Abstract
Background
Genomic variants identified by genome-wide association studies (GWAS) explain <20% of heritability of coronary artery disease (CAD), thus many risk variants remain missing for CAD. Identification of new variants may unravel new biological pathways and genetic mechanisms for CAD. To identify new variants associated with CAD, we developed a candidate pathway-based GWAS by integrating expression quantitative loci (eQTL) analysis and mining of GWAS data with variants in a candidate pathway.
Methods and Results
Mining of GWAS data was performed to analyze variants in 32 complement system genes for positive association with CAD. Functional variants in genes showing positive association were then identified by searching existing expression quantitative loci databases and validated by RT-PCR. A follow-up case control design was then used to determine whether the functional variants are associated with CAD in two independent GeneID Chinese populations. Candidate pathway-based GWAS identified positive association between variants in C3AR1 and C6 and CAD. Two functional variants, rs7842 in C3AR1 and rs4400166 in C6, were found to be associated with expression levels of C3AR1 and C6, respectively. Significant association was identified between rs7842 and CAD (P=3.99×10−6, OR=1.47) and between rs4400166 and CAD (P=9.30×10−3, OR=1.24) in the validation cohort. The significant findings were confirmed in the replication cohort (P=1.53×10−5, OR=1.37 for rs7842; P=8.41×10−3, OR=1.21 for rs4400166.
Conclusions
Integration of GWAS with biological pathways and eQTL is effective in identifying new risk variants for CAD. Functional variants increasing C3AR1 and C6 expression were shown to confer significant risk of CAD for the first time.
Keywords: coronary artery disease, genome-wide association study (GWAS), variants, single nucleotide polymorphisms, complement system
Coronary artery disease (CAD) and its major complication myocardial infarction (MI) are the major cause of morbidity and mortality in the Western world as well as in China.1; 2 The pathogenesis of CAD is a complex process mediated by many genetic and environmental factors and their interactions.3 CAD is known to be influenced by abnormal lipid metabolism, smoking, diabetes, hypertension, obesity, physical inactivity, alcohol intake, and psychosocial situation.4 Heritability of CAD was estimated to be 40%–60%, indicating that genetic factors contribute significantly to the development of this disease.5 Identification of novel genetic variants may reveal new biological pathways and mechanisms of CAD and facilitate improved diagnosis, prevention and treatment.
Genome-wide association studies (GWAS) and their meta-analyses (Meta-GWAS) have identified about 50 risk variants associated with CAD and MI (Catalog of Published GWAS: http://www.genome.gov/26525384).6; 7 However, GWAS identify mostly common variants and all variants for CAD and MI identified by GWAS account for 10.60% of heritability in aggregate.8 Therefore, a large portion of heritability remains missing for CAD and MI (referred to as missing heritability), and much effort is needed to identify new risk variants for CAD and MI to account for missing heritability.
Most GWAS for CAD and MI were completed in European ancestry populations, but recently two GWAS were reported in the Chinese population. We reported the first GWAS for CAD in the Chinese Han population using a large GeneID database in 2011 and identified the C6orf105 gene (now known as ADTRP) as a susceptibility gene for CAD specifically in the Chinese population.9 The finding was replicated in two independent reports.10; 11 In 2012, another GWAS identified four new loci for CAD in or near TTC32-WDR35, GUCY1A3, C6orf10-BTNL2, and ATP2B1.12 However, these identified variants may explain little heredity of CAD in the Chinese population.
In order to identify new genomic variants associated with CAD and MI in the Chinese population, we focused on mining of GWAS genotyping data for variants in individual pathways for positive association with CAD without adjusting for multiple testing (P<0.01), followed by identification of functional variants by eQTL analysis and association analysis between functional variants and CAD. We refer this strategy as candidate pathway-based GWAS. Using this approach, we found that functional SNPs in two genes in the complement system, C3AR1 encoding complement component 3a receptor and the C6 encoding complement component 6, were significantly associated with risk of CAD.
Subjects and Methods
Study populations
The study subjects involved in this study were selected from the GeneID population, which is a large ongoing database with clinical data and tissue samples from more than 80,000 Chinese patients and controls. The major aim of GeneID is to identify genes for cardiovascular and cerebrolvascular diseases in the Chinese Han population.9 The study subjects are of the ethnic Han origin by self-description. This study was approved by appropriate local institutional review boards on human subject research and conformed to the guidelines set forth by the Declaration of Helsinki. Written informed consent was obtained from all study subjects. The details on the diagnosis of CAD, MI, hypertension, and diabetes and controls were described in the Data Supplement.
SNP genotyping
SNP genotyping was carried out as described9 and in detail in the Data Supplement.
eQTL analysis, SNP selection, and LD analysis
We searched the SNP express database (http://compute1.lsrc.duke.edu/softwares/SNPExpress/) and Genevar 3.3 (http://www.sanger.ac.uk/resources/software/genevar/) to identify the expression quantitative loci for the C3AR1 and C6 genes.13
To determine whether the GWAS variants and the variants with eQTLs are in the same linkage disequilibrium (LD) block, we computed the r2 values using data from the HapMap and 1000genomes databases and investigated the genomic region for the recombination rate covering these variants using Locuszoom (http://csg.sph.umich.edu/locuszoom/).14
Real-time quantitative RT-PCR analysis
Quantitative real-time PCR analysis was carried out according to the MIQE guidelines as described previoulsy15 and in detail in the Data Supplement.
Statistical analysis
Genotyping data were analyzed for allelic and genotypic association using Pearson’s 2×2 or 2×3 contingency tables Chi-square tests as implemented in PLINK version 1.06, respectively. P values and corresponding odds ratios (ORs) with 95% confidential intervals were computed for each SNP using PLINK version 1.06. Statistical analyses for eQTLs and power analysis were performed as reported previously16 and in detail in the Data Supplement.
