Abstract
A fundamental challenge in treating disease is identifying molecular states that affect cellular responses to drugs. Here, we focus on GSK-3, a key regulator for many of the hallmark behaviors of cancer cells. We alter GSK-3 activity in colon epithelial cells to test its role in modulating drug response. We find that GSK-3 activity broadly affects the cellular sensitivities to a panel of oncology drugs and kinase inhibitors. Specifically, inhibition of GSK-3 activity can strongly desensitize or sensitize cells to kinase inhibitors (e.g. mTOR or PLK1 inhibitors, respectively). Additionally, colorectal cancer cell lines, in which GSK-3 function is commonly suppressed, are resistant to mTOR inhibitors and yet highly sensitive to PLK1 inhibitors and this is further exacerbated by additional GSK-3 inhibition. Finally, by conducting a kinome-wide RNAi screen, we find that GSK-3 modulates the cell proliferative phenotype of a significant fraction (~35%) of the kinome, which includes ~50% of current, clinically relevant kinase-targeted drugs. Our results highlight an under-appreciated interplay of GSK-3 with therapeutically important kinases and suggest strategies for identifying disease-specific molecular profiles that can guide optimal selection of drug treatment.
Introduction
A fundamental challenge in drug discovery and personalized medicine is the identification of molecular drivers of sensitivity or resistance to therapy. Common approaches focus on a specific drug and investigate how its efficacy is altered by various signaling components. An complementary approach—which we take here—is to focus on a specific signaling component and investigate how its state can alter the efficacy of a broad spectrum of drugs. The identification of key signaling components whose states modify cellular responses to a broad spectrum of drugs, will help provide strategies for optimal selection of individualized drug treatments.
We focused our study on the serine/threonine protein kinase Glycogen Synthase Kinase 3 (GSK-3) as a broad modulator of drug potency for four key reasons. First, GSK-3 is a highly networked kinase; GSK-3 regulates the function of tens, if not hundreds, of proteins through binding and/or enzymatic modification1,2. Second, GSK-3 is a downstream signaling conduit for multiple growth factor pathways, including Receptor Tyrosine Kinase (RTK), Hedgehog (HH), and Wnt signaling pathways3; when these growth factor pathways are activated, GSK-3 activity towards pathway-specific substrates is typically decreased2. Third, GSK-3 generally functions to regulate cell proliferation and differentiation in many tissues1,2; active GSK-3 suppresses pro-proliferation substrates, e.g. β-catenin, Myc, Jun, Snail, and enhances pro-differentiation substrates, e.g. p53, Rb, PTEN, TSC1/24. Fourth, GSK-3 activity is often down regulated5-9 during tumor progression, although GSK-3 is rarely mutated itself. In fact, the three most common mutations in highly aggressive, drug-resistant colorectal cancer, (APC, KRAS, and PI3K), can perturb GSK-3’s function, typically leading to decreased phosphorylation of GSK-3 substrates10. Together, we hypothesized that GSK-3 is positioned to act as a key player in the cellular response to drugs.
Here we modulated GSK-3 activity, using small molecule and genetic perturbations, to uncover its role in drug response. We found that loss of GSK-3 activity significantly alters cellular responses to a wide array of oncology drugs and kinase inhibitors. Specifically, we found that inhibition of GSK-3 desensitizes cells to mTOR inhibitors, but sensitizes cells to PLK1 inhibitors. We confirmed our results for mTOR and PLK1 inhibitors in multiple colorectal cancer cell lines of diverse genetic backgrounds. Finally, we performed a GSK-3 modifier screen across the known human kinome and found that ~35% of kinases interact with GSK-3, a subset of which are the targets of ~50% of current, clinically relevant kinase-inhibitors listed in DrugBank11 (Supplementary Results, Supplementary Data set 1). Our study suggests that GSK-3 is a gatekeeper for therapeutically important kinases—its activity state can strongly alter the potency of drug treatment—and suggests strategies for predicting and improving kinase-targeted drug potency.
Results
GSK-3 activity affects response to oncology drugs and kinase inhibitors
To investigate how GSK-3 influences the landscape of cellular response to drugs, we chose to make use of human colonic epithelial cells (HCECs) in our large-scale screens for two reasons. First, HCECs are clonally derived from healthy patient tissue and are diploid and genetically stable12; thus, HCECs serve as a model cell line for rapidly proliferating epithelial cells. Second, HCECs do not contain the genetic alterations of cancer cell lines; thus, HCECs provides a clean genetic background for understanding the unique contribution of GSK-3 to drug sensitivity in human epithelial cells. We then made use of a panel of colorectal cancer cell lines to test our key findings. To modulate the activity of GSK-3, we used the potent and specific GSK-3 inhibitor CHIR99021 (“CHIR”) (Fig. 1a). In humans, GSK-3 is encoded by two genes, GSK-3α and GSK-3 β (double knockout of both genes is lethal13), and CHIR99021 (“CHIR”) blocks both GSK-3α and GSK-3β activity14. We chose a concentration (3 μM) that showed measurable effects on multiple GSK-3 substrates yet had no discernible effect on cell proliferation or cell cycle phasing (Supplementary Fig. 1). This allowed us to identify drug effects that were not due simply to cell cycle arrest.
