 2009;84(5):651–657. doi: 10.1016/j.ajhg.2009.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye K, Schulz MH, Long Q, Apweiler R, Ning Z. Pindel: a pattern growth approach to detect break points of large deletions and medium sized insertions from paired-end short reads. Bioinformatics. 2009;25(21):2865–2871. doi: 10.1093/bioinformatics/btp394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu HC, Sloan JL, Scharer G, et al. An X-linked cobalamin disorder caused by mutations in transcriptional coregulator HCFC1. Am J Hum Genet. 2013;93(3):506–514. doi: 10.1016/j.ajhg.2013.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Cobalamin - Text & Tables -revised with track changes.docx (DOCX 153 KB)