Results
Description of a candidate pathway-based GWAS strategy for association studies for common disease
We previously performed genome-wide genotyping of 44,0794 SNPs using Genome-wide Human SNP 5.0 arrays in two independent case-control discovery cohorts for CAD from GeneID.9 SNPs showing positive association for CAD with P of <0.01 in both cohorts were selected for follow-up validation and multiple replication studies, which led to the identification of association between an ADTRP variant and CAD and MI.9 To further explore the GWAS data, we developed a candidate pathway-based GWAS strategy, which consists of three steps. First, we mine the GWAS data by focusing on a specific candidate biological pathway, e.g. the complement system in the present study, to identify variants that show nominal significance with CAD without adjustment for multiple testing (P<0.01) to reduce false negatives as in many GWAS for CAD. Second, considering that most variants identified by GWAS are simply genetic markers and have low effects on the risk of disease, we identify functional variants in close proximity to the positive GWAS variants by eQTL and real-time RT-PCR analyses. The functional variants may be causatively associated with the disease, and thus have a larger effect, which is expected to increase the power of statistically analysis. Third, the functional variants are evaluated for association with the disease in a case control cohort (validation cohort) and significant association is then further assessed in an independent replication cohort. We tested the candidate pathway-GWAS strategy in this study and successfully established the genetic association between the complement system and CAD. Atherosclerosis is a chronic inflammatory process, and abnormalities of the immune system may play an important role in the initiation and development of the vascular atherosclerotic lesions.17 The complement system is a major component of the innate immune system, and was suggested to be involved in the pathogenesis of atherosclerosis.18 Moreover, few human genetic studies for CAD investigated the complement system. Therefore, in our pilot study of the candidate pathway-GWAS strategy, we focused on 32 genes of the complement system.
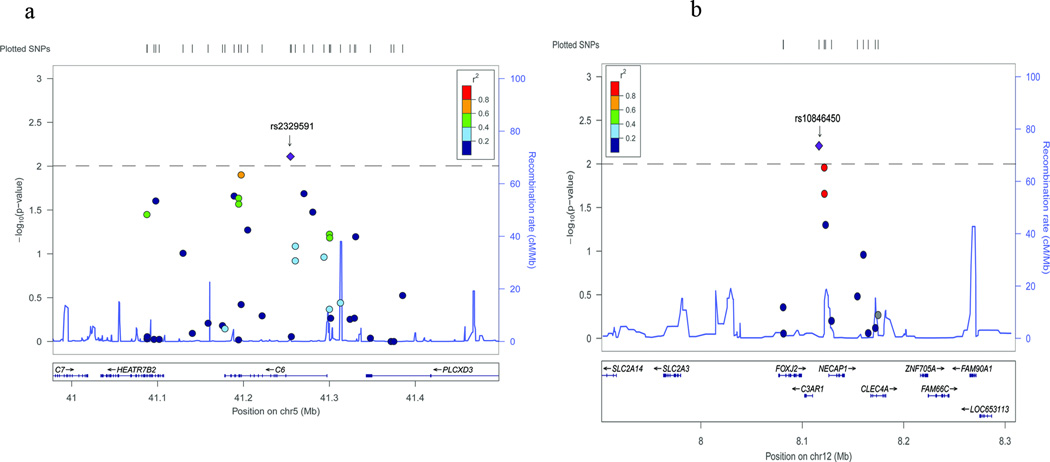
Mining of GWAS data identified two SNP hits (rs10846450 and rs2329591) from 668 variants in the complement system showing nominal association with CAD
We assessed SNPs in all genes of the complement system in exons, introns, 5-UTR, 3’-UTR, and genomic regions 100 kb upstream or downstream of each gene for potential association with CAD using our combined GWAS genotyping data from 230 CAD patients and 230 controls. In total, 668 variants in 32 genes in the complement pathway were on Genome-Wide Human SNP 5.0 arrays. In the association analysis of the 668 SNPs for CAD, only two SNPs achieved a P value of <0.01, including rs10846450 located 12 kb upstream of C3AR1 and rs2329591 in C6. Minor allele A of C3AR1 variant rs10846450 showed a positive association with CAD (P =6.20×10−3, OR= 1.88) (Table 1 and Figure 1). Minor allele A of C6 variant rs2329591 also a positive association with CAD (P = 7.75×10−3, OR=2.11) (Table 1 and Figure 1).
Table 1.
Association analysis of 32 genes in the complement system using GWAS data from a Chinese Han GeneID population
| Gene Symbol | SNPs | Top SNPa | P value | Gene Symbol | SNPs | Top SNPa | P value |
|---|---|---|---|---|---|---|---|
| C1 | 25 | rs34962575 | 0.54 | C7 | 27 | rs404223 | 0.15 |
| C1QA | 7 | rs209693 | 0.083 | C8a | 52 | rs1620855 | 0.022 |
| C1QB | 13 | rs2869513 | 0.054 | C9 | 45 | rs1421094 | 0.03 |
| C1S | 5 | rs7306673 | 0.40 | CFB | 5 | rs522162 | 0.44 |
| C1R | 5 | rs2110073 | 0.31 | CFI | 20 | rs6815517 | 0.071 |
| C1RL | 7 | rs7297327 | 0.09 | CRP | 26 | rs12068753 | 0.074 |
| C2 | 7 | rs9267673 | 0.13 | CFH | 23 | rs424535 | 0.22 |
| C3 | 20 | rs11569536 | 0.16 | CFHR1 | 10 | rs7413265 | 0.081 |
| C3AR1 | 12 | rs10846450 | 6.20×10−3 | CR2 | 14 | rs311299 | 0.33 |
| C4A | 6 | rs550513 | 0.36 | CR1 | 14 | rs677066 | 0.30 |
| C4B | 13 | rs204991 | 0.24 | CD59 | 12 | rs831608 | 0.28 |
| C4BPB | 28 | rs11120211 | 0.10 | CD93 | 33 | rs6082986 | 0.08 |
| C4BPA | 47 | rs2012296 | 0.46 | CD55 | 27 | rs6658718 | 0.24 |
| C5 | 27 | rs1468672 | 0.14 | SERING1 | 16 | rs2649662 | 0.04 |
| C5AR1 | 6 | rs10853782 | 0.79 | MASP2 | 19 | rs37303981 | 0.037 |
| C6 | 37 | rs2329591 | 7.75×10−3 | MBL2 | 117 | rs7095636 | 0.06 |
The GWAS data were from 230 CAD patients and 230 controls as reported previously by Wang et al.9
Top SNP: a SNP in a specific gene with the lowest P value for association with CAD.