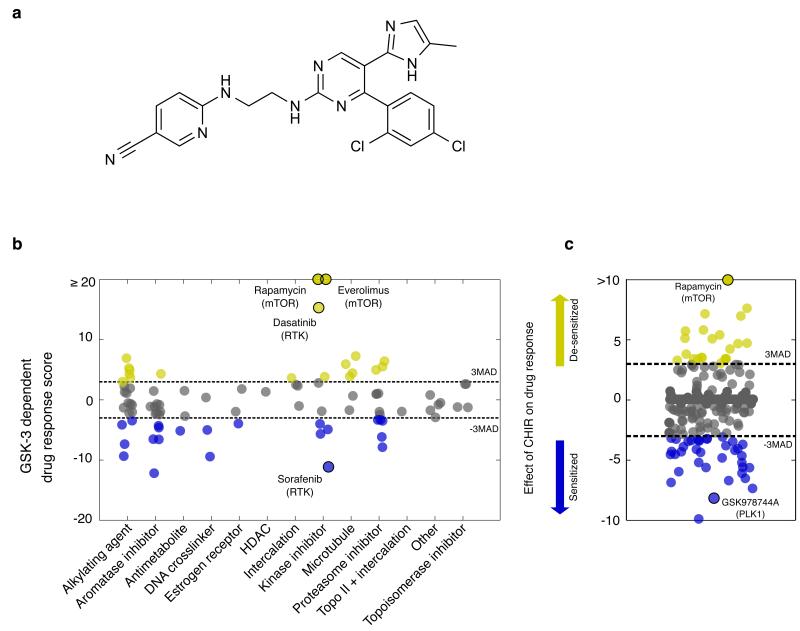
Figure 1. Decreased GSK-3 activity alters cellular response to oncology drugs and kinase inhibitors.
(a) Chemical structure of CHIR99021 (b) The effects of inhibiting GSK-3 (via CHIR) on cellular responses of the 89 drugs in the NCI Approved Oncology Drug Set II are quantified (Supplementary Note). Kinase inhibitors exhibit the widest range of GSK-3 dependent response scores, with mTOR inhibitors (rapamycin and everolimus) completely losing activity under GSK-3 inhibition. (c) A screen of a kinase inhibitor set reveals that inhibiting GSK-3 activity can either strongly enhance (n= 37) or suppress (n = 22) kinase inhibitor activities, e.g. PLK1 and mTOR inhibitors. Compounds labeled in yellow or blue reflect score ≥ 3MAD or score ≤ - 3MAD, respectively (Supplementary Data Set 2, 3).
To investigate the effect of GSK-3 activity on cellular responses to cytotoxic and cytostatic compounds, we first performed a “modifier” screen to determine how modulating the activity of GSK-3 affects potency of the NCI Approved Oncology Drug Set15, a collection of 89 current and diverse FDA-approved anticancer drugs (Supplementary Data set 2). HCEC cells were treated with compounds at 6 concentrations, covering a 1000-fold concentration range, with or without CHIR present. Cell viability was evaluated by measuring cellular ATP concentrations, and dose-response curves were fit to each drug treatment. A “Drug-Response Score,” indicating the effect of GSK-3 inhibition on each drug response, was calculated by transforming the area between dose-response curves of each drug, with or without CHIR, into a robust z-score (Supplementary Note, Supplementary Fig. 2).
We found that GSK-3 inhibition significantly affected nearly half (46%) of the drugs, across many mechanisms of action (Fig. 1b). Notably, the kinase-inhibitor category had a wide range of responses, suggesting that targeted therapies may be significantly perturbed by the activity of GSK-3. mTOR inhibitors were the most strongly perturbed by GSK-3 inhibition; rapamycin and its analog, everolimus, showed a complete loss of growth-inhibitory effects under CHIR treatment. To a lesser extent, GSK-3 inhibition desensitized cells to the receptor tyrosine kinase (RTK) inhibitors dasatinib, erlotinib and gefitinib.
The effects of GSK-3 on the NCI library’s small set of kinase-targeted inhibitors prompted us to test a larger and more diverse set of kinase inhibitors. We used a small molecule library of ~370 published kinase-inhibitors (Supplementary Data Set 3). As before, we measured six doses of each compound in a viability assay in the presence and absence of CHIR and calculated the drug-response score. GSK-3 inhibition strongly affected the response curves of 89 out of 367 compounds (24%), with cells being either sensitized or desensitized to drug treatment. Notable among the extremes, mTOR inhibitors were again suppressed and a PLK1 inhibitor was enhanced (Fig. 1c). In light of the poly-pharmacological signature of many compounds in our kinase-inhibitor set, we sought to validate our observed interactions between GSK-3 activity and mTOR and PLK1 inhibitors.
GSK-3 inhibition desensitizes cells to mTOR inhibitors
First, we explored the effect of GSK-3 inhibition on the desensitization of cells to mTOR inhibitors. We confirmed that CHIR’s effect is specific to GSK-3 by testing another GSK-3 inhibitor, BIO, as well as siRNA for GSK-3 α/β, both of which desensitized cells to rapamycin (Fig. 2a, Supplementary Fig. 3a). We then tested several mTOR-pathway independent and dependent mechanisms that could explain how inhibition of GSK-3 could lead to rapamycin insensitivity. To investigate mTOR-pathway independent mechanisms, we tested the possibilities that GSK-3 inhibition could lead to stabilization of the transcriptional activators β-catenin or Myc, which are both well known to directly activate pro-growth and survival pathways. However, we found that β-catenin or Myc loss-of-function (using either RNAi or small molecule inhibition) did not prevent GSK-3’s effect on the cellular response to rapamycin (Supplementary Fig. 4)16,17.