Figure 1.
Analysis of SNPs in and close to C3AR1 (a) and C6 (b) for association with CAD in Chinese Han GeneID populations using GWAS data. Regional association plots of the GWAS data are shown for C3AR1 (a) and C6 (b). Each panel shows SNPs plotted by their positions on the corresponding chromosome against −log10 P. Estimated recombination rates from 1000 genomes (JPT+CHB populations) were plotted in blue to reflect the local linkage disequilibrium (LD) structure on a secondary y axis. The most significant lead SNP (diamond) is annotated with its observed P value, and flanking SNPs (circle) are color-coded to represent the pairwise r2 measure of LD with the lead SNP: red, r2 ≥ 0·8; orange, 0·6 ≤ r2 < 0·8; green, 0·4 ≤ r2<0·6; light blue, 0·2 ≤ r2< 0·4; blue, r2 < 0·2. All positions are based on hg18. These plots were generated by Locuszoom (https://statgen.sph.umich.edu/locuszoom/)
eQTLs analysis identified two functional variants, rs7842 and rs4400166 associated with the expression levels of C3AR1 and C6, respectively
We examined whether C3AR1 variant rs10846450 and C6 variant rs2329591 showing nominal association with CAD in GWAS data may be associated the expression levels of corresponding genes. Mining of the SNP express database and Genevar 3.3 showed that rs10846450 and rs2329591 were not associated with the expression levels of C3AR1 and C6. By contrast, we found that variant rs7842 located in the 3’UTR of C3AR1 was associated increased expression of C3AR1 (P=5.47×10−5) (Figure S1 in the Data Supplement). We also found that variant rs4400166 in intron 13 of C6 showed significant association with increased expression of the C6 gene (P=3.73×10−4) (Figure S1 in the Data Supplement).
Both rs7842 in C3AR1 and rs4400166 in C6 were not on the GWAS chips. We analyzed LD for nominally-associated variants rs10846450 and rs2329591 and variants with eQTLs (rs7842 and rs4400166) by computing the r2 values using the HapMap and newly released 1000genomes data(hg19/ 1000genomes Mar 2012 data) and the recombination rates using Locuszoom with the 1000genomes data. We found that for C3AR1, rs7842 and rs10846450 were not located in the same LD block because of a low r2 value of 0.01 in the HapMap database and 0.08 in the 1000 genomes database and a recombination rate of 4.6% in Asian populations. Similarly, for the C6 gene, rs4400166 and rs2329591 were not located in the same LD block due to a low r2 value of 0.06 in the HapMap database and 0.14 in the 1000 genomes database and a recombination rate of 0.8% in Asian populations (Figure S2 in the Data Supplement).
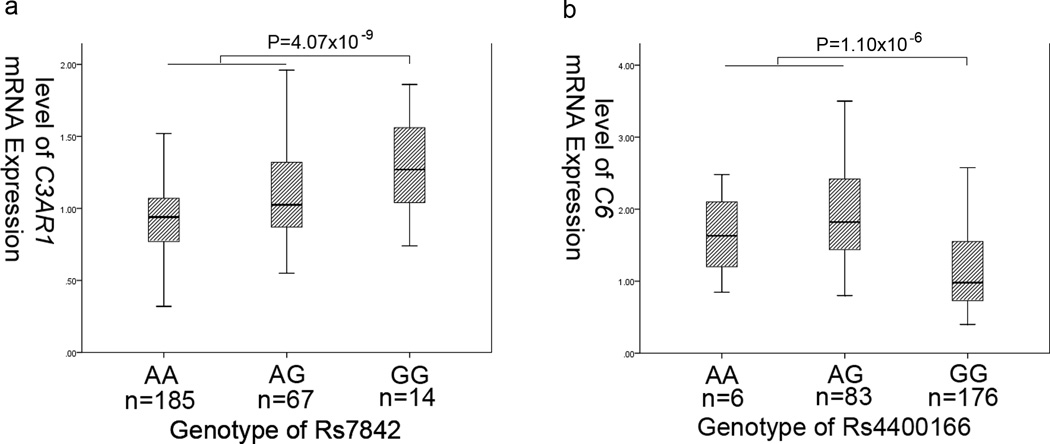
The significant associations between rs7842 and C3AR1 expression and between rs4400166 and C6 expression were validated experimentally in the Chinese Han population. We measured the expression levels of C3AR1 and C6 in leukocytes from 266 people randomly selected from the general population by real-time qRT-PCR analysis and genotyped these individuals with rs7842 and rs4400166. For rs7842, the mean relative expression levels of C3AR1 were 1.42±0.24 in 14 GG genotype carriers, 1.10±0.23 in 67 AG genotype carriers and 0.92±0.18 in 185 AA genotype carriers. Statistical analysis with general linear modeling showed that minor allele G of rs7842 was significantly associated with a higher C3AR1 mRNA expression level under a dominant model with a standardized coefficients (β) of 0.35 (mean expression levels: 1.13±0.30 for GG+GA vs. 0.92±0.18 in AA, P=4.07×10−9) (Figure 2a).
Figure 2.