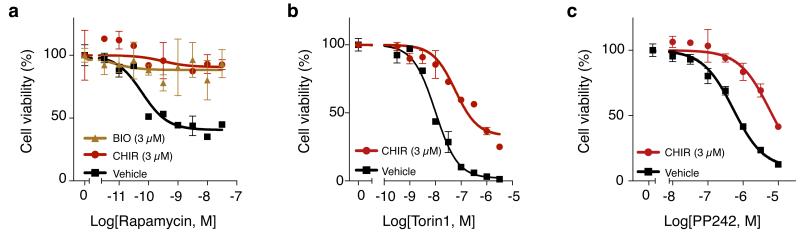
Figure 2. GSK-3 inhibition desensitizes cells to mTOR inhibitors.
Inhibition of GSK-3 activity via CHIR or BIO desensitizes cells to mTOR inhibitors rapamycin (a), Torin1 (b), and PP242 (c). (Experiment conditions for all panels were performed in duplicate, data represent mean values ± s.d.)
To investigate mTOR-pathway dependent mechanisms, we tested whether inactivation of GSK-3 could lead (directly or indirectly) to phosphorylation of the immediate downstream substrates of drug-inhibited mTORC1 or mTORC2 complexes. Rapamycin and its analogs (rapalogs) work by causing FKBP12 to allosterically inhibit the kinase activity of mTOR in the mTORC1 complex, but rapalogs have little effect on the partially-redundant mTORC2 complex18,19. Much of the effect that mTOR has on cell growth is mediated by phosphorylation of 4EBP and S6K by the mTORC1 complex; thus, a possible explanation for CHIR-induced desensitization to rapamycin is that GSK-3 inhibition prevents FKBP12 from blocking mTORC1 signaling. However, we found that GSK-3 inhibition did not reverse rapamycin’s inhibition of 4EBP or S6K phosphorylation (Supplementary Fig. 5). Alternatively, it has been speculated that lack of efficacy of rapalogs in some settings could be due to compensatory mTORC2 signaling or incomplete inhibition of mTORC120-22. Thus, a possible explanation for CHIR-induced desensitization is parallel signaling by the mTORC2 complex8,20,23-25. Potent ATP-site inhibitors of mTOR that block both mTORC1 and mTORC2 complex activities have begun to emerge, of which torin1 and PP242 are examples26,27. Here, we found that torin1 and PP242, like rapamycin, lost effectiveness in the presence of CHIR (Fig. 2b,c). However, the phosphorylation of AKT at serine 473, a known mTORC2 substrate, was not affected by CHIR treatment (Supplementary Fig. 5). Taken together, our results suggest that GSK-3 inhibition desensitizes cells to mTOR inhibitors through a mechanism that bypasses the obvious candidates of β-catenin, c-Myc or mTORC1/2 signaling. Given GSK-3’s high connectivity within the proteome24,25, an intriguing possibility is that rapamycin resistance emerges from the small effects from many interactions rather than a single dominant interaction.
GSK-3 inhibition sensitizes cells to PLK1 inhibitors
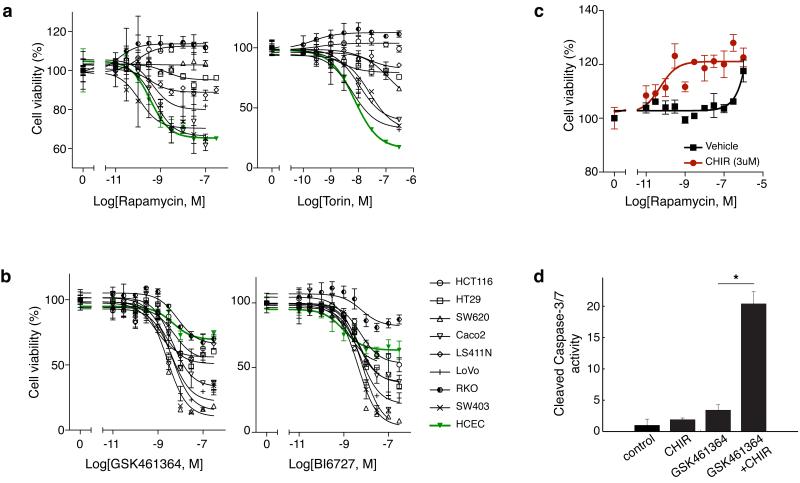
Next, we explored the effects of GSK-3 inhibition on the sensitization of cells to PLK1 inhibitors. PLK1 plays a critical role in mitotic progression and chromosome stability28; suppression of PLK1 activity leads to mitotic arrest in prometaphase and, eventually, to mitotic catastrophe, fragmented nuclei (Fig. 3a), and cell death28. We made use of PLK1 RNAi and the potent PLK1 inhibitor GSK461364, which has a strong specificity profile and is currently being tested in the clinic29,30. We quantified the number of fragmented nuclei after GSK461364 (Fig. 3b) or PLK1 RNAi (Supplementary Fig. 3b) and found that CHIR inhibition of GSK-3 greatly increased PLK1-induced DNA fragmentation. We also measured apoptosis through cleaved caspase 3 staining (Fig. 3c) and found it also was increased by combining inhibition of GSK-3 with GSK461364 (Fig. 3d) or PLK1 RNAi (Supplementary Fig. 3c) treatment. Our results suggest that the potency of PLK1 inhibitors could be enhanced by GSK-3 inhibitors. Recently, small molecule inhibitors of PLK1 have received much attention from the pharmaceutical industry, where they have moved into clinical trials and are beginning to show promise as anti-cancer agents30,31. Our results suggest that pathologically low GSK-3 activity found in various cancers5-7,9,32 could be partially responsible for the potency of PLK1 inhibitors in preclinical cancer studies.