Association of SNPs rs7842 and rs4400166 with mRNA expression levels of C3AR1 (a) and C6 (b)
For SNP rs4400166, the mean relative expression levels of C6 were 1.17±0.58 for 176 subjects with the GG genotype and 1.88±0.64 for 83 subjects with the GA genotype combined with 6 AA genotype carriers (1.90±0.64 for the GA genotype and 1.65±0.60 for the AA genotype, respectively). Minor allele A of rs4400166 showed a highly significant association with an increased C6 mRNA expression level under a dominant model (β=0.46, P=1.10×10−6) (Figure 2b).
Significant association between rs7842 in C3AR1 and CAD
The studies above showed that C3AR1 variant rs10846450 and C6 variant rs2329591 from GWAS data mining were not associated with the expression levels of their respective genes, thus we did not pursue the two GWAS variants any further. Interestingly, significant association was demonstrated between rs7842 in C3AR1 and rs4400166 in C6 and the expression levels of C3AR1 and C6, respectively. Therefore, we performed case control association analyses for SNPs rs7842 and rs4400166 for their association with CAD next.
Table 2 showed the details of basic demographical and clinical characteristics of the two cohorts used in our case control association studies. The association analysis was first carried out in a case control cohort with 924 CAD cases and 904 non-CAD controls from the GeneID Chinese database (validation cohort). Significant association from the validation cohort was then replicated in another independent cohort with 1,065 cases and 1,398 controls, also from the GeneID Chinese database (replication cohort).
Table 2.
Basic demographic and clinical characteristics of CAD patients and controls involved in the present study
| Characteristic | Validation cohort† | Replication cohort† | ||||
|---|---|---|---|---|---|---|
| Cases (n=924) |
Controls (n=904) |
P§ | Cases (n=1,065) |
Controls (n=1,398) |
P§ | |
| Age*, mean±SD (years) | 61.33±11.41 | 62.33±8.64 | 0.68 | 62.92±11.07 | 62.61±9.83 | 0.46 |
| Sex, % of females | 37.66% | 41.15% | 0.13 | 38.21% | 39.34% | 0.57 |
| Hypertension, No.% | 61.26% | 60.51% | 0.74 | 61.05% | 58.44% | 0.22 |
| Diabetes, No.% | 16.56% | 15.15% | 0.41 | 16.62% | 14.74% | 0.07 |
| Total cholesterol, mean±SD (mmol/L) | 4.48±1.10 | 4.46±1.17 | 0.88 | 4.49±1.11 | 4.43±0.84 | 0.03 |
| Triglyceride, mean±SD (mmol/L) | 1.50±0.84 | 1.42±0.89 | 0.04 | 1.52±1.29 | 1.45±1.43 | 0.04 |
| HDL-cholesterol, mean±SD (mmol/L) | 1.21±0.44 | 1.25±0.41 | 0.06 | 1.16±0.38 | 1.21±0.34 | 0.01 |
| LDL-cholesterol, mean±SD (mmol/L) | 2.59±0.85 | 2.48±0.83 | 0.01 | 2.64±0.85 | 2.55±0.87 | 0.01 |
| Smoker, No.% | 33.44% | 32.08% | 0.77 | 34.74% | 31.47% | 0.12 |
Data are shown as mean +/− standard deviation (SD) for quantitative variables and percent (%) for qualitative variables.
Age at the first diagnosis of the disease in CAD case and at enrollment of CAD controls.
These samples are independent from GWAS samples.
P values for comparison of means between cases and controls.
The power estimation was performed based on assumptions that SNPs rs7842 can confer a risk of CAD with an OR of >1.3 and a minor allelic frequency (MAF) of 0.16 in the Chinese population in the HapMap database. Both cohorts can provide sufficient statistical power to detect the association between rs7842 and CAD with a type I error of 0.05 (86% power for rs7842 in the first validation cohort; 94% for rs7842 in the replication cohort).
Genotyping data for variant rs7842 in C3AR1 did not deviate from the Hardy-Weinberg equilibrium in the controls in the first cohort (P=0.92). The allelic frequencies of rs7842 were significantly different between CAD cases and controls (Table 3). Significant association was identified with an OR of 1.47 and a P value (P-obs) of 3.99×10−6. After adjusting for potential confounders including age, gender, smoking, hypertension, diabetes mellitus and lipid concentrations (Tch, TG, HDL-c and LDL-c), the association remained significant (OR=1.43 with an adjusted P or P-adj=4.94 ×10−4).
Table 3.
Analysis of allelic association of SNP rs7842 with CAD in the Chinese Han population
| Cohort (n, case/control) |
Risk Allele |
Frequency (case/control) |
Without Adjustment | With Adjustment* | ||
|---|---|---|---|---|---|---|
| P-obs | OR (95% CI) | P-adj | OR (95% CI) | |||
| Validation cohort (924/904) | G | 0.23 / 0.17 | 3.99×10−6 | 1.47(1.25–1.75) | 4.94×10−4 | 1.43(1.17–1.75) |
| Replication cohort (1,065/1,398) | G | 0.22 / 0.17 | 1.53×10−5 | 1.37(1.19–1.58) | 8.87×10−3 | 1.28(1.07–1.58) |
| Combined (1,989/2,302) | G | 0.23/0.170 | 1.89 ×10−10 | 1.41(1.21–1.57) | 2.87 ×10−4 | 1.31(1.13–1.52) |
P-obs: P value observed; P-adj: P value after adjustment for covariates; OR: odds ratio.
Adjusted P value by multivariate logistic regression analysis for potential confounders including age, gender, smoking, hypertension, diabetes mellitus and lipid concentrations (total cholesterol, triglyceride, HDL-cholesterol and LDL-cholesterol).
Genotyping data for rs7842 did not deviate from the Hardy-Weinberg equilibrium in the controls in the replication cohort (P=0.09). The G allele of rs7842 conferred significant risk of CAD in the replication cohort (P-obs=1.53×10−5, OR=1.37; P-adj=8.87×10−3, OR=1.28) (Table 3).
We performed a meta-analysis by combining two cohorts together, which generated a large cohort of 1,989 cases and 2,302 controls. SNP rs7842 showed highly significant association with CAD in the combined cohort (P-obs=1.89×10−10, OR=1.41). After adjustment for covariates of age, gender, hypertension, diabetes, smoking and lipids levels, the association remained significant (P-adj=2.87×10−4, OR=1.31). The association remained significant after Bonferroni correction for multiple testing of two SNPs. These results suggest that SNP rs7842 in the C3AR1 gene confers a significant risk of CAD in the Chinese population.
We also carried out genotypic association analysis, which allows assessment of the risk of rs7842 for CAD under different genetic models. Highly significant genotypic association was identified for CAD in both cohorts under all three models (dominant, recessive or additive) (Table 1S in the Data Supplement).
Significant association between rs4400166 in C6 and CAD
Similar power estimation as for rs7842 showed that the validation and replication cohorts can provide sufficient statistical power to detect the association between rs4400166 and CAD with a type I error of 0.05 (89% power in the first validation cohort; 96% in the replication cohort; MAF of 0.19). Genotyping data for variant rs4400166 did not deviate from the Hardy-Weinberg equilibrium in the controls in the first cohort (P>0.05).
Significant association was found between rs4400166 and CAD (P-obs=9.30×10−3, OR=1.24) (Table 4). After adjusting for potential confounders, the association remained significant (P-adj=0.03, OR=1.20) (Table 4). The significant association was successfully confirmed in the replication cohort. The A allele of rs4400166 showed significant association with CAD in the replication cohort (P-obs=8.41×10−3, OR=1.21; P-adj=0.01, OR=1.18) (Table 4). In the combined cohort with 1,989 cases and 2,302 controls, rs4400166 showed highly significant association with CAD (P-obs of 1.69×10−4, OR=1.23). After adjustment for covariates, the association remained significant for rs4400166 with an adjusted P-adj of 4.43×10−3 and an OR of 1.18 (Table 4). Significant genotypic association was also found between rs4400166 and CAD under a dominant model (P-obs=1.95×10−4, OR=1.27; P-adj=3.23×10−3, OR=1.26) (Table S1 in the Data Supplement). The association remained significant after Bonferroni correction for multiple testing of two SNPs. These data suggest that rs4400166 in C6 confers a risk of CAD in the Chinese Han population.
Table 4.
Analysis of allelic association of SNP rs4400166 with CAD in Chinese Han population
| Cohort (n, case/control) |
Risk Allele |
Frequency (case/control) |
Without Adjustment | With Adjustment* | ||
|---|---|---|---|---|---|---|
| P-obs | OR (95% CI) | P-adj | OR (95% CI) | |||
| Validation cohort (924/904) | A | 0.23/0.19 | 9.30×10−3 | 1.24(1.05–1.45) | 0.03 | 1.20(1.01–1.43) |
| Replication cohort (1065/1398) | A | 0.22/0.19 | 8.41×10−3 | 1.21(1.05–1.40) | 0.01 | 1.18(1.02–1.41) |
| Combined (1989/2302) | A | 0.22/0.19 | 1.69 ×10−4 | 1.23(1.10–1.36) | 4.43×10−3 | 1.18(1.02–1.40) |
P-obs: P value observed; P-adj: P value after adjustment for covariates; OR: odds ratio.
Adjusted P value by multivariate logistic regression analysis for potential confounders including age, gender, smoking, hypertension, diabetes mellitus and lipid concentrations (total cholesterol, triglyceride, HDL-cholesterol and LDL-cholesterol).
Discussion
Initial small-sized GWAS and follow-up meta-GWAS with larger sample sizes pooled together by several groups owning GWAS data have identified about 50 variants associated with CAD and MI.19 But, these variants explain only 10.6% of the disease heritability.8 How to identify the hidden missing heritability of CAD is the most challenging issue for the genetic field of CAD and MI and other common diseases. The latest meta-GWAS for CAD used 63,746 cases and 130,681 controls.8 Identification of new variants for CAD by further expansion of sample sizes for meta-GWAS is intimidating and may not be easily feasible. In this study, we demonstrate that a new candidate pathway-based candidate gene strategy for CAD is effective in identifying some remaining heritability (new genetic variants for CAD).
In this study, we systematically analyzed the association of 668 SNPs in 32 genes regulating the complement system with CAD from our GeneID GWAS data. Through this candidate pathway-based GWAS and follow-up case control association analyses, we provide strong evidence to demonstrate that SNPs in two genes of the complement system, C3AR1 and C6, were significantly associated with CAD and MI in the Chinese Han GeneID population. First, by analyzing 32 genes in the complement system with the GWAS data, nominal positive association was found between CAD and C3AR1 or C6. Second, two cis-acting variants that affect the expression of C3AR1 and C6 were identified and validated independently using the GeneID samples, including rs7842 in the 3’UTR of C3AR1 and rs4400166 in intron 13 of C6. Finally, in two independent case control cohorts, rs7842 and rs4400166 were found to be significantly associated with CAD in the Chinese Han GeneID population. We found that rs7842 contributed 0.85% and rs4400166 contributed 0.31% to the heritability of CAD.
To the best of our knowledge, this is the first systematic analysis of the genetic association between the complement system and CAD, which identified novel associations between variants in C3AR1 and C6 and risk of CAD. Several studies evaluated the association between variant rs3746731 in CD93 and CAD in a small case control study with 340 cases and 300 controls, a P value of 0.02 was achieved, but became non-significant after multiple testing of 45 SNPs studied.20 In a separate study with 2,145 familial hypercholesterolemia patients, rs3746731 showed a P value of 0.01 for association with CAD, which became non-significant after correction of testing of 10 SNPs.21 Several studies showed association between the Y402H variant in complement factor H associated with age-related macular degeneration and CAD, but recent meta-analysis with 48,000 patients showed that this variant was not associated with CAD.22–24 Similarly, a recent large scale study showed that variants in CRP were not associated with risk of CAD.25 In our GWAS data, there was one variant in CRP, rs12068753, which did not show positive association with CAD (P=0.07). In a small case control study with 217 CAD patients and 217 controls from American Indian communities identified association between composite genotypes from three SNPs and one promoter variant in the MBL gene (mannose-binding lectin) and CAD.26 Although interesting, the study was small and not replicated. On the other hand, our study with two independent, relatively large populations provides strong genetic evidence that variants in genes in the complement system confer significant risk of CAD in the Chinese population.