Figure 3. GSK-3 inhibition sensitizes cells to PLK1 inhibitors.
Inhibition of GSK-3 activity via CHIR sensitizes cells to PLK1 inhibition with PLK1 RNAi (a,c) the small molecule GSK461364 (b,d). We measured either fragmented nuclei after 72 hours treatment (a,b) or cleaved caspase-3/7 activity after 48 hours of treatment (c,d). Data points were normalized to the control; each point represents mean values ± s.d. from two biological replicates. Statistical significance was determined using a K-S test (Supplementary Note); * indicates p-values < 0.01. (Scale bars, 25 μm.)
CRC cells are resistant to mTOR inhibitors and sensitive to PLK1 inhibitors
To what degree our findings in HCECs apply more broadly to epithelial cells derived from colorectal cancer? GSK-3 kinase activity is commonly suppressed in colorectal cancer (CRC) due to mutations in PI3K, KRAS and/or Wnt signaling upstream of GSK-310. Thus we speculated that most cancer lines would be more resistant to mTOR inhibitors and potentially more sensitive to PLK1 inhibitors when compared to non-oncogenic HCECs. Indeed, we conducted cell viability dose-response curves for mTOR inhibitors, (rapamycin, torin1) and Caspase assays for PLK1 inhibitor (GSK461364) across 8 CRC lines and found the majority were significantly less responsive to mTOR inhibitors and more responsive to PLK1 inhibitors (Fig. 4a,b). To test if drug responsiveness could yet be further perturbed by GSK-3 inhibition, we tested drug response with or without CHIR. We found most cell lines could be desensitized to mTOR inhibition or sensitized to PLK1 inhibition (Fig. 4c,d for SW620, Supplementary Fig. 6 for the rest of the CRC panel). Interestingly, rapamycin treatment alone increased basal cell proliferation rates of several CRC lines, and these rates increased even more dramatically with combined mTOR and GSK-3 inhibition (e.g. SW620, HCT116, RKO). Combined inhibition of these kinases in certain oncogenic backgrounds could allow for a surprisingly significant growth advantage for tumor tissue over healthy epithelium. Overall, we find that alteration in GSK-3 activity has a consistent role in modulating drug response in diverse colorectal cancer cell lines and HCECs. Together, our results suggest that in colon cancer the therapeutic window for mTOR inhibitors might be poor, whereas the therapeutic window for PLK1 inhibitors could be favorable.
Figure 4. Colorectal cancer cell lines are intrinsically more resistant to mTOR inhibitors and sensitive to PLK1 inhibitors.
A panel of eight colorectal cancer cell lines were treated with (a) mTOR inhibitors: rapamycin (left panel), torin1 (right panel), and (b) PLK1 inhibitors: GSK461364 (left panel), and BI6727 (right panel) for 72 hours and viability observed by measuring cellular ATP levels. Non-transformed HCECs are shown in green for comparison. GSK-3 inhibition further (c) desensitizes SW620 to rapamycin (72 hours treatment) and (d) sensitizes to PLK1 inhibitor GSK4611364 (30nM; 48 hours treatment, cleaved caspase-3/7 activity was observed). Normalization and significance were determined as in Figure 3.
GSK-3 interacts with the kinome to control cell proliferation
In light of GSK-3’s broad modification of kinase-inhibitor activity, we sought to expand our understanding of GSK-3’s relationship with kinases. A characteristic of GSK-3 is that priming phosphorylation events (via other kinases) on its targets can be a prerequisite for interaction33-35. Thus, GSK-3 acts in concert with other members of the kinome. Additionally, a caveat to screening kinase small molecule inhibitor libraries is that they often exhibit poly-pharmacological profiles and thus are rarely specific and may inhibit many kinases simultaneously, making interpretation of GSK-3-kinase interactions a challenge. To achieve a gene-by-gene understanding of which kinases interact with GSK-3 in controlling cell proliferation, we conducted a modifier RNAi screen of the entire human kinome in the presence or absence of CHIR. We applied siRNA pools of 3-4 oligos that target individual kinases to HCECs, followed by treatment of CHIR or vehicle control. After 72 hours, we fixed and stained for DNA and actin and imaged the plates by automated microscopy. We processed images to quantify intact nuclear and cellular boundaries as measures of cell viability (Supplementary Note, Supplementary Fig. 7). By examining cell counts, we identified kinases whose loss-of-function effects on cell viability were significantly different in CHIR and non-CHIR treatments (Fig. 5a, Supplementary Fig. 8).
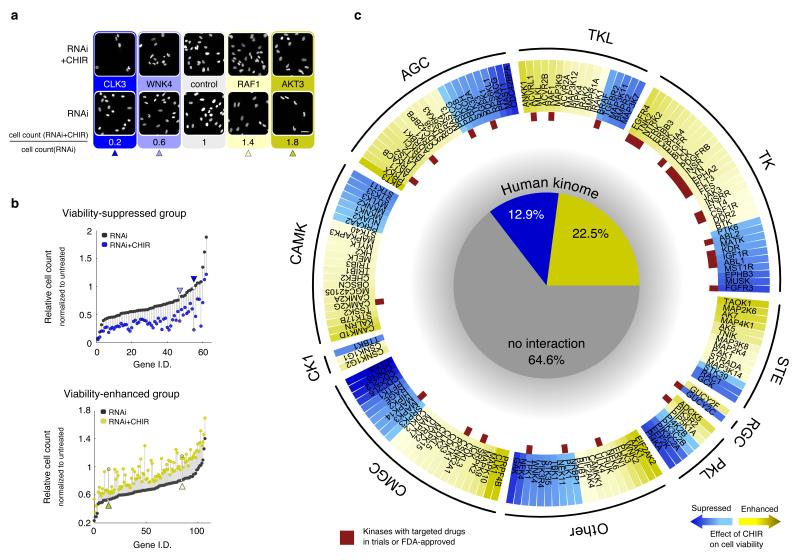
Figure 5. GSK-3 interacts with the kinome to control cell proliferation.
A screen of the human kinome with RNAi revealed that GSK-3 interacts with over one third of known human kinases. Yellow /blue are kinase knockdowns whose effects on cell viability were enhanced/suppressed in the presence of CHIR. (a) Representative examples of nuclear stained images from kinome screening (scale bar, 50μm). (b) Top panel: cell viability-suppressed group (~60 kinases) with and without CHIR treatment. Bottom panel: cell viability-enhanced group (~115 kinases) with and without CHIR treatment. Y-axis shows cell counts normalized to untreated (i.e. without CHIR or RNAi) and then sorted by the effect of RNAi alone. Vertical lines indicate the size of the synthetic effect of CHIR with RNAi. Arrowheads indicate genes listed in (a) with the same shading. (c) A breakdown of kinase families and kinase inhibitor drugs that interact with GSK-3. Darker shade indicates stronger CHIR effects (see Supplementary Information). Red rectangles mark kinases with targeted drugs that are FDA-approved or in clinical trials.
In agreement with the role of GSK-3 as major signaling hub, inhibition of GSK-3 affected cell viability of a large percentage of the kinome. CHIR enhanced viability for ~23% of kinases and suppressed viability for 13% (Fig. 5b and pie chart in Fig. 5c). Thus, 35% of the kinome shows an interaction with GSK-3 for cell viability in colonic epithelial cells. In line with its broad role in cell physiology, GSK-3 showed interactions with subsets of kinases from all the major kinase groups with no obvious enrichment for any one group or kinase subfamily (Fig. 5c). Many kinases known to genetically or biochemically interact with GSK-3 were identified in the screen, including AKT, PI3K, BRAF, and tyrosine kinase receptors for IGF, EGF, FGF. Kinases known to prime substrates for GSK-3 were also found (CDK, CK1, AKT). Additionally, many new or unappreciated interactions were identified. These include clinically important kinases such as TNK2, LCK, ABL1 and PRKCB (Fig. 5c, 6, and Supplementary Data Set 4). mTOR was not identified from this RNAi screen due to its subtle effect in cell number (as measured by our image-based screen) and major effect on size (Supplementary Fig. 9); our drug studies using ATP as a readout could not distinguish the two. However, cell viability from silencing of multiple mitotic kinases, including PLK1, was altered by CHIR treatment (consistent with the various mitotic poisons, such as microtubule drugs or topoisomerase inhibitors, found in our NCI library; Fig. 1a, Supplementary Data Set 2.). Though our choice of 3 μM CHIR had minimal effect on the cell cycle (compared with 10 μM CHIR, which has previously been shown to delay cell cycle36), it is possible that the mitotic phase of the cell cycle is subtly destabilized and thereby alters the potency of mitotic poisons. Finally, GSK-3 inhibition also altered cell viability in response to silencing members of the tyrosine kinase family (also in agreement with our drug screens; Fig. 1, Supplementary Data Set 2, 3.), an important emerging class of targeted therapies. Further, of the 196 kinase-targeted drugs currently in clinical trials or FDA-approved according to DrugBank11, GSK-3 interacts with ~50% (96) of them (Supplementary Note, Supplementary Data Set 1). Taken together, the state of GSK-3 activity has the ability to dictate cellular response to inhibiting a large fraction of the human kinome.
Figure 6. Summary of drug-targeted kinases affected by GSK-3 activity.
Schematic shows the kinase targets for FDA-approved or clinical trial drugs identified from the GSK-3 interaction RNAi screen. Kinases in the yellow box are likely to be more responsive to their inhibitors when GSK-3 activity is high. Kinases in the blue box are likely to be more responsive to their inhibitors when GSK-3 activity is low.
Discussion
Our findings reveal a GSK-3-mediated mechanism that can switch cells between sensitivity and insensitivity to kinase-targeted drugs-the second-most drugged protein targets after GPCRs. The observation that GSK-3 can affect cellular responses to such a wide array of kinases suggests that GSK-3 signaling could be predictive for drug response, and modifiers of GSK-3 signaling could be important for devising new combination therapies that can overcome drug resistance. For example, GSK-3 inhibitors could be used to enhance the efficacy of PLK1 inhibitors. Additionally, if GSK-3 activity were increased or reactivated by a targeted small molecule, cells would become re-sensitized to drugs from diverse drug classes, from nucleotide analogs to tyrosine kinase inhibitors. A few examples of kinase activation by small molecules exist and are currently being validated in pre-clinical studies37-39. Due to GSK-3’s vast number of protein substrates and cellular functions, it is likely that the mechanism by which GSK-3 sensitizes or desensitizes cells to any single drug is unique to the drug’s mode of action. Taken together, our pharmacological and genetic studies suggest that the state of GSK-3 in a cellular population can strongly affect the outcome of drug treatment.