There are several key points for the pathway-based GWAS. First, the focus is on a specific biological pathway and a fixed set of genes, which reduces the number of SNPs to be analyzed. Second, the transition from positive SNPs in the GWAS data to functional variants in the same genes is most important. Most variants identified by GWAS are genetic markers with low effects, especially those variants showing weak association not passing the stringent multiple testing corrections. In this case, the possibilities of false positives and a difficulty of replication increased, thus, in the present study, we did not perform the replication study between CAD and the SNPs which showed positive association with CAD in the initial GWAS data. Instead, we tried to find the possible causal variants with larger effects by eQTL and real-time RT-PCR analysis. The functional variants are then pursued in the validation and replication stages of the study. It is interesting to note that the initial positive SNPs identified by GWAS mining (rs10846451, rs2329591) are not in LD with the respective functional eQTL SNPs analyzed further (rs7842, rs4400166) (R2=0.01–0.08 for rs7842 and rs10846451 and 0.06–0.14 for rs4400166 and rs2329591). This transition from SNPs with a weak effect identified by GWAS mining to functional SNPs in different LD blocks and with a potentially larger effect may be a key to the success of candidate pathway-based GWAS. Our research successfully identified and validated two cis-acting variants, which affect the expression of C3AR1 and C6 and confer risk to CAD, and demonstrated that our pathway-based GWAS strategy is effective identifying causal or larger effect variants of common disease.
After completion of the study, we went back and genotyped the two SNPs, rs2329591 and rs10846450, identified initially from the GWAS data in the validation and replication cohorts. In the combined population, minor allele A of C3AR1 variant rs10846450 showed significant association with CAD (P=0.02, OR= 1.12). After adjustment for age, gender, hypertension, diabetes, smoking and lipids levels, the association remained significant for rs10846450 with an adjusted P-adj of 0.04 and an OR of 1.08 (Table S2 in the Data Supplement), but became non-significant after multiple testing correction for two SNPs studied. Significant genotypic association was also found between rs10846450 and CAD under an additive model (P-obs=0.03; P-adj=0.05, OR=1.10) (Table S3 in the Data Supplement), but became non-significant after multiple testing correction. SNP rs2329591 in C6 did not show any significant association with CAD (P>0.05) (Table S2 in the Data Supplement). Therefore, in the candidate pathway-based GWAS, there is no need to validate or replicate the initial positive SNP hits from the initial GWAS data mining.
Inflammation is central to the development and progression of CAD and MI.18 The complement system is a part of the innate immune system and has been implicated in the pathogenesis of atherosclerosis. A number of studies reported detection of various complement activation products, regulatory proteins, and complement receptors in human atherosclerotic lesions, which may directly cause local cell damage and indirectly attract and activate immune cells.19; 27 Therefore, genes regulating the complement system are candidates for CAD and MI. It is possible that variants in the complement system genes have a small effect on CAD and MI, and their potential association with CAD and MI may be too modest to be detected in GWAS due to a low power to achieve a high significant level. Our candidate pathway-based approach integrates a priori knowledge on atherosclerosis together with eQTL analysis to identify potentially functional cis-acting variants, which may amplify the effect of the candidate genes and provide an explanation for the successful identification of SNPs in C3AR1 and C6 conferring risk of CAD. Future studies are needed to further assess whether this novel candidate pathway-based strategy is effective in identifying new susceptibility genes for CAD in other biological pathways and other complex diseases in general.
The complement system is composed of three activation pathways, including the alternative, classical, and lectin pathways.28 All three major complement activation pathways lead to cleavage of C3, which generates C3a and C3b fragments.29 C3a is one of the most potent pro-inflammatory anaphylatoxins generated during complement pathway activation, and involved in activation of leukocytes and leukocyte recruitment to inflammatory sites in the vessel wall, which are important processes in atherosclerosis.30; 31 C3AR1 is a Gi-coupled G protein–coupled receptor for C3a.32 In the present study, we found that minor allele G of rs7842 in the 3’-UTR of C3AR1 was associated with an increased expression level of C3AR1 and increased risk of CAD (Figure 2 and Table 3). These data suggest that increased C3AR1 expression augments the risk of CAD. Our results are supported by the finding that knockout mice deficient in C3AR1 was found to significantly decrease the aortic lesion size at the ApoE−/− background and by the finding that expression of the C3AR1 protein was increased 5-fold in coronary atherosclerotic plaques in humans.33; 34
C3b leads to the formation of the C5 convertase, which cleaves C5 into the C5a and C5b fragments.28 C5b then forms a complex with C6, C7, C8α, C8β, C8γ, and C9 on the cell surface, resulting in the membrane attack complex (MAC).35 MAC causes swelling and lysis of the target cells and the release of inflammatory molecules.35 Here we show that minor allele A of rs4400166 in the C6 gene was associated with increased C6 expression and risk of CAD (Figure 2 and Table 4), suggesting that increased C6 expression increases risk of CAD. This conclusion is supported by the finding that C6-deficient rabbits and C6 knockout mice showed a lack of MAC formation, less atherosclerotic lesions, and reduced plaque areas than controls under Apoe−/− background.36; 37
One limitation of the present study is that the initial sample size for GWAS is small. However, it was effective in identifying the 9p21 and 6p24 variants that confer risk to CAD.9 It is highly likely that the power for the candidate pathway GWAS may be significantly increased if large GWAS or huge meta-GWAS databases are mined.