A key challenge in designing optimized therapies is identifying molecular states that affect cellular responses to drugs. Knowing the intrinsic state of GSK-3 within a diseased tissue may be essential information when predicting drug potencies and selecting therapeutic strategies. Our studies with GSK-3 may serve as a prototype for identifying other highly-networked signaling molecules that broadly modulate cellular response to therapeutics.
Methods
Antibodies, drugs and reagents
Immunoblotting was performed using the following antibodies: α - β -catenin (BD Transduction Labs); α -p4EBP, α -4EBP, α -pS6, α -S6, c-Myc, CREB, Snail (Cell Signaling); α -actin (ImmunO); rabbit α -GSK-3 (Santa Cruz). Reagents used include Torin1, PP 242, BIO, CHIR 99021 (Tocris, >98% purity); BI 6727, GSK461364 (Selleck, >97% purity); LiCl, Rapamycin, Everolimus (Sigma, >95% purity); DMSO was used as vehicle in all experiments unless otherwise stated.
Cell lines and cell viability assay
Human Colonic Epithelial Cells12 and cancer cell lines were kind gifts from Jerry Shay (UT Southwestern). HCECs and cancer cell lines were maintained in DMEM with 10% FBS and antibiotics, 37degC and 5% C02. Cell viability was obtained by measuring cellular ATP as a proxy for cell number using CellTiter-Glo (Promega) following manufacturer’s instructions. For all growth curves, cells were seeded and grown in log-growth phase for the entire course of drug treatment.
GSK kinase inhibitor screen
The Published Kinase Inhibitor Set (PKIS) was acquired from GlaxoSmithKline by the University of Texas Southwestern High Throughput Screen facility. We modified the PKIS by adding torin1 and rapamycin into our screen. 100μl HCEC (1,000 cells/ml) cells were seeded into each wells and let adhere O/N. Half the plates received fresh media with 1% DMSO, the other half received fresh media with 300 μM BIO. Library compounds and controls were added using Echo liquid handler (Labcyte). 6 doses of each drug at 10,000 3,000, 1000, 300, 100, 30 nM were tested. Cells were then incubated for 72 hrs at 37°C and 5% CO2. Next, media was removed and 25 μl of CellTiter-Glo diluted 1:5 with passive lysis buffer (Promega) was added. Plates were incubated for 10 mins at RT with shaking and read on Envision (Perkin Elmer).
Kinome RNAi screen
Darmacon siRNA library was used for kinome screening. First, 0.5 μl of oligo (5μM) was diluted in 20 μl of Optimum and 0.2 μl of RNAi Max in 20 μl of Optimum in master plates. 20 μl of oligo mix was added to each well followed by 20 μl of Lipid mix. Plates were incubated for 20 mins at room temperature. Next 100 μl HCEC (1,000 cells/ml) cells were seeded into each wells and let adhere for 4 hours. Then, 3 μM of CHIR99021 was added with an Echo liquid handler and plates were place in incubators for 72 hours. Cells were then fixed by the addition of 37 μl PFA and incubated for 10 mins. Then cells were permeabilized and stained by discarding fixative and adding 50 μl of PBS with 0.2% triton containing Hoechst and phalloidin alexa 546 for 30 minutes. Staining solution was discarded and 100 μl of PBS was added for imaging. Plates (96-well BD Falcon Microplates) were then imaged on Nikon Ti-Eclipse with automated stage (see detail below).
Image acquisition
Fluorescence images were acquired using a Nikon Ti-E inverted-microscope equipped with integrated Perfect-Focus System (PFS), Nikon CFI S Flour 10× objective lens, and 14-bit CoolSnap HQ2 CCD camera (Photometrics). Images were acquired 16 frames per well at 15× (10 × 1.5× zoom) controlled by NIS-Elements software version 4.0. Uneven-illumination was corrected and 0.1 quantile of pixel values was considered as background and subtracted out from the distribution resulting in a flat, background subtracted foreground40.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Melanie Cobb and members of the Altschuler and Wu lab for critical reading of the manuscript; Jerry Shay for providing the HCEC cell line and guidance on culture techniques; Michael Rodriguez for experimental support; and Jane Life for help with HTS experiments. This research was supported by a Cancer Biology Training Grant T32 CA124334 (C.A.T.), an American Cancer Society-Lakeshore Division Postdoctoral Fellowship (C.A.T), the National Institute of Health grants K99 DK103126-01 (C.A.T.), CA133253 (S.J.A.), CA184984 (L.F.W.), R01 GM081549 (L.F.W.), CPRIT RP10900 (L.F.W), and the Welch Foundation I-1619 (S.J.A.) and I-1644 (L.F.W.).
Footnotes
CONTRIBUTIONS
C.A.T. designed, performed, and analyzed experiments. C.W. designed experiments and performed analysis of data. A.D.C. aided in analysis of data. B.A.P. designed HTS experiments and provided critical reagents. C.A.T., C.W., L.F.W. and S.J.A. wrote the manuscript with advice from all authors. L.F.W. and S.J.A. guided all aspects of this study.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
BIBLIOGRAGHY
- 1.Doble BW, Woodgett JR. GSK-3: tricks of the trade for a multi-tasking kinase. Journal of cell science. 2003;116:1175–1186. doi: 10.1242/jcs.00384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kim L, Kimmel AR. GSK3, a master switch regulating cell-fate specification and tumorigenesis. Current opinion in genetics & development. 2000;10:508–514. doi: 10.1016/s0959-437x(00)00120-9. [DOI] [PubMed] [Google Scholar]
- 3.Pap M, Cooper GM. Role of glycogen synthase kinase-3 in the phosphatidylinositol 3-Kinase/Akt cell survival pathway. The Journal of biological chemistry. 1998;273:19929–19932. doi: 10.1074/jbc.273.32.19929. [DOI] [PubMed] [Google Scholar]
- 4.Sutherland C. What Are the bona fide GSK3 Substrates? International journal of Alzheimer’s disease. 2011;2011:505607. doi: 10.4061/2011/505607. doi:10.4061/2011/505607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Farago M, et al. Kinase-inactive glycogen synthase kinase 3beta promotes Wnt signaling and mammary tumorigenesis. Cancer research. 2005;65:5792–5801. doi: 10.1158/0008-5472.CAN-05-1021. doi:10.1158/0008-5472.CAN-05-1021. [DOI] [PubMed] [Google Scholar]
- 6.Kang T, et al. GSK-3 beta targets Cdc25A for ubiquitin-mediated proteolysis, and GSK-3 beta inactivation correlates with Cdc25A overproduction in human cancers. Cancer cell. 2008;13:36–47. doi: 10.1016/j.ccr.2007.12.002. doi:10.1016/j.ccr.2007.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Leis H, Segrelles C, Ruiz S, Santos M, Paramio JM. Expression, localization, and activity of glycogen synthase kinase 3beta during mouse skin tumorigenesis. Molecular carcinogenesis. 2002;35:180–185. doi: 10.1002/mc.10087. doi:10.1002/mc.10087. [DOI] [PubMed] [Google Scholar]
- 8.Zhang HH, Lipovsky AI, Dibble CC, Sahin M, Manning BD. S6K1 regulates GSK3 under conditions of mTOR-dependent feedback inhibition of Akt. Molecular cell. 2006;24:185–197. doi: 10.1016/j.molcel.2006.09.019. doi:10.1016/j.molcel.2006.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhou BP, et al. Dual regulation of Snail by GSK-3beta-mediated phosphorylation in control of epithelial-mesenchymal transition. Nature cell biology. 2004;6:931–940. doi: 10.1038/ncb1173. doi:10.1038/ncb1173. [DOI] [PubMed] [Google Scholar]
- 10.Cancer Genome Atlas N. Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487:330–337. doi: 10.1038/nature11252. doi:10.1038/nature11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Knox C, et al. DrugBank 3.0: a comprehensive resource for ‘omics’ research on drugs. Nucleic acids research. 2011;39 doi: 10.1093/nar/gkq1126. D1035-1041, doi:10.1093/nar/gkq1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Roig AI, et al. Immortalized epithelial cells derived from human colon biopsies express stem cell markers and differentiate in vitro. Gastroenterology. 2010;138:1012–1021. doi: 10.1053/j.gastro.2009.11.052. e1011-1015, doi:10.1053/j.gastro.2009.11.052. [DOI] [PubMed] [Google Scholar]
- 13.Doble BW, Patel S, Wood GA, Kockeritz LK, Woodgett JR. Functional redundancy of GSK-3alpha and GSK-3beta in Wnt/beta-catenin signaling shown by using an allelic series of embryonic stem cell lines. Developmental cell. 2007;12:957–971. doi: 10.1016/j.devcel.2007.04.001. doi:10.1016/j.devcel.2007.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bennett CN, et al. Regulation of Wnt signaling during adipogenesis. The Journal of biological chemistry. 2002;277:30998–31004. doi: 10.1074/jbc.M204527200. doi:10.1074/jbc.M204527200. [DOI] [PubMed] [Google Scholar]
- 15. http://dtp.nci.nih.gov/branches/dscb/oncology_drugset_explanation.html.
- 16.Yost C, et al. The axis-inducing activity, stability, and subcellular distribution of beta-catenin is regulated in Xenopus embryos by glycogen synthase kinase 3. Genes & development. 1996;10:1443–1454. doi: 10.1101/gad.10.12.1443. [DOI] [PubMed] [Google Scholar]
- 17.Liu C, et al. Control of beta-catenin phosphorylation/degradation by a dual-kinase mechanism. Cell. 2002;108:837–847. doi: 10.1016/s0092-8674(02)00685-2. [DOI] [PubMed] [Google Scholar]
- 18.Jacinto E, et al. Mammalian TOR complex 2 controls the actin cytoskeleton and is rapamycin insensitive. Nature cell biology. 2004;6:1122–1128. doi: 10.1038/ncb1183. doi:10.1038/ncb1183. [DOI] [PubMed] [Google Scholar]
- 19.Sarbassov DD, et al. Rictor, a novel binding partner of mTOR, defines a rapamycin-insensitive and raptor-independent pathway that regulates the cytoskeleton. Current biology: CB. 2004;14:1296–1302. doi: 10.1016/j.cub.2004.06.054. doi:10.1016/j.cub.2004.06.054. [DOI] [PubMed] [Google Scholar]