In conclusion, we employed a novel candidate pathway-based GWAS strategy that combines mining of GWAS data for variants in a candidate pathway and eQTL analysis to identify new risk variants associated with CAD. Here we performed the first systematic analysis of the genetic association between all genes in the complement system and CAD. We provided strong genetic evidence to demonstrate that SNPs in two genes in the complement pathway, C3AR1 and C6, are significantly associated with CAD in the Chinese Han GeneID population. These results show that the two most important terminal signals for complement activation, including C3a-C3aR signaling and C6-mediated MAC formation, may confer risk in the pathogenesis of CAD. Therefore, restriction of complement activation may be a novel potential scheme for the treatment of CAD. Future studies are needed to determine the detailed molecular mechanism by which C3AR1 and C6 mediates atherosclerosis.
Supplementary Material
Acknowledgments
Funding Sources: Chinese National Basic Research Programs (973 Programs 2013CB531101 and 2012CB517801), grants from the National Natural Science Foundation of China(31430047, 81200073, 81270163), the Hubei Province’s Outstanding Medical Academic Leader Program, China Postdoctoral Science Foundation (2012T50644), NIH/NHLBI grant R01 HL121358, Specialized Research Fund for the Doctoral Program of Higher Education from the Ministry of Education, the “Innovative Development of New Drugs” Key Scientific Project (2011ZX09307-001-09), and Program for New Century Excellent Talents in University of China (NCET-11-0181).
Footnotes
Conflict of Interest Disclosures: None
References
- 1.Lozano R, Naghavi M, Foreman K, Lim S, Shibuya K, Aboyans V, et al. Global and regional mortality from 235 causes of death for 20 age groups in 1990 and 2010: A systematic analysis for the global burden of disease study 2010. Lancet. 2013;380:2095–2128. doi: 10.1016/S0140-6736(12)61728-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhang X, Lu Z, Liu L. Coronary heart disease in china. Heart. 2008;94:1126–1131. doi: 10.1136/hrt.2007.132423. [DOI] [PubMed] [Google Scholar]
- 3.Wang Q. Molecular genetics of coronary artery disease. Curr Opin Cardiol. 2005;20:182. doi: 10.1097/01.hco.0000160373.77190.f1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Torpy JM, Burke AE, Glass RM. Coronary heart disease risk factors. JAMA. 2009;302:2388–2388. doi: 10.1001/jama.302.21.2388. [DOI] [PubMed] [Google Scholar]
- 5.Watkins H, Farrall M. Genetic susceptibility to coronary artery disease: From promise to progress. Nat Rev Genet. 2006;7:163–173. doi: 10.1038/nrg1805. [DOI] [PubMed] [Google Scholar]
- 6.Kessler T, Erdmann J, Schunkert H. Genetics of coronary artery disease and myocardial infarction-2013. Curr Cardiol Rep. 2013;15:368–368. doi: 10.1007/s11886-013-0368-0. [DOI] [PubMed] [Google Scholar]
- 7.Hindorff LA, Sethupathy P, Junkins HA, Ramos EM, Mehta JP. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. P Natl Acad Sci USA. 2009;106:9362–9367. doi: 10.1073/pnas.0903103106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Deloukas P, Kanoni S, Willenborg C, Farrall M, Assimes TL, Thompson JR, et al. Large-scale association analysis identifies new risk loci for coronary artery disease. Nat Genet. 2013;45:25–33. doi: 10.1038/ng.2480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang F, Xu CQ, He Q, Cai JP, Li XC, Wang D, et al. Genome-wide association identifies a susceptibility locus for coronary artery disease in the chinese han population. Nat Genet. 2011;43:345–349. doi: 10.1038/ng.783. [DOI] [PubMed] [Google Scholar]
- 10.Tayebi N, Ke T, Foo JN, Friedlander Y, Liu J, Heng C-K. Association of single nucleotide polymorphism rs6903956 on chromosome 6p24. 1 with coronary artery disease and lipid levels in different ethnic groups of the singaporean population. Clin Biochem. 2013;46:755–759. doi: 10.1016/j.clinbiochem.2013.01.004. [DOI] [PubMed] [Google Scholar]
- 11.Guo CY, Gu Y, Li L, Jia EZ, Li CJ, Wang LS, et al. Association of snp rs6903956 on chromosome 6p24.1 with angiographical characteristics of coronary atherosclerosis in a chinese population. PLoS One. 2012;7:e43732. doi: 10.1371/journal.pone.0043732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lu X, Wang L, Chen S, He L, Yang X, Shi Y, et al. Genome-wide association study in han chinese identifies four new susceptibility loci for coronary artery disease. Nat Genet. 2012;44:890–894. doi: 10.1038/ng.2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sanders MA, Verhaak RG, Geertsma-Kleinekoort WM, Abbas S, Horsman S, van der Spek PJ, et al. Snpexpress: Integrated visualization of genome-wide genotypes, copy numbers and gene expression levels. BMC Genomics. 2008;9:41. doi: 10.1186/1471-2164-9-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xiong X, Xu C, Zhang Y, Li X, Wang B, Wang F, et al. Brg1 variant rs1122608 on chromosome 19p13. 2 confers protection against stroke and regulates expression of pre-mrna-splicing factor sfrs3. Hum Genet. 2013:1–10. doi: 10.1007/s00439-013-1389-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xu C, Wang F, Wang B, Li X, Li C, Wang D, et al. Minor allele c of chromosome 1p32 single nucleotide polymorphism rs11206510 confers risk of ischemic stroke in the chinese han population. Stroke. 2010;41:1587–1592. doi: 10.1161/STROKEAHA.110.583096. [DOI] [PubMed] [Google Scholar]