- 20.Efeyan A, Sabatini DM. mTOR and cancer: many loops in one pathway. Current opinion in cell biology. 2010;22:169–176. doi: 10.1016/j.ceb.2009.10.007. doi:10.1016/j.ceb.2009.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sabatini DM. mTOR and cancer: insights into a complex relationship. Nature reviews. Cancer. 2006;6:729–734. doi: 10.1038/nrc1974. doi:10.1038/nrc1974. [DOI] [PubMed] [Google Scholar]
- 22.Thoreen CC, Sabatini DM. Rapamycin inhibits mTORC1, but not completely. Autophagy. 2009;5:725–726. doi: 10.4161/auto.5.5.8504. [DOI] [PubMed] [Google Scholar]
- 23.O’Reilly KE, et al. mTOR inhibition induces upstream receptor tyrosine kinase signaling and activates Akt. Cancer research. 2006;66:1500–1508. doi: 10.1158/0008-5472.CAN-05-2925. doi:10.1158/0008-5472.CAN-05-2925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen CH, et al. ER stress inhibits mTORC2 and Akt signaling through GSK-3beta-mediated phosphorylation of rictor. Science signaling. 2011;4:ra10. doi: 10.1126/scisignal.2001731. doi:10.1126/scisignal.2001731. [DOI] [PubMed] [Google Scholar]
- 25.Huang J, Manning BD. A complex interplay between Akt, TSC2 and the two mTOR complexes. Biochemical Society transactions. 2009;37:217–222. doi: 10.1042/BST0370217. doi:10.1042/BST0370217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Thoreen CC, et al. An ATP-competitive mammalian target of rapamycin inhibitor reveals rapamycin-resistant functions of mTORC1. The Journal of biological chemistry. 2009;284:8023–8032. doi: 10.1074/jbc.M900301200. doi:10.1074/jbc.M900301200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Feldman ME, et al. Active-site inhibitors of mTOR target rapamycin-resistant outputs of mTORC1 and mTORC2. PLoS biology. 2009;7:e38. doi: 10.1371/journal.pbio.1000038. doi:10.1371/journal.pbio.1000038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Petronczki M, Lenart P, Peters JM. Polo on the Rise-from Mitotic Entry to Cytokinesis with Plk1. Developmental cell. 2008;14:646–659. doi: 10.1016/j.devcel.2008.04.014. doi:10.1016/j.devcel.2008.04.014. [DOI] [PubMed] [Google Scholar]
- 29.Rudolph D, et al. BI 6727, a Polo-like kinase inhibitor with improved pharmacokinetic profile and broad antitumor activity. Clinical cancer research: an official journal of the American Association for Cancer Research. 2009;15:3094–3102. doi: 10.1158/1078-0432.CCR-08-2445. doi:10.1158/1078-0432.CCR-08-2445. [DOI] [PubMed] [Google Scholar]
- 30.Olmos D, et al. Phase I study of GSK461364, a specific and competitive Polo-like kinase 1 inhibitor, in patients with advanced solid malignancies. Clinical cancer research: an official journal of the American Association for Cancer Research. 2011;17:3420–3430. doi: 10.1158/1078-0432.CCR-10-2946. doi:10.1158/1078-0432.CCR-10-2946. [DOI] [PubMed] [Google Scholar]
- 31.Schoffski P. Polo-like kinase (PLK) inhibitors in preclinical and early clinical development in oncology. The oncologist. 2009;14:559–570. doi: 10.1634/theoncologist.2009-0010. doi:10.1634/theoncologist.2009-0010. [DOI] [PubMed] [Google Scholar]
- 32.Zheng H, Saito H, Masuda S, Yang X, Takano Y. Phosphorylated GSK3beta-ser9 and EGFR are good prognostic factors for lung carcinomas. Anticancer research. 2007;27:3561–3569. [PubMed] [Google Scholar]
- 33.Fiol CJ, et al. Phosphoserine as a recognition determinant for glycogen synthase kinase-3: phosphorylation of a synthetic peptide based on the G-component of protein phosphatase-1. Archives of biochemistry and biophysics. 1988;267:797–802. doi: 10.1016/0003-9861(88)90089-6. [DOI] [PubMed] [Google Scholar]
- 34.Frame S, Cohen P, Biondi RM. A common phosphate binding site explains the unique substrate specificity of GSK3 and its inactivation by phosphorylation. Molecular cell. 2001;7:1321–1327. doi: 10.1016/s1097-2765(01)00253-2. [DOI] [PubMed] [Google Scholar]
- 35.Thomas GM, et al. A GSK3-binding peptide from FRAT1 selectively inhibits the GSK3-catalysed phosphorylation of axin and beta-catenin. FEBS letters. 1999;458:247–251. doi: 10.1016/s0014-5793(99)01161-8. [DOI] [PubMed] [Google Scholar]
- 36.Tighe A, Ray-Sinha A, Staples OD, Taylor SS. GSK-3 inhibitors induce chromosome instability. BMC cell biology. 2007;8:34. doi: 10.1186/1471-2121-8-34. doi:10.1186/1471-2121-8-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hertz NT, et al. A neo-substrate that amplifies catalytic activity of parkinson’s-disease-related kinase PINK1. Cell. 2013;154:737–747. doi: 10.1016/j.cell.2013.07.030. doi:10.1016/j.cell.2013.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Simpson GL, Hughes JA, Washio Y, Bertrand SM. Direct small-molecule kinase activation: Novel approaches for a new era of drug discovery. Current opinion in drug discovery & development. 2009;12:585–596. [PubMed] [Google Scholar]
- 39.Thorne CA, et al. Small-molecule inhibition of Wnt signaling through activation of casein kinase 1alpha. Nature chemical biology. 2010;6:829–836. doi: 10.1038/nchembio.453. doi:10.1038/nchembio.453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Coster AD, Wichaidit C, Rajaram S, Altschuler SJ, Wu LF. A simple image correction method for high-throughput microscopy. Nature methods. 2014;11:602. doi: 10.1038/nmeth.2971. doi:10.1038/nmeth.2971. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.