- 16.Lieb W, Vasan RS. Genetics of coronary artery disease. Circulation. 2013;128:1131–1138. doi: 10.1161/CIRCULATIONAHA.113.005350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kane JP, Aouizerat BE, Luke MM, Shiffman D, Iakoubova O, Liu D, et al. Novel genetic markers for structural coronary artery disease, myocardial infarction, and familial combined hyperlipidemia: Candidate and genome scans of functional snps. International Congress Series. 2004;1262:309–312. [Google Scholar]
- 18.van der Net JB, Oosterveer DM, Versmissen J, Defesche JC, Yazdanpanah M, Aouizerat BE, et al. Replication study of 10 genetic polymorphisms associated with coronary heart disease in a specific high-risk population with familial hypercholesterolemia. Eur Heart J. 2008;29:2195–2201. doi: 10.1093/eurheartj/ehn303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kardys I, Klaver CC, Despriet DD, Bergen AA, Uitterlinden AG, Hofman A, et al. A common polymorphism in the complement factor h gene is associated with increased risk of myocardial infarction the rotterdam study. J Am Coll Cardiol. 2006;47:1568–1575. doi: 10.1016/j.jacc.2005.11.076. [DOI] [PubMed] [Google Scholar]
- 20.Sofat R, Casas JP, Kumari M, Talmud PJ, Ireland H, Kivimaki M, et al. Genetic variation in complement factor h and risk of coronary heart disease: Eight new studies and a meta-analysis of around 48,000 individuals. Atherosclerosis. 2010;213:184–190. doi: 10.1016/j.atherosclerosis.2010.07.021. [DOI] [PubMed] [Google Scholar]
- 21.Jylhävä J, Eklund C, Pessi T, Raitakari O, Juonala M, Kähönen M, et al. Genetics of c-reactive protein and complement factor h have an epistatic effect on carotid artery compliance: The cardiovascular risk in young finns study. Clin Exp Immunol. 2009;155:53–58. doi: 10.1111/j.1365-2249.2008.03752.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Collaboration CRPCHDG. Association between c reactive protein and coronary heart disease: Mendelian randomisation analysis based on individual participant data. Brit Med J. 2011;342:d548. doi: 10.1136/bmj.d548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Best LG, Davidson M, North KE, MacCluer JW, Zhang Y, Lee ET, et al. Prospective analysis of mannose-binding lectin genotypes and coronary artery disease in american indians the strong heart study. Circulation. 2004;109:471–475. doi: 10.1161/01.CIR.0000109757.95461.10. [DOI] [PubMed] [Google Scholar]
- 24.Libby P, Ridker PM, Hansson GK. Inflammation in atherosclerosis: From pathophysiology to practice. J Am Coll Cardiol. 2009;54:2129–2138. doi: 10.1016/j.jacc.2009.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Speidl WS, Kastl SP, Huber K, Wojta J. Complement in atherosclerosis: Friend or foe? J Thromb Haemost. 2011;9:428–440. doi: 10.1111/j.1538-7836.2010.04172.x. [DOI] [PubMed] [Google Scholar]
- 26.Széplaki G, Varga L, Füst G, Prohászka Z. Role of complement in the pathomechanism of atherosclerotic vascular diseases. Mol Immunol. 2009;46:2784–2793. doi: 10.1016/j.molimm.2009.04.028. [DOI] [PubMed] [Google Scholar]
- 27.Carroll MC. The complement system in regulation of adaptive immunity. Nat Immunol. 2004;5:981–986. doi: 10.1038/ni1113. [DOI] [PubMed] [Google Scholar]
- 28.Sarma JV, Ward PA. The complement system. Cell Tissue Res. 2011;343:227–235. doi: 10.1007/s00441-010-1034-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hertle E, van Greevenbroek MMJ, Stehouwer CDA. Complement c3: An emerging risk factor in cardiometabolic disease. Diabetologia. 2012;55:881–884. doi: 10.1007/s00125-012-2462-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Veneskoski M, Turunen SP, Kummu O, Nissinen A, Rannikko S, Levonen A-L, et al. Specific recognition of malondialdehyde and malondialdehyde acetaldehyde adducts on oxidized ldl and apoptotic cells by complement anaphylatoxin c3a. Free Radical Bio Med. 2011;51:834–843. doi: 10.1016/j.freeradbiomed.2011.05.029. [DOI] [PubMed] [Google Scholar]
- 31.Guo Q, Subramanian H, Gupta K, Ali H. Regulation of c3a receptor signaling in human mast cells by g protein coupled receptor kinases. PLoS One. 2011;6:e22559. doi: 10.1371/journal.pone.0022559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang X, Peterson L, Thieringer R, Deignan JL, Wang X, Zhu J, et al. Identification and validation of genes affecting aortic lesions in mice. J Clin Invest. 2010;120:2414–2422. doi: 10.1172/JCI42742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Oksjoki R, Laine P, Helske S, Vehmaan-Kreula P, Mayranpaa MI, Gasque P, et al. Receptors for the anaphylatoxins c3a and c5a are expressed in human atherosclerotic coronary plaques. Atherosclerosis. 2007;195:90–99. doi: 10.1016/j.atherosclerosis.2006.12.016. [DOI] [PubMed] [Google Scholar]
- 34.Tegla CA, Cudrici C, Patel S, Trippe R, III, Rus V, Niculescu F, et al. Membrane attack by complement: The assembly and biology of terminal complement complexes. Immunol Res. 2011;51:45–60. doi: 10.1007/s12026-011-8239-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ricklin D, Hajishengallis G, Yang K, Lambris JD. Complement: A key system for immune surveillance and homeostasis. Nat Immunol. 2010;11:785–797. doi: 10.1038/ni.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lewis RD, Jackson CL, Morgan BP, Hughes TR. The membrane attack complex of complement drives the progression of atherosclerosis in apolipoprotein e knockout mice. Mol Immunol. 2010;47:1098–1105. doi: 10.1016/j.molimm.2009.10.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schmiedt W, Kinscherf R, Deigner H-P, Kamencic H, Nauen O, Kilo J, et al. Complement c6 deficiency protects against diet-induced atherosclerosis in rabbits. Arterioscl Thromb Vas. Biol. 1998;18:1790–1795. doi: 10.1161/01.atv.18.11.1790. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